Abstract
Symbiotic associations with microorganisms are pivotal in many insects. Yet, the functional roles of obligate symbionts have been difficult to study because it has not been possible to cultivate these organisms in vitro. The medically important tsetse fly (Diptera: Glossinidae) relies on its obligate endosymbiont, Wigglesworthia glossinidia, a member of the Enterobacteriaceae, closely related to Escherichia coli, for fertility and possibly nutrition. We show here that the intracellular Wigglesworthia has a reduced genome size smaller than 770 kb. In an attempt to understand the composition of its genome, we used the gene arrays developed for E. coli. We were able to identify 650 orthologous genes in Wigglesworthia corresponding to ≈85% of its genome. The arrays were also applied for expression analysis using Wigglesworthia cDNA and 61 gene products were detected, presumably coding for some of its most abundant products. Overall, genes involved in cell processes, DNA replication, transcription, and translation were found largely retained in the small genome of Wigglesworthia. In addition, genes coding for transport proteins, chaperones, biosynthesis of cofactors, and some amino acids were found to comprise a significant portion, suggesting an important role for these proteins in its symbiotic life. Based on its expression profile, we predict that Wigglesworthia may be a facultative anaerobic organism that utilizes ammonia as its major source of nitrogen. We present an application of E. coli gene arrays to obtain broad genome information for a closely related organism in the absence of complete genome sequence data.
Keywords: insect, Glossina, symbiosis, Wigglesworthia, expression profile
Different microorganisms have established obligate endosymbiotic associations with a range of arthropods including aphids, psyllids, whiteflies, mealybugs, termites, ants, reduviid bugs, and tsetse flies (1). These associations are thought to provide nutritional supplements to the insect host, which in each case relies on a single food source such as plant phloem, wood, or blood during all developmental stages of their life cycle. In addition to harboring symbionts, some of these insects are involved in the transmission of pathogens that cause devastating diseases in humans, animals, or plants. Tsetse flies are important insect vectors that exclusively feed on vertebrate blood and in the feeding process transmit African trypanosomes, the causative agents of sleeping sickness disease in humans and nagana in animals. Because these insects are dependent on symbiotic associations for viability, their microorganisms have long been of interest as potential means to genetically manipulate vector populations. It has not been possible to cultivate most endosymbionts in vitro and hence their correct taxonomic positioning has been difficult. Recently, phylogenetic analyses based on symbiont gene sequences have made it possible to compare these organisms and have shown that various microorganisms in distant clades of the Eubacteria have been able to enter and fulfill such mutualistic functions.
Microorganisms with different ultrastructural characteristics have been reported from various tissues of tsetse (2–6). Molecular analyses have now confirmed that tsetse harbors three different symbionts (7). Two of these organisms are members of the Enterobacteriaceae: the primary (P)-symbiont (genus Wigglesworthia; refs. 8 and 9) and the secondary (S)-symbiont (genus Sodalis; refs. 8, 10, and 11). The primary symbiont Wigglesworthia lives intracellularly in specialized epithelial cells (bacteriocytes) that make up the bacteriome-tissue in the anterior midgut, whereas Sodalis is harbored both inter- and intracellularly, principally in the midgut tissue (10). The third symbiont belongs to the genus Wolbachia and is present in some tsetse species.
The phylogenetic characterization of Wigglesworthia from distant tsetse species has shown that they form a distinct clade in the Enterobacteriaceae (7) and display concordant evolution with their host species (12). This finding suggests that a tsetse ancestor had been infected with a bacterium some 50–80 million years ago, and from this ancestral pair extant species of tsetse and associated Wigglesworthia strains radiated without horizontal transfer events between species. The bacteriome-associated symbionts of other insect taxa, such as aphids (13), whiteflies (14), psyllids (15), mealybugs (16), cockroaches (17), and carpenter ants (18), have similarly been found to represent distinct lineages that parallel the evolutionary histories of their host insect species.
Most of the obligate intracellular bacteria in insects are strictly vertically transmitted to the progeny. In tsetse, Wigglesworthia is maternally inherited by the intrauterine larva, which receives nutrients along with Wigglesworthia from its mother via milk-gland secretions (19, 20). It has been difficult to study the functional role of obligate symbionts in insects including tsetse because attempts to eliminate them have resulted in retarded growth of the insect and a decrease in egg production, preventing the ability of the aposymbiotic host to reproduce (21–23). However, the ability to reproduce could be partially restored in tsetse when the aposymbiotic flies were given a bloodmeal supplemented with B-complex vitamins (thiamine, pantothenic acid, pyridoxine, folic acid, and biotin), suggesting that the endosymbionts might play a role in metabolism that involves these compounds (24).
