Abstract
Cellular and plasma lipid levels are tightly controlled by complex gene regulatory mechanisms. Elevated plasma lipid content, or hyperlipidemia, is a significant risk factor for cardiovascular morbidity and mortality. MicroRNAs (miRNAs) are posttranscriptional regulators of gene expression and have emerged as important modulators of lipid homeostasis, but the extent of their role has not been systematically investigated. In this study, we performed high-throughput small RNA sequencing and detected approximately 150 miRNAs in mouse liver. We then employed an unbiased, in silico strategy to identify miRNA regulatory hubs in lipid metabolism, and miR-27b was identified as the strongest such hub in human and mouse liver. In addition, hepatic miR-27b levels were determined to be sensitive to plasma hyperlipidemia, as evidenced by its ~3-fold up-regulation in the liver of mice on a high-fat diet (42% calories from fat). Further, we showed in a human hepatocyte cell line (Huh7) that miR-27b regulates the expression (mRNA and protein) of several key lipid-metabolism genes, including Angptl3 and Gpam. Finally, we demonstrated that hepatic miR-27b and its target genes are inversely altered in a mouse model of dyslipidemia and atherosclerosis.
Conclusion
miR-27b is responsive to lipid levels, and controls multiple genes critical to dyslipidemia.
Keywords: miRNA, lipid, triglyceride, dyslipidemia, atherosclerosis, GPAM, ANGPTL3
Cellular and plasma lipid levels are tightly controlled by complex feed-back and feed-forward mechanisms, which regulate the expression and activity of key metabolic genes(1) at both the transcriptional and post-transcriptional levels(2, 3). Dysregulation of lipid metabolism can lead to hyperlipidemia, a major risk factor for cardiovascular disease(4). Several key process for regulating cellular and systemic lipid levels have been identified(5); however, post-transcriptional mechanisms remain less well characterized.
MicroRNAs (miRNAs) are short (~22 nt) non-coding RNAs that regulate gene expression at the post-transcriptional level(6, 7). They serve as stable plasma biomarkers for various disorders(8), are important factors in the pathogenesis of several diseases(9, 10), and are promising targets of novel therapeutic strategies(11, 12). In regard to lipid metabolic control, miRNAs have recently been found to modulate cholesterol homeostasis(13). In vivo inhibition of a liver-specific miRNA, miR-122, significantly lowers plasma cholesterol levels in both mice and non-human primates(14–16). In addition, miR-33, which is encoded within an intron of SREBF2, is co-transcribed with SREBF2 and regulates the expression of the ATP-binding cassette transfer protein (ABCA1), a critical player in reverse cholesterol transport and in lipoprotein biogenesis(17–19). miR-33 has also been shown to regulate fatty acid oxidation in hepatic cell lines(20). Nevertheless, despite these important advances, the full extent of post-transcriptional control of lipid metabolism by miRNAs remains incompletely understood and has not been systematically investigated(21).
Using a novel in silico approach, which should be generally applicable toward the identification of key regulatory miRNAs in any biological process, we predicted miR-27b as a regulatory hub in lipid metabolism. Furthermore, we demonstrate that hepatic miR-27b is responsive to lipid levels and regulates the expression (mRNA and protein) of key metabolic genes, including angiopoietin-like 3 (ANGPTL3) and glycerol-3-phosphate acyltransferase 1 (GPAM), which have been implicated previously in the pathobiology of lipid-related disorders.
EXPERIMENTAL PROCEDURES
Animal studies
Eight-week old wild-type C57BL/6J mice were placed on either normal chow diet (4% fat, NIH-31 open chow, Zeigler Brothers, Gardners, PA) or high-fat Western diet (21% fat, 42% calories from fat, ad libitum, TD88137, Harlan-Teklad, Frederick, MD) for three weeks (19–21 days). Adult (8–10 wk) female apolipoprotein E null mice (Apoe−/−, C57BL/6J background, The Jackson Laboratory, Bar Harbor, ME) were placed on either normal chow or Cocoa butter diet with sodium cholate (16% fat, 37% calories from fat, 1.25% cholesterol, 0.125% choline chloride, 0.5% sodium cholate, TD90221, Harlan-Teklad, Frederick, MD) for four weeks (28 days). Mouse livers were excised and homogenized (100 mg) in Qiazol Total RNA extraction buffer. All mice were housed, and the relevant studies were completed, under active protocols approved by the National Institutes of Health, National Heart, Lung and Blood Institute Animal Care and Use Committee. All protocols complied with, and all animals received humane care according to, the criteria outlined in the NIH "Guide for the Care and Use of Laboratory Animals".
