Abstract
We conducted a case-control study to investigate whether vascular endothelial growth factor (VEGF −2578, −1154, −634, and 936) and kinase insert domain containing receptor (KDR −604, 1192, and 1719) polymorphisms are associated with moyamoya disease. Korean patients with moyamoya disease (n = 107, mean age, 20.9±15.9 years; 66.4% female) and 243 healthy control subjects (mean age, 23.0±16.1 years; 56.8% female) were included. The subjects were divided into pediatric and adult groups. Among the 64 surgical patients, we evaluated collateral vessel formation after 2 years and divided patients into good (collateral grade A) or poor (collateral grade B and C) groups. The frequencies and distributions of four VEGF (−2578, −1154, −634, and 936) and KDR (−604, 1192, and 1719) polymorphisms were assessed from patients with moyamoya disease and compared to the control group. No differences were observed in VEGF −2578, −1154, −634, and 936 or KDR −604, 1192, and 1719 polymorphisms between the control group and moyamoya disease group. However, we found the −634CC genotype occurred less frequently in the pediatric moyamoya group (p = 0.040) whereas the KDR −604C/1192A/1719T haplotype increased the risk of pediatric moyamoya (p = 0.024). Patients with the CC genotype of VEGF −634 had better collateral vessel formation after surgery. Our results suggest that the VEGF −634G allele is associated with pediatric moyamoya disease and poor collateral vessel formation.
Introduction
Although many studies have investigated the etiology of moyamoya disease, unsatisfactory progress has been made. Moyamoya disease etiology may be idiopathic, environmental, or genetic. The search for genetic loci linked to moyamoya disease has uncovered associations with chromosomes 3, 6, 7, 8, and 17 and the HLA haplotype [1]–[8], but relevant genes have not been identified [9]. Recent linkage analyses from East Asian families with moyamoya disease demonstrated a linkage of 17q25.3 with the disease at a locus that is −1480 bp from the transcription site of the Raptor gene. However, an allele of this gene was not detected in samples from Caucasian patients with this disease [10].
Vascular endothelial growth factor (VEGF) is involved in vasculogenesis and vascular permeability in various intracranial lesions [11]. In ischemic disease, cerebral angiogenesis is caused by the release of VEGF [12], [13]. VEGF affects vasculogenesis, endothelial cell proliferation and migration, vascular permeability, and stromal degradation through the activation of proteolytic enzymes that are involved in angiogenesis [14], [15]. VEGF binds its receptor tyrosine kinases, VEGF receptor-1 and VEGF receptor-2 (also known as kinase insert domain containing receptor, or KDR) but KDR is the key receptor mediating angiogenesis [16] and is essential for endothelial cell survival and integrity [17].
Although excess VEGF in moyamoya disease has been demonstrated [11], [18] and the association is convincing, the specific role for VEGF remains unclear. Therefore, we studied the relationship of VEGF and KDR polymorphisms and moyamoya disease. VEGF is a major angiogenic factor and a prime regulator of endothelial cell proliferation [19]. The gene that encodes VEGF is located on chromosome 6 and is comprised of a 14-kb coding region with eight exons and seven introns [20]. VEGF is activated transcriptionally and posttranscriptionally by hypoxia in tumor necrosis and in various models of ischemia [21], [22]. Ischemia stimulates VEGF expression in the brain suggesting that it may be important for the vascular response to cerebral ischemia [23], [24], [25]. Several single nucleotide polymorphisms (SNPs) have been described in the VEGF gene (National Center for Biotechnology Information, Gene association no: NT 007592). The VEGF gene includes at least 4 relatively common polymorphisms that may influence VEGF expression: −2578C>A (rs699947), −1154G>A (rs1570360), −634G>C (rs2010963), and 936C>T (rs3025039) [26], [27], [28].
Three of these polymorphisms are located in the promoter region at −2578, −1154, and −634 relative to the translation start site. The −2578A, −1154A, and −634G alleles are all associated with decreased VEGF expression [26], [27]. In addition to promoter region polymorphisms, the T allele of the common 936C>T polymorphism in the 3′-untranslated region is also associated with significantly decreased serum VEGF levels [28]. Recently, several SNPs of the VEGF gene have been associated with cancer risk and prognosis [29], as well as coronary arterial disease [30]. Moreover, KDR −604C, 1192A, and 1719A alleles of chromosome 4 were associated with decreased VEGF binding activity and coronary artery disease [31]. These results indicate the importance of VEGF-KDR signaling in human disease.
To our knowledge, no previous study has evaluated both KDR and VEGF polymorphisms with moyamoya disease and collateral vessel formation after surgery. The aim of present study was to evaluate the frequencies of the VEGF −2578C>A, −1154G>A, −634G>C, and 936C>T and KDR −604T>C (rs2071559), 1192G>A (rs2305948), and 1719T>A (rs1870377) polymorphisms in Korean patients with moyamoya disease in an effort to determine the relationship of these polymorphisms with moyamoya disease.
Methods
Subjects
One hundred seven consecutive Korean patients with moyamoya disease [mean age, 20.9±15.9 years; 71 females (66.4%), 36 males (33.6%)] were recruited for this study. Moyamoya disease was defined as the presence of clinical ischemic or hemorrhagic symptoms in combination with vascular lesions in magnetic resonance imaging (MRI) or magnetic resonance angiography (MRA).
The control group was comprised of 243 healthy subjects [mean age, 23.0±16.1 years; 138 female (56.8%); 105 male (43.2%)] from the same geographic region as the moyamoya patients. These age- and sex-matched subjects were recruited from outpatient clinics at Severance Hospital (Seoul, Korea) and CHA Bundang Medical Center (Seongnam, Korea).
Moyamoya disease has a bimodal pattern of incidence so we divided the patients into pediatric (<18 years) and adult (≥18 years) groups. We further divided the moyamoya patients into ischemic or hemorrhagic groups based on clinical and MRI findings. We performed indirect bypass surgery in 64 patients and direct superficial temporal artery to middle cerebral artery bypass plus encephalo-duro-arterio-myo-synangiosis (STA-MCA plus EDAMS) in one patient. We graded newly developed collateral vessels according to the method of Matsushima et al. [32]. Briefly, Grade A represented synangiosis-induced filling of greater than two-thirds of MCA circulation, Grade B represented between one-third and two-thirds, and Grade C represented less than one-third. We further divided the 64 indirect bypass surgical patients by collateral vessel formation after 2 years into good (collateral grade A) and poor (collateral grade B, C) using MRA. Table 1 shows the demographic characteristics of the moyamoya patients and control subjects.