Here we report the characterization of the size and the overall composition of the Wigglesworthia genome. To understand the potential coding capacity of Wigglesworthia, we have hybridized its DNA to a heterologous gene array that contains all of the protein coding genes of the phylogenetically related enteric bacterium, Escherichia coli. Wigglesworthia cDNA was also hybridized to the arrays to gain insight into its most abundant gene products. We present the potential genome contents of Wigglesworthia and speculate on its putative role in tsetse biology based on some of its expressed gene products. These findings are discussed in comparison to the genomes characterized from free living bacteria, as well as from other intracellular and symbiotic organisms. We present an application of the gene-array technology that can be used to gain preliminary insights into the biology of closely related taxa in the absence of full genome sequence information.
Materials and Methods
Insects and Source of Symbionts.
The Glossina brevipalpis, Glossina pallidipes, and Glossina palpalis colonies are maintained in the insectary at Yale University Laboratory of Epidemiology and Public Health at 24–26°C with 65% humidity and are fed daily on defibrinated bovine blood (Crane Laboratories, Syracuse, NY), using an artificial membrane system (25). Puparia of colonized G. brevipalpis, G. pallidipes, and G. palpalis have been kindly supplied by the Seibersdorf Agricultural Research Laboratory, Vienna, Austria. For analysis of Wigglesworthia, bacteriome structures were dissected in PBS from pregnant females 3 weeks of age and the bacteria were released by gentle homogenization of the tissue.
Analysis of Wigglesworthia Genome Size.
Agarose plugs were prepared by resuspending bacterial cells from 200 bacteriomes in 100 μl TE buffer (10 mM Tris/1 mM EDTA, pH 8.0) and mixing with an equal volume of 1% low-melting agarose. The plugs were treated overnight in EC solution (6 mM Tris⋅HCl, pH 7.6, 100 mM EDTA, 1 M NaCl, 0.5% Brij 58, 0.2% deoxycholate, 0.5% N-lauroylsarcosine in the presence of 1 mg/ml lysozyme and 20 μg/ml RNase) at 37°C, as described for Buchnera (26). The EC solution was replaced with ESP (0.5 M EDTA, pH 8, 1% N-lauroylsarcosine, 1 mg/ml proteinase K) and incubated at 50°C for 2 days. Undigested DNA plugs were first electrophoresed by using pulse conditions of 50–200 s at 200 volts for 24 h. Our preliminary experiments have shown that under these conditions, contaminating extrachromosomal or degraded eukaryotic DNA enters into the gel while the high-molecular-weight intact bacterial DNA remains in the plugs. The plugs were then recovered from the wells and digested with restriction enzymes SmaI and ApaI and analyzed by contour-clamped homogeneous electric fields (CHEF) gel electrophoresis by using the conditions 5–40 s at 200 volts for 24 h.
Analysis of Gene Arrays.
E. coli macroarrays (Panorama E. coli gene arrays from Genosys, The Woodlands, TX) were used to identify common loci in Wigglesworthia obtained from G. pallidipes. Both Wigglesworthia DNA and RNA were labeled for hybridization analysis. Bacterial DNA isolated according to standard protocols was labeled with 32P-dGTP or 32P-dGTP and 32PdTTP by nick translation using DNA Pol I/Dnase I (Nick translation system from GIBCO/BRL). Hybridization conditions were changed from 42°C to 45°C, in the presence of buffers containing 45% formamide, 5× Denhardt's (0.02% polyvinylpyrrolidone/0.02% Ficoll/0.02% BSA), 5× SSC, and 0.5% SDS. Arrays were washed to a final stringency of 0.1× SSC/0.5% SDS at 42°C to 45°C. The use of a double isotope in the DNA labeling protocol enhanced the level of detection by approximately 3-fold. Total RNA was isolated from bacteriome tissues by the RNeasy kit (Qiagen) and 32P-dCTP labeled cDNA was synthesized by using the random primers in a multiprime DNA labeling system (Amersham Pharmacia). RNA was denatured for 2 min at 90°C, and ramped down to 42°C in 20 min to allow the hexamer primers to anneal. cDNA was synthesized by using the SuperscriptII reverse transcriptase at 42°C for 3 h. The labeled cDNA was purified and used as a probe in array hybridization at similar conditions as described for DNA.
Arrays were exposed to Maximum Resolution Films (Eastman Kodak, BMR) and signals were scored as strong (44%), medium (48%), or weak (8%). There were no cases where duplicate spots gave contradictory results.
Confirmation of DNA Methylation.
Wigglesworthia DNA was incubated overnight with BstNI and EcoRII, Sau3AI, MboI at 60°C and 37°C, respectively, and analyzed by conventional gel electrophoresis. Sodalis genomic DNA was used as a control to confirm the efficacy of the restriction enzymes (data not shown).
