Abstract
Purpose
To quantitatively measure tCho levels in healthy breasts using Proton-Echo-Planar-Spectroscopic-Imaging (PEPSI).
Material and Methods
The 2-dimensional mapping of tCho at 3 Tesla across an entire breast slice using PEPSI and a hybrid spectral quantification method based on LCModel fitting and integration of tCho using the fitted spectrum were developed. This method was validated in 19 healthy females and compared with single voxel spectroscopy (SVS) and with PRESS prelocalized conventional Magnetic Resonance Spectroscopic Imaging (MRSI) using identical voxel size (8 cc) and similar scan times (~7 min).
Results
A tCho peak with a signal to noise ratio larger than 2 was detected in 10 subjects using both PEPSI and SVS. The average tCho concentration in these subjects was 0.45 ± 0.2 mmol/kg using PEPSI and 0.48±0.3 mmol/kg using SVS. Comparable results were obtained in 2 subjects using conventional MRSI. High lipid content in the spectra of 9 tCho negative subjects was associated with spectral line broadening of more than 26 Hz, which made tCho detection impossible. Conventional MRSI with PRESS prelocalization in glandular tissue in two of these subjects yielded tCho concentrations comparable to PEPSI.
Conclusion
The detection sensitivity of PEPSI is comparable to SVS and conventional PRESS-MRSI. PEPSI can be potentially used in the evaluation of tCho in breast cancer. A tCho threshold concentration value of ~0.7mmol/kg might be used to differentiate between cancerous and healthy (or benign) breast tissues based on this work and previous studies.
Keywords: MR spectroscopic imaging, breast spectral quantification, spectral modeling, echo-planar spectroscopic imaging, prelocalization
Introduction
The overall specificity of MRI in breast cancer diagnostics has been low, with the result that a considerable number of biopsies have been performed on lesions subsequently determined to be benign. It has recently been found (1–3) that adding the results from quantitative MR spectroscopic (MRS) measurements of total choline-containing compounds (tCho) to dynamic contrast enhanced (DCE) MRI exams improved the sensitivity, specificity, accuracy, and inter-observer agreement of breast cancer diagnosis. Several studies have observed an increased tCho concentration in breast cancer patients compared to healthy control subjects (1,4,5). A second promising application of breast MRS involves predicting the individual response to treatment (6–8). A decrease in the concentration of tCho within 24 hours after the first dose of chemotherapy may serve as an early indicator of the clinical response for locally advanced breast cancer (9,10). A working hypothesis behind these findings is that an elevated tCho concentration, which can be measured by MRS, is an indicator of the increased cellular proliferation, membrane breakdown, and cell density that characterizes the neoplasic state (11). In order for this change in Choline metabolism to be used clinically, the normal concentration of tCho, and its range of variation, must be known so that alterations in tCho can be accurately interpreted.
Conventional single-voxel spectroscopy (SVS) does not allow characterization of lesion heterogeneity. MR spectroscopic imaging (MRSI) is a more desirable technique for clinical applications, since it offers opportunities for whole breast examination, the demonstration of lesion heterogeneity, and the efficient characterization of multi-focal lesions. Furthermore, spectroscopic imaging allows for retrospective adjustment of the voxel position along the imaging dimensions, reducing the need for accurate voxel placement during the data acquisition.
The feasibility of mapping tCho, water and lipids in breast cancer patients using single slice MRSI at 1.5 T was demonstrated by Jacobs et al. (12), who reported (13) that the tCho signal to noise ratio (SNR) in malignant tissues was significantly higher than that found in benign breast abnormalities. One marker for cancer diagnosis could therefore be the tCho SNR. Baek et al. (14) showed that proper adjustment of the tCho SNR threshold based on receiver operating characteristic (ROC) curves, resulted in a total accuracy of 81% in differentiating malignant tissues from benign tissues in a group of 36 patients who underwent an MRSI exam. However, the tCho SNR depends on measurement sensitivity and therefore varies with field strength, coil design, voxel size, and signal averaging.
MRSI in the breast currently suffers from two important limitations. The first is that conventional, phase-encoded MRSI methods perform spatial encoding over long time durations, which increases their sensitivity to motion and frequency shifts due to respiration, and prevents 3D mapping in the clinical setting. This also makes quantification using water as an internal reference peak prohibitively time consuming. The slow encoding speed also prevents implementation of TE averaging, which has been shown to reduce gradient sideband artifacts (15), and of 2D NMR spectroscopy, which has been shown to improve identification of tCho in the presence of confounding lipid signals (16–18). The second limitation is that PRESS prelocalization has been needed in all MRSI studies to prevent the spatial leakage of overwhelming lipid signals from adipose tissue outside the region of interest, but placement of the PRESS volume is time consuming and prone to operator errors. Mapping the entire breast without prelocalization is desirable in order to study multi-centric and multi-focal breast cancer and to avoid PRESS volume placement errors.
High-speed proton MRSI with interleaved spatial-spectral gradient encoding, which is based on earlier work by Mansfield and others (19–31), is increasingly becoming a practical alternative to conventional phase-encoded spectroscopic imaging techniques. It is possible to integrate this MRSI with parallel imaging to further accelerate encoding speed. These high-speed MRSI techniques offer more than an order-of-magnitude temporal acceleration while also providing the high spectral resolution, adequate spectral widths, the feasibility of short echo times and the excellent sensitivity per unit time and volume, of conventional spectroscopic imaging techniques (30). In our former work, the proton-echo-planar-spectroscopic-imaging (PEPSI) technique with high spatial resolution and in-line data reconstruction has been developed(31) on clinical scanners. To allow for very short echo time acquisition for water referencing, single spin echo with slice selection, instead of double spin echo with PRESS box selection, was implemented. Adaptation of this methodology for breast studies in which data are acquired under more challenging conditions, such as off-center volume selection and considerable magnetic field inhomogeneity across the volume of interest, is desirable.
MRSI across large parts of the breast without volume prelocalization is technically more challenging than SVS. The definition of the baseline underlying the tCho peak suffers from partial volume contamination from lipid signals, as well as from inhomogeneous line broadening and from Gibbs ringing from lipid signals in adjacent voxels. The difficulty of shimming in the presence of dominating lipid signals as well as inaccurate B1 calibration over a larger imaging volume further impairs MRSI measurements of tCho in the breast. Spectral fitting with linear interpolation of the baseline, which has been developed for single voxel MRS(1), has not yet been adapted for MRSI. The LCModel software package, which is widely used to quantify brain spectra (32), has recently been extended to quantify breast spectra(33). However, in our experience with MRSI data, both in vitro and in vivo, LCModel fitting of breast spectra is very challenging, since the distorted lipid peaks in lipid-suppressed breast MRSI spectra are insufficiently represented in the standard LCModel basis set. The residual lipid peaks near 2.8, 2.3, 2.0, 1.3, and 0.9 ppm are broad and their line shapes vary from pixel to pixel, which requires manual adaptation of the LCModel fitting parameters on a voxel-by-voxel basis. In addition, the proximal 2.8 ppm lipid peak distorts the baseline in the vicinity of the tCho signals at 3.22 ppm. In regions with a low tCho concentration this can lead to overfitting of the baseline with a high order polynomial curve, resulting in wrong estimation of the tCho peak area. It is desirable to automate the quantification of MRSI data sets acquired without volume prelocalization in a manner that is insensitive to considerable lipid contamination.
In this study we demonstrate the feasibility of tCho measurements using 2-dimensional high-speed Proton-Echo-Planar-Spectroscopic-Imaging (PEPSI) without in-plane volume prelocalization(31). We used whole-slice PEPSI, with weighted averaging, to quantitatively map total Choline (tCho) with respect to tissue water in 19 healthy subjects. These results were compared with tCho quantification by means of both PRESS-CSI and single voxel spectroscopy. Automated spectral quantification of tCho with respect to tissue water was performed using a combination of LCModel fitting with a custom-designed spectral model and spectral integration of tCho in the fitted spectrum. This approach using a noise-free representation of the spectrum enabled reliable modeling of a linear baseline underneath the tCho peak and was found to be robust with respect to considerable lipid contamination. A secondary aim of this study was to establish the mean, standard deviation and an upper limit of the normal tCho concentration in healthy controls as a reference for volumetric mapping of tCho in patients with breast cancer, which is currently under way in our laboratory.