Table 1. Demographic characteristics between controls and moyamoya patients.
| Characteristic | Control (n = 243) | Moyamoya(n = 107) | P * | Ischemicmoyamoya(n = 92) | Hemorrhagic moyamoya(n = 15) | P * |
| Number of subjects | ||||||
| <18 years | 102 (42.0) | 56 (52.3) | 54 (58.7) | 2 (13.3) | ||
| ≥18 years | 141 (58.0) | 51 (47.7) | 38 (41.3) | 13 (86.7) | ||
| Age (means±SD) | ||||||
| <18 years | 7.71±4.05 | 7.98±4.13 | 0.920 | 8.11±4.12 | 4.50±3.54 | NA |
| ≥18 years | 36.72±10.05 | 34.98±11.29 | 0.248 | 34.63±11.45 | 36.00±11.21 | 0.689 |
| Sex [male, n(%)] | ||||||
| <18 years | 54 (52.9) | 22 (39.3) | 0.100 | 21 (38.9) | 1 (50.0) | 1.000† |
| ≥18 years | 51 (21.0) | 14 (27.5) | 0.260 | 12 (31.6) | 2 (15.4) | 0.472† |
| Collateral vessel formation score (n = 64) | ||||||
| 0 | – | 2 | ||||
| 1 | – | 22 | ||||
| 2 | – | 40 |
P values were calculated using the Mann-Whitney test for continuous data and χ2-test for categorical data.
Fisher's exact test. NA; not applicable.
All participants gave informed written consent prior to enrollment in the study. The institutional review boards of Severance Hospital (4-2008-0308) and CHA Bundang Medical Center (PBC09-103) approved this study.
VEGF Genotyping
We investigated four relevant single nucleotide polymorphism (SNP) candidates in the VEGF gene. We used the G-DEX blood extraction kit (iNtRON Biotechnology, Inc., Seongnam, South Korea) according to the manufacturer’s instructions for DNA extraction. We obtained all SNP sequences from the HapMap database (www.hapmap.org) [33]. The VEGF -2578C>A, -1154G>A, -634G>C, and 936C>T polymorphisms were analyzed by polymerase chain reaction–restriction fragment length polymorphism (PCR-RFLP) method.
We used following primers: VEGF −2578C>A polymorphism, forward 5′-GGA TGG GGC TGA CTA GGT AAG-3′ and reverse 5′-AGC CCC CTT TTC CTC CAA C-3′, that amplifies a 308 bp (C allele) or 326 bp (A allele) product; VEGF −1154G>A polymorphism, forward 5′-CGC GTG TCT CTG GAC AGA GTT TCC-3′ and reverse 5′-CGG GGA CAG GCG AGC TTC AG-3′, that amplifies a 173 bp product; VEGF −634G>C polymorphism, forward 5′-CAG GTC ACT CAC TTT GCC CCG GTC-3′ and reverse 5′-GCT TGC CAT TCC CCA CTT GAA TCG-3′, that amplifies a 204 bp product; and VEGF 936C>T polymorphism, forward 5′-AAG GAA GAG GAG ACT CTG CGC AGA GC-3′ and reverse 5′-TAA ATG TAT GTA TGT GGG TGG GTG TGT CTA CAG G-3′, that amplifies a 208 bp fragment.
The VEGF −2578C>A and −634G>C polymorphisms were identified by digesting the PCR product with the restriction endonuclease AvaII (New England Biolabs, Beverly, MA, USA). The VEGF −1154G>A polymorphism was identified by digesting the PCR product with the restriction endonuclease MnlI (New England Biolabs). The VEGF 936C>T polymorphism was identified by digesting the PCR product with the restriction endonuclease NlaIII (New England Biolabs). All restriction digests were performed at 37°C for 16 h.
KDR Genotyping
We identified 3 well-known SNPs in the KDR gene, including one in the promoter region (−604) and two in the coding region (1192 and 1719). All SNP sequences were obtained from the HapMap database (http://www.hapmap.org) [33]. We used previously described primers and PCR-RFLP conditions for KDR polymorphism analyses [31]. For SNP −604, two DNA fragments (174 bp and 116 bp) were observed for the C allele and one band (290 bp) was produced for the T allele. For the 1192 polymorphism, two DNA fragments (30 bp and 232 bp) indicated the A allele and one band (252 bp) was produced for the G allele. For SNP 1719, two DNA fragments (191 bp and 213 bp) were observed for the A allele and one band (404 bp) was produced for the T allele. The genotyping reproducibility was confirmed by bi-directional sequencing of 400 randomly selected samples.
Measurement of Plasma Total Homocysteine (tHcy), Folic Acid (FA), and Vitamin B12 (VB12)
Blood was collected from moyamoya patients into a tube containing anticoagulant 12 hours after a meal. The tube was centrifuged for 15 min at 1000 × g, and the plasma was separated. The concentration of tHcy (n = 37, 10.18±2.81 µmol/L) in the plasma was measured by fluorescent polarizing immunoassay (FPIA) with IMx (Abbott Laboratories, Chicago, IL, USA). The plasma concentration of FA (n = 30, 10.25±3.50 ng/ml) and VB12 (n = 35, 848.06±319.22 pg/ml) was determined using a radioassay kit (ACS 180; Bayer, Tarrytown, NY, USA).
Measurement of Whole Blood Nitric Oxide (NO)
Blood was collected from moyamoya patients into a tube containing anticoagulant 12 hours after a meal. NO production was evaluated by determining the circulating levels of nitrosyl-hemoglobin complexes. In the present study, the paramagnetic properties of nitrosyl-heme adducts were used to detect whole blood NO-hemoglobin derivatives (n = 33, 5.30±6.93 arbitrary unit [AU]) by electron paramagnetic resonance (EPR) spectroscopy.
Statistical Analyses
To analyze the demographic characteristics of moyamoya disease, we used the Mann–Whitney and chi-square (χ2) tests for continuous and categorical data, respectively. The associations among pediatric and adult patients were estimated by computing the odds ratios (ORs) and 95% confidence intervals (CIs) using Fisher’s exact test. The adjusted odds ratios (AORs) for VEGF and KDR polymorphisms were calculated using multiple logistic regression analyses using gender and age. The genotype distribution of each polymorphism was expected under Hardy-Weinberg equilibrium. Statistical analyses were performed using GraphPad Prism 4.0 (GraphPad Software, Inc., San Diego, CA, USA) and StatsDirect software (version 2.4.4; StatsDirect Ltd., Altrincham, UK). Haplotype analyses were performed using HAPSTAT (version 3.0; University of North Carolina, Chapel Hill, NC, USA) and Haploview 4.2 (Broad Institute, Cambridge, MA, USA). StatsDirect Statistical Software (Version 2.4.4; StatsDirect Ltd, Altrincham, UK) was used to calculate the adjusted OR and 95% CI. The linkage disequilibrium between loci was measured using the absolute value of Lewontin’s D [34].
Results
Figure 1 shows the linkage disequilibrium of VEGF and KDR polymorphisms from the present study. A comparison of genotype frequencies between moyamoya patients and control subjects of the VEGF −2578C>A, −1154G>A, −634G>C, and 936C>T polymorphisms and the KDR −604T>C, 1192G>A, and 1719T>A polymorphisms is shown in Table 2. There were no statistically significant differences between moyamoya patients and controls in any of the polymorphisms evaluated (Tables 2 and 3).
Figure 1. Linkage disequilibrium(LD) of VEGF and KDR polymorphisms.