Confirmation of AmtB Expression by Western Blot Analysis.
Equal numbers of bacteriomes were dissected from female and male flies. In addition, muscle tissue was collected as a control. Total proteins from bacteriomes, muscle tissue, and in vitro cultured Sodalis were prepared in triple-detergent lysis buffer (50 mM Tris⋅Cl, pH 8.0, 150 mM NaCl, 100 μg/ml PMSF, 0.1% SDS, 1% Nonidet P-40, 0.25% deoxycholate). The samples were resolved by SDS/8% PAGE analysis and blots were reacted to AmtB (E. coli-specific) and GroEL (Wigglesworthia-specific; ref. 27) antibodies. AmtB peptide antibodies were kindly provided by S. Kustu (University of California, Berkeley, CA). The peptide sequence used to generate these antibodies was PEEQEREGLDVNSHGENAYN.
Genomic Library Preparation and Screening.
For genomic DNA library construction, Wigglesworthia chromosomal DNA was digested with Sau3AI restriction enzyme and 5–10-kb-sized fragments were ligated to Zap Express predigested lambda vector (Stratagene). This library was screened with 32P-dCTP-labeled Wigglesworthia cDNA probe as described above. Recombinant phages with positive signals were picked and plated for hybridization with gene-specific probes. The genes groEL, tktA, gshA, atoB, and dnaN were PCR amplified from E. coli by using gene-specific oligonucleotide primers (Genosys) and were radioactively labeled with 32P-dCTP to be used as individual probes for the second screening.
Results and Discussion
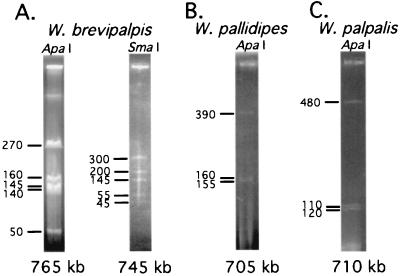
The genome size of Wigglesworthia was first characterized from Glossina brevipalpis, a species in the fusca subgenus. Although it has not been possible to cultivate the obligate intracellular symbionts of insects, we were able to obtain enough quantities of Wigglesworthia by microscopically dissecting the bacteriome tissue from tsetse guts. Bacterial DNA was embedded in agarose blocks, digested, and subjected to contour-clamped homogeneous electric fields (CHEF) gel electrophoresis analysis (Fig. 1). The size of the Wigglesworthia brevipalpis genome was found to be smaller than 770 kb (Fig. 1A), about one-sixth of that of the related free-living E. coli (4.6 Mb). Genome size reductions have also been observed for intracellular pathogens such as Chlamydia trachomatis (1.04 Mb), Treponema pallidum (1.14 Mb), Mycoplasma genitalium (0.58 Mb; ref. 28) and Rickettsia prowazekii (1.1 Mb). Recently, the genome of the obligate endosymbiont of aphids, Buchnera, another member of the Enterobacteriaceae, has been completely sequenced and shown to be 640 kb in size, comparable to that of Wigglesworthia (26, 29). The genome sizes of these mutualists, Buchnera and Wigglesworthia, are apparently reduced and approaching that of M. genitalium, the smallest bacterial genome reported so far. Similar genome size characterization of Wigglesworthia from tsetse flies in different subgenera were conducted, including G. pallidipes in the morsitans subgenus and G. palpalis in the palpalis subgenus. The genomes of their Wigglesworthia symbionts were found to have undergone similar reductions to about 705 kb and 710 kb, respectively (Fig. 1 B and C). A molecular clock analysis between G. brevipalpis and Glossina morsitans flies had indicated that they are separated by about 50–80 million years (12). When the genome sizes of Buchnera symbionts from distant aphids were determined and compared they were found to be similar in size, indicating that the symbiont genome reductions do not correlate with the evolutionary age of the symbiotic associations (30). The genome reductions in intracellular bacteria in each case imply gene and presumably related function loss and reflect the increased exploitation and dependence of these intracellular organisms on their host cells. The comparative analysis of intracellular symbiotic and pathogenic bacterial genomes might reveal the functional aspects of mutualistic versus parasitic nature of these associations.
Figure 1.
Genome size characterization of Wigglesworthia by contour-clamped homogeneous electric fields (CHEF) gel electrophoresis. (A) W. brevipalpis chromosome digested with ApaI and SmaI restriction enzymes were run at pulse times ranging from 5–40 s for 27 h and 20–100 s for 24 h, respectively. (B) Wigglesworthia pallidipes and (C) Wigglesworthia palpalis chromosomes were digested with ApaI restriction enzyme and run at pulse times ranging from 3–60 s for 48 h and 5–30 s at 23 h, respectively.
