Abstract
Autophagy is a tightly regulated lysosomal self-digestion process that can both promote and impede tumorigenesis. Here, we utilize a three-dimensional (3D) culture model to address how interactions between autophagy and the PI3K/Akt/mTOR pathway impact the malignant behavior of cells carrying a tumor-derived, activating mutation in PI3K (PI3K-H1047R). In this model, autophagy simultaneously mediates tumor suppressive and promoting functions within individual glandular structures. In 3D culture, constitutive PI3K activation overcomes proliferation arrest and promotes resistance to anoikis in the luminal space, resulting in aberrant structures with filled lumen. Inhibiting autophagy in PI3K-H1047R structures triggers luminal cell apoptosis, resulting in lumen clearance. At the same time, ATG depletion strongly enhances PI3K-H1047R cell proliferation during 3D morphogenesis, revealing an unexpected role for autophagy in restricting proliferation driven by PI3K activation. Intriguingly, over-expression of the autophagy cargo receptor p62/SQSTM1 in PI3K-H1047R cells is sufficient to enhance cell proliferation, activate the ERK/MAPK pathway, and to promote EGF-independent proliferation in 3D culture. Overall, these results indicate that autophagy antagonizes specific aspects of oncogenic PI3K transformation, with the loss of autophagy promoting proliferation.
Keywords: Autophagy, oncogenic PI3K, proliferation, 3D culture
INTRODUCTION
Autophagy is an evolutionarily conserved lysosomal degradation process that serves as an important mechanism for protein turnover and organelle homeostasis (1). Autophagy is activated in response to multiple stresses relevant for cancer progression, including nutrient starvation, hypoxia, the unfolded protein response (ER stress), and extracellular matrix detachment as well as upon treatment with a wide spectrum of cytotoxic and targeted chemotherapeutic agents (2). Because autophagy commonly functions as a survival and fitness mechanism in response to these diverse stresses, significant interest has emerged in inhibiting autophagy as a therapeutic strategy to kill or prevent the expansion of tumor cells. Notably, the lysosomal inhibitor hydroxychloroquine which inhibits the final maturation steps during autophagy, has gained special attention as an anti-cancer agent because of its long history of use as an anti-malarial agent and in diseases such as rheumatoid arthritis; as a result, multiple clinical trials using hydroxychloroquine as a sensitizing agent in combination with standard cancer therapies are under evaluation in different cancers (3).
Despite this enthusiasm for autophagy inhibition as a potential anti-cancer strategy, one cannot ignore genetic evidence supporting that autophagy can exert important tumor suppressor functions. The ability of autophagy to prevent tumorigenesis was initially broached through studies of Beclin1/ATG6, which is allelically deleted in a large number of human breast, ovarian and prostate cancers (4). Moreover, mice heterozygous for beclin1 show increased rates of spontaneous tumor development (5, 6). More recently, mice with systemic mosaic deletion of atg5 and liver-specific atg7−/− mice were found to develop hyperproliferative liver adenomas, further corroborating a genetic role for autophagy as a suppressor of spontaneous tumorigenesis (7, 8). In addition, studies demonstrate that autophagy serves as an important mechanism for tumor suppression; defects in autophagy lead to activation of DNA damage responses, genomic instability, and gene amplification (9, 10). Importantly, ATG deficiency elicits high levels of tumor cell proliferation in these animal models, suggesting that autophagy plays an important role in proliferative suppression in vivo. However, the exact mechanisms through which ATG deficiency causes an increase in cell proliferation remain unclear. Furthermore, it remains uncertain whether autophagy similarly augments cell proliferation downstream of oncogene activation. Here, utilizing a threedimensional (3D) culture system, we interrogate how autophagy deficiency modulates oncogenic Phosphatidylinositol 3-kinase (PI3K) mediated transformation of mammary epithelial cells.
The aberrant activation of the PI3K/Akt/mTOR signaling pathway is commonly observed in many cancers; the class I PI3K plays a central role in cell growth, proliferation, differentiation, and metabolism. PIK3CA, which encodes the p110α catalytic subunit of class I PI3K, is frequently mutated in cancers. In breast cancer, more than 25% of tumors harbor somatic mutations in PIK3CA, and commonly involve a hotspot mutation in the C-terminal kinase domain (H1047R) (11). Cells expressing mutant PIK3CA H1047R exhibit increased AKT and mTOR activity and have several phenotypic changes associated with oncogenic transformation, including adhesion- and growth factor-independent proliferation, protection from anoikis, and abnormal morphogenesis in 3D culture (12, 13).
The aforementioned studies motivated us to scrutinize how autophagy impacts the growth and morphogenesis of mammary epithelial structures expressing PI3K-H1047R. We demonstrate that inhibiting autophagy sensitizes PI3K-H1047R cells occupying the 3D luminal space to undergo apoptosis. At the same time, we uncover an important role for autophagy in restricting cell proliferation driven by oncogenic PI3K activation. Accordingly, defective autophagy in PI3K-transformed epithelial cells results in enhanced levels of proliferation in 3D culture. Moreover, in response to rapamycin-mediated mTORC1 inhibition, we demonstrate that autophagy contributes to the suppression of proliferation in PI3K-transformed cells. Finally, we uncover that accumulation of the autophagy substrate p62/SQSTM1 is sufficient to activate the extracellular signal related kinase/mitogen activated protein kinase (ERK/MAPK) pathway and to enhance cell proliferation in oncogenic PI3K expressing structures. Overall, these results point to important tumor suppressive functions for autophagy in the context of oncogenic PI3K activation, with the loss of autophagy promoting proliferation in 3D organotypic culture.
RESULTS
Autophagy inhibition concomitantly enhances luminal apoptosis and proliferation during 3D morphogenesis driven by the oncogenic PIK3CA mutant H1047R
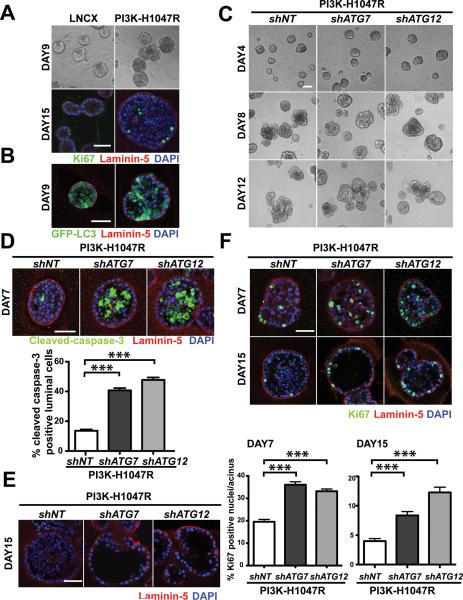
We utilized an established 3D culture system using MCF10A, a non-transformed human mammary epithelial cell line, to investigate PI3K-H1047R driven transformation in a tissue-relevant context (14). MCF10A cells stably expressing empty vector (LNCX) or PI3K-H1047R were cultured on a reconstituted laminin-rich basement membrane. Consistent with previous work (13, 15), MCF10A cells expressing LNCX formed hollow gland-like structures that underwent proliferative arrest, evidenced by the near complete absence of the proliferation marker Ki-67 following 15 days in 3D culture (Figure 1A). In contrast, over this timecourse, PI3K-H1047R expressing cells formed larger acini, and over longer periods, multi-acinar clusters. Importantly, these structures exhibited increased numbers of Ki67 positive cells and prominent luminal filling, indicating the expression of PI3K-H1047R enabled cells to both escape from proliferative suppression in 3D culture as well as survive in the luminal space (Figure 1A).