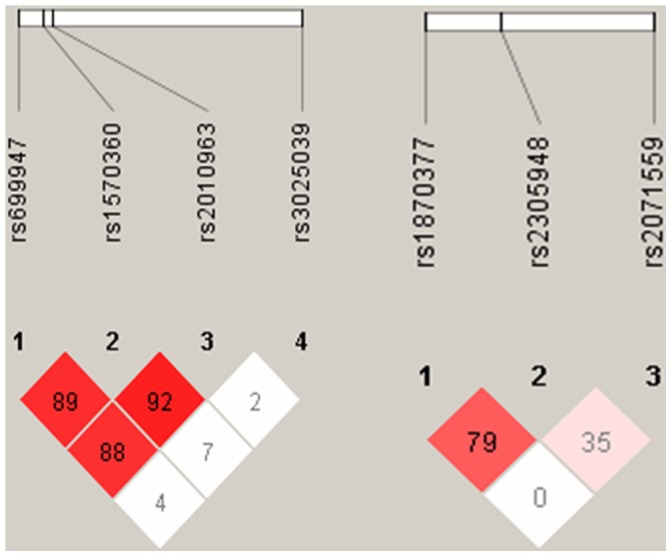
There was strong LD between −1154G>A (rs 1570460) and −634G>C (rs2010963) (D′ = 0.92), −2578C>A (rs 699947)and 1154G>A (rs1570360) (D′ = 0.89). There was strong LD between 1719T>A(rs1870377) and 1192G>A (rs2305948) (D′ = 0.79).
Table 2. The frequencies of the VEGF −2578C>A, −1154G>A, −634G>C, and 936C>T polymorphisms between control subjects and patients with moyamoya disease.
| Characteristics | Controls (n = 243, %) | Moyamoya patients (n = 107, %) | AOR (95% CI) | P * |
| VEGF −2578C>A | ||||
| CC | 128 (52.7) | 62 (57.9) | 1.000 (reference) | |
| CA | 99 (40.7) | 36 (33.6) | 0.769 (0.470–1.258) | 0.296 |
| AA | 16 (6.6) | 9 (8.4) | 1.188 (0.491–2.873) | 0.702 |
| Dominant (CC vs. CA+AA) | 0.827 (0.520–1.315) | 0.422 | ||
| Recessive (CC+CA vs. AA) | 1.296 (0.547–3.072) | 0.556 | ||
| VEGF–1154G>A | ||||
| GG | 173 (71.2) | 80 (74.8) | 1.000 (reference) | |
| GA | 62 (25.5) | 25 (23.4) | 0.873 (0.510–1.496) | 0.622 |
| AA | 8 (3.3) | 2 (1.9) | 0.517 (0.106–2.519) | 0.414 |
| Dominant (GG vs. GA+AA) | 0.834 (0.495–1.405) | 0.495 | ||
| Recessive (GG+GA vs. AA) | 0.546 (0.113–2.653) | 0.454 | ||
| VEGF −634G>C | ||||
| GG | 90 (37.0) | 37 (34.6) | 1.000 (reference) | |
| GC | 103(42.4) | 48 (44.9) | 1.127 (0.666–1.906) | 0.655 |
| CC | 50 (20.6) | 22 (20.6) | 1.083 (0.575–2.042) | 0.804 |
| Dominant (GG vs. GC+CC) | 1.124 (0.695–1.817) | 0.634 | ||
| Recessive (GG+GC vs. CC) | 1.024 (0.579–1.811) | 0.934 | ||
| VEGF 936C>T | ||||
| CC | 172 (70.8) | 67 (62.6) | 1.000 (reference) | |
| CT | 64 (26.3) | 37 (34.6) | 1.530 (0.926–2.529) | 0.097 |
| TT | 7 (2.9) | 3 (2.8) | 1.230 (0.301–5.027) | 0.773 |
| Dominant (CC vs. CT+TT) | 1.484 (0.912–2.416) | 0.112 | ||
| Recessive (CC+CT vs. TT) | 1.079 (0.270–4.313) | 0.915 | ||
Adjusted by age and gender. AOR, adjusted odds ratio. CI, confidence interval.
Table 3. The frequencies of the KDR −604T>C, 1192G>A, and 1719T>A polymorphisms between control subjects and patients with moyamoya disease.
| Characteristics | Controls (n = 243, %) | Moyamoya patients (n = 107, %) | AOR (95% CI) | P * |
| KDR −604T>C | ||||
| TT | 113 (46.5) | 47 (43.9) | 1.000 (reference) | |
| TC | 108 (44.4) | 50 (46.7) | 1.131 (0.698–1.833) | 0.617 |
| CC | 22 (9.1) | 10 (9.3) | 1.102 (0.482–2.517) | 0.818 |
| Dominant (CC vs. CA+AA) | 1.128 (0.710–1.792) | 0.609 | ||
| Recessive (CC+CA vs. AA) | 1.039 (0.470–2.296) | 0.925 | ||
| KDR 1192G>A | ||||
| GG | 186 (76.5) | 80 (74.8) | 1.000 (reference) | |
| GA | 52 (21.4) | 22 (20.6) | 0.994 (0.563–1.754) | 0.983 |
| AA | 5 (2.1) | 5 (4.7) | 2.140 (0.596–7.693) | 0.244 |
| Dominant (GG vs. GA+AA) | 1.101 (0.646–1.875) | 0.725 | ||
| Recessive (GG+GA vs. AA) | 2.139 (0.597–7.665) | 0.243 | ||
| KDR 1719T>A | ||||
| TT | 46 (18.9) | 26 (24.3) | 1.000 (reference) | |
| TA | 114 (46.9) | 45 (42.1) | 0.764 (0.417–1.401) | 0.384 |
| AA | 83 (34.2) | 36 (33.6) | 0.799 (0.427–1.495) | 0.482 |
| Dominant (TT vs. TA+AA) | 0.775 (0.446–1.346) | 0.365 | ||
| Recessive (TT+TA vs. AA) | 0.997 (0.613–1.620) | 0.989 | ||
Adjusted by age and gender. AOR, adjusted odds ratio. CI, confidence interval.
In subgroup analyses (Tables 4 and 5), the CC genotype of the VEGF −634 was less frequent in pediatric moyamoya disease (p = 0.040; CC vs. GG) and comparison with the GG+GC genotype was also significantly different in pediatric moyamoya patients. In the adult subgroup, the VEGF −634CC genotype was more frequent in moyamoya disease (p = 0.024; CC vs. GG). The frequencies of the KDR polymorphisms in both the pediatric and adult subgroups were not significantly different.