Another hallmark of intracellular genomes is the A + T bias in their coding sequences. The Buchnera genome is found to be 74% A + T rich (29) and the overall A + T contents of the genomes of R. prowazekii, M. genitalium, and C. trachomatis are 71, 68, and 59%, respectively. Analysis of the coding capacity of these reduced genomes has indicated that loci encoding for DNA repair and recombination functions have been lost or limited in many (31). It is thought that this loss of the repair functions might have led to the high A + T bias observed in these intracellular genomes (32). Our preliminary analysis with Wigglesworthia gene sequences also indicates a similar A + T bias. The genes coding for GroEL and FtsZ are found to be 63 and 66% A + T rich, respectively (GenBank accession nos. AF326971 and AF012886, respectively). The groEL and ftsZ genes of Wigglesworthia are 70 and 65% identical in nucleotide sequences to those of their corresponding orthologs in E. coli, respectively.
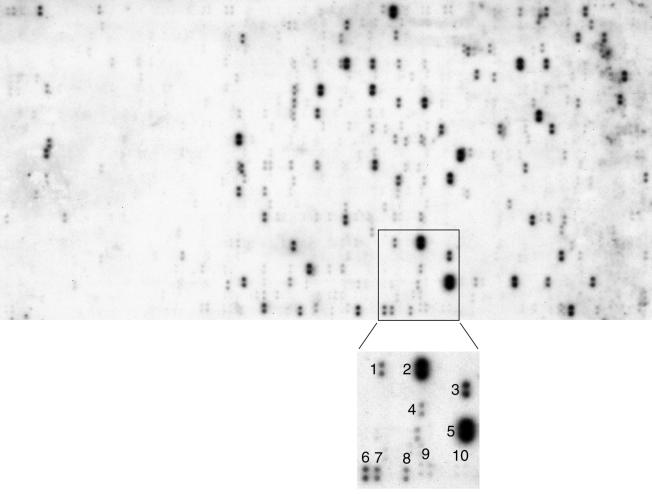
Gene array technology has become a popular tool for expression analysis and in addition has been successfully applied for screening of polymorphisms and evolutionary dynamics between different bacterial strains (33, 34). Here, we have applied this technology to understand the broad genomic aspects of an enteric bacterium of a different taxon in the absence of genome sequence information. Hybridization experiments were performed with E. coli gene arrays that contain 4,290 PCR-amplified ORFs corresponding to all E. coli protein-coding genes (Fig. 2). Although loci unique to Wigglesworthia or those significantly different from E. coli could not be detected by using this approach, 650 orthologs (see Table 1, which is published as supplemental data on the PNAS web site, www.pnas.org) were identified that represent about 85% of its genome assuming an average size of 1 kb per gene (29). Of the genes detected, known functional roles were assigned to 457 (70%) in E. coli, whereas 64 (9.8%) genes were reported to code for putative enzymes and 129 (19.8%) corresponded to hypothetical proteins with no known functions. As anticipated, among the genes detected with strong signals were those coding for conserved housekeeping proteins such as GroEL and HslU (chaperonins), TufA (elongation factor) and DnaN (DNA polymerase) as shown in Fig. 2. However, the signal intensities reflect not only sequence homology but also the length of genes. Accordingly, some of the genes detected with weaker signals were found to code for conserved but small proteins (e.g., GroES, ribosomal subunits).
Figure 2.
Gene array hybridization analysis. The autoradiogram shows the signals detected by hybridization of W. pallidipes DNA to Panorama E. coli macroarrays under stringent conditions. Each gene is spotted in duplicate and 4,290 genes are distributed over three panels. The enlarged insert shows some of the genes (and their products in parenthesis) identified in this region: 1, fusA (protein elongation factor G); 2, tufA (elongation factor EF-Tu); 3, hslU (heat shock protein); 4, prlC (oligopeptidase A); 5, groEL (chaperonin); 6, b0669 (function unknown); 7, lipA (lipoic acid synthetase); 8, b0667 (function unknown); 9 and 10, quadruple control spots containing 10 and 5 ng of E. coli genomic DNA, respectively.