Figure 1.
Autophagy inhibition concomitantly enhances luminal apoptosis and proliferation in cells expressing the oncogenic PIK3CA mutant H1047R. (A) MCF10A cells stably expressing empty vector (LNCX) or PI3K H1047R were cultured on laminin-rich basement membrane (Matrigel™). Top: Phase images of day 9 structures. Bottom: Confocal images of day 15 acini stained for Ki67 (green), laminin-5 (red) and DAPI (blue). Bar, 50μm. (B) Confocal images of day 9 acini derived from LNCX or PI3K-H1047R cells expressing GFP-LC3. (C) Phase time course of 3D cultures of the indicated cell types. Bar, 100μm. (D) Day 7 3D cultures of the indicated cell types were immunostained for cleaved-caspase 3 (green), laminin-5 (red), and counterstained with DAPI (blue). To assess luminal apoptosis, the percentage of cleaved-caspase-3 positive luminal cells was quantified from 150 acini of each cell type obtained from three independent experiments (mean ± SEM). *** p< 0.001, Student's t test. Bar, 50μm. (E) Day 15 acini of the indicated cell types were immunostained for laminin-5 (red) and counterstained with DAPI (blue). Bar, 50μm. (F) Day 7 and day 15 acini derived from the indicated cell types were immunostained for Ki67 (green), laminin-5 (red), and DAPI counterstained (blue). Percentage of Ki67 positive nuclei per acinus was quantified on day 7 and day 15. For each cell type, a total of 150 acini from three independent experiments were analyzed. Results represent mean ± SEM. *** p< 0.001, Student's t test. Bar, 50μm.
Although previous studies suggest that activation of PI3K pathway suppresses autophagy induction via activating mTOR (16), we observed autophagosome formation (punctate GFP-LC3) in acini derived from both LNCX and PI3K-H1047R cells stably expressing a GFP-LC3 reporter (Figure 1B). To corroborate this result, we assessed the induction of autophagy in response to ECM detachment. MCF10A cells expressing LNCX or PI3K-H1047R were grown attached (Att) or in suspension (Sus) for 24h, upon which autophagy was assessed by monitoring the turnover of lipidated LC3/ATG8 (LC3-II) in the absence versus presence of the lysosomal cathepsin inhibitors, E64D and pepstatin A (E/P) (17). LNCX and PI3K-H1047R cells showed equivalent levels of autophagic flux following ECM detachment (Supplementary Figure1A). Furthermore, both LNCX and PI3K-H1047R cells stably expressing a GFP-LC3 reporter both showed a robust induction of autophagosomes upon ECM detachment (Supplementary Figure 1A). Overall, these results indicate that the activation of PI3K/Akt pathway does not suppress autophagy during 3D morphogenesis or in response to ECM detachment.
Our recent work demonstrated that autophagy functions as a survival pathway during luminal clearance in MCF10A acini grown in 3D culture (18). To assess whether autophagy contributes to the luminal filling in PI3K-H1047R expressing cells derived 3D structures, we transduced these cells with lentiviral constructs encoding short hairpin RNAs (shRNA) against ATG7 (shATG7) or ATG12 (shATG12), two essential autophagy genes (ATGs). Stable ATG7 or ATG12 knockdown reduced LC3-II turnover (autophagic flux) during ECM detachment to approximately 50% of controls (Supplementary Figure1B). Notably, ATG knockdown did not alter PI3K activity as assessed by serine 473 phosphorylation of the downstream effector, AKT (Supplementary Figure1C). Although ATG depletion did not significantly alter the overall morphology of PI3K-H1047R structures (Figure 1C), these knockdowns resulted in up to a 3-fold increase in luminal apoptosis, evidenced by increased cleaved caspase-3 positive cells occupying the luminal space of day 7 structures (Figure 1D). Importantly, these apoptotic cells lacked direct contact with the underlying basement membrane, indicating that they were dying via anoikis (15). By day 15, the majority of shATG7 and shATG12 cell-derived acini exhibited luminal space clearance, in contrast to PI3K-H1047R shNT acini, which exhibited filled lumen (Figure 1E). In addition, in soft agar transformation assays, adhesion-independent growth of PI3K-H1047R cells decreased by 61.8% and 75.8% upon ATG7 or ATG12 depletion, respectively (Supplementary Figure 1D). Overall, these results demonstrate that autophagy is critical for the survival of PI3K-H1047R-expressing cells lacking ECM contact.
Although ATG depleted acini exhibited enhanced luminal apoptosis, the total number of cells recovered from day 7 3D culture of shATG7 and shATG12 cells was only slightly decreased compared to controls (Supplementary Figure 2A). Given these modest effects, we assess whether ATG depletion concomitantly affected cell proliferation driven by oncogenic PI3K in 3D culture. Both PI3K-H1047R-shATG7 and shATG12 structures exhibited an almost 2-fold increase in Ki67 activity compared to control cells. This hyper-proliferative phenotype in ATG-depleted structures was observed in both early (day 7) and late (day 15) stages of 3D culture (Figure 1F). These results were corroborated using a second proliferation marker, phosphorylated Rb (pRb-S780) (Supplementary Figure 2B). Increased proliferation was not observed in ATG depleted cells cultured in monolayer (Supplementary Figure 2C), indicating that autophagy inhibition does not enhance proliferative capacity of PI3K-H1047R cells grown in 2D conditions; rather, it enhances the ability of these cells to escape proliferative suppression in 3D culture.