Materials and Methods
Human Subjects and Scanners
This study was approved by the institutional review boards at both participating institutions. Written informed consent was obtained before participation. In vivo experiments in 19 healthy volunteers were performed at two sites, both using a 3T MR scanner (Tim Trio, Siemens, Erlangen, Germany). One site (UMN) was equipped with 4-channel breast coil from Siemens (Erlangen, Germany) and the other site (Mind Research Network) was equipped with an 8-channel breast coil (Sentinelle Medical Inc, Canada). Second order automated shimming was performed across the entire breast region encompassed within the PEPSI slice.
Phantom
A breast phantom used for in vitro tests at both sites consisted of a cylindrical, 2 liter bottle filled with canola oil. A 40mm diameter sphere, containing 1mM phosphocholine chloride, 10mM trimethylsilyl-propionic acid (TSP) and 0.2mM Gd-DTPA in 0.9% saline, was placed within and below the center of the cylinder (34).
MR Imaging and MR Spectroscopic Imaging
Breast anatomy was imaged using a bilateral localizers and 3D axial MRI scans. Then, a 2D high resolution multi-slice FLASH scan was performed using: TR/TE = 156 ms/2.75 ms, FOV = 280×280 mm, slice thickness = 3 mm, number of slices = 8, TA = 1 minute. Water and lipid suppressed (WS) and non-water suppressed (NWS) PEPSI scans were acquired using the same slice orientation as the 2D FLASH scan with the following parameters: TR/TE=1500 ms/12 5ms, 32×32 matrix size, FOV = 640 mm×640 mm, slice thickness = 20 mm, and a voxel size of 8 cc. This relatively large voxel was used in order to match the voxel size typically used for SVS in the human breast and to provide adequate sensitivity for detecting the expected low tCho concentration in healthy breast tissue.
The PEPSI pulse sequence consisted of the following modules: WET water suppression (35), spin-echo RF excitation with MEGA lipid suppression (36), and echo-planar readout with 2048 readout gradients. The reconstructed spectral width after even-odd echo editing was 1087 Hz, with 1 Hz digital spectral resolution. Sampling on the readout gradient ramps was performed to maximize sensitivity. Two-fold oversampling and regridding to correct ramp sampling were used in the readout direction. For the WS PEPSI scans, weighted averaging was used in the phase encoding (PE) direction to further decrease acquisition time. The acquisition time consisted of 6.3 minutes for the WS scan and 48 seconds for the single NWS reference scan, resulting in a total acquisition time of 7.1 minutes.
SVS was performed with PRESS localization and MEGA lipid suppression, using TR/TE = 3 s/125 ms and a 2×2×2 cm voxel. SVS water reference scan was acquired in the voxel using TR = 3s, and TE = 50ms or 125ms, with 4 dummy scans. The single voxel was positioned in the center of glandular tissue. PRESS-CSI was performed with MEGA lipid suppression using identical TR/TE, slice selection and voxel size as the PEPSI scan and a 4×4 cm2 PRESS area selection. The PRESS-CSI water reference scan was performed with identical parameters but with water and lipid suppression disabled, requiring an acquisition time of 6.3 minutes. The scan times used for water-suppressed SVS and PRESS-CSI in vivo were similar to that for PEPSI, i.e., 6.3 minutes, whereas the total scan time including water reference acquisition was 6.5 minutes for SVS, and 12.6 minutes for PRESS-CSI.
Data Reconstruction
Reconstruction of PEPSI data was implemented on the scanner in the Integrated Computing Environment (ICE), which is part of the scanner operating system (Siemens, Erlangen, Germany). Following online regridding and downsampling of the oversampled ADC readout signal, data were saved in a data buffer according to coil, spectral and k-space dimensions, and the signals were averaged online. A Hamming filter (50% window) was applied across the k-space dimensions before performing a Fast Fourier Transform (FFT) and coil combination. For each voxel, the first measured data point on the NWS spectrum was used as the coil sensitivity metric. Coil combination of the WS and NWS spectra was performed using weighted averaging, with the weighting factors being the conjugate of the coil sensitivity data but normalized by the their sum of square (37). Even and odd gradient echo data were processed separately and averaged to maintain SNR and to avoid spectral ghosting. ICE reconstruction enabled display of the reconstructed spectra on the scanner immediately after the acquisition. The reconstructed spectral array data in DICOM format were then sent to a customized Matlab program for offline processing. Manual inversion of some of the spectra was necessary due to the phase correction using the oversuppressed water signal. The spectrum at each pixel was extracted and converted into LCModel format for spectral quantification. Coil combination and spatial reconstruction of SVS and PRESS-CSI data was performed using the manufacturer’s software.
Spectral Quantification
To illustrate the challenges of fitting breast MRSI data we examined sample data sets. LCModel software using the standard breast-3 basis set is unable to adequately fit the residual lipid peaks near 2.8, 2.3, 2.0, 1.3, and 0.9 in breast MRSI data, which often have broad and irregular line shapes. Fig1a demonstrates a non-ideal LCModel fitting result of a SVS spectrum. LCModel fitting of CSI breast data is even more challenging due to the stronger lipid contamination and line shape distortion as a result of Gibb’s ringing. The large residual lipid signal tends to bias the parameterization of LCModel fitting, which may lead to incorrect estimation of the tCho concentration. A modified weighting factor for lipid peaks might be used to solve this issue, but this is beyond the scope of the present study.
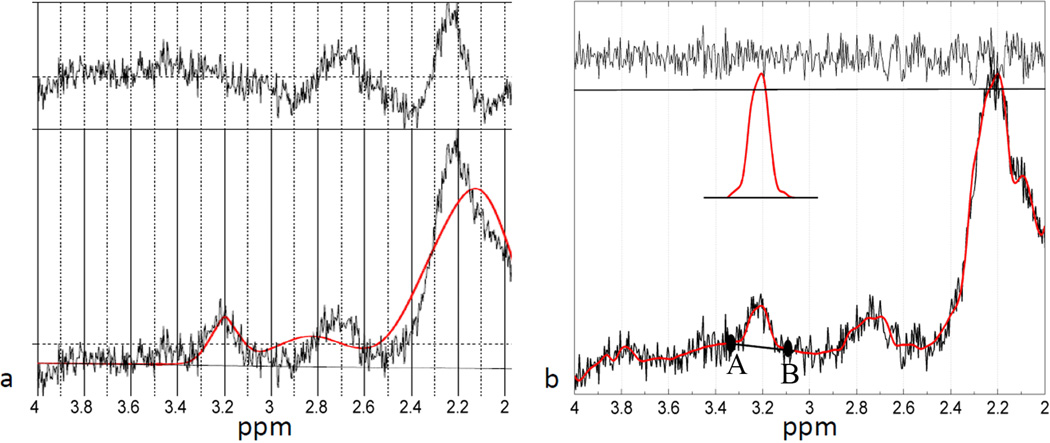
Figure 1.
Display of the hybrid tCho quantification procedure on a SVS spectrum. (a) LCModel fitting using the default basis-set shows incapability of fitting the ppm range where residual lipid resides. (b) Demonstration of the LCModel fitting using the customized basis-set over the [4.0 – 2.0] ppm range. The LCModel fit is shown as smooth curve overlaid on the original noisy spectrum. In the fitted spectrum, the baseline underlying the tCho peak is detected by finding the local minima at points A and B, to the right and left of 3.22ppm within a 0.15ppm distance. The tCho peak with a three time magnification is shown above the spectrum. Also shown is the residual noise on top of the plot.