When the contents of the completely sequenced pathogenic intracellular genomes were analyzed, genes involved in cell structure, replication, transcription, and translation were found to be retained, whereas the genes for amino acid biosynthesis, intermediate metabolism, and biosynthetic pathways were found to be largely lost (31). An example is the genome of M. genitalium, which has only a single gene related to amino acid biosynthesis as it exploits its host genome for nutrients. In contrast to intracellular pathogens, however, the small genome of the mutualist Buchnera has 55 genes involved in amino acid biosynthetic pathways, some of which have been found amplified on extra-chromosomal plasmids. Thus, Buchnera has retained the ability to synthesize all amino acids essential for its aphid host, whereas genes coding for nonessential amino acids are missing from its reduced genome (29). Indeed, nutritional studies have demonstrated that among the essential amino acids Buchnera provides to its aphid host are tryptophan, cysteine, and methionine (35, 36). Wigglesworthia apparently has the orthologs of the genes that encode for many essential proteins involved in DNA replication, transcription, and translation, in addition to 37 orthologs encoding for amino acid biosynthetic enzymes. We were unable to detect many of the genes involved in the biosynthetic pathways for proline, histidine, phenylalanine, tryptophan, serine, and leucine. However, specific transporters for proline, histidine, phenylalanine, and serine, as well as some amino acid ABC-transporter orthologs, could be detected and hence they may be involved in the uptake of these amino acids from the host diet. In Buchnera, retention of the essential amino acid genes reflects its mutualistic nature where both the bacteria and the insect host benefit from the provision of some of the amino acids. Considering the dietary differences between tsetse and aphid (i.e., blood versus plant sap), further characterization of amino acid biosynthetic pathways in Wigglesworthia may similarly reveal an interplay related to its obligate association with tsetse.
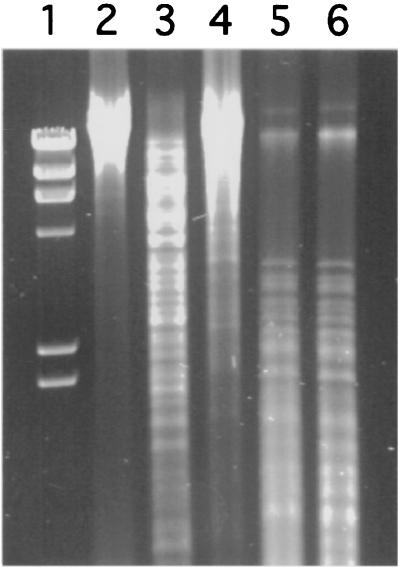
DNA methylation is a ubiquitous biological process that occurs in diverse organisms. DNA methylation in bacteria is most often thought of in its role to protect DNA from restriction endonucleases. Given the intracellular protected environment of the endosymbiotic bacteria, they are unlikely to methylate their DNA for protection. Indeed, the genome of Buchnera does not contain any methylase coding sequences. However, the gene for DNA cytosine methyltransferase (dcm) was detected in Wigglesworthia by array hybridization analysis, whereas the genes coding for two other DNA methyltransferases, dam and hsd, could not be found. To verify the functional presence of Dcm, Wigglesworthia DNA was digested with a pair of isoschizomers with identical recognition sequences but different methylation requirements. The enzyme pair BstNI and EcoRII is diagnostic for Dcm methylation status: BstNI can cleave DNA regardless of Dcm methylation, whereas its isoschizomer EcoRII requires unmethylated DNA at the same recognition site. When genomic DNA was subjected to digestion with these enzymes, it could be cleaved with BstNI (Fig. 3, lane 3), but not with EcoRII (Fig. 3, lane 4), indicating that Wigglesworthia DNA contains 5-methylcytosine modifications. Under the same conditions, Sodalis control DNA could be cleaved with EcoRII (data not shown). Similarly, the enzyme pair Sau3AI and MboI is diagnostic for Dam methylation status. These two enzymes generated similar restriction patterns, indicating that Wigglesworthia DNA is not subjected to adenine methylation, confirming the absence of the Dam locus (Fig. 3, Lanes 5 and 6, respectively). Whether Dcm methylation in Wigglesworthia might reflect regulatory function(s) for gene expression or cell division is unknown.
Figure 3.
DNA methylation status of W. pallidipes. Lane 1, λ molecular weight marker; lane 2, Wigglesworthia total DNA uncut; lanes 3–6, Wigglesworthia DNA digested with restriction enzymes BstNI, EcoRII, Sau3AI, and MboI, respectively.
When Wigglesworthia cDNA was hybridized to the E. coli array, 61 genes were detected, 37 of which had attributed functional roles in E. coli. Among these 37, some were detected with strong signals such as groEL, hslU, tktA, and dnaN, which had shown strong signals on genomic DNA hybridization as well, as mentioned above (data not shown). On the other hand, some of the genes with weak signals on genomic DNA hybridization were also detected with high signal intensities in the expression analysis (e.g., amtB, gshA, atoB, dapE, deaD, and pta). This observation suggests that the strong signal intensities in the expression analysis represent abundant expression levels and not necessarily high sequence conservation. When the same cDNA probe was hybridized to a Wigglesworthia genomic DNA library, approximately 100 clones could be identified with strong signals. Partial characterization of these clones by hybridization to E. coli gene-specific DNA probes, such as groEL, dnaN, gshA, atoB, and tktA, confirmed their abundant expression (data not shown).