Autophagy is not essential for the growth-suppressive effects of rapamycin in PI3K-H1047R 3D cultures
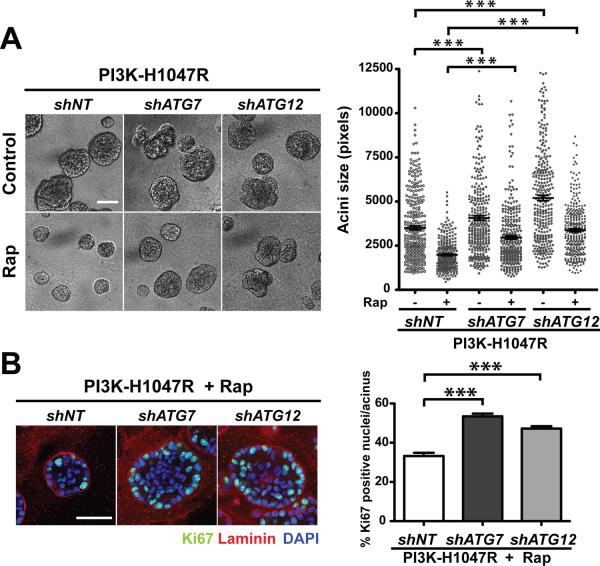
Previous work indicates that mTORC1 activity is required for the phenotypes mediated by PI3K/Akt pathway activation in 3D culture; the allosteric mTORC1 inhibitor rapamycin suppresses the development of large, distorted structures mediated by PI3-KH1047R (13). Because rapamycin-mediated inhibition of mTORC1 induces autophagy in cells (19), we assessed the functional impact of autophagy inhibition on rapamycin-mediated growth suppression. Rapamycin treatment reduced acinar size of the PI3-KH1047R-derived cells by 43.5% (Figure 2A). In contrast, rapamycin elicited a 26.9% and 35% reduction in structure size of PI3K-H1047R-shATG7 and shATG12 cells, respectively. Although these results suggest that autophagy may partly contribute to the reduction of PI3K-H1047R structure size during rapamycin treatment, autophagy is not essential for rapamycin-mediated growth suppression in this model. Furthermore, we observed significantly increased Ki67 activity in ATG knockdown cells treated with rapamycin, indicating that autophagy continues to restrict proliferation in response to rapamycin treatment (Figure 2B)
Figure 2.
Autophagy is not essential for rapamycin-induced growth suppression of PI3K-H1047R 3D structures. (A) Left: Day 7 phase images of the indicated cell types grown in the presence or absence of 25nM rapamycin (Rap). Right: Acinar size distribution on day 7 of 3D culture. A total of 300 acini from three independent experiments were measured for each condition. Results represent mean ± SEM. *** p<0.001, Student's t test. Bar, 100μm. (B) Indicated cell types were grown in 3D for 7 days in the presence of 25nM Rap. Left: Acini were immunostained for Ki67 (green), laminin-5 (red), counterstained with DAPI (blue). Right: The percentage of Ki67 positive nuclei per acinus was quantified. For each cell type and condition, a total of 150 acini from three independent experiments were analyzed. Results represent the mean ± SEM *** p<0.001, Student's t test. Bar, 50μm.
To further corroborate these results, we examined whether autophagy deficiency similarly affected 3D morphogenesis of cells expressing oncogenic AKT. In MCF10A cells expressing a conditionally activated variant of AKT (ER-AKT), treatment of cells with 4-hydroxytamoxifen (4-OHT) activates AKT and induces proliferation in 3D culture, resulting in large, misshapen structures. Similar to PI3K-H1047R, rapamycin-mediated mTORC1 inhibition reverts these phenotypes associated with AKT activation (Supplementary Figure 3A and B) (20). In ER-AKT cells, ATG7 knockdown did not significantly affect AKT activation induced by 4-OHT treatment (Supplementary Figure 3A); in the presence of 4-OHT, shATG7 cells formed larger, distorted acini in 3D culture resembling non-targeting controls (ER-AKT-shNT) (Supplementary Figure 3B). Upon rapamycin treatment, ER-AKT-shNT cells formed small, spherical acini that resembled ethanol (ETOH) treated controls; ERAKT cells with ATG7 depletion also exhibited a significant reduction in structure size in response to rapamycin (Supplementary Figure 3B). However, while rapamycin treatment decreased Ki67 activity in AKT-activated structures, knockdown of either ATG7 or ATG5, another essential autophagy gene, enabled higher rates of proliferation in ER-AKT cells in the presence of rapamycin (Supplementary Figure 3C). Overall, these results corroborate that autophagy continues to restrict oncogenic PI3K/Akt-driven proliferation in response to rapamycin-mediated mTORC1 inhibition.
Chloroquine-mediated lysosomal inhibition enhances ERK/MAPK pathway activation and induces the proliferative outgrowth of PI3K-H1047R subpopulations in 3D culture
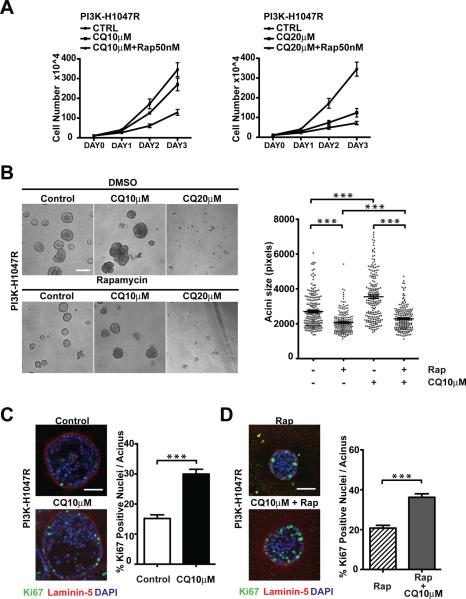
The anti-malarial chloroquine (CQ) is a weak base that can neutralize acidic intracellular compartments such as late endosomes and lysosomes (21). Both CQ and its derivative, hydroxychloroquine, are currently being used in numerous human cancer clinical trials as autophagy inhibitor to block the late stages of autophagy (22). We therefore evaluated how CQ treatment impacted PI3K-H1047R driven morphogenesis. Based on the accumulation of LC3-II, we established that a range of concentrations of CQ were able to effectively inhibit autophagic degradation with maximal inhibition obtained at doses of 20μM or higher (Supplementary Figure 4A). In monolayer culture, CQ suppressed PI3K-H1047R cell proliferation in a dose-dependent manner that became even more pronounced when combined with rapamycin (Figure 3A). In 3D culture, treating cells with 20μM CQ alone or in combination with rapamycin fully suppressed the outgrowth of PI3K-H1047R acini (Figure 3B). In contrast, upon treatment with a lower dose of 10μM, a heterogeneous cell population was observed. While certain CQ treated individual cells were unable to grow out in these cultures, a significant subpopulation of cells exhibited enhanced proliferative capacity compared to untreated controls, resulting in the formation of large cell structures that were highly Ki67 positive (Figure 3B and C). Notably, numerous hyperproliferative outgrowths were also observed in cultures concomitantly treated with CQ and rapamycin, although the size of the outgrowths was reduced in comparison to cultures treated with CQ alone (Figure 3B); furthermore, structures treated with rapamycin plus 10μM CQ exhibited higher Ki67 activity compared to those treated with rapamycin alone (Figure 3D).