Table 4. The frequency of the VEGF polymorphisms according to age.
| Age <18 years | Age ≥18 years | |||||||
| Characteristics | Controls (n = 102, %) | Moyamoya (n = 56, %) | AOR (95% CI) | P * | Controls (n = 141, %) | Moyamoya (n = 51, %) | AOR (95% CI) | P * |
| VEGF −2578C>A | ||||||||
| CC | 57 (55.9) | 29 (51.8) | 1.000 (reference) | 71 (50.4) | 33 (64.7) | 1.000 (reference) | ||
| CA | 42 (41.2) | 22 (39.3) | 1.100 (0.548–2.206) | 0.789 | 57 (40.4) | 14 (27.5) | 0.539 (0.261–1.113) | 0.095 |
| AA | 3 (2.9) | 5 (8.9) | 3.191 (0.696–14.631) | 0.135 | 13 (9.2) | 4 (7.8) | 0.669 (0.200–2.242) | 0.514 |
| Dominant (CC vs. CA+AA) | 1.246 (0.638–2.433) | 0.519 | 0.543 (0.277–1.063) | 0.075 | ||||
| Recessive (CC+CA vs. AA) | 3.044 (0.686–13.499) | 0.143 | 0.777 (0.239–2.525) | 0.674 | ||||
| VEGF −1154G>A | ||||||||
| GG | 77 (75.5) | 44 (78.6) | 1.000 (reference) | 109 (77.3) | 36 (70.6) | 1.000 (reference) | ||
| GA | 22 (21.6) | 9 (16.1) | 1.614 (0.755–3.449) | 0.217 | 30 (21.3) | 13 (25.5) | 0.391 (0.160–0.957) | 0.040 |
| AA | 3 (2.9) | 3 (5.4) | 0.695 (0.067–7.200) | 0.760 | 2 (1.4) | 2 (3.9) | 0.445 (0.049–4.029) | 0.472 |
| Dominant (GG vs. GA+AA) | 1.522 (0.729–3.178) | 0.264 | 0.395 (0.169–0.923) | 0.032 | ||||
| Recessive (GG+GA vs. AA) | 0.608 (0.061–6.044) | 0.671 | 0.517 (0.058–4.589) | 0.554 | ||||
| VEGF −634G>C | ||||||||
| GG | 34 (33.3) | 24 (42.9) | 1.000 (reference) | 56 (39.7) | 13 (25.5) | 1.000 (reference) | ||
| GC | 46 (45.1) | 27 (48.2) | 0.813 (0.398–1.664) | 0.571 | 57 (40.4) | 21 (41.2) | 1.715 (0.771–3.816) | 0.186 |
| CC | 22 (21.6) | 5 (8.9) | 0.301 (0.096–0.946) | 0.040 | 28 (19.9) | 17 (33.3) | 2.698 (1.137–6.403) | 0.024 |
| Dominant (GG vs. GC+CC) | 0.644 (0.236–1.273) | 0.206 | 2.016 (0.979–4.152) | 0.057 | ||||
| Recessive (GG+GC vs. CC) | 0.334 (0.117–0.955) | 0.041 | 2.087 (1.014–4.295) | 0.046 | ||||
| VEGF 936C>T | ||||||||
| CC | 76 (74.5) | 38 (67.9) | 1.000 (reference) | 96 (68.1) | 31 (60.8) | 1.000 (reference) | ||
| CT | 24 (23.5) | 18 (32.1) | 1.580 (0.756–3.303) | 0.224 | 41 (29.1) | 17 (33.3) | 1.322 (0.652–2.681) | 0.438 |
| TT | 2 (2.0) | 0 (0.0) | NA | NA | 4 (2.8) | 3 (5.9) | 2.562 (0.525–12.506) | 0.245 |
| Dominant (CC vs. CT+TT) | 1.405 (0.681–2.900) | 0.357 | 1.378 (0.702–2.702) | 0.351 | ||||
| Recessive (CC+CT vs. TT) | 2.207 (0.472–10.328) | 0.315 | ||||||
Adjusted by age and gender. NA; not applicable. AOR, adjusted odds ratio. CI, confidence interval.
Table 5. The frequency of the KDR polymorphisms according to age.
| Age <18 years | Age ≥18 years | ||||||||
| Characteristics | Controls (n = 102, %) | Moyamoya (n = 56, %) | AOR (95% CI) | P * | Controls (n = 141, %) | Moyamoya (n = 51, %) | AOR (95% CI) | P * | |
| KDR −604T>C | |||||||||
| CC | 54 (52.9) | 25 (44.6) | 1.000 (reference) | 59 (41.8) | 22 (43.1) | 1.000 (reference) | |||
| CA | 40 (39.2) | 27 (48.2) | 1.413(0.711–2.808) | 0.324 | 68 (48.2) | 23 (45.1) | 0.929 (0.467–1.847) | 0.833 | |
| AA | 8 (7.8) | 4 (7.1) | 1.096(0.298–4.031) | 0.890 | 14 (9.9) | 6 (11.8) | 1.141 (0.388–3.356) | 0.811 | |
| Dominant (CC vs. CA+AA) | 1.350(0.697–2.616) | 0.374 | 0.951 (0.495–1.826) | 0.879 | |||||
| Recessive (CC+CA vs. AA) | 0.889(0.251–3.143) | 0.855 | 1.198 (0.433–3.320) | 0.728 | |||||
| KDR 1192G>A | |||||||||
| GG | 20 (19.6) | 16 (28.6) | 1.000 (reference) | 26 (18.4) | 10 (19.6) | 1.000 (reference) | |||
| GA | 47 (46.1) | 22 (39.3) | 0.572 (0.246–1.323) | 0.193 | 67 (47.5) | 23 (45.1) | 1.082 (0.440–2.664) | 0.864 | |
| AA | 35 (34.2) | 18 (32.1) | 0.654 (0.232–1.572) | 0.342 | 48 (34.0) | 18 (35.3) | 0.965 (0.382–2.435) | 0.940 | |
| Dominant (GG vs. GA+AA) | 0.611 (0.284–1.313) | 0.207 | 1.003 (0.440–2.286) | 0.994 | |||||
| Recessive (GG+GA vs. AA) | 0.927 (0.460–1.869) | 0.833 | 1.069 (0.543–2.103) | 0.848 | |||||
| KDR 1719T>A | |||||||||
| TT | 77 (75.5) | 44 (78.6) | 1.000 (reference) | 109 (77.3) | 36 (70.6) | 1.000 (reference) | |||
| TA | 22 (21.6) | 9 (16.1) | 0.739 (0.310–1.762) | 0.495 | 30 (21.3) | 13 (25.5) | 1.283 (0.602–2.734) | 0.518 | |
| AA | 3 (2.9) | 3 (5.4) | 1.848 (0.349–9.773) | 0.470 | 2 (1.4) | 2 (3.9) | 3.003 (0.403–22.390) | 0.283 | |
| Dominant (TT vs. TA+AA) | 0.877 (0.398–1.934) | 0.746 | 1.378 (0.667–2.846) | 0.386 | |||||
| Recessive (TT+TA vs. AA) | 2.026 (0.382–10.736) | 0.407 | 2.675 (0.362–19.768) | 0.335 | |||||
Adjusted by age and gender. NA; not applicable. AOR, adjusted odds ratio. CI, confidence interval.
We conducted haplotype analyses of the VEGF and KDR polymorphisms (Table 6). The C-G-C-C haplotype (VEGF −2578/−1154/−634/936) in pediatric moyamoya patients was significantly different. In addition, the C-A-T haplotype (KDR −604/1192/1719) increased the risk of pediatric moyamoya. In adult moyamoya, the A-A-G-C haplotype (VEGF −2578/−1154/−634) was significantly different whereas other haplotypes were not. We have estimated and provided several haplotype frequencies in Table 6.