In this work, we used a two-step, hybrid approach to calculate the tCho area in the WS spectrum. In the first step, we obtained a noise-free representation of the raw data using LCModel. In principal, any curve fitting algorithm, such as b-spline fitting, can be adopted to produce a smooth spectrum, as long as it preserves spectral details of the spectrum. Here, we utilized the superior fitting capability of LCModel using a customized basis set that contains 11 Lorentzian singlet peaks representing tCho (δ = 3.22 ppm) and 10 peaks that accommodate broad and irregular line shapes of residual lipid signals within the 2.0 – 2.9 ppm range. The basis set was optimized empirically by fitting different number of simulated Lorentzian peaks in the 2.0 to 2.9 ppm range with the goal of minimizing excessive modulation of the fitted baseline that negatively impacts the quantification of tCho. Further increase of the number of basis functions did not improve the fit quality.
In the second step, the outputs of LCModel, including the original and the fitted spectra were further processed using custom software written in Matlab (Mathworks 2008a) to calculate the tCho area and the residual noise level. The tCho peak was identified in the fitted WS spectra by finding the local maximum in the vicinity of 3.22ppm. Then, the two data points with local minimum values in the 3.07 – 3.22 and the 3.22 – 3.37 ppm ranges were selected in the fitted spectrum. The baseline underlying the tCho peak was modeled by linear interpolation using these two points. The tCho signal was integrated above this baseline to yield the tCho area. Residual noise was the difference between the fitted spectrum and the original spectrum. The tCho SNR values were calculated based on tCho peak amplitudes and the standard deviation of the residual noise. The hybrid approach performed on the same SVS spectrum as in Fig1a provides a strongly improved estimate of the tCho area (Fig. 1b).
In the NWS spectra, water and total lipid areas were calculated by integrating the ranges from 4.0 to 5.0 ppm, and 0.4 to 2.6 ppm, respectively. The millimolal concentration of tCho (millimoles of tCho per kilogram of water) was calculated using equation (1):
| [1] |
where ηwater = 2 and ηCho = 9 are number of protons per molecule of water and tCho, respectively. MWwater = 18 g/mol is the molecular weight of water. Consistent with our previous studies, molal instead of molar concentration was used because the density of MR-visible water is unknown for each voxel, due to the variable partial volume contribution of the lipids(1).
The relaxation parameters of water and tCho in breast tissues were taken from previous studies (1,5,14,38–41). Table 1 shows that the results vary between healthy and cancerous tissue. The reported T1 times of water in healthy fibroglandular tissue (5,40,41) are ~40% longer than in cancerous tissue (1,5,14,38,39). On the other hand, these studies showed that the water T1 values in each tissue type, healthy or cancerous, are relatively consistent among studies. Due to the low concentration of tCho in healthy tissue, the direct measurement of the T1 of tCho is challenging in individual subjects and was not performed in this study. In keeping with previous studies (e.g. (5)) we thus assumed that the T1 value of tCho is at the same level as the water T1 value in healthy breast tissue (1,5), therefore neglecting the T1 weighting in equation 1. To correct for transverse relaxation, we used T2,water = 60ms, T2,Cho=399ms, consistent with previous studies(1). For the in vivo experiments, SNR=2 was used as the threshold to determine whether the tCho signal was detectable or not.
Table 1.
Relaxation times (ms) reported in Breast MRS studies
| Sample | Reference | T2 Cho [ms] |
T2 H2O [ms] |
T1 Cho [ms] |
T1H2O [ms] |
Field Strength [T] |
tCho Concentration |
|---|---|---|---|---|---|---|---|
| Cancer Patients | Bolan 2003 | 399 | 60 | 870 | 4 | 1.38–10 mmol/kg | |
| Baik 2006, 2008 | 269 ± 61 | 97 ± 10 | 1513 ± 156 | 746 ± 118 | 1.5 | 0.76–21.20 mmol/kg | |
| Sijens 2010 | 305† | 97 | 1513 | 746 | 1.5 | 2.8 & 3.4 mM | |
| Bakken 2001 | 340 | 1.5 | |||||
| Healthy Controls | Rakow-Penner 2006 | 57.51 | 1136, 1266* | 1.5 | |||
| Rakow-Penner 2006 | 54.36 | 1324, 1445* | 3 | ||||
| Graham 1997Δ | 40 | 1301 | 1.5 | ||||
| Sijens 2010 | 311 | 49◊ | 1240 | 1317 | 1.5 | 0.3 & 0.6 mM | |
| Phantom | Baik 2006 | 485–664 | 1122–1282 | 2301–2415 | 2574–2619 | 1.5 |
Average value based on Baik 2006 and Bakken 2011.
Values measured using two techniques.
Values measured in ex vivo breast tissue.
Average value based on Graham 1997 and Penner 2006.
Results
Method validation in Breast Phantom
Comparison of the application of conventional PRESS-CSI and PEPSI sequence in a breast phantom showed that each sequence produced comparable spectral quality with similar lipid contamination. The PEPSI sequence was slightly modified to incorporate double spin echo excitation with PRESS selection to best match the product design of CSI to facilitate comparison. The line width of the resonances (mean ± standard deviation) observed using the MRSI techniques was found to be slightly larger (6.5 ± 2.1 Hz) than that obtained from the SVS technique (5.4 Hz) due to the larger shim volume needed for MRSI. The line-shapes and line-widths obtained from PEPSI and CSI, on the other hand, are very close, which indicates that the eddy current effect introduced by the echo planar acquisition is negligible. In addition, the tCho SNRs per unit acquisition time and unit volume measured with PEPSI and CSI were also very comparable, as expected. The choline concentration measured with all 3 methods was within ~10% of the true phantom concentration of 1.0 mM: 0.95 mM for SVS, 0.92 ± 0.23 mM for PRESS-PEPSI, and 1.02 ± 0.20 mM for PRESS-CSI.
In vivo measurements
PEPSI and SVS data from fibro-glandular tissue were collected from 19 subjects. A tCho resonance with a signal to noise ratio greater than two was detected in 11 of these 19 subjects using SVS. Among these 11 subjects, 10 subjects showed detectable tCho (SNR > 2) on PEPSI scans. The group of 11 subjects in whom tCho was detected in SVS scans is referred to as the choline-positive (Cho+) group. The remaining group of 8 subjects with non-detectable tCho levels is referred to as the choline-negative (Cho−) group. By visual inspection, the SVS spectra exhibited less baseline distortion due to lipid contamination than the PEPSI data. Across the 19 subjects in whom both SVS and PEPSI scans were performed, the NWS SVS spectra showed narrower water lines (21.8 ± 8.6 Hz) than the NWS PEPSI spectra (33.9 ± 12.6Hz), whereas the linewidths of the lipid methylene peaks at δ = 1.3 ppm obtained using the two techniques were comparable (33.5 ± 8.0 Hz vs. 33.2 ± 4.1 Hz). The narrower water linewidth obtained using SVS was due to the smaller shim volume used in SVS compared to the PEPSI scans. However, the water linewidth measured in the PEPSI data showed a positive correlation (p < 0.003) with the water linewidth measured in SVS data across subjects (Fig. 2). For PEPSI spectra alone, Cho+ water linewidth (22.7 ± 7.4Hz) is much smaller (p < 0.005) than Cho− water linewidth (41.3 ± 6.9Hz). Similarly, for SVS water spectra, Cho+ water linewidth (15.8 ± 6.7Hz) presented significantly smaller (p<0.005) water linewidth than that of the Cho− group (26.5 ± 7.3Hz). Furthermore, subjects in the Cho− group presented a much larger lipid signal in both SVS and PEPSI scans than those in the Cho+ group.
Figure 2.
Characterization of water line width. Subjects in Cho- group presented a much larger water line-width than the Cho+ group. A positive cross correlation between the water line-width values obtained using the two techniques is clearly seen (p<0.003).