One of the genes found in the expression array analysis was ushA ortholog, which encodes for UDP-sugar hydrolase 5′-nucleotidase, suggesting that Wigglesworthia might be scavenging exogenous nucleotides. In support of this, we were unable to detect a complete set of genes for de novo synthesis of nucleic acids, indicating that Wigglesworthia may be incapable of synthesizing all of its own nucleotides, but might instead depend on tsetse, using the salvage pathways. Genes for transketolases (tktA, tktB) were also found expressed at high levels, suggesting that the nonoxidative branch of the pentose-P pathway might be used preferentially to generate pyruvate in glycolysis. Pyruvate can be anaeorobically disseminated to acetyl-coA and formate by the action of Pyruvate formate lyase (pfl).
Wigglesworthia might be able to grow anaerobically by using the formate-dependent respiratory nitrite reductase enzymes encoded by nrfD and E products using ubiquinones (Q). Indeed, the gene coding for NrfE, which is involved in nitrite reduction, was among the overexpressed genes in Wigglesworthia. Formate could be the electron donor using the enzyme formate dehydrogenase (FdnG, H). Typically, formate dehydrogenases contain selenocysteine, which is incorporated cotranslationally by selA-D products, some of which were found present in the Wigglesworthia genome.
A role for vitamin B metabolites has been suggested for tsetse symbionts based on artificial diets where the diet of aposymbiotic female tsetse flies has been supplemented with vitamins and its impact on fertility has been documented (24). Our gene array analysis showed that Wigglesworthia has various gene homologs for the synthesis of riboflavin (ribH), folate, and dihydrofolate (folA, C, D, K), as well as genes whose products are involved in the biosynthesis of thiazole leading to the synthesis of thiamin (vitamin B1). In addition, genes involved in the synthesis of lipoic acid (lipA, B) and the intermediates involved in biotin synthesis were also detected. The tktA gene detected in expression analysis has also been implicated in pyridoxine (vitamin B6) synthesis (37). Although the presence of genes and specific pathways will be confirmed by the ongoing Wigglesworthia genome sequencing project, our preliminary findings further support the notion that vitamin biosynthetic pathways play a significant role in the functional biology of tsetse-Wigglesworthia symbiosis.
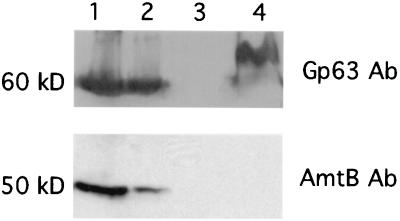
The ammonium/methylammonium transporter gene (amtB) was also found expressed by cDNA profiling in Wigglesworthia. The expression of amtB was further demonstrated by Western blot analysis using a peptide antibody raised against the E. coli AmtB (Fig. 4). A protein of expected size (50 kDa) was detected only in the bacteriome tissue (Fig. 4, Lane 1 and 2), whereas no specific signals were detected in either muscle tissue (Lane 3) or Sodalis (Lane 4). When the same blot was hybridized to GroEL (Gp63) antibody, both Wigglesworthia and Sodalis were found to overexpress this chaperonin as had been reported (27). The ammonium/methylammonium transporter is involved in the uptake of ammonium, which can be converted to glutamine by the action of glutamine synthetase (GS). Glutamine can serve as the cellular nitrogen source for the biosynthesis of amino acids. The gene coding for GS has also been detected in Wigglesworthia genome by this array analysis. Nitrogen regulatory protein C (NtrC) has been shown in E. coli to activate the transcription of genes whose products enhance growth under nitrogen-limiting conditions, including amtB (38). Although ntrC was detected in Wigglesworthia genome, it remains to be seen whether it plays a similar regulatory role in amtB expression in the event of host starvation.
Figure 4.
AmtB expression in bacteriome tissue. The Western blot was probed with antibodies against Gp63 (GroEL) (Top) and AmtB (Bottom), successively. Lane 1, female bacteriome; lane 2, male bacteriome; lane 3, muscle tissue; lane 4, protein extract from an in vitro culture of Sodalis.