Table 6. Haplotype frequencies of the VEGF and KDR polymorphisms according to age.
| Age <18 years | Age ≥18 years | |||||||
| Characteristics | Controls (n = 102, %) | Moyamoya (n = 56, %) | OR (95% CI) | P * | Controls (n = 141, %) | Moyamoya (n = 51, %) | OR (95% CI) | P * |
| VEGF −2578/−1154/−634/936 | ||||||||
| CGCC | 0.3502 | 0.2331 | 0.566 (0.335–0.957) | 0.041 | 0.3251 | 0.4350 | 1.594 (1.001–2.540) | 0.053 |
| CGGC | 0.2894 | 0.3561 | 1.365 (0.836–2.231) | 0.254 | 0.2360 | 0.2047 | 0.842 (0.484–1.465) | 0.584 |
| AAGC | 0.1234 | 0.1361 | 1.107 (0.557–2.199) | 0.860 | 0.1471 | 0.0406 | 0.250 (0.087–0.718) | 0.006 |
| CGCT | 0.0910 | 0.0578 | 0.551 (0.213–1.423) | 0.277 | 0.0708 | 0.0741 | 0.976 (0.400–2.382) | 1.000 |
| AGGC | 0.0840 | 0.0648 | 0.733 (0.295–1.826) | 0.658 | 0.1085 | 0.0587 | 0.511 (0.207–1.265) | 0.171 |
| CGGT | 0.0233 | 0.0372 | 1.474 (0.388–5.606) | 0.726 | 0.0656 | 0.0620 | 0.926 (0.357–2.403) | 1.000 |
| AGGT | 0.0152 | 0.0532 | 3.792 (0.930–15.470) | 0.073 | 0.0189 | 0.0310 | 1.696 (0.398–7.231) | 0.440 |
| CAGC | 0.0109 | 0.0221 | 1.836 (0.255–13.220) | 0.617 | NA | NA | NA | NA |
| AAGT | NA | NA | NA | NA | 0.0149 | 0.0368 | 2.866 (0.703–11.680) | 0.216 |
| KDR −604/1192/1719 | ||||||||
| TGA | 0.4141 | 0.3786 | 0.857 (0.534–1.376) | 0.550 | 0.3624 | 0.3736 | 1.048 (0.655–1.675) | 0.845 |
| TGT | 0.2187 | 0.2688 | 1.293 (0.758–2.204) | 0.407 | 0.2551 | 0.2337 | 0.897 (0.528–1.525) | 0.689 |
| CGA | 0.1325 | 0.1393 | 1.093 (0.561–2.128) | 0.864 | 0.1990 | 0.1919 | 0.984 (0.557–1.740) | 0.957 |
| CGT | 0.0974 | 0.0794 | 0.804 (0.353–1.831) | 0.687 | 0.0630 | 0.0341 | 0.444 (0.128–1.542) | 0.190 |
| TAT | 0.0773 | 0.0402 | 0.549 (0.196–1.541) | 0.346 | 0.0421 | 0.0495 | 1.160 (0.398–3.378) | 0.786 |
| CAT | 0.0330 | 0.0938 | 3.065 (1.153–8.148) | 0.024 | 0.0618 | 0.1042 | 1.884 (0.851–4.173) | 0.113 |
P-values after 10,000 permutation test. NA; not applicable. OR, odds ratio. CI, confidence interval.
We performed indirect bypass surgery in 64 patients. The genotypes containing the VEGF −634C allele had better collateral vessel formation after surgery whereas −634GG was associated with poor collateral grade (Table 7). We also investigated the haplotype frequency differences related to collateral grades. However, there were no statistical differences (Table S1). Our results potentially implicate the VEGF −634G>C polymorphism in the development of collateral vessel formation in moyamoya disease. The other VEGF and KDR polymorphisms we studied did not exhibit statistically significant differences in collateral vessel formation.
Table 7. Comparison of the VEGF and KDR genotype frequencies according to collateral vessel formation score.
| Characteristics | Collateral grade A (n = 40) | Collateral grade B and C (n = 24) | AOR (95% CI) | P * |
| VEGF −2578C>A | ||||
| CC | 20 (50.0) | 13 (54.2) | 1.000 (reference) | |
| CA | 18 (45.0) | 7 (29.2) | 0.615 (0.200–1.893) | 0.397 |
| AA | 2 (5.0) | 4 (16.7) | 3.060 (0.473–19.816) | 0.241 |
| Dominant (CC vs. CA+AA) | 0.867 (0.313–2.408) | 0.785 | ||
| Recessive (CC+CA vs. AA) | 3.757 (0.625–22.590) | 0.148 | ||
| VEGF −1154G>A | ||||
| GG | 27 (67.5) | 17 (70.8) | 1.000 (reference) | |
| GA | 12 (30.0) | 6 (25.0) | 0.841 (0.253–2.796) | 0.777 |
| AA | 1 (2.5) | 1 (4.2) | 1.475 (0.082–26.410) | 0.792 |
| Dominant (GG vs. GA+AA) | 0.926 (0.297–2.886) | 0.895 | ||
| Recessive (GG+GA vs. AA) | 1.565 (0.090–27.244) | 0.759 | ||
| VEGF −634G>C | ||||
| GG | 11 (27.5) | 13 (54.2) | 1.000 (reference) | |
| GC | 22 (55.0) | 7 (29.2) | 0.252 (0.076–0.837) | 0.024 |
| CC | 7 (17.5) | 4 (16.7) | 0.284 (0.045–1.780) | 0.179 |
| Dominant (GG vs. GC+CC) | 0.286 (0.095–0.859) | 0.026 | ||
| Recessive (GG+GC vs. CC) | 0.857 (0.215–3.412) | 0.827 | ||
| VEGF 936C>T | ||||
| CC | 27 (67.5) | 16 (66.7) | 1.000 (reference) | |
| CT | 13 (32.5) | 8 (33.3) | 1.069 (0.357–3.206) | 0.905 |
| TT | 0 (0.0) | 0 (0.0) | NA | NA |
| Dominant (CC vs. CT+TT) | 1.069 (0.357–3.206) | 0.905 | ||
| Recessive (CC+CT vs. TT) | NA | NA | ||
| KDR −604T>C | ||||
| TT | 17 (42.5) | 11 (45.8) | 1.000 (reference) | |
| TC | 19 (47.5) | 12 (50.0) | 0.950 (0.325–2.778) | 0.926 |
| CC | 4 (10.0) | 1 (4.2) | 0.370 (0.035–3.962) | 0.411 |
| Dominant (TT vs. TC+CC) | 0.840 (0.298–2.367) | 0.741 | ||
| Recessive (TT+TC vs. CC) | 0.346 (0.035–3.409) | 0.363 | ||
| KDR 1192G>A | ||||
| GG | 30 (75.0) | 16 (66.7) | 1.000 (reference) | |
| GA | 8 (20.0) | 7 (29.2) | 1.381 (0.401–4.758) | 0.609 |
| AA | 2 (5.0) | 1 (4.2) | 0.843 (0.063–11.263) | 0.897 |
| Dominant (GG vs. GA+AA) | 1.355 (0.420–4.375) | 0.612 | ||
| Recessive (GG+GA vs. AA) | 0.664 (0.053–8.390) | 0.752 | ||
| KDR 1719T>A | ||||
| TT | 10 (25.0) | 7 (29.2) | 1.000 (reference) | |
| TA | 18 (45.0) | 9 (37.5) | 0.786 (0.219–2.824) | 0.712 |
| AA | 12 (30.0) | 8 (33.3) | 0.886 (0.227–3.462) | 0.861 |
| Dominant (TT vs. TA+AA) | 0.825 (0.259–2.626) | 0.745 | ||
| Recessive (TT+TA vs. AA) | 1.195 (0.398–3.593) | 0.751 | ||
Adjusted by age and gender. NA; not applicable. AOR, adjusted odds ratio. CI, confidence interval.