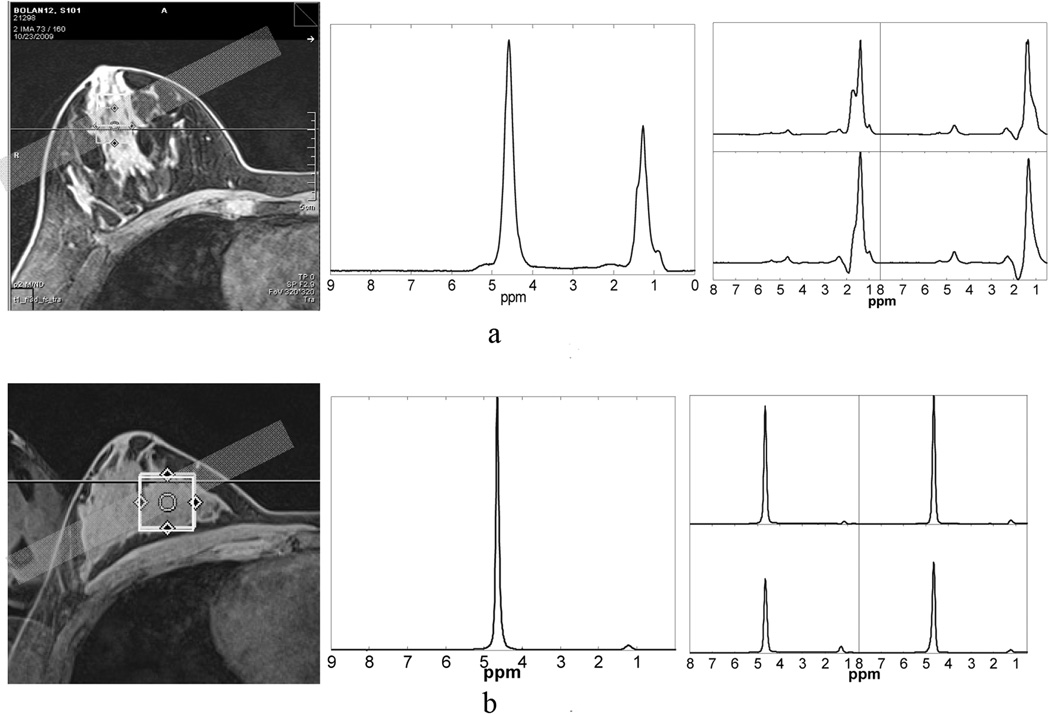
The heterogeneous distribution of water and lipid across breast tissue influences the spectral quality, and hence the detection of tCho, as illustrated in Fig. 3, where images are shown from axial scans from breasts with both high and low lipid contents. The PEPSI and SVS positions are indicated along with the SVS NWS and PEPSI NWS spectra. Fig. 3a shows data from a breast with large glandular lipid regions and considerable tissue heterogeneity. The water linewidths in the NWS spectra were 31.3 Hz for SVS and 44.2 ±3.4 Hz for PEPSI. A tCho resonance was not detectable in this subject. In contrast, Fig. 3b shows an MR image of a more homogeneous breast with denser fibro-glandular tissue (subject 8 in Table 2). In this case, the spectra exhibited much narrower water linewidths for SVS and PEPSI (15.6 Hz and 26.9±3.4 Hz, respectively) and we observed a detectable tCho peak at 3.22ppm, which yielded measured tCho concentrations using SVS and PEPSI of 0.61 mmol/kg and 0.44±0.38 mmol/kg, respectively.
Figure 3.
Influence of lipid content on tCho detection sensitivity. From left to right the axial MRI, NWS SVS and NWS PEPSI spectra obtained from 4 voxels in (a) a breast with high lipid content and (b) a breast with low lipid content are shown. For the subject in (a), tCho was not detectable. PEPSI spectra show small water peaks due to the large lipid content in the 4 voxels, while the SVS voxel was carefully positioned over the glandular tissue, yielding a larger water signal. The subject in (b) exhibited narrower water and lipid line-width and tCho was detected with measured concentrations of 0.61mmol/kg in SVS data and 0.44mmol/kg (mean) in the PEPSI data (subject 10 in Table 2).
Table 2.
Quantification of tCho concentration and SNR in vivo in the tCho+ group using SVS, PRESS-CSI and PEPSI.
| Coil | Subject No. |
Age | tCho Concentration (mmol/kg) and SNR | |||||
|---|---|---|---|---|---|---|---|---|
| SVS | PEPSI | CSI | ||||||
| [tCho] | SNR | [tCho] | SNR | [tCho] | SNR | |||
| 4-Channel | 1 | 20 | 0.17 | 2.62 | 0.65 ± 0.16 (n=11) | 4.88 ± 1.20 | ◊ | |
| 2 | 20 | 0.71 | 5.54 | 0.62 ± 0.19 (n=5) | 2.61 ± 0.32 | |||
| 3 | 25 | 0.25 | 2.58 | 0.68 ± 0.08 (n=2) | 2.89 ± 0.78 | |||
| 4 | 22 | 0.26 | 5.94 | 0.23 ± 0.19 (n=4) | 2.30 ± 0.23 | |||
| 5 | 21 | 0.18 | 4.95 | 0.32 (n=1) | 2.01 | |||
| 6 | 26 | 0.98 | 4.88 | 0.56 ± 0.23 (n=4) | 2.95 ± 0.43 | |||
| 8-Channel | 7 | 20 | 0.37 | 3.16 | 0.68 ± 0.33 (n=6) | 9.88 ± 3.85 | 0.53 ± 0.37 (n=4) | 11.9 ± 5.14 |
| 8 | 20 | 0.61 | 3.98 | 0.44 ± 0.38 (n=11) | 10.3 ± 6.70 | ◊ | ||
| 9 | 26 | 0.91 | 4.49 | 0.15 ± 0.04 (n=4) | 4.74 ± 0.92 | 0.15 ± 0.02 (n=4) | 5.22 ± 1.39 | |
| 10 | 22 | 0.74 | 2.76 | * | * | |||
| 11 | 29 | 0.14 | 2.66 | 0.17 ± 0.04 (n=6) | 4.30 ± 0.81 | ◊ | ||
| Mean | 22.8±3.1 (n=11) | 0.48 ± 0.3 (n=11) | 3.96 ± 1.3 (n=11) | 0.45 ± 0.21 (n=10) | 4.69 ± 3.02 (n=10) | 0.34 ± 0.3 (n=2) | 8.56 ± 4.7 (n=2) | |
Experiments not performed or completed due to time limitations or subject discomfort.
No tCho detected.