Figure 3.
Effects of chloroquine-mediated lysosomal inhibition on 3D morphogenesis driven by oncogenic PI3K. (A) Monolayer growth curves of PI3K-H1047R cells treated with 10μM (left) or 20μM chloroquine (CQ) (right) in the presence or absence of 50nM Rapamycin (Rap). (B) Left: Phase images of day 7 3D cultures of PI3K-H1047R cells subject to the indicated treatments. Bar, 100μm. Right: Acinar size distribution on day 7 of 3D culture. A total of 200 acini from two independent experiments were measured for each condition. (C) Left: PI3K-H1047R cells were cultured in the presence of absence of 10μM CQ for 7 days. The outgrowths were immunostained with antibodies against Ki67 (green), laminin-5 (red), and DAPI-counterstained (blue). Bar, 50μm. Right: the percentage of Ki67 positive nuclei per acinus was quantified. For each condition, a total of 75 acini from three independent experiments were analyzed. (D) Left: PI3K-H1047R cells were treated with 25nM rapamycin alone or in combination of 10μM CQ. Day 7 outgrowths were immunostained with antibodies against Ki67 (green), laminin-5 (red), and DAPI-counterstained (blue). Bar, 50μm. Right: The percentage of Ki67 positive nuclei per acinus was quantified. For each condition, a total of 75 acini from three independent experiments were analyzed. Results represent the mean ± SEM *** p<0.001, Student's t test.
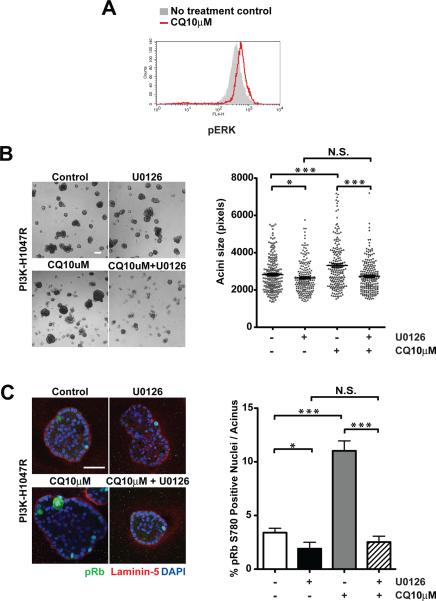
Because the ERK/MAPK pathway is an important driver of proliferation in MCF10A 3D culture (15, 23), we further interrogated whether CQ treatment impacted activation of ERK/MAPK pathway. FACS analysis revealed that cells treated with low-dose CQ exhibited enhanced ERK activation, as delineated by increased phospho-ERK1/2 (pERK) staining (Figure 4A). We then tested whether the outgrowth of PI3K-H1047R cells in the presence of low-dose CQ is sensitive to ERK/MAPK pathway inhibition using U0126, a highly selective and potent inhibitor of the ERK/MAPK signaling cascade via inhibition of the immediate upstream activators, MAP Kinase Kinase 1 and 2 (MAPKK1/2, also known as MEK1/2). We treated 3D cultures with U0126 alone or in combination with low-dose CQ for 8 days. Treatment with U0126 alone was found to have modest effects on PI3K-H1047R structure size in 3D culture. Nevertheless, U0126 robustly suppressed the outgrowth of PI3K-H1047R cells in the presence of 10μM CQ (Figure 4B). Furthermore, staining for the proliferation marker pRb780 revealed that ERK/MAPK inhibition dramatically decreased cell proliferation in these structures (Figure 4C). Together, these results indicate that although CQ can effectively inhibit autophagy over a range of concentrations, its ultimate outcome on 3D growth and proliferation driven by PI3K-H1047R is dose dependent; at lower doses, CQ treatment promotes the outgrowth of a highly proliferative PI3K-H1047R subpopulation, which appears to be at least partly arise due to the activation of ERK/MAPK pathway in these cells.
Figure 4.
Chloroquine-treatment promotes outgrowth of PI3KH1047R cells in 3D culture through activation of ERK/MAPK pathway. (A) PI3K-H1047R cells were cultured in the presence of absence 10μM CQ. FACS analysis of activation specific ERK phosphorylation (pERK) was performed in cells recovered from day 11 3D cultures. (B) Left: Phase images of day 8 3D cultures treated with 10μM U0126, 10μM CQ, or 10μM U0126+10μM CQ as indicated. Bar, 100μm. Right: Acinar size distribution on day 7 of 3D culture. A total of 200 acini from two independent experiments were measured for each condition. (C) Left: Day 7 outgrowths were fixed and immunostained with antibodies against pRb780 (green), laminin-5 (red), and DAPI-counterstained (blue). Bar, 50μm. Right: The percentage of pRb780 positive nuclei per acinus was quantified. For each cell type and condition, a total of 100 acini from two independent experiments were analyzed. Results represent the mean ± SEM, * p< 0.05 *** p<0.001, Student's t test. N.S. not significant.