To assess the clinical significance of the VEGF and KDR polymorphisms, we surveyed the association between the studied polymorphisms and various vascular risk factors (tHcy, FA, VB12, and NO). Blood tHcy, FA, VB12, and NO are essential for vascular homeostasis regulation [35]. There were no significant distinctions according to VEGF polymorphisms but the KDR −604C (p = 0.017) and 1192A (p = 0.032) alleles were linked to decreased NO levels (Table S2). The association of the KDR −604 and 1192 polymorphisms with NO levels suggests that KDR haplotypes containing −604C or 1192A may adversely affect vascular homeostasis.
Discussion
In this study, we found VEGF or KDR polymorphisms influence moyamoya disease in subgroup analyses as well as the formation of revascularization after bypass surgery. VEGF is involved in vasculogenesis in different intracranial lesions [11], is an endothelial cell mitogen that induces transient vascular leakage, and a potent angiogenic factor [36]. VEGF promotes angiogenesis in cerebral ischemia [11], [37] and causes pathologic vessel formation [12].
Moyamoya disease is characterized by the angiographic findings of arterial stenosis and occlusion of the circle of Willis [38]. It can lead to transient ischemic attacks or a cerebral infarction pattern in juveniles [39] and a hemorrhagic stroke pattern in adults [40], [41]. Takekawa et al. [18] reported increased VEGF expression in autopsy specimens from adults with moyamoya disease and Sakamoto et al. reported that the total meningeal cellularity and VEGF expression in the moyamoya dura was significantly higher in moyamoya patients compared to controls [11]. Although increased VEGF concentrations have been demonstrated in moyamoya disease [11], [18], the specific role of VEGF in moyamoya genetics remains unclear. We therefore reasoned that mutations and genetic polymorphisms of the VEGF gene may cause cerebral ischemia in moyamoya disease.
Vascular endothelial growth factor A (VEGF-A) is a disulfide-bonded dimeric glycoprotein that is a member of a protein family that includes VEGF-B, VEGF-C, VEGF-D, and placental growth factor (PGF). The gene that encodes VEGF-A is located on human chromosome 6 and comprises a 14-kb coding region with eight exons. VEGF cellular signaling activity depends on specific membrane receptors. The receptors include fms like tyrosine kinase-1 (Flt-1 or VEGFR-1), KDR (also known as VEGFR-2) [42], [43], and Flt-4 (also known as VEGFR-3) [44], [45]. VEGF acts on endothelial cells particularly through Flt-1 and KDR [46], [47]. Epidermal growth factor (EGF) arising from hypoxia stimulates Flt-1 expression and inhibits KDR expression [48].
VEGF binding to KDR activates multiple signaling cascades that affect angiogenesis as well as endothelial cell survival, proliferation, and migration. Recently, several SNPs in the VEGF gene have been linked to cancer risk and prognosis [29] and coronary arterial disease [30], indicating the importance of the VEGF-KDR signaling pathway in human disease. Oh et al. reported the SNP 1719T allele conferred ischemic stroke risk in a dose dependent manner [49].
Revascularization after bypass surgery and the formation of new pial vessels may play a role in moyamoya disease. Several studies have suggested that endogenous VEGF production mediates compensatory revascularization during various physiological and pathological processes [37], [50], [51], [52].
In addition, as suggested by the data in our study, VEGF −634 G>C may be a possible prognostic biomarker after bypass surgery. Taken together, we can speculate that VEGF polymorphisms influence moyamoya disease as well as the formation of synangiosis-induced collateral vessel after bypass surgery. Although revascularization with pial synangiosis helps ameliorate ischemic symptoms, some patients progress to cerebral infarction or hemorrhage, even after surgery. This likely reflects the degree of synangiosis and is dependent on the genetic characteristics of the patient. Therefore, VEGF or KDR polymorphisms can be used as prognostic factors after revascularization surgery.
There are some limitations in correlating VEGF or KDR polymorphisms with our clinical findings. Our data should be interpreted with caution because of the relatively small sample size. Patients require long-term follow up to assess clinical outcomes and variation in the clinical characteristics of moyamoya disease makes it difficult to identify specific genes that are associated with the disease. In addition, moyamoya disease is characterized by genetic heterogeneity and complex interactions between genes and other factors. Although there are limitations with sample size and long-term follow-up clinical findings, it is important to determine the relationship of VEGF or KDR polymorphisms with moyamoya disease and collateral vessel formation after surgery.
In summary, no differences in VEGF −2578, −1154, −634, and 936 or KDR −604, 1192, and 1719 polymorphisms were observed between total moyamoya disease patients and control subjects. However, in subgroup analyses, we found that the CC genotype in VEGF −634 occurred less frequently in pediatric patients (p = 0.040) and occurred more often in adult moyamoya patients (p = 0.024). The genotypes including the VEGF −634C allele had better collateral vessel formation after surgery. In addition, the C-G-C-C (VEGF −2578/−1154/−634/936) haplotype and the C-A-T (KDR -604/1192/1719) haplotype in pediatric patients, as well as the A-A-G-C (VEGF −2578/−1154/−634/936) in adult moyamoya patients, had significant differences. Therefore, these results suggest that VEGF or KDR polymorphisms influence moyamoya disease as well as the formation of synangiosis-induced collateral vessel after bypass surgery.
Supporting Information
Haplotype analyses of VEGF and KDR polymorphisms according to collateral score. *P-values after 10,000 permutation test.
(DOC)
Association of VEGF and KDR polymorphisms with vascular risk factors. *P-values between major and minor alleles of each polymorphism analyzed by Mann-Whitney test. AU; arbitrary units.
(DOC)
Acknowledgments
We thank all the people who give the help for this study.
Funding Statement
This work was supported by the National Research Foundation of Korea (2010-0023851). The funders had no role in the study design or decision to publish.