Quantification of tCho in vivo
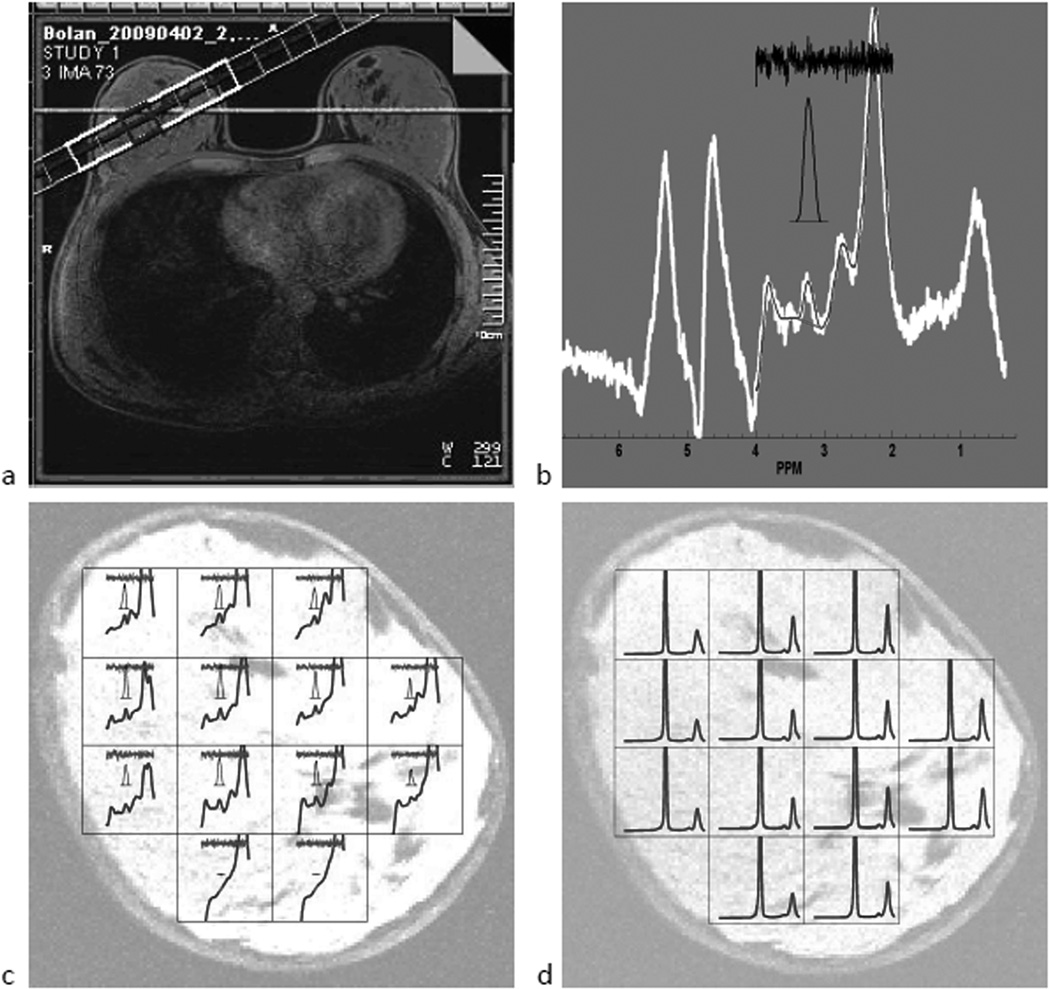
The hybrid approach for tCho measurement in PEPSI data is illustrated in Fig. 4 for subject 1 in Table 2. An oblique 2D coronal PEPSI slice (Fig. 4a, left) was overlaid on the axial breast image and the PEPSI spectral grid was placed on the oblique image (Fig. 4a, right). An example spectrum from a central voxel (Fig. 4b) shows the results of the hybrid fitting procedure. The raw spectrum is displayed here in white, while the fitted spectrum, over the 2 – 4 ppm range is shown as a narrow black line. The residual noise is shown on top in black (Fig. 4b) and the 3-fold magnified tCho peak is displayed between the residual noise and the fitted spectrum. In this spectrum, a tCho peak with a SNR of 4.7 was detected at 3.22 ppm. The WS spectra from the entire 13-voxel PEPSI array within the breast are overlaid onto a breast MRI with the same slice orientation and position as the PEPSI scan in Fig. 4c. At each grid point, from top to bottom, the residual noise, the detected tCho peak with 3-fold magnification, and the fitted spectrum across the 2 – 4 ppm range are displayed. In this particular subject, a total of 11 voxels showed tCho signals with SNR greater than 3. For these 11 voxels, the average SNR was 4.9 ± 1.1. The corresponding NWS spectral array is shown in Fig. 4d. In this subject the water and 1.3 ppm lipid peak linewidths (mean ± standard deviation) were 24.8 ± 7.1 and 51.3 ± 13.0, Hz respectively. The average tCho/water area ratio was (3.1 ± 0.78)×10−4. The millimolal concentration (millimoles of solute per kilogram of water) of tCho was found to be 0.65 ± 0.16 mmol/kg.
Figure 4.
Example of 2D spectral array obtained from a subject with small lipid to water ratio (no. 1 in Table 2) (a) PEPSI slice positioning overlaid on axial and oblique coronal MRIs. (b) A single spectrum showing the tCho and residual lipid peaks. (c) WS spectral array overlaid onto the MRI image. (d) water reference spectral array.
Overall, the average tCho concentrations in the Cho+ group, in healthy breasts measured using SVS and PEPSI, were found to be 0.48 ± 0.31 mmol/kg (n = 11) and 0.45 ± 0.21 mmol/kg (n = 10), respectively. The individual tCho quantification results obtained from SVS and PEPSI measurements for the Cho+ group are given in Table 2. In this study, measurement of the tCho concentration in 2 subjects using conventional CSI with PRESS prelocalization in fibro-glandular tissue, yielded a value of 0.34 ± 0.27 mmol/kg. The tCho concentration obtained using PEPSI in healthy breast tissue was found to be not significantly different from that determined using either SVS or PRESS-CSI sequences.
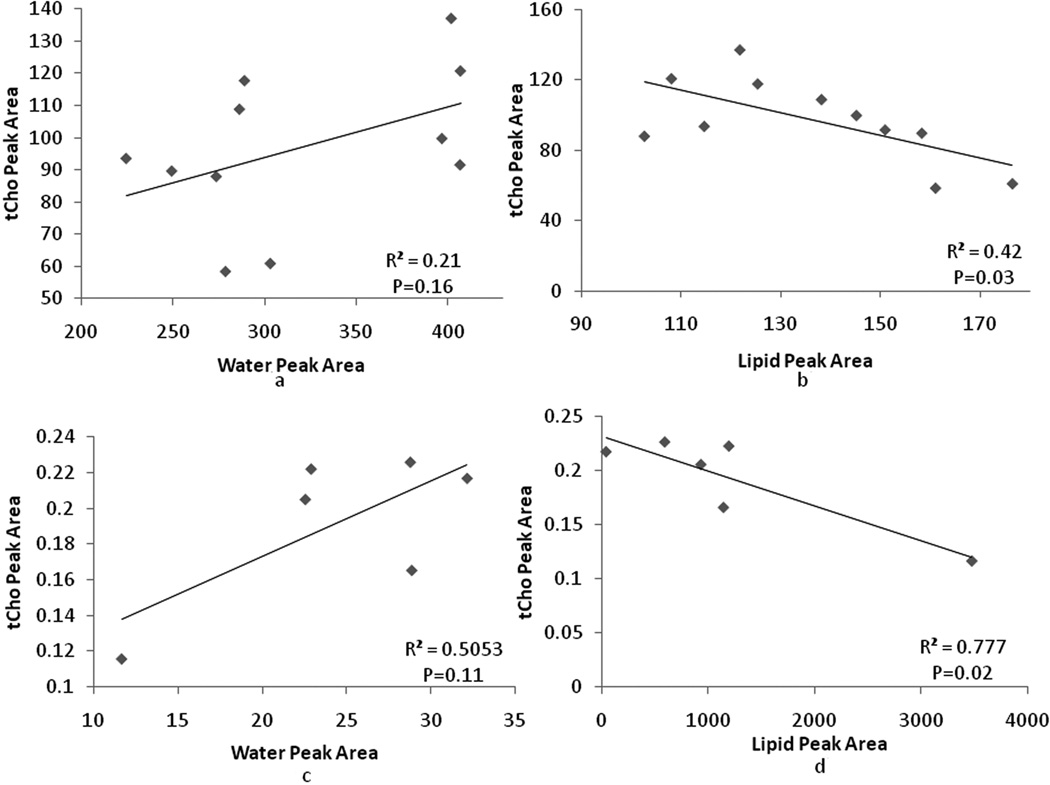
An interesting quality check on the methods used is to determine the relationship between the measured tCho integrals and those from the water and lipid signals. Choline-containing compounds are water, and not lipid, soluble. Therefore, one expects to find a direct correlation between the choline and water signal integrals, and an inverse correlation between the choline and lipid signal integrals in each voxel in each subject. Linear regressions between water, lipid and tCho peaks across voxels were performed for each of the four subjects who had 6 or more voxels with detectable tCho peaks. For subject 8, a significant positive correlation between the tCho and water peak areas was observed (p<0.05). For subject 1 and 11, a significant negative correlation between tCho and lipids were observed (p<0.05). For subject 7, the correlation between tCho and lipids showed a trend with a p-value of 0.08. Other regression tests between, tCho and water, tCho and lipids peak areas were not statistically significant and only showed trends. As examples, the tCho-water and tCho-lipid area correlations for subject 1 and 11 are shown in Fig. 5. The observed trends between tcho and water, tCho and lipids were consistent in all data sets with 6 or more tCho positive pixels. These observed correlations between tCho and water or lipids also indicate that lipid contamination in the quantification of tCho was limited. Voxels with larger tCho signals contained more cellular water, and less lipid.