p62/SQSTM1 cooperates with PI3K-H1047R to enhance cell proliferation in 3D culture
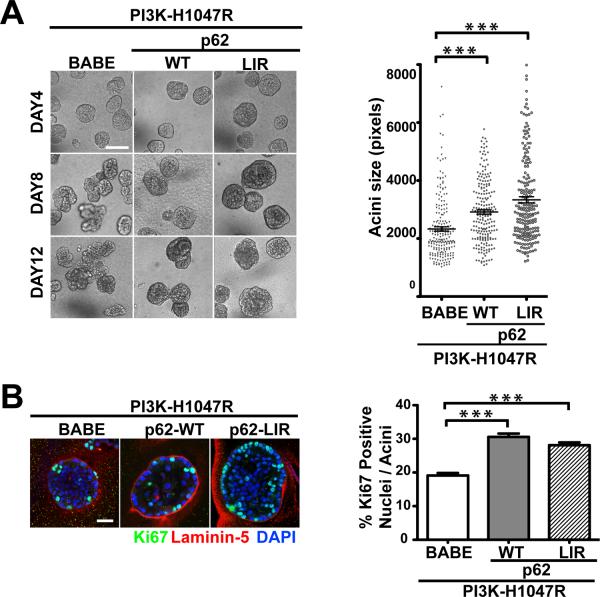
p62/SQSTM1 is a signaling adaptor protein that is selectively degraded via autophagic pathway (24). Genetic deletion of autophagy genes results in spontaneous tumor formation and p62 accumulation in tumor cells (7, 8), while p62 overexpression enhances tumorigenicity of autophagy-deficient cells in vivo (25). These studies support that p62 accumulation plays an important role in the tumorigenesis mediated by autophagy deficiency. In this 3D culture system, PI3K-H1047R cells treated with CQ alone or in combination with rapamycin exhibited significantly increased p62 levels (Supplementary Figure 4B), which motivated us to interrogate whether p62 accumulation is sufficient to enhance cell proliferation associated with autophagy deficiency in 3D culture. We stably expressed HA-tagged wild type p62 (p62-WT) or a mutant p62 (p62-LIR) in control cells (LNCX) and PI3K-H1047R expressing cells. The p62-LIR contains a mutation (W338A) within the LC3-interacting region (LIR), rendering it resistant to degradation via autophagy (26). As shown in Supplementary Figure 5A, wild-type p62 but not p62-LIR was degraded in response to nutrient starvation (HBSS treatment) in both LNCX and PI3K-H1047R cells. In 3D culture, p62-WT or p62-LIR expression had minimal effects on acini size in LNCX cells (Supplementary Figure 5B). In contrast, when co-expressed with PI3K-H1047R, p62-WT expression caused a 25% increase in average acini size; this phenotype was further enhanced in PI3K-H1047R cells expressing p62-LIR, which produced a 43% increase in average acini size (Figure 5A). Additionally, we observed significantly increased Ki67 activity in p62-WT and p62-LIR expressing PI3K-H1047R acini (Figure 5B). Similar to our results with ATG depletion, we did not observe enhanced cell proliferation in p62 over-expressing PI3K-H1047R cells grown in monolayer culture (Supplementary Figure 5C); rather, the proliferative phenotype was only observed in 3D culture conditions. Overall, these results indicate that while the accumulation of p62 alone is not sufficient to enhance growth and proliferation in wild type (LNCX) MCF10A acini, it can enhance proliferation and growth capacity in cooperation with oncogenic PI3K activation in 3D culture.
Figure 5.
p62/SQSTM1 cooperates with oncogenic PI3K to enhance cell proliferation in 3D culture. (A) Left: Phase timecourse of the indicated cell types in 3D culture. Right: Acinar size distribution of day 7 3D cultures of the indicated cell types. For each cell type, a total of 200 acini from three independent experiments were measured. Results represent mean ± SEM. *** p<0.001, Student's t test. Bar, 100μm. (B) Left: Day 7 acini derived from the indicated cell types were immunostained with antibodies against Ki67 (green), laminin-5 (red), and counterstained with DAPI (blue). Right: Percentage of Ki67 positive nuclei per acinus was quantified. For each cell type, a total of 150 acini from three independent experiments were counted. *** p<0.001, Student's t test. Bar, 50μm.
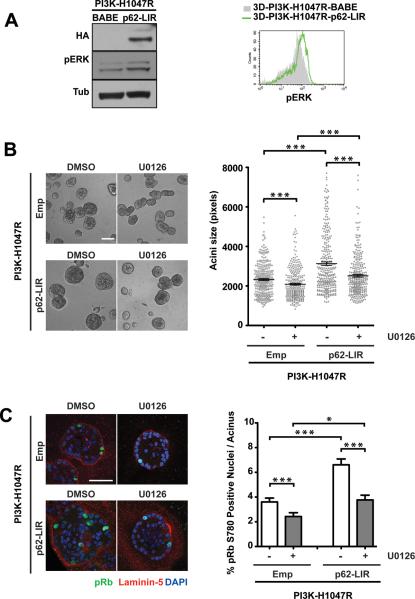
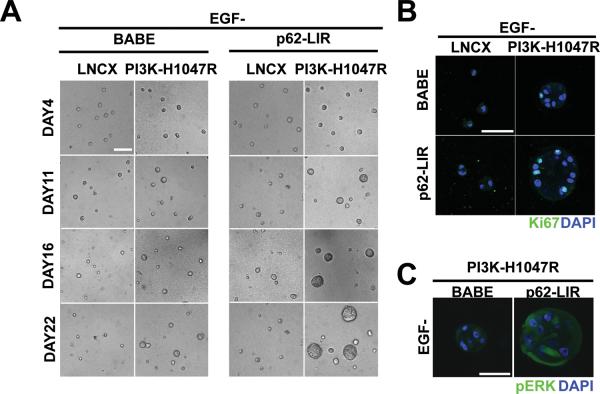
Furthermore, we also detected a stronger pERK signal in PI3K-p62-LIR cells collected from day 7 3D cultures (Figure 6A, Left). Flow cytometric quantification corroborated a 40% increase in pERK signal intensity in PI3K-p62-LIR cells harvested on day12 compared to controls (Figure 6A, Right). Treatment of PI3K-p62-LIR cells with U0126 significantly decreased structure size and cell proliferation compared to untreated cultures, indicating that ERK/MAPK pathway activation is an important contributor to p62-LIR-induced hyper-proliferation in PI3K-H1047R cells (Figure 6B and C). Because of these effects of p62 on ERK/MAPK activation, we subsequently evaluated how p62 accumulation influenced proliferation in 3D cultures lacking epidermal growth factor (EGF). Whereas proliferation of wild type MCF10A cells is strictly EGF-dependent, PI3K-H1047R-expressing cells exhibit low-level proliferation in the absence of exogenously provided EGF (13). Consistent with previous results, PI3K-H1047R, but not vector control (LNCX) cells, formed small structures after 22 days of culture (Figure 7A, left). The expression of p62-LIR significantly enhanced the EGF-independent growth of PI3K-H1047R cells in 3D culture (Figure 7A, right). Whereas LNCX-p62-LIR cells showed extremely limited proliferation capacity in the absence of exogenous EGF, PI3K-H1047R-p62-LIR cultures contained multiple cell clusters that exhibited increased Ki67 activity compared to controls (Figure 7B). Furthermore, a stronger pERK signal was detected in PI3K-p62-LIR acini grown in the absence of EGF in comparison to PI3K-BABE controls (Figure 7C). Overall, these results suggest that p62 accumulation cooperates with oncogenic PI3K to enhance cell proliferation in MCF10A 3D culture, both in the presence or absence of EGF; moreover, this proliferative phenotype correlates with enhanced activation of the ERK/MAPK pathway.
Figure 6.
p62/SQSTM1-mediated proliferation of PI3K-H1047R cells in 3D culture is associated with activation of ERK/MAPK pathway. (A) In 3D cultures of the indicated cell types grown in standard assay media, phosphorylated ERK (pERK) levels were determined by immunoblotting of cell lysates (day 7) or flow cytometry (day 12). (B) Left: Phase images of day 7 3D cultures treated with DMSO or 10μM U0126 as indicated. Bar, 100μm. Right: Acinar size distribution on day 7 of 3D culture. A total of 300 acini from three independent experiments were measured for each condition. Results represent mean ± SEM. *** p<0.001, Student's t test. (C) Left: Day 7 3D structures were fixed and immunostained with antibodies against pRb780 (green), laminin-5 (red), and DAPI-counterstained (blue). Bar, 50μm. Right: the percentage of pRb780 positive nuclei per acinus was quantified. For each cell type and condition, a total of 150 acini from three independent experiments were analyzed. Results represent the mean ± SEM * p< 0.05, *** p<0.001, Student's t test.