High-throughput small RNA sequencing
miRNA isolation and Illumina sequencing were completed as previously reported(22). Details are provided in Supplementary Methods.
Bioinformatics
Target sites (seed, centered) were predicted for miR-27b in both the 3’ UTRs and the open reading frames of the 151 lipid metabolism genes. Details of target site prediction and the identification of candidate miRNA regulatory hubs by Monte Carlo simulations are provided in Supplementary Methods.
Cell Culture
Human hepatocytes (Huh7) were cultured in F12 DMEM, supplemented with 10% fetal bovine serum, penicillin (100 U/mL), and streptomycin (100 µg/mL), and maintained at 37°C with 5% CO2. For transfection, Huh7 cells were plated at 1 × 105 cells/mL for 24 h prior to transient transfection (DharmaFECT4, Dharmacon, Lafayette, CO) with 10 nM miR-27b miRIDIAN mimic (C-300589-05) or 10/100 nM HI-27b miRIDIAN hairpin inhibitor (IH-300589-07) for 48 h.
Gene expression studies
Gene expression in Huh7 cells was assayed by microarray (Affymetrix GeneChIP Human Genome U133A 2.0), real time PCR (Applied Biosystems TaqMan assays), and ELISA according to standard protocols. Details of the methodology and reagents used are provided in Supplementary Methods.
Reporter Gene (Luciferase) Assays
Reporter gene assays were conducted with a truncated 3’ UTR (bases 1-806) of human GPAM (NM_020918.3), which was cloned downstream of firefly luciferase in a pEZXMT01 vector (GeneCopoeia). Site-directed mutagenesis (QuickChange II XL, Stratagene), using custom primers (Supplementary Table S4), was performed to alter the predicted miR-27b target site at base 329 (G>A). Transformation, DNA extraction, transient transfections, and Luciferase activity measurements were conducted according to standard protocols, which are described in detail in the Supplementary Methods.
Lipid analysis
Murine plasma and hepatic lipid levels were measured according to standard enzymatic quantification (Roche Diagnostics). Details of blood collection, tissue extraction, and reagents used are provided in Supplementary Methods.
Statistics
When comparing two groups, Mann-Whitney non-parametric tests (two-tailed) were used unless otherwise stated. For all tests, P-values ≤ 0.05 were considered significant. All results are expressed as means +/− standard error, and n = derives from independent experiments.
RESULTS
High-throughput small RNA sequencing detects at least 150 miRNAs in mouse liver
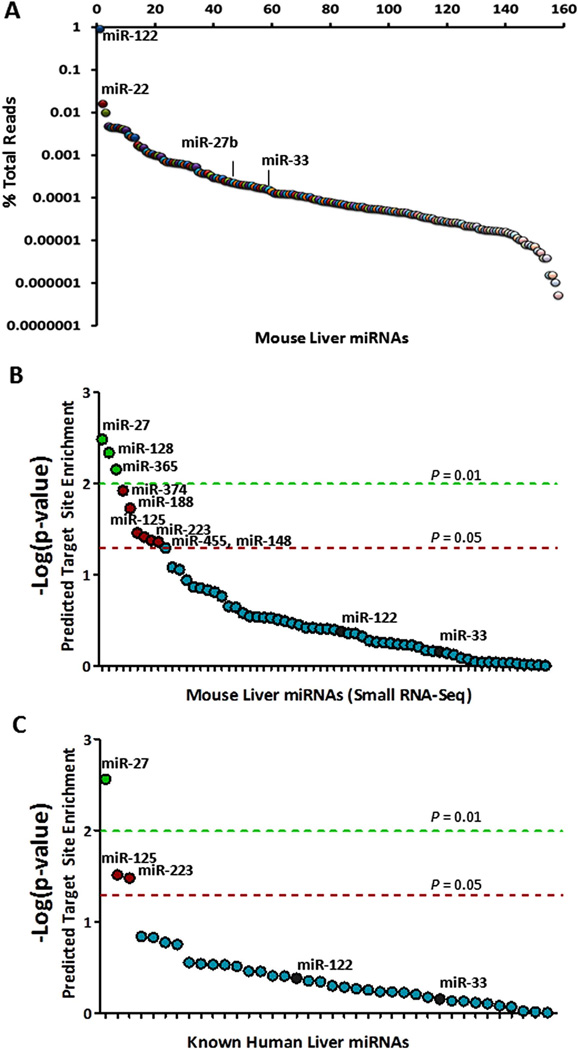
To characterize mouse liver miRNAs, we performed high-throughput sequencing on a small RNA library generated from mouse liver and obtained ~9.9 million small RNA reads (Methods). Using an in-house bioinformatic strategy, we determined that ~40% (~3.9 million) of the reads matched exactly (no mismatches) to 160 annotated mouse miRNAs in miRBase. Almost all of these miRNAs (n = 157) were represented by ≥ 3 exactlymatching sequence reads and were thus identified as hepatic miRNAs (Figure 1A; Supplementary Table S1). The diversity and number of hepatic miRNAs is consistent with results from the few other previously published small RNA sequencing studies performed in other murine tissues(23, 24). The most highly abundant miRNA, miR-122, accounts for ~90% of the miRNA-related sequence reads in the mouse liver (Figure 1A). Nevertheless, many of the less abundant miRNAs have been shown to regulate important processes in the liver, such as miR-33 (cholesterol homeostasis, fatty acid oxidation)(20, 25), miR-22 (hepatocyte proliferation)(26), miR-125a-5p (lipid uptake)(27), miR-30 (hepatobiliary development)(28), and miR-29b (liver fibrosis)(29).
Figure 1. miR-27b is a strong candidate regulatory hub in lipid metabolism.
(A) For each murine hepatic miRNA (x-axis), the read depth from small RNA-seq (y-axis; logscale) is shown. (B) For each murine hepatic miRNA (x-axis), the negative logarithm of the empirical p-value (y-axis) for the level of enrichment of predicted target sites in lipid-related genes is shown. (C) For each known human hepatic miRNA (x-axis), the negative logarithm of the empirical p-value (y-axis) for the level of enrichment of predicted target sites in lipid-related genes is shown. Dashed lines indicate empirical P = 0.01 (green) and empirical P = 0.05 (red).
miR-27b is a regulatory hub in lipid metabolism
A post-transcriptional “miRNA hub” in lipid metabolism was defined as a miRNA that is predicted to target more lipid metabolism-associated genes than expected by chance(30). To identify lipid metabolic miRNA hubs, we assembled a high-confidence list of 151 known lipid metabolism associated genes (Supplementary Table S2) from three high-throughput screens: [1] a large-scale hepatic gene expression analysis (microarray) of transgenic mice over-expressing SREBF1 or SREBF2(31), [2] a systematic siRNA screen for lipid-regulating genes assayed by quantitative analysis of cellular cholesterol levels(32), and [3] a genome-wide screen for common genetic variants associated with plasma lipid levels(33).
Typically, the most effective miRNA target sites occur within 3’ untranslated regions (3’ UTRs) of mRNAs and have perfect base pairing with the “seed” region of the miRNA (nucleotides 2 through 7 from the 5’-end of the miRNA)(34). For each of the 157 hepatic miRNAs identified by small RNA sequencing, we scanned the 3’ UTRs of the 151 known lipid metabolism-associated genes for seed-based target sites, and the number of genes with at least one such predicted site was scored. We then performed Monte-Carlo simulations to obtain the expected number of genes predicted to be targeted by chance for each miRNA. Target sites for three hepatic miRNAs, namely miR-27b, miR-128, and miR-365, were significantly over-represented (empirical uncorrected P < 0.01) in the 151 known lipid metabolism genes. Among all mouse liver miRNAs, miR-27b had the most predicted lipid metabolism gene targets (n=27) (empirical uncorrected P = 0.003) (Figure 1B). We repeated this analysis for those miRNAs previously detected in human liver tissue by small RNA cloning(35), and again miR-27 was the most significant (Figure 1C).
miR-27b levels are sensitive to plasma and hepatic lipid content
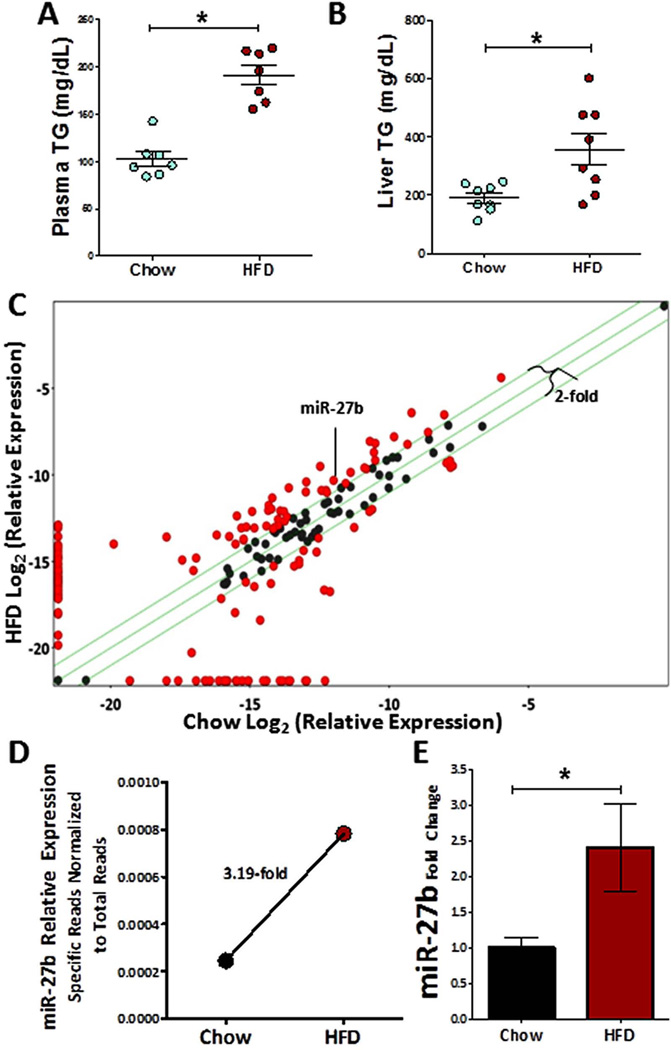
To identify lipid-responsive hepatic miRNAs, we used high-throughput small RNA sequencing to quantify and compare miRNA expression in the livers of C57BL/6J mice on a normal chow diet and on a high-fat “Western” diet (HFD, 42% calories from fat). After 3 weeks, triglyceride levels (mg/dL) were significantly increased in the plasma (1.86-fold, P = 0.0006) (Figure 2A) and liver (1.87-fold, P = 0.01) (Figure 2B) of HFD mice compared to mice fed a normal chow diet. Analysis of the small RNA sequence data revealed that at least 50 miRNAs were ≥ 2-fold more abundant (percent of total reads) in HFD mouse liver (Figure 2C, Supplementary Table S1), including miR-27b, which was up-regulated 3.2-fold (Figure 2D). To confirm this observation, we performed real-time PCR using individual TaqMan assays, and found miR-27b to be significantly increased (2.4-fold, P = 0.03) in HFD livers compared to normal mouse livers (Figure 2E). However, interestingly, levels of the primary transcript of miR-27b (pri-miR-27b) were not increased (Supplementary Figure S1).
Figure 2. miR-27b is sensitive to hyperlipidemia.
(A) Triglyceride levels (mg/dL) are significantly increased in the mouse plasma after three weeks of HFD (chow n=7, high-fat diet (HFD) n=7). (B) Triglyceride levels (mg/dL) are significantly increased in the mouse liver after three weeks of HFD (chow n=8, HFD n=8). (C) Differential abundance of normalized mouse liver miRNA reads (Small RNA-seq) on high fat diet is shown. Green lines represent 2-fold change. X-axis: Log2(percent total reads chow diet), Y-axis: Log2(percent total reads HFD). Red circles indicate ≥ 2-fold change. (D) Hepatic miR-27b expression levels, as determined by small RNA-seq, are significantly increased in HFD (2995 reads / 3,825,273 total reads) relative to chow (955 reads / 3,902,443 total reads). (E) Hepatic miR-27b expression levels, as determined by real-time PCR, are significantly increased in HFD relative to chow. Relative quantitative value (RQV) is represented as fold change to chow diet mean (chow, n=9; HFD, n=3). *, Mann-Whitney non-parametric test P < 0.05.
miR-27b represses critical regulators of lipid homeostasis
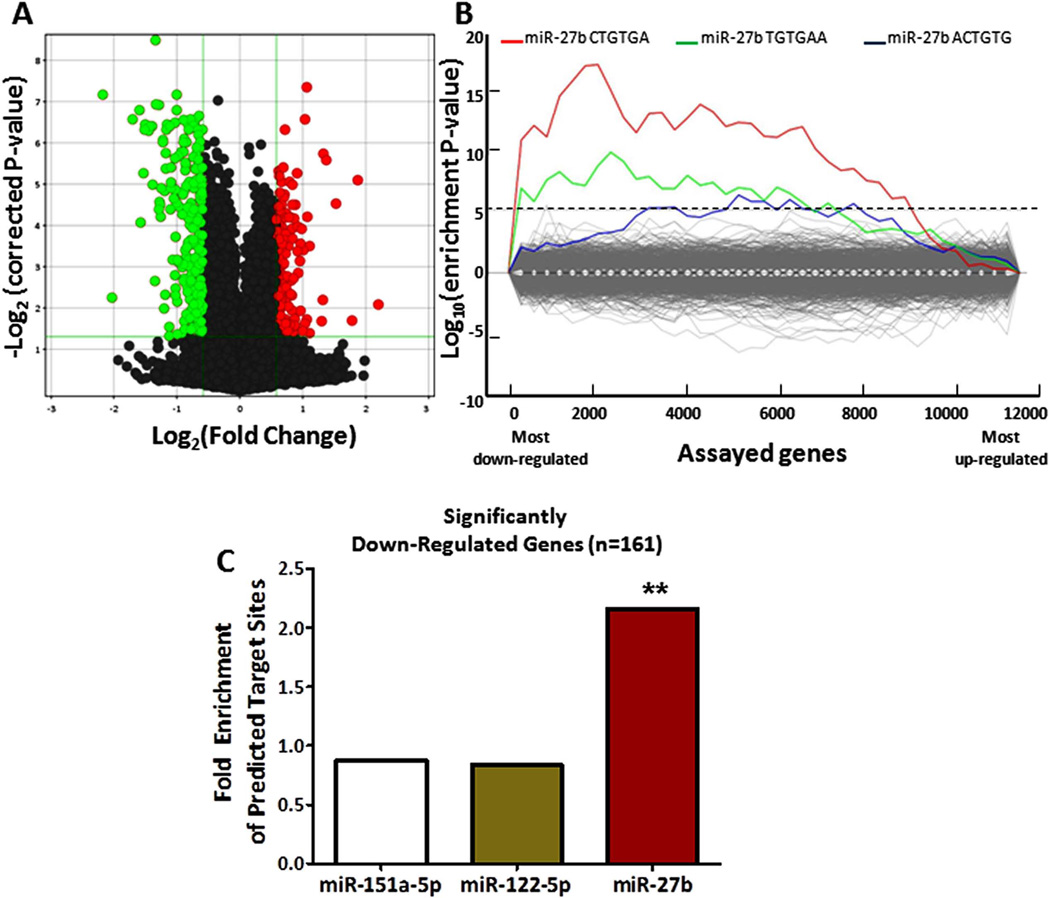
To validate miR-27b targeting of lipid metabolism genes experimentally, we transfected miR-27b in human hepatocytes (Huh7 cells) and performed whole-genome microarray expression analysis. Of the 13,785 unique genes assayed on the array, 1,318 were down-regulated at FDR < 0.05, including ~10% of the original set of 151 known lipid metabolism genes. Of these 1,318 genes, 173 were down-regulated by a fold-change < - 1.5 (Figure 3A; Supplementary Table S3). We applied the Sylamer algorithm(36) to calculate enrichment scores for all possible 6-mer motifs in the 3’ UTRs of the 13,785 genes, sorted according to their level of down-regulation upon miR-27b over-expression. The most enriched 6-mer motif among down-regulated genes is CTGTGA (Figure 3B), which is the exact reverse complement of the miR-27b “canonical seed” region (nucleotides 2–7 from the 5’-end of the miRNA) (34). The two next-most enriched motifs, TGTGAA and ACTGTG, are reverse complements of the miR-27b seed region shifted by 1-nucleotide on either side (nucleotides 1–6 and 3–8 from the 5’-end of miR-27b, respectively). We next implemented an algorithm that identifies “seed” target sites. Among the significantly down-regulated genes with annotated 3’ UTR sequence (n=161), ~73% (n=118) have canonical miR-27b seed sites, which represents a 2.2-fold enrichment compared to background expectation (Fisher’s exact test P < 0.0001) (Figure 3C). Non-canonical “shifted seed” sites are present in an additional ~9% (n=14) of the down-regulated genes, leaving ~18% (n=29) of the genes without any predicted 3’ UTR target site.
Figure 3. miR-27b over-expression in human hepatocytes significantly down-regulates 173 genes.
(A) Results of a microarray analysis upon miR-27b over-expression (10 nM) in hepatocytes are shown. 173 genes (green) are significantly down-regulated (Fold Change < −1.5, FDR < 0.05) and 128 genes (red) are significantly up-regulated (Fold Change > 1.5, FDR < 0.05). (B) A sylamer plot of motif enrichment in the 3’ UTRs of the genes assayed on the microarray is shown. The six-nucleotide motif matching the miR-27b “seed” region is the most significantly enriched among down-regulated genes (left-most portion of graph). (C) Canonical miR-27b seed-match sites are significantly overrepresented in the set of 161 most down-regulated genes, whereas predicted seed-match sites for two control miRNAs, miR-151a-5p and miR-122-5p, are not. **, Fisher’s exact test P < 0.0001.
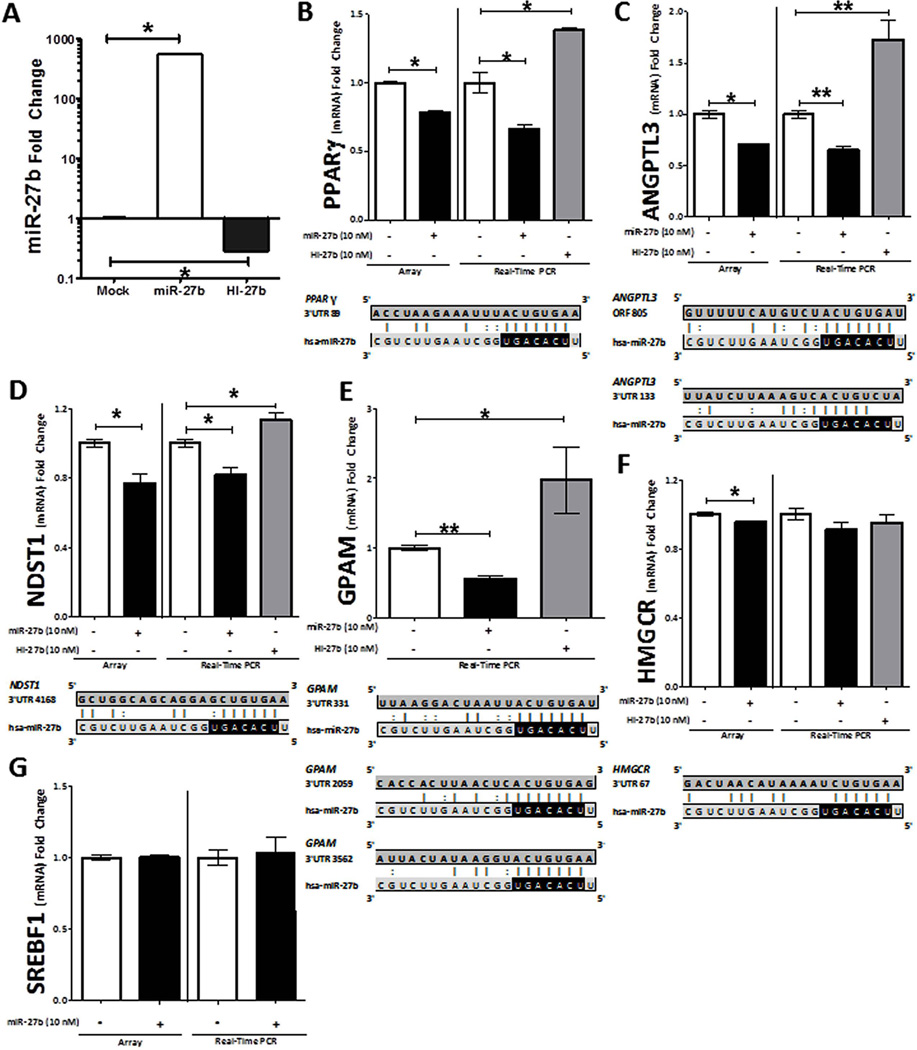
To further validate miR-27b-mediated regulation of lipid metabolism genes, we introduced miR-27b mimics or inhibitors (antagomiRs) into Huh7 cells by transient transfection. Over-expression of miR-27b mimics resulted in a significant increase (552-fold, P = 0.02) in intracellular miR-27b levels, and inhibition of endogenous miR-27b resulted in a significant decrease (71% loss, P = 0.02) in intracellular miR-27b levels (Figure 4A). We then assayed by real-time quantitative PCR the mRNA levels of six genes: Peroxisome proliferator-activated receptor gamma (PPARG), Angiopoietin-like 3 (ANGPTL3), N-deacetylase/N-sulfotransferase 1 (NDST1), 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR), Glycerol-3-phosphate acyltransferase 1, mitochondrial (GPAM), and Sterol regulatory element binding factor 1 (SREBF1). These six genes were selected on the basis of their well-established relevance to lipid metabolism (Supplementary Methods).
Figure 4. miR-27b represses key lipid metabolism genes.
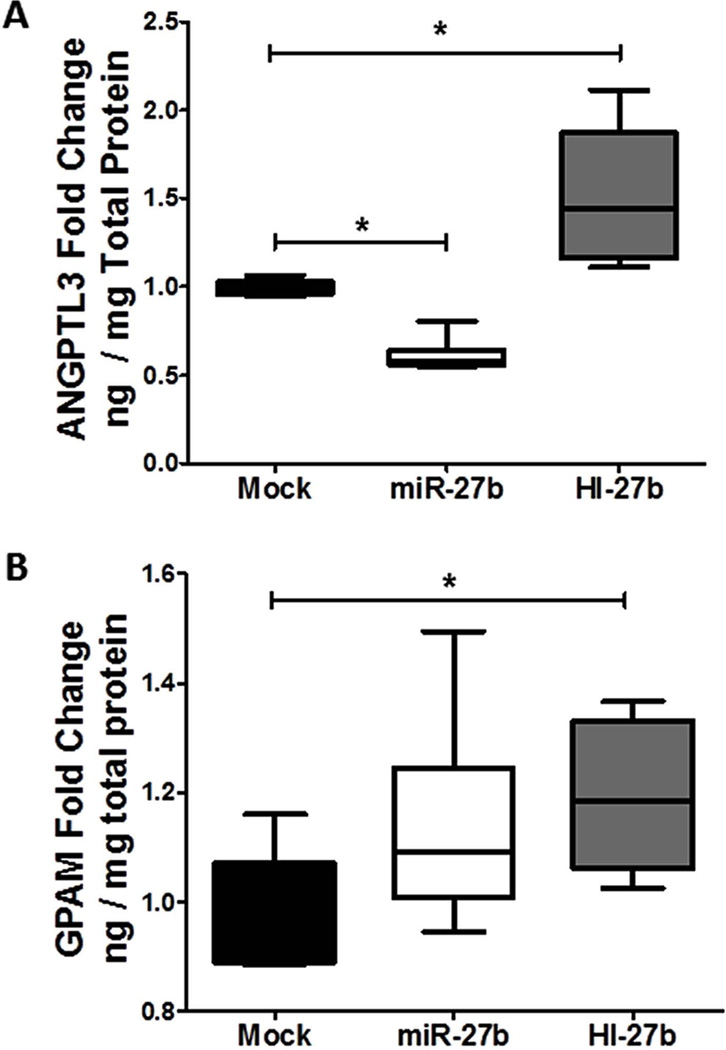
(A) Quantification by real-time PCR of intracellular mature miR-27b levels after transient transfection (48 h) of miR-27b or HI-27b (anti-miR-27b) is shown. (B–G) Results of microarray and real-time quantitative PCR (RT-qPCR) analysis upon miR-27b and HI-27b over-expression (10 nM) in hepatocytes are shown. Microarray data: (B–D, F, G) Each array group (+/−miR-27b), n=6. (E) No microarray data is shown for GPAM, because probes for this gene were not present on the microarray. Real-time PCR data: (B) PPARG: mock, n=7; miR-27b, n=7; and HI-27b, n=3. (C) ANGPTL3: mock, n=17; miR-27b, n=13; and HI-27b, n=14. (D) NDST1: mock, n=13; miR-27b, n=9; and HI-27b, n=11. (E) GPAM: mock, n=16; miR-27b, n=19; and HI-27b, n=12. (F) HMGCR: mock, n=10; miR-27b, n=10; and HI-27b, n=6. (F) SREBF1: mock, n=10; miR-27b, n=10. The mRNA levels of four genes (PPARG, ANGPTL3, NDST1, and GPAM) are significantly reduced upon miR-27b over-expression (black) and elevated upon HI-27b over-expression (shaded). PPARG, NDST1, HMGCR, and GPAM each have at least one predicted “seed-based” 3’ UTR target site for miR-27b. ANGPTL3 has both a predicted “centered” 3’ UTR site and a “seed-based” coding-region site for miR-27b. P-value computations are based on the Mann-Whitney non-parametric test. *, P < 0.05; **, P < 0.0001.
Four of the six genes were significantly down-regulated by miR-27b over-expression (PPARG, P=0.0006; ANGPTL3, P<0.0001; NDST1, P=0.0008; and GPAM, P<0.0001; Figure 4B–E) and one (HMGCR, P=0.06) was just outside of significance at the 5% threshold (Figure 4F). Inhibition of endogenous miR-27b significantly up-regulated the same four genes (PPARG, P=0.01; ANGPTL3, P<0.0001; NDST1, P=0.02; and GPAM, P=0.004; Figure 4B–E). SREBF1 was not affected by miR-27b over-expression (Figure 4G).
To assess the influence of miR-27b targeting on protein levels of these key lipid metabolism genes, secreted (ANGPTL3) and cellular (GPAM) protein levels were quantified by ELISA. Inhibition of endogenous miR-27b by transient transfection of Huh7 cells with antagomiRs significantly (P=0.002) increased secreted ANGPTL3 protein levels in the media after 48 hrs (Figure 5A). miR-27b inhibition also resulted in significantly (P=0.04) increased cellular GPAM levels (Figure 5B). To determine if miR-27b modulates PPARG transcriptional activity, we performed PPARG binding assays with nuclear extracts from transfected Huh7 cells (Supplementary Methods). Inhibition of endogenous miR-27b resulted in a significant (P=0.01) increase in PPARG binding to immobilized response elements (Supplementary Figure S2). It should be noted that while over-expression of miR-27b significantly reduced (39% loss, P=0.002) secreted ANGPTL3 levels (Figure 5A) after 48 hrs, cellular GPAM protein levels and PPARG transcriptional activity were not affected (Figure 5B, Supplementary Figure S2). These observations are likely explained at least in part by the stability and temporal dynamics of each protein.
Figure 5. Inhibition of miR-27b increases cellular GPAM and secreted ANGPTL3 protein levels.
Results of ELISA from hepatocyte (Huh7) culture media or cellular extracts upon miR-27b and HI-27b (anti-miR-27b) over-expression (10 nM) are shown. (A) ANGPTL3 protein levels in hepatocyte culture media, normalized to total cellular protein (ng/mg), is shown (n=6). (B) GPAM protein levels in cellular extracts, normalized to total cellular protein (ng/mg), is shown (n=6). P-value computations are based on the Mann-Whitney non-parametric test. *, P < 0.05.
Next we searched for canonical 3’ UTR seed-based miR-27b target sites within each of the six genes (Methods). As expected, SREBF1, which did not change (mRNA level) in response to over-expression of either miR-27b mimic or its antagomiR, did not harbor any canonical miR-27b seed sites (Figure 4F). Three out of the five genes that were repressed by miR-27b (PPARG, NDST1, and GPAM) contained one or more seed sites within their 3’ UTRs (Figure 4A–E). GPAM harbors two highly conserved and one moderately conserved miR-27b target site within its 3’ UTR (Figure 4). To determine if miR-27b directly targets GPAM through one of these predicted sites, we performed reporter gene (luciferase) assays. A portion of the GPAM 3’ UTR, containing one putative miR-27b site, was cloned downstream of firefly luciferase (Methods). Dual transfection with miR-27b in HEK293 cells significantly (P=0.001) reduced firefly luciferase activity (Supplementary Figure S3). After site-directed mutagenesis to eliminate the putative miR-27b site (Methods), miR-27b failed to knock-down firefly luciferase activity, indicating that the site is directly involved in miR-27b mediated regulation of GPAM (Supplementary Figure S3).
ANGPTL3 was the only down-regulated gene that did not harbor any miR-27b seed sites in its 3’ UTR. To further investigate the observed strong miR-27b-mediated regulation of ANGPTL3 (Figure 4C, 5A), we expanded the search to two recently discovered classes of target sites: [1] 3’ UTR centered sites and [2] open reading frame (ORF) sites (Methods). As its name implies, 3’ UTR centered sites base pair to the center of the miRNA sequence(37), as opposed to the 5'-end seed region. Functional ORF sites are typically preceded by a stretch of rare codons(38), which can cause ribosomal pausing(39), thereby allowing miRNA silencing complexes to form stable interactions with the target site without ribosomal interference. We developed and implemented computational strategies to predict 3’ UTR centered sites based on the strength of base pairing to the center of a miRNA, and ORF seed sites based on a metric that evaluates codon rarity in the preceding sequence (Methods). Of the six genes examined, only ANGPTL3 was predicted to have both a 3’ UTR centered site and a high-confidence ORF target site for miR-27b (Figure 4C). Furthermore, no other hepatic miRNA besides miR-27 was predicted to have high-confidence ORF or 3’ UTR sites in ANGPTL3.
Hepatic miR-27b is up-regulated in a mouse model of dyslipidemia and atherosclerosis