Figure 5.
Cross correlation between water, lipid and tCho peak areas across voxels in which tCho was detected. Examples from two of the subjects in Table 2 are shown: (a) and (b) shows regression between tCho and water/lipid peak area for subject 1. (c) and (d) shows the regressions for subject 11.
Discussion
The concentration of tCho in the healthy breast
The results presented here show that choline compounds are present in healthy breasts at concentrations detectable using MRS on clinical instruments. Ten out of 19 (53%) subjects showed a measureable tCho signal using PEPSI. The mean tCho concentration was 0.45 ± 0.21 mmol/kg. For each individual subject, the tCho level across the breast varied only moderately, as indicated by the deviation of tCho concentration for each subject in Table 2, indicating a relatively homogeneous distribution of tCho concentration in normal fibroglandular breast tissue.
A previous study at 4 Tesla found that tCho levels of more than 1.0 mmol/kg distinguished malignant lesions from benign lesions and normal tissue(1). In the same study, four of five normal subjects with a detectable tCho peak exhibited tCho levels of 0.3 – 0.7 mmol/kg. In a recent CSI study in breast cancer, an average tCho concentration of 0.45 mmol/kg was reported(5) in one healthy control subject. Another breast MRS study at 7 T revealed a tCho concentration of 0.7 mmol/kg in healthy control subjects (4). Thus, the tCho concentration in healthy female subjects measured in our study was consistent with previously reported values. At 1.5 T, a previous study reported that 3 of 40 healthy (mean age 44 years) non-lactating females demonstrated a composite signal at 3.22 ppm on MRS examination(42). The relatively high lipid content in post-menopausal females and the low signal sensitivity at 1.5 T may have contributed to the low detection rate for tCho in that study. In breast cancer patients, greater tCho concentration values with a considerable larger range (0.7 ~ 21.2 mmol/kg) have been reported(1,4,5,8,38). Therefore, a tCho concentration of greater than 0.7 mmol/kg might be a threshold for breast cancer diagnosis. The elevated tCho concentrations greater than 0.7mmol/kg might be in part due to lipid contamination resulting from insufficient lipid suppression of the MEGA pulses.
In this study, the average age of the volunteers was 23. A control group around this age may not be representative for females who are normally older and have a higher risk of breast cancer, which was a limitation of the present study. Young females tend to have denser breast tissue and the measurement of tCho concentration is relatively easier than in older individuals. Furthermore the tCho concentration may vary with age. Nevertheless, the obtained tCho concentrations may serve as a reproducible reference data set for setting thresholds in breast cancer diagnosis.
The identity of the tCho peak in healthy controls requires further characterization. One study at 1.5T reported that in healthy volunteers, both lactating and non-lactating females, a resonance peak located at 3.28ppm might be a composite peak resulting from myo-inositol/taurine/GPC (42). A recent in vivo study using localized COSY at 3T reported a peak at 3.4ppm from taurine in healthy breast tissue and much lower Cho/Fat3 ratio in healthy controls compared to ductal and lobular carcinoma (43). Further studies are needed to confirm these findings. Further characterization of the tCho peak in breast tissue may be achieved using 2D MRS approaches such as J-resolved spectroscopy (15) and COSY (18) integrated into PEPSI, however, this is beyond the scope of the present study.
While PEPSI has the same SNR per unit acquisition time as conventional MRSI techniques, its spatial encoding time is much shorter. To achieve equivalent SNR the overall acquisition time is the same with both PEPSI and conventional MRSI, but the PEPSI approach would average over multiple fully-encoded k-spaces. These distinct averages can be used to integrate well-established MRS techniques, such as TE averaging and 2D NMR spectroscopy, into the MRSI data acquisition to improve lipid suppression and unambiguous tCho detection in a clinical setting. This study shows that PEPSI offers comparable sensitivity to conventional CSI. Furthermore, PEPSI can feasibly be extended to 3D spectroscopic imaging in the breast; conventional CSI is not feasible for 3D acquisition due to the long acquisition time required. In principle, a 3D WS breast spectroscopic imaging can be performed using PEPSI in the same acquisition time as that used in this work (i.e. 6.3 minutes) without sacrificing the SNR.
The method for tCho quantification
In this work we assume that the observed tCho signal is only originating from tissue compartments containing tissue water. Because the MR-visible water content per unit volume of tissue is unknown for each voxel due to the variable partial volume contribution of the lipids, molal concentration was used. Further work is required to investigate the distribution of tCho in healthy breast tissue compartments.
Due to the variation in intensity, line-width and line-shape of residual lipid resonances across voxels, the quantification of tCho in water-, and lipid-suppressed breast spectra was not straightforward. The commonly-used polynomial fitting of the baseline was not sufficient in this case. Using a consistent parameter set for LCModel using the breast basis set (parameter SPTYPE = 'breast-3') to process all voxels automatically in bulk mode was not possible. Manual adjustments were frequently needed to quantify the breast spectra on a voxel by voxel basis. Therefore, a hybrid tCho quantification method was developed that used clusters of singlet peaks to fit the spurious residual lipid peaks around 2.3 and 2.8 ppm. This allowed fitting over the larger spectral range of 2 ppm compared to the standard LCModel fitting, which only worked well over a 0.5 ppm range around the tCho peak in the WS spectra. This customized basis set offered spectral quantification with reduced residuals, and enabled automated spectral array data processing. In this hybrid approach, the curve fitting functionality of LCModel was used only for the water suppressed spectra, whereas tCho quantification was performed based on a tCho-water area ratio calculations using spectral integration. The quantification results using the proposed hybrid method were compared with the results obtained from standard LCModel fitting over a 0.5 ppm spectral range around the tCho peak. The results were comparable for tCho peaks with a high SNR and a flat baseline. For tCho peaks with a large residual lipid peak at around 2.8 ppm, standard LCModel fitting consistently overestimated tCho due to instability of estimating the baseline with a high order polynomial function. In addition, the detection of the residual lipid peak around 2.3 and 2.8 ppm may have provided useful information for tCho baseline detection by compensating distortions caused by the slopes from lipid peaks in the vicinity of the tCho resonance.
The observed correlations between tCho and water or lipids (Fig. 5) indicated that lipid contamination in the quantification of tCho was limited. Voxels with larger tCho signals contained more cellular water, and less lipid. Nevertheless, lipid sidebands may have contributed to the composite signal at 3.22 ppm, and caused variation of the tCho concentration evaluation across the breast area for each individual subject. To remove this potential influence of the spurious lipid side band, TE averaging will be incorporated into the PEPSI sequence in a future technical implementation.
The effect of tissue heterogeneity on spectral quality
It was found that higher lipid content in breast tissues increased tissue heterogeneity and decreased spectral quality, making tCho detection in the high lipid-content healthy breast challenging. This conclusion was supported by the finding that the water linewidths measured using PEPSI and SVS sequences in the same subject’s voxels were positively correlated (Fig. 2), an indication that the residual magnetic field inhomogeneity after shimming was largely dependent on the intrinsic tissue heterogeneity. Further evidence for this effect of tissue heterogeneity on spectral quality was found from a comparison of the water linewidths between the Cho+ and Cho− populations. The width of the water line in the Cho+ group was consistently smaller than that found in the Cho− group.