Figure 7.
p62/SQSTM1 promotes EGF-independent proliferation of PI3K-H1047R structures. (A) Phase timecourse of indicated cell types grown in 3D in the absence of EGF. Bar, 100μm. (B) Day 22 structures derived from the indicated cell types in the absence of EGF were immunostained with antibodies against Ki67 (green) and DAPI-counterstained (blue). Bar, 50μm. (C) Structures derived from the indicated cell types in the absence of EGF (day 22) were immunostained with antibodies against pERK (green) and DAPI-counterstained (blue). Bar, 50μm.
ATG depletion enhances ERK/MAPK pathway activation in PI3K-H1047R structures
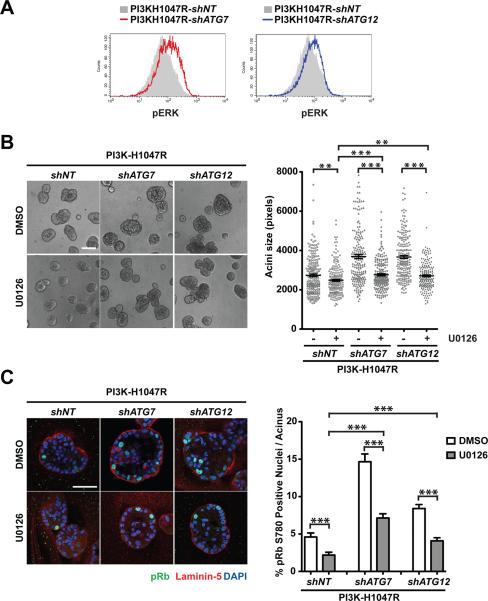
Lastly, we assessed the affects of ATG depletion on ERK/MAPK pathway activation in PI3K-H1047R cells in 3D culture. FACS analysis revealed PI3K-shATG7 and –shATG12 cells exhibited increased pERK activity compared to control cells (Figure 8A). U0126 treatment significantly decreased the structure size of ATG depleted PI3K-H1047R cells; upon U0126 treatment, structure size was reduced by 9% in shNT cells, 25.2% in shATG7, and 26.1% in shATG12 cells (Figure 8B). ERK/MAPK pathway inhibition also decreased ATG depletion-mediated hyperproliferation of PI3K-H1047R cells (Figure 8C). Taken together, these results support that the activation of the ERK/MAPK pathway serves as one important mechanism by which autophagy deficiency enhances the proliferation of PI3K-H1047R cells in 3D culture.
Figure 8.
ATG depletion enhances ERK/MAPK activation in 3D cultures of PI3K-H1047R cells. (A) In 3D cultures of the indicated cell types grown in standard assay media, phosphorylated ERK (pERK) levels were determined by flow cytometry (day 11). (B) Left: Phase images of day 7 3D cultures treated with DMSO or 10μM U0126 as indicated. Bar, 100μm. Right: Acinar size distribution on day 7 of 3D culture. A total of 200 acini from two independent experiments were measured for each condition. Results represent mean ± SEM. ** p<0.01, *** p<0.001, Student's t test. (C) Left: Day 7 3D structures were fixed and immunostained with antibodies against pRb780 (green), laminin-5 (red), and DAPI-counterstained (blue). Bar, 50μm. Right: the percentage of pRb780 positive nuclei per acinus was quantified. For each cell type and condition, a total of 100 acini from two independent experiments were analyzed. Results represent the mean ± SEM *** p<0.001, Student's t test.
DISCUSSION
Growing evidence reveals multiple context-dependent roles for autophagy in cancer (3). Genetic studies reveal that autophagy promotes genome integrity and serves as a barrier to limit cancer initiation. However, in established tumors, autophagy functions as a survival and fitness mechanism that is critical for tumor progression and maintenance (27). Here, we utilize a 3D epithelial culture model to demonstrate that in response to oncogenic PI3K activation, autophagy can simultaneously mediate both tumor suppressive and promoting functions within the same glandular structure. We uncover that ATG depletion strongly enhances cell proliferation within PI3K-H1047R structures, suggesting that autophagy restricts the proliferation of oncogenic PI3K expressing cells in 3D culture. At the same time, ATG knockdown results in increased luminal apoptosis and luminal space clearance, indicating that autophagy is crucial for the survival of oncogenic PI3K expressing cells deprived of ECM contact. Importantly, we find that cell-ECM contact dictates the manifestation of these two phenotypes within these structures. Increased levels of proliferation are observed throughout the structure, both detached cells occupying the lumen as well as in outer cells that directly contact the surrounding ECM. In contrast, the increase in apoptosis is restricted only to the centrally located cells deprived of ECM contact. These findings illustrate that although both tumor suppressive and promoting functions of autophagy may co-exist, the precise effect of autophagy inhibition on cell fate is ultimately determined by environmental constraints imposed on the oncogene-expressing cells.
Increased cell proliferation has been observed in several autophagy-deficient tissues in vivo. However, in monolayer cultures, autophagy inhibition often decreases or has no effect on cell proliferation in vitro (28–30). Here, we demonstrate that the enhanced cell proliferation arising from autophagy deficiency, pharmacological lysosomal inhibition, and p62 accumulation is only evident in 3D culture conditions; no such phenotypes are present in PI3K-H1047R cells grown in monolayer. We propose that the enhanced rates of proliferation resulting from defective autophagy only become manifest in more tissue relevant settings, such as those provided in 3D culture models. Hence, 3D systems may serve as useful tools to dissect how autophagy impacts proliferation control and cell fate during oncogenic transformation (31).
Currently, multiple clinical trials using hydroxychloroquine (HCQ) are in progress to evaluate whether autophagy inhibition is able to sensitize tumor cells to undergo death, either alone or in combination with anti-cancer therapy. At the same time, there is immense clinical interest in mTOR inhibition as a therapeutic strategy against tumors (32). Because mTOR serves as a key negative regulator of autophagy, an important question is whether autophagy contributes to the anti-cancer effects of pharmacological mTOR inhibitors, or alternatively, if this process promotes drug resistance in response to such agents (33). Our results using rapamycin-mediated allosteric mTORC1 inhibition in cells transformed by oncogenic PI3K (or Akt) support that autophagy continues to contribute to efficient proliferative suppression in 3D culture; however, autophagy is not critical for the ability of rapamycin to suppress the growth of PI3K-H1047R 3D structures. In addition, although the lysosomal inhibitor chloroquine (CQ) is able to suppress the proliferation of PI3K-H1047R cells propagated in standard monolayer culture, we demonstrate that a subpopulation of hyperproliferative structures readily emerge in 3D culture conditions, especially when CQ is utilized at lower doses. Since CQ and HCQ are currently being used in clinical trials as a treatment against a wide spectrum of cancers, our findings point to the importance of optimal dosage of these agents in clinical oncology settings.