The expression of the ammonium transporter in Wigglesworthia might represent an adaptation to the digestive physiology of its host. Wigglesworthia may use the transported ammonia to synthesize nitrogenous compounds, which may supplement the amino acid-based energy metabolism of its tsetse host. The ammonium uptake may further reduce the burden of large amounts of nitrogenous compounds, which tsetse must excrete rapidly to avoid water loss and desiccation. It has been shown that uric acid is the main excretory product in the fly representing the final product of nitrogen metabolism. Indeed, it makes up more than 60% of the dry weight of tsetse faeces. It has been estimated that tsetse flies use more than 50% of the energy gained by bloodmeal for synthesis and excretion of uric acid (39). Expression of the ammonium transporter may be particularly important for the tsetse intrauterine larva, which receives its nutrition from its mother through an intricate network of milk glands. Glutamic acid has been shown to be the main energy source in larva (40, 41), and Wigglesworthia in larva might be contributing to its synthesis. Indeed, the longevity of adult tsetse does not appear to be affected when Wigglesworthia is eliminated by antibiotic treatment; however, its absence adversely affects tsetse fecundity. Because it has not been feasible to obtain larva from symbiont-cured flies, it has not been possible for us to evaluate the functional significance of AmtB in the nitrogen metabolism of intrauterine larva.
Gene arrays are being used widely for quantitative expression analysis. Here we show an additional application of this technology where macroarrays can be successfully used to obtain information on the genome coding capacity of unexplored taxa in the absence of genome sequencing data. Although the lower sequence homology and the existence of paralogous genes or pseudogenes may potentially result in false negative/positive findings, the simultaneous utilization of arrays from related organisms would increase the confidence levels in similar analyses of unknown genomes. Alternatively, composite arrays that contain the genes for biosynthetic pathways of interest from different organisms can be constructed to explore specific pathways. The availability of gene arrays from different organisms and their similar application stand to provide a cost-effective approach to rapidly expand our knowledge base on the overall biology of microorganisms.
Supplementary Material
Acknowledgments
We are grateful to Drs. Scott L. O'Neill and K. P.-Chang for their critical reading of the manuscript. We are also grateful to the members of the Seibersdorf Agricultural Research Laboratory, Austria, for supplying tsetse pupae and to Drs. S. Kustu and E. Soupene for providing the E. coli AmtB antibody and technical advice. This work was supported by National Institutes of Health/National Institute of Allergy and Infectious Diseases Grant AI-34033 (to S.A.). L.A. is funded through the James Hudson Brown–Alexander Brown Coxe postdoctoral fellowship.
Footnotes
References
- 1.Moran N, Baumann P. Curr Opin Microbiol. 2000;3:270–275. doi: 10.1016/s1369-5274(00)00088-6. [DOI] [PubMed] [Google Scholar]
- 2.Pinnock D E, Hess R T. Acta Trop. 1974;31:70–79. [PubMed] [Google Scholar]
- 3.Roubaud B. Ann Inst Pasteur (Paris) 1919;33:489–537. [PubMed] [Google Scholar]
- 4.Shaw M K, Moloo S K. Parasitology. 1991;102:193–199. doi: 10.1017/s003118200006248x. [DOI] [PubMed] [Google Scholar]
- 5.Stuhlman F. Arb Gesundh Amte (Berlin) 1907;26:301–308. [Google Scholar]
- 6.Wigglesworth V B. Parasitology. 1929;21:288–321. [Google Scholar]
- 7.Aksoy S. Parasitol Today. 2000;16:114–119. doi: 10.1016/s0169-4758(99)01606-3. [DOI] [PubMed] [Google Scholar]
- 8.Aksoy S, Pourhosseini A A, Chow A. Insect Mol Biol. 1995;4:15–22. doi: 10.1111/j.1365-2583.1995.tb00003.x. [DOI] [PubMed] [Google Scholar]
- 9.Aksoy S. Int J Syst Bacteriol. 1995;45:848–851. doi: 10.1099/00207713-45-4-848. [DOI] [PubMed] [Google Scholar]
- 10.Cheng Q, Aksoy S. Insect Mol Biol. 1999;8:125–132. doi: 10.1046/j.1365-2583.1999.810125.x. [DOI] [PubMed] [Google Scholar]