References
- 1. Kang HS, Kim SK, Cho BK, Kim YY, Hwang YS, et al. (2006) Single nucleotide polymorphisms of tissue inhibitor of metalloproteinase genes in familial moyamoya disease. Neurosurgery 58: 1074–1080. [DOI] [PubMed] [Google Scholar]
- 2. Ikeda H, Sasaki T, Yoshimoto T, Fukui M, Arinami T (1999) Mapping of a familial moyamoya disease gene to chromosome 3p24.2–p26. Am J Hum Genet 64: 533–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Inoue TK, Ikezaki K, Sasazuki T, Matsushima T, Fukui M (2000) J Child Neurol. 15: 179–182. [DOI] [PubMed] [Google Scholar]
- 4. Sakurai K, Horiuchi Y, Ikeda H, Ikezaki K, Yoshimoto T, et al. (2004) J Hum Genet. 49: 278–281. [DOI] [PubMed] [Google Scholar]
- 5. Yamauchi T, Tada M, Houkin K, Tanaka T, Nakamura Y, et al. (2000) Linkage of familial moyamoya disease (spontaneous occlusion of the circle of Willis) to chromosome 17q25. Stroke 31: 930–935. [DOI] [PubMed] [Google Scholar]
- 6. Han H, Pyo CW, Yoo DS, Huh PW, Cho KS, et al. (2003) Associations of Moyamoya patients with HLA class I and class II alleles in the Korean population. J Korean Med Sci 18: 876–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mineharu Y, Liu W, Inoue K, Matsuura N, Inoue S, et al. (2008) Autosomal dominant moyamoya disease maps to chromosome 17q25.3. Neurology 70: 2357–2363. [DOI] [PubMed] [Google Scholar]
- 8. Park YS, Min KT, Kim TG, Lee YH, Cheong HJ, et al. (2011) Age-specific eNOS polymorphisms in moyamoya disease. Childs Nerv Syst 27: 1919–1926. [DOI] [PubMed] [Google Scholar]
- 9. Nanba R, Tada M, Kuroda S, Houkin K, Iwasaki Y (2005) Sequence analysis and bioinformatics analysis of chromosome 17q25 in familial moyamoya disease. Childs Nerv Syst 21: 62–68. [DOI] [PubMed] [Google Scholar]
- 10. Liu W, Hashikata H, Inoue K, Matsuura N, Mineharu Y, et al. (2010) A rare Asian founder polymorphism of Raptor may explain the high prevalence of Moyamoya disease among East Asians and its low prevalence among Caucasians. Environ Health Prev Med 15: 94–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sakamoto S, Kiura Y, Yamasaki F, Shibukawa M, Ohba S, et al.. (2008) Expression of vascular endothelial growth factor in dura mater of patients with moyamoya disease. Neurosurg Rev 31: 77–81; discussion 81. [DOI] [PubMed]
- 12. Cao Y, Hong A, Schulten H, Post MJ (2005) Update on therapeutic neovascularization. Cardiovasc Res 65: 639–648. [DOI] [PubMed] [Google Scholar]
- 13. Jin KL, Mao XO, Nagayama T, Goldsmith PC, Greenberg DA (2000) Induction of vascular endothelial growth factor and hypoxia-inducible factor-1alpha by global ischemia in rat brain. Neuroscience 99: 577–585. [DOI] [PubMed] [Google Scholar]
- 14. Mukhopadhyay D, Tsiokas L, Zhou XM, Foster D, Brugge JS, et al. (1995) Hypoxic induction of human vascular endothelial growth factor expression through c-Src activation. Nature 375: 577–581. [DOI] [PubMed] [Google Scholar]
- 15. Ikeda E, Achen MG, Breier G, Risau W (1995) Hypoxia-induced transcriptional activation and increased mRNA stability of vascular endothelial growth factor in C6 glioma cells. J Biol Chem 270: 19761–19766. [DOI] [PubMed] [Google Scholar]
- 16. Waltenberger J, Claesson-Welsh L, Siegbahn A, Shibuya M, Heldin CH (1994) Different signal transduction properties of KDR and Flt1, two receptors for vascular endothelial growth factor. J Biol Chem 269: 26988–26995. [PubMed] [Google Scholar]
- 17. Gerber HP, McMurtrey A, Kowalski J, Yan M, Keyt BA, et al. (1998) Vascular endothelial growth factor regulates endothelial cell survival through the phosphatidylinositol 3'-kinase/Akt signal transduction pathway. Requirement for Flk-1/KDR activation. J Biol Chem 273: 30336–30343. [DOI] [PubMed] [Google Scholar]
- 18. Takekawa Y, Umezawa T, Ueno Y, Sawada T, Kobayashi M (2004) Pathological and immunohistochemical findings of an autopsy case of adult moyamoya disease. Neuropathology 24: 236–242. [DOI] [PubMed] [Google Scholar]
- 19. La Rosa S, Uccella S, Finzi G, Albarello L, Sessa F, et al. (2003) Localization of vascular endothelial growth factor and its receptors in digestive endocrine tumors: correlation with microvessel density and clinicopathologic features. Hum Pathol 34: 18–27. [DOI] [PubMed] [Google Scholar]
- 20. Vincenti V, Cassano C, Rocchi M, Persico G (1996) Assignment of the vascular endothelial growth factor gene to human chromosome 6p21.3. Circulation 93: 1493–1495. [DOI] [PubMed] [Google Scholar]
- 21. Leung DW, Cachianes G, Kuang WJ, Goeddel DV, Ferrara N (1989) Vascular endothelial growth factor is a secreted angiogenic mitogen. Science 246: 1306–1309. [DOI] [PubMed] [Google Scholar]
- 22. Shweiki D, Itin A, Soffer D, Keshet E (1992) Vascular endothelial growth factor induced by hypoxia may mediate hypoxia-initiated angiogenesis. Nature 359: 843–845. [DOI] [PubMed] [Google Scholar]
- 23.Kovacs Z, Ikezaki K, Samoto K, Inamura T, Fukui M (1996) VEGF and flt. Expression time kinetics in rat brain infarct. Stroke 27: 1865–1872; discussion 1872–1863. [DOI] [PubMed]
- 24. Hayashi T, Abe K, Suzuki H, Itoyama Y (1997) Rapid induction of vascular endothelial growth factor gene expression after transient middle cerebral artery occlusion in rats. Stroke 28: 2039–2044. [DOI] [PubMed] [Google Scholar]
- 25. Lennmyr F, Ata KA, Funa K, Olsson Y, Terent A (1998) Expression of vascular endothelial growth factor (VEGF) and its receptors (Flt-1 and Flk-1) following permanent and transient occlusion of the middle cerebral artery in the rat. J Neuropathol Exp Neurol 57: 874–882. [DOI] [PubMed] [Google Scholar]
- 26. Brogan IJ, Khan N, Isaac K, Hutchinson JA, Pravica V, et al. (1999) Novel polymorphisms in the promoter and 5' UTR regions of the human vascular endothelial growth factor gene. Hum Immunol 60: 1245–1249. [DOI] [PubMed] [Google Scholar]
- 27. Awata T, Inoue K, Kurihara S, Ohkubo T, Watanabe M, et al. (2002) A common polymorphism in the 5'-untranslated region of the VEGF gene is associated with diabetic retinopathy in type 2 diabetes. Diabetes 51: 1635–1639. [DOI] [PubMed] [Google Scholar]