Finally, we demonstrate that accumulation of the autophagy substrate and cargo receptor, p62/SQSTM1, is sufficient to enhance proliferation in PI3K-H1047R cells. Interestingly, our results indicate that p62 overexpression is not sufficient as an isolated insult to overcome 3D proliferative suppression, nor is it able to promote EGF independent proliferation in 3D culture. These results broach the possibility that the ability of p62 to drive proliferation in autophagy-deficient cells may be significantly amplified in combination with additional oncogenic insults that activate the PI3K/Akt signaling axis.
As a adaptor protein, p62/SQSTM1 binds to proteins involved in several signaling pathways (34). In this model, the expression of an autophagy-resistant form of p62 (p62-LIR) enhances ERK/MAPK pathway activation, which cooperates with oncogenic PI3K to promote cell proliferation. Similarly, both CQ-mediated autophagy inhibition and ATG depletion in PI3K-H1047R cells are able to promote ERK/MAPK activation in 3D culture. Furthermore, the MEK inhibitor U0126 attenuates the enhanced growth and proliferation observed upon autophagy inhibition or p62-LIR overexpression in 3D culture. Altogether, these data support that autophagy restricts proliferation in PI3K-H1047R cells at least partly via downregulation of the ERK/MAPK mitogenic pathway. However, it is important to recognize that despite this sensitivity of ATG deficient and p62-LIR expressing cells to MEK inhibition in 3D culture, the U0126-treated structures derived from these cells do remain significantly larger and more proliferative than U0126-treated control cells (Figure 6C and 8C). These results broach the possibility that autophagy and p62/SQSTM1 direct additional pathways, such as the previously reported NF-κB and Nfr2 pathways, which contribute to their ability to suppress tumor cell growth and proliferation.
In summary, our data delineate diverse biological functions for autophagy during 3D morphogenesis driven by oncogenic PI3K. Importantly, autophagy inhibition promotes the apoptosis of oncogenic cells under duress, in this case, due to ECM deprivation. Nevertheless, inhibiting autophagy leads to enhanced cell proliferation and activation of ERK/MAPK pathway in this 3D culture model. Currently, there is great interest in targeting autophagy as an anti-cancer strategy. Our study suggests that the potential untoward effects of autophagy inhibition on cell proliferation should be considered when applying autophagy inhibition as a clinical treatment against cancer.
MATERIAL AND METHODS
Cell lines and culture conditions
MCF10A derived cell lines were generated and cultured as previously described (14). Stable pools were generated by selection with 2μg/ml Puromycin or 300μg/ml G418.
3D morphogenesis assays
3D culture of MCF10A derived cell lines was performed as previously described (14). Culture media was refreshed every 4 days, unless otherwise indicated. When indicated, the following reagents were added on day 1 of culture and refreshed every 4 days: 25nM rapamycin (Rap, Calbiochem), 10μM or 20μM chloroquine diphosphate salt (CQ, Sigma), 1μM 4-Hydroxytamoxifen (4OHT, Sigma). For proliferation assays, 10μM U0126 (Merck) was added on day 1, refreshed on day 4 and 24h prior to analysis.
Antibodies
Commercial antibodies include: α-Ki67 (Invitrogen), α-phospho-Akt (Ser473), α-AKT, anti-cleaved caspase-3, α-phospho-Rb (Ser780), α-phospho-p44/42 MAPK (Erk1/2)-Alexa Fluor® 488 Conjugate, α-phospho-p44/42 MAPK (Erk1/2)-Alexa Fluor® 647 Conjugate, α-ATG12 (Cell Signaling Technology), α-phospho-ERK1/2 (Biosource), α-ATG7 (Santa Cruz Biotechnology), α-laminin 5 (Millipore), α-p62 (Progen Biotechnik), α-HA (HA.11, Covance), α-α-tubulin (Sigma), Anti-LC3 antibody has been previously described (18).
Retroviral vectors and virus production
The retroviral vector pLNCX-neo containing PI3K-H1047R was kindly provided by Dr. W. Weiss. pBABEpuro GFP-LC3 and pWZLneo ER-Akt have been previously described (20, 30). Human p62 cDNA was a kind gift from Dr. T. Johansen. To generate pBABEpuro-p62-WT, p62 was amplified and cloned into a pBABEpuro vector containing a 5' HA-tag. To generate pBABEpuro-p62-LIR, a point mutation at position 338 (p62W338A) in pBABEpuro-p62-WT was introduced using the QuickChange XL Site-Directed Mutagenesis kit (Stratagene). For retroviral transduction, VSV-G-pseudotyped retroviruses were generated, and cells were infected and selected as previously described (14).
shRNA and siRNA
pLKO.1 lentiviral expression vectors containing short hairpin RNAs against ATG7 and ATG12 were purchased from Sigma (Mission shRNA). The target sequences for hairpins directed against human ATG7 (NM_006395) is: shATG7 (TRCN0000007584): GCCTGCTGAGGAGCTCTCCA; directed against human ATG12 (NM_004707) is: shATG12 (TRCN0000007394): TGGAACTCTCTATGAGTGTTT. For siRNA-mediated knockdown of autophagy genes, siGenome SMARTpool siRNAs against human ATG7 (M-020112-01) and human ATG5 (M-004374-04) were purchased from Dharmacon and cells were transfected as previously described (18).
Phase and immunofluorescence image acquisition and analysis
Phase images of 3D culture were acquired on an Axiovert 200 microscope (Carl Zeiss) equipped with a Spot RT camera (Diagnostic Instruments). For confocal analyses, 3D cultures were fixed and stained as previously described (14). Images were acquired using a C1Si confocal laser-scanning microscope (Nikon) and analyzed using EZ-C1 software (v3.20). Acini area was measured using MetaMorph software (v6.0).
Flow cytometry
Cells were harvested from 3D cultures and fixed in 4% paraformaldehyde followed by resuspension in cold 90% methanol for 30 minutes on ice. Cells were then subject to antibody staining according to the manufacturer's protocol. For flow cytometric detection of pERK activity, assay media for 3D cultures was not replenished prior to harvest.