This effect of tissue heterogeneity was also seen in previous studies, which set a threshold for the lipid to water ratio of 0.33 above which the data were considered unsuitable for fitting(1). Partial volume effects and susceptibility differences due to high lipid content in a voxel introduced line broadening, baseline distortion and spectral contamination, making tCho detection difficult. Spectral contamination from lipids in adipose tissue may propagate into voxels centered on fibroglandular tissue. Therefore, effective lipid suppression was critical during tCho measurement in the breast. MEGA lipid suppression substantially reduced the lipid peak, but residual B0 and B1 inhomogeneity limited the lipid suppression efficiency. Lipid suppression strategies under development, such as frequency selective refocusing(44) and inversion recovery nulling of the lipid signals, were beyond the scope of the current study.
Overall, the water linewidth measured in PEPSI spectra was ~50 % larger than that seen in the SVS spectra, indicating the challenge of shimming across the whole slice for the PEPSI scan. Tissue heterogeneity in the large shim volume and macroscopic magnetic field distortion at tissue edges likely contributed to the larger linewidth. Lipid contamination in SVS data was considerably smaller than that found with the whole-slice PEPSI technique, which was limited by Gibbs ringing associated with the Fourier Transform.
In conclusion, although the tCho level in healthy breasts was relatively low, it was feasible to measure its 2D concentration distribution across the entire breast using a short (7.1 min) PEPSI protocol at 3 Tesla. The mean concentration of tCho, measured using PEPSI in healthy females, was 0.45 mmol/kg, which was comparable to the value measured using SVS and conventional PRESS-CSI techniques. In breast MRS, the water linewidth and lipid to water signal ratio in the reference scan were important factors determining the detectability of tCho within the imaging volume. The application of PEPSI in the breast offered the advantage of minimizing the encoding time required for the acquisition of a water-suppressed data set and a non-water-suppressed reference data set compared to conventional CSI. This would enable integration of well-established MRS techniques, such as TE averaging and 2D NMR spectroscopy, into the MRSI data acquisition to improve data quality and spectral specificity in a clinical setting. The feasibility of studying tCho in healthy glandular tissue supported the potential of using PEPSI for the evaluation of tCho in breast cancer, where the concentration of tCho is considerably higher.
Acknowledgments
Grant Support: We acknowledge funding by grants NCI 1RC1EB010617-01, NIH P41 RR08079, R01 CA120509, NCI R01 CA123194, DoD postdoc award BC093217, and support from UNM cancer center. We thank Dr. Linda Casey and Dr. Robert Rosenberg for reading the breast MRI scans.
References
- 1.Bolan PJ, Meisamy S, Baker EH, et al. In vivo quantification of choline compounds in the breast with 1H MR spectroscopy. Magn Reson Med. 2003;50(6):1134–1143. doi: 10.1002/mrm.10654. [DOI] [PubMed] [Google Scholar]
- 2.Huang W, Fisher PR, Dulaimy K, Tudorica LA, O'Hea B, Button TM. Detection of breast malignancy: diagnostic MR protocol for improved specificity. Radiology. 2004;232(2):585–591. doi: 10.1148/radiol.2322030547. [DOI] [PubMed] [Google Scholar]
- 3.Meisamy S, Bolan PJ, Baker EH, et al. Adding in vivo quantitative 1H MR spectroscopy to improve diagnostic accuracy of breast MR imaging: preliminary results of observer performance study at 4.0 T. Radiology. 2005;236(2):465–475. doi: 10.1148/radiol.2362040836. [DOI] [PubMed] [Google Scholar]
- 4.Klomp DW, van de Bank BL, Raaijmakers A, et al. (31) P MRSI and (1) H MRS at 7 T: initial results in human breast cancer. NMR Biomed. 2011 doi: 10.1002/nbm.1696. [DOI] [PubMed] [Google Scholar]
- 5.Sijens PE, Dorrius MD, Kappert P, Baron P, Pijnappel RM, Oudkerk M. Quantitative multivoxel proton chemical shift imaging of the breast. Magn Reson Imaging. 28(3):314–319. doi: 10.1016/j.mri.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 6.Danishad KK, Sharma U, Sah RG, Seenu V, Parshad R, Jagannathan NR. Assessment of therapeutic response of locally advanced breast cancer (LABC) patients undergoing neoadjuvant chemotherapy (NACT) monitored using sequential magnetic resonance spectroscopic imaging (MRSI) NMR Biomed. 2010;23(3):233–241. doi: 10.1002/nbm.1436. [DOI] [PubMed] [Google Scholar]
- 7.Jagannathan NR, Kumar M, Seenu V, et al. Evaluation of total choline from in-vivo volume localized proton MR spectroscopy and its response to neoadjuvant chemotherapy in locally advanced breast cancer. Br J Cancer. 2001;84(8):1016–1022. doi: 10.1054/bjoc.2000.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Korteweg MA, Veldhuis WB, Visser F, et al. Feasibility of 7 Tesla Breast Magnetic Resonance Imaging Determination of Intrinsic Sensitivity and High-Resolution Magnetic Resonance Imaging, Diffusion-Weighted Imaging, and 1H-Magnetic Resonance Spectroscopy of Breast Cancer Patients Receiving Neoadjuvant Therapy. Invest Radiol. 2011;46(6):370–376. doi: 10.1097/RLI.0b013e31820df706. [DOI] [PubMed] [Google Scholar]
- 9.Haddadin IS, McIntosh A, Meisamy S, et al. Metabolite quantification and high-field MRS in breast cancer. NMR Biomed. 2009;22(1):65–76. doi: 10.1002/nbm.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meisamy S, Bolan PJ, Baker EH, et al. Neoadjuvant chemotherapy of locally advanced breast cancer: predicting response with in vivo (1)H MR spectroscopy--a pilot study at 4 T. Radiology. 2004;233(2):424–431. doi: 10.1148/radiol.2332031285. [DOI] [PubMed] [Google Scholar]
- 11.Bolan PJ, Nelson MT, Yee D, Garwood M. Imaging in breast cancer: Magnetic resonance spectroscopy. Breast Cancer Res. 2005;7(4):149–152. doi: 10.1186/bcr1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacobs MA, Barker PB, Argani P, Ouwerkerk R, Bhujwalla ZM, Bluemke DA. Combined dynamic contrast enhanced breast MR and proton spectroscopic imaging: a feasibility study. J Magn Reson Imaging. 2005;21(1):23–28. doi: 10.1002/jmri.20239. [DOI] [PubMed] [Google Scholar]
- 13.Jacobs MA, Barker PB, Bottomley PA, Bhujwalla Z, Bluemke DA. Proton magnetic resonance spectroscopic imaging of human breast cancer: a preliminary study. J Magn Reson Imaging. 2004;19(1):68–75. doi: 10.1002/jmri.10427. [DOI] [PubMed] [Google Scholar]