- 11.Dale C, Maudlin I. Int J Syst Bacteriol. 1999;49:267–275. doi: 10.1099/00207713-49-1-267. [DOI] [PubMed] [Google Scholar]
- 12.Chen X, Song L, Aksoy S. J Mol Evol. 1999;48:49–58. doi: 10.1007/pl00006444. [DOI] [PubMed] [Google Scholar]
- 13.Munson M, Baumann P, Kinsey M. Int J Syst Bacteriol. 1991;41:566–568. [Google Scholar]
- 14.Campbell B C. Curr Microbiol. 1993;26:129–132. doi: 10.1007/BF01577365. [DOI] [PubMed] [Google Scholar]
- 15.Thao M L, Moran N A, Abbot P, Brennan E B, Burckhardt D H, Baumann P. Appl Environ Microbiol. 2000;66:2898–2905. doi: 10.1128/aem.66.7.2898-2905.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Munson M A, Baumann P, Moran N A. Mol Phylogenet Evol. 1992;1:26–30. doi: 10.1016/1055-7903(92)90032-c. [DOI] [PubMed] [Google Scholar]
- 17.Bandi C, Damiani G, Magrassi L, Grigolo A, Fani R, Sacchi L. Proc R Soc London Ser B. 1994;257:43–48. doi: 10.1098/rspb.1994.0092. [DOI] [PubMed] [Google Scholar]
- 18.Schroder D, Deppisch H, Obermayer M, Krohne G, Stackebrandt E, Holldobler B, Goebel W, Gross R. Mol Microbiol. 1996;21:479–489. doi: 10.1111/j.1365-2958.1996.tb02557.x. [DOI] [PubMed] [Google Scholar]
- 19.Ma W-C, Denlinger D L. Nature (London) 1974;247:301–303. [Google Scholar]
- 20.Aksoy S, Chen X, Hypsa V. Insect Mol Biol. 1997;6:183–190. doi: 10.1111/j.1365-2583.1997.tb00086.x. [DOI] [PubMed] [Google Scholar]
- 21.Nogge G. Experientia. 1976;32:995–996. doi: 10.1007/BF01933932. [DOI] [PubMed] [Google Scholar]
- 22.Southwood T R, Khalaf S, Sinden R E. Acta Trop. 1975;32:259–266. [PubMed] [Google Scholar]
- 23.Hill P D S, Campbell J A. Trans R Soc Trop Med Hyg. 1973;67:727–728. doi: 10.1016/0035-9203(73)90051-5. [DOI] [PubMed] [Google Scholar]
- 24.Nogge G. Parasitology. 1981;82:101–104. [Google Scholar]
- 25.Moloo S K. Parasitology. 1971;63:507–512. doi: 10.1017/s0031182000080021. [DOI] [PubMed] [Google Scholar]
- 26.Charles H, Ishikawa H. J Mol Evol. 1999;48:142–150. doi: 10.1007/pl00006452. [DOI] [PubMed] [Google Scholar]
- 27.Aksoy S. Insect Mol Biol. 1995;4:23–29. doi: 10.1111/j.1365-2583.1995.tb00004.x. [DOI] [PubMed] [Google Scholar]
- 28.Fraser C, Gocayne J, White O, Adams M D, Clayton R A, Fleischmann R D, Bult C J, Kerlavage A R, Sutton G, Kelley J M, et al. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 29.Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. Nature (London) 2000;407:81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- 30.Wernegreen J J, Ochman H, Jones I B, Moran N A. J Bacteriol. 2000;182:3867–3869. doi: 10.1128/jb.182.13.3867-3869.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moran N, Wernegreen J. Trends Ecol Evol. 2000;15:321–326. doi: 10.1016/s0169-5347(00)01902-9. [DOI] [PubMed] [Google Scholar]
- 32.Moran N A. Proc Natl Acad Sci USA. 1996;93:2873–2878. doi: 10.1073/pnas.93.7.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ochman H, Jones I B. EMBO J. 2000;19:6637–6643. doi: 10.1093/emboj/19.24.6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Troesch A, Nguyen H, Miyada C G, Desvarenne S, Gingeras T R, Kaplan P M, Cros P, Mabilat C. J Clin Microbiol. 1999;37:49–55. doi: 10.1128/jcm.37.1.49-55.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baumann P, Baumann L, Lai C Y, Rouhbakhsh D, Moran N A, Clark M A. Annu Rev Microbiol. 1995;49:55–94. doi: 10.1146/annurev.mi.49.100195.000415. [DOI] [PubMed] [Google Scholar]
- 36.Douglas A E, Prosser W A. J Insect Physiol. 1992;38:565–568. [Google Scholar]
- 37.Zhao G, Winkler M. J Bacteriol. 1994;176:6134–6138. doi: 10.1128/jb.176.19.6134-6138.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zimmer D P, Soupene E, Lee H L, Wendisch V F, Khodursky A B, Peter B J, Bender R A, Kustu S. Proc Natl Acad Sci USA. 2000;97:14674–14679. doi: 10.1073/pnas.97.26.14674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bursell E. J Insect Physiol. 1965;11:993–1001. doi: 10.1016/0022-1910(65)90202-7. [DOI] [PubMed] [Google Scholar]
- 40.Moloo S K. J Insect Physiol. 1976;22:563–567. doi: 10.1016/0022-1910(76)90177-3. [DOI] [PubMed] [Google Scholar]
- 41.Moloo S K. Acta Trop. 1976;33:133–142. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.