- 28. Renner W, Kotschan S, Hoffmann C, Obermayer-Pietsch B, Pilger E (2000) A common 936 C/T mutation in the gene for vascular endothelial growth factor is associated with vascular endothelial growth factor plasma levels. J Vasc Res 37: 443–448. [DOI] [PubMed] [Google Scholar]
- 29. Schneider BP, Wang M, Radovich M, Sledge GW, Badve S, et al. (2008) Association of vascular endothelial growth factor and vascular endothelial growth factor receptor-2 genetic polymorphisms with outcome in a trial of paclitaxel compared with paclitaxel plus bevacizumab in advanced breast cancer: ECOG 2100. J Clin Oncol 26: 4672–4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Petrovic D, Verhovec R, Globocnik Petrovic M, Osredkar J, et al. (2007) Association of vascular endothelial growth factor gene polymorphism with myocardial infarction in patients with type 2 diabetes. Cardiology 107: 291–295. [DOI] [PubMed] [Google Scholar]
- 31. Wang Y, Zheng Y, Zhang W, Yu H, Lou K, et al. (2007) Polymorphisms of KDR gene are associated with coronary heart disease. J Am Coll Cardiol 50: 760–767. [DOI] [PubMed] [Google Scholar]
- 32. Matsushima T, Inoue T, Suzuki SO, Fujii K, Fukui M, et al. (1992) Surgical treatment of moyamoya disease in pediatric patients–comparison between the results of indirect and direct revascularization procedures. Neurosurgery 31: 401–405. [DOI] [PubMed] [Google Scholar]
- 33. Consortium IH (2003) The international HapMap Project. Nature 426: 789–796. [DOI] [PubMed] [Google Scholar]
- 34. Hedrick PW (1987) Gametic disequilibrium measures: proceed with caution. Genetics 117: 331–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fabian E, Kickinger A, Wagner KH, Elmadfa I (2011) Homocysteine and asymmetric dimethylarginine in relation to B vitamins in elderly people. Wien Klin Wochenschr 123: 496–501. [DOI] [PubMed] [Google Scholar]
- 36. Thomas KA (1996) Vascular endothelial growth factor, a potent and selective angiogenic agent. J Biol Chem 271: 603–606. [DOI] [PubMed] [Google Scholar]
- 37. Issa R, Krupinski J, Bujny T, Kumar S, Kaluza J, et al. (1999) Vascular endothelial growth factor and its receptor, KDR, in human brain tissue after ischemic stroke. Lab Invest 79: 417–425. [PubMed] [Google Scholar]
- 38. Suzuki J, Takaku A (1969) Cerebrovascular "Moyamoya" disease. Disease showing abnormal net-like vessels in base of brain. Arch Neurol 20: 288–299. [DOI] [PubMed] [Google Scholar]
- 39. Fukui M (1997) Current state of study on moyamoya disease in Japan. Surg Neurol 47: 138–143. [DOI] [PubMed] [Google Scholar]
- 40. Kuroda S, Hashimoto N, Yoshimoto T, Iwasaki Y (2007) Radiological findings, clinical course, and outcome in asymptomatic moyamoya disease: results of multicenter survey in Japan. Stroke 38: 1430–1435. [DOI] [PubMed] [Google Scholar]
- 41. Mineharu Y, Liu W, Inoue K, Matsuura N, Inoue S, et al. (2008) Autosomal dominant moyamoya disease maps to chromosome 17q25.3. Neurology 70: 2357–2363. [DOI] [PubMed] [Google Scholar]
- 42. Takahashi T, Shibuya M (1997) The 230 kDa mature form of KDR/Flk-1 (VEGF receptor-2) activates the PLC-gamma pathway and partially induces mitotic signals in NIH3T3 fibroblasts. Oncogene 14: 2079–2089. [DOI] [PubMed] [Google Scholar]
- 43. Soker S, Takashima S, Miao HQ, Neufeld G, Klagsbrun M (1998) Neuropilin-1 is expressed by endothelial and tumor cells as an isoform-specific receptor for vascular endothelial growth factor. Cell 92: 735–745. [DOI] [PubMed] [Google Scholar]
- 44. Harry LE, Paleolog EM (2003) From the cradle to the clinic: VEGF in developmental, physiological, and pathological angiogenesis. Birth Defects Res C Embryo Today 69: 363–374. [DOI] [PubMed] [Google Scholar]
- 45. Ferrara N, Gerber HP, LeCouter J (2003) The biology of VEGF and its receptors. Nat Med 9: 669–676. [DOI] [PubMed] [Google Scholar]
- 46. Rahimi N, Dayanir V, Lashkari K (2000) Receptor chimeras indicate that the vascular endothelial growth factor receptor-1 (VEGFR-1) modulates mitogenic activity of VEGFR-2 in endothelial cells. J Biol Chem 275: 16986–16992. [DOI] [PubMed] [Google Scholar]
- 47. Marchand GS, Noiseux N, Tanguay JF, Sirois MG (2002) Blockade of in vivo VEGF-mediated angiogenesis by antisense gene therapy: role of Flk-1 and Flt-1 receptors. Am J Physiol Heart Circ Physiol 282: H194–204. [DOI] [PubMed] [Google Scholar]
- 48. Okuda Y, Tsurumaru K, Suzuki S, Miyauchi T, Asano M, et al. (1998) Hypoxia and endothelin-1 induce VEGF production in human vascular smooth muscle cells. Life Sci 63: 477–484. [DOI] [PubMed] [Google Scholar]
- 49. Oh SH, Min KT, Jeon YJ, Kim MH, Moon JS, et al. (2011) Association between kinase insert domain-containing receptor gene polymorphism and haplotypes and ischemic stroke. J Neurol Sci 308: 62–66. [DOI] [PubMed] [Google Scholar]
- 50. Marti HJ, Bernaudin M, Bellail A, Schoch H, Euler M, et al. (2000) Hypoxia-induced vascular endothelial growth factor expression precedes neovascularization after cerebral ischemia. Am J Pathol 156: 965–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kuo NT, Benhayon D, Przybylski RJ, Martin RJ, LaManna JC (1999) Prolonged hypoxia increases vascular endothelial growth factor mRNA and protein in adult mouse brain. J Appl Physiol 86: 260–264. [DOI] [PubMed] [Google Scholar]
- 52. Dvorak HF, Brown LF, Detmar M, Dvorak AM (1995) Vascular permeability factor/vascular endothelial growth factor, microvascular hyperpermeability, and angiogenesis. Am J Pathol 146: 1029–1039. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Haplotype analyses of VEGF and KDR polymorphisms according to collateral score. *P-values after 10,000 permutation test.
(DOC)
Association of VEGF and KDR polymorphisms with vascular risk factors. *P-values between major and minor alleles of each polymorphism analyzed by Mann-Whitney test. AU; arbitrary units.
(DOC)