- 14.Baek HM, Chen JH, Yu HJ, Mehta R, Nalcioglu O, Su MY. Detection of choline signal in human breast lesions with chemical-shift imaging. J Magn Reson Imaging. 2008;27(5):1114–1121. doi: 10.1002/jmri.21309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bolan PJ, DelaBarre L, Baker EH, et al. Eliminating spurious lipid sidebands in H-1 MRS of breast lesions. Magnetic Resonance in Medicine. 2002;48(2):215–222. doi: 10.1002/mrm.10224. [DOI] [PubMed] [Google Scholar]
- 16.Mountford C, Ramadan S, Stanwell P, Malycha P. Proton MRS of the breast in the clinical setting. NMR Biomed. 2009;22(1):54–64. doi: 10.1002/nbm.1301. [DOI] [PubMed] [Google Scholar]
- 17.Thomas MA, Binesh N, Yue K, DeBruhl N. Volume-localized two-dimensional correlated magnetic resonance spectroscopy of human breast cancer. J Magn Reson Imaging. 2001;14(2):181–186. doi: 10.1002/jmri.1170. [DOI] [PubMed] [Google Scholar]
- 18.Thomas MA, Lipnick S, Velan SS, et al. Investigation of breast cancer using two-dimensional MRS. NMR Biomed. 2009;22(1):77–91. doi: 10.1002/nbm.1310. [DOI] [PubMed] [Google Scholar]
- 19.Mansfield P. Spatial mapping of the chemical shift in NMR. Magn Reson Med. 1984;1(3):370–386. doi: 10.1002/mrm.1910010308. [DOI] [PubMed] [Google Scholar]
- 20.Guilfoyle DN, Mansfield P. Chemical-shift imaging. Magn Reson Med. 1985;2(5):479–489. doi: 10.1002/mrm.1910020507. [DOI] [PubMed] [Google Scholar]
- 21.Matsui S, Sekihara K, Kohno H. High-speed spatially resolved high-resolution NMR spectroscopy. J Am Chem Soc. 1985;107(9):2817–2819. [Google Scholar]
- 22.Guilfoyle DN, Blamire A, Chapman B, Ordidge RJ, Mansfield P. PEEP--a rapid chemical-shift imaging method. Magn Reson Med. 1989;10(2):282–287. doi: 10.1002/mrm.1910100213. [DOI] [PubMed] [Google Scholar]
- 23.Webb P, Spielman D, Macovski A. A fast spectroscopic imaging method using a blipped phase encode gradient. Magn Reson Med. 1989;12(3):306–315. doi: 10.1002/mrm.1910120303. [DOI] [PubMed] [Google Scholar]
- 24.Twieg DB. Multiple-output chemical shift imaging (MOCSI): a practical technique for rapid spectroscopic imaging. Magn Reson Med. 1989;12(1):64–73. doi: 10.1002/mrm.1910120108. [DOI] [PubMed] [Google Scholar]
- 25.Posse S, DeCarli C, Le Bihan D. Three-dimensional echo-planar MR spectroscopic imaging at short echo times in the human brain. Radiology. 1994;192(3):733–738. doi: 10.1148/radiology.192.3.8058941. [DOI] [PubMed] [Google Scholar]
- 26.Posse S, Tedeschi G, Risinger R, Ogg R, Le Bihan D. High speed 1H spectroscopic imaging in human brain by echo planar spatial-spectral encoding. Magn Reson Med. 1995;33(1):34–40. doi: 10.1002/mrm.1910330106. [DOI] [PubMed] [Google Scholar]
- 27.Adalsteinsson E, Irarrazabal P, Spielman DM, Macovski A. Three-dimensional spectroscopic imaging with time-varying gradients. Magn Reson Med. 1995;33(4):461–466. doi: 10.1002/mrm.1910330402. [DOI] [PubMed] [Google Scholar]
- 28.Ebel A, Soher BJ, Maudsley AA. Assessment of 3D proton MR echo-planar spectroscopic imaging using automated spectral analysis. Magn Reson Med. 2001;46(6):1072–1078. doi: 10.1002/mrm.1301. [DOI] [PubMed] [Google Scholar]
- 29.Maudsley AA, Domenig C, Govind V, et al. Mapping of Brain Metabolite Distributions by Volumetric Proton MR Spectroscopic Imaging (MRSI) Magnetic Resonance in Medicine. 2009;61(3):548–559. doi: 10.1002/mrm.21875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pohmann R, von Kienlin M, Haase A. Theoretical evaluation and comparison of fast chemical shift imaging methods. J Magn Reson. 1997;129(2):145–160. doi: 10.1006/jmre.1997.1245. [DOI] [PubMed] [Google Scholar]
- 31.Posse S, Otazo R, Caprihan A, et al. Proton echo-planar spectroscopic imaging of J-coupled resonances in human brain at 3 and 4 Tesla. Magn Reson Med. 2007;58(2):236–244. doi: 10.1002/mrm.21287. [DOI] [PubMed] [Google Scholar]
- 32.Provencher SW. Estimation of metabolite concentrations from localized in vivo proton NMR spectra. Magn Reson Med. 1993;30(6):672–679. doi: 10.1002/mrm.1910300604. [DOI] [PubMed] [Google Scholar]
- 33.Provencher S. http://s-provencher.com/pages/lcmodel.shtml. [Google Scholar]
- 34.Bolan P, Garwood M, Rosen M, et al. Design of Quality Control Measures for a Multi-Site Clinical Trial of Breast MRS - ACRIN 6657. Proceedings of the 16th Annual Meeting ISMRM; Toronto, Canada. 2008. [Google Scholar]
- 35.Ogg RJ, Kingsley PB, Taylor JS. Wet, a T-1-Insensitive and B-1-Insensitive Water-Suppression Method for in-Vivo Localized H-1-Nmr Spectroscopy. Journal of Magnetic Resonance Series B. 1994;104(1):1–10. doi: 10.1006/jmrb.1994.1048. [DOI] [PubMed] [Google Scholar]
- 36.Mescher M, Merkle H, Kirsch J, Garwood M, Gruetter R. Simultaneous in vivo spectral editing and water suppression. NMR Biomed. 1998;11(6):266–272. doi: 10.1002/(sici)1099-1492(199810)11:6<266::aid-nbm530>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 37.Brown MA. Time-Domain Combination of MR pectroscopy Data Acquired Using Phased-Array Coils. Magnetic Resonance in Medicine. 2004;52(5):1207–1213. doi: 10.1002/mrm.20244. [DOI] [PubMed] [Google Scholar]
- 38.Baik HM, Su MY, Yu H, Mehta R, Nalcioglu O. Quantification of choline-containing compounds in malignant breast tumors by 1H MR spectroscopy using water as an internal reference at 1.5 T. MAGMA. 2006;19(2):96–104. doi: 10.1007/s10334-006-0032-4. [DOI] [PubMed] [Google Scholar]
- 39.Bakken IJ, Gribbestad IS, Singstad TE, Kvistad KA. External standard method for the in vivo quantification of choline-containing compounds in breast tumors by proton MR spectroscopy at 1.5 Tesla. Magn Reson Med. 2001;46(1):189–192. doi: 10.1002/mrm.1175. [DOI] [PubMed] [Google Scholar]
- 40.Graham SJ, Ness S, Hamilton BS, Bronskill MJ. Magnetic resonance properties of ex vivo breast tissue at 1.5 T. Magn Reson Med. 1997;38(4):669–677. doi: 10.1002/mrm.1910380422. [DOI] [PubMed] [Google Scholar]
- 41.Rakow-Penner R, Daniel B, Yu H, Sawyer-Glover A, Glover GH. Relaxation times of breast tissue at 1.5T and 3T measured using IDEAL. J Magn Reson Imaging. 2006;23(1):87–91. doi: 10.1002/jmri.20469. [DOI] [PubMed] [Google Scholar]
- 42.Stanwell P, Gluch L, Clark D, et al. Specificity of choline metabolites for in vivo diagnosis of breast cancer using 1H MRS at 1.5 T. Eur Radiol. 2005;15(5):1037–1043. doi: 10.1007/s00330-004-2475-1. [DOI] [PubMed] [Google Scholar]
- 43.Ramadan S, Box HN, Baltzer P, et al. Distinction of Invasive Lobular Carcinoma, Invasive Ductal Carcinoma, and Healthy Breast Tissue In Vivo With L-COSY at 3T. ISMRM 19th Annual Meeting & Exhibition; Montréal, Québec, Canada. 2011. [Google Scholar]
- 44.Choi C, Coupland NJ, Bhardwaj PP, Malykhin N, Gheorghiu D, Allen PS. Measurement of brain glutamate and glutamine by spectrally-selective refocusing at 3 tesla. Magnetic Resonance in Medicine. 2006;55(5):997–1005. doi: 10.1002/mrm.20875. [DOI] [PubMed] [Google Scholar]