Statistical analysis
Graphs represent the average values from three independent experiments with error bars reflecting the standard error. GraphPad Prism software (v5.0b) was used for statistical analysis. P values were determined by unpaired Student's t-test with p<0.05 considered significant.
Supplementary Material
Acknowledgements
We thank Drs. William Weiss, Qiwen Fan, and members of the Debnath lab for critically reading the manuscript, and Drs. William Weiss and Terge Johansen for generously providing constructs. Grant support to JD includes the NIH (RO1 CA126792 and ARRA supplement CA126792-S1) and the DOD BCRP (W81XWH-11-1-0130). NE received a Short Term Stay Fellowship from the Spanish Ministry of Health. RL is a DOD BCRP Predoctoral Scholar (W81XWH-08-1-0759).
Footnotes
Conflict of interest The authors declare no conflict of interest.
REFERENCES
- 1.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kondo Y, Kanzawa T, Sawaya R, Kondo S. The role of autophagy in cancer development and response to therapy. Nat Rev Cancer. 2005 Sep;5(9):726–34. doi: 10.1038/nrc1692. [DOI] [PubMed] [Google Scholar]
- 3.Chen N, Debnath J. Autophagy and tumorigenesis. FEBS Lett. 2010 Apr 2;584(7):1427–35. doi: 10.1016/j.febslet.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999 Dec 9;402(6762):672–6. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 5.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. The Journal of clinical investigation. 2003 Dec;112(12):1809–20. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003 Dec 9;100(25):15077–82. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Inami Y, Waguri S, Sakamoto A, Kouno T, Nakada K, Hino O, et al. Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells. J Cell Biol. 2011 Apr 18;193(2):275–84. doi: 10.1083/jcb.201102031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S, et al. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011 Apr 15;25(8):795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karantza-Wadsworth V, Patel S, Kravchuk O, Chen G, Mathew R, Jin S, et al. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 2007 Jul 1;21(13):1621–35. doi: 10.1101/gad.1565707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mathew R, Kongara S, Beaudoin B, Karp CM, Bray K, Degenhardt K, et al. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007 Jun 1;21(11):1367–81. doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004 Apr 23;304(5670):554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 12.Kang S, Bader AG, Vogt PK. Phosphatidylinositol 3-kinase mutations identified in human cancer are oncogenic. Proc Natl Acad Sci U S A. 2005 Jan 18;102(3):802–7. doi: 10.1073/pnas.0408864102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Isakoff SJ, Engelman JA, Irie HY, Luo J, Brachmann SM, Pearline RV, et al. Breast cancer-associated PIK3CA mutations are oncogenic in mammary epithelial cells. Cancer Res. 2005 Dec 1;65(23):10992–1000. doi: 10.1158/0008-5472.CAN-05-2612. [DOI] [PubMed] [Google Scholar]
- 14.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003 Jul;30(3):256–68. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 15.Debnath J, Mills KR, Collins NL, Reginato MJ, Muthuswamy SK, Brugge JS. The role of apoptosis in creating and maintaining luminal space within normal and oncogene-expressing mammary acini. Cell. 2002 Oct 4;111(1):29–40. doi: 10.1016/s0092-8674(02)01001-2. [DOI] [PubMed] [Google Scholar]
- 16.Petiot A, Ogier-Denis E, Blommaart EF, Meijer AJ, Codogno P. Distinct classes of phosphatidylinositol 3'-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J Biol Chem. 2000 Jan 14;275(2):992–8. doi: 10.1074/jbc.275.2.992. [DOI] [PubMed] [Google Scholar]
- 17.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008 Feb;4(2):151–75. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fung C, Lock R, Gao S, Salas E, Debnath J. Induction of Autophagy during Extracellular Matrix Detachment Promotes Cell Survival. Mol Biol Cell. 2008 Mar;19(3):797–806. doi: 10.1091/mbc.E07-10-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanazawa T, Taneike I, Akaishi R, Yoshizawa F, Furuya N, Fujimura S, et al. Amino acids and insulin control autophagic proteolysis through different signaling pathways in relation to mTOR in isolated rat hepatocytes. J Biol Chem. 2004 Feb 27;279(9):8452–9. doi: 10.1074/jbc.M306337200. [DOI] [PubMed] [Google Scholar]
- 20.Debnath J, Walker SJ, Brugge JS. Akt activation disrupts mammary acinar architecture and enhances proliferation in an mTOR-dependent manner. J Cell Biol. 2003 Oct 27;163(2):315–26. doi: 10.1083/jcb.200304159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luiken JJ, Aerts JM, Meijer AJ. The role of the intralysosomal pH in the control of autophagic proteolytic flux in rat hepatocytes. Eur J Biochem. 1996 Feb 1;235(3):564–73. doi: 10.1111/j.1432-1033.1996.00564.x-i2. [DOI] [PubMed] [Google Scholar]
- 22.Amaravadi RK, Lippincott-Schwartz J, Yin XM, Weiss WA, Takebe N, Timmer W, et al. Principles and current strategies for targeting autophagy for cancer treatment. Clin Cancer Res. 2011 Feb 15;17(4):654–66. doi: 10.1158/1078-0432.CCR-10-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Debnath J, Brugge JS. Modelling glandular epithelial cancers in three-dimensional cultures. Nat Rev Cancer. 2005 Sep;5(9):675–88. doi: 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- 24.Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005 Nov 21;171(4):603–14. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009 Jun 12;137(6):1062–75. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007 Aug 17;282(33):24131–45. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 27.Kimmelman AC. The dynamic nature of autophagy in cancer. Genes Dev. 2011 Oct 1;25(19):1999–2010. doi: 10.1101/gad.17558811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011 Mar 1;25(5):460–70. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011 Apr 1;25(7):717–29. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lock R, Roy S, Kenific CM, Su JS, Salas E, Ronen SM, et al. Autophagy facilitates glycolysis during Ras-mediated oncogenic transformation. Mol Biol Cell. 2011 Jan;22(2):165–78. doi: 10.1091/mbc.E10-06-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Debnath J. Detachment-induced autophagy in three-dimensional epithelial cell cultures. Methods Enzymol. 2009;452:423–39. doi: 10.1016/S0076-6879(08)03625-2. [DOI] [PubMed] [Google Scholar]
- 32.Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer. 2009 Aug;9(8):550–62. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- 33.Janku F, McConkey DJ, Hong DS, Kurzrock R. Autophagy as a target for anticancer therapy. Nat Rev Clin Oncol. 2011;8(9):528–39. doi: 10.1038/nrclinonc.2011.71. [DOI] [PubMed] [Google Scholar]
- 34.Moscat J, Diaz-Meco MT. Feedback on Fat: p62-mTORC1-Autophagy Connections. Cell. 2011 Nov 11;147(4):724–7. doi: 10.1016/j.cell.2011.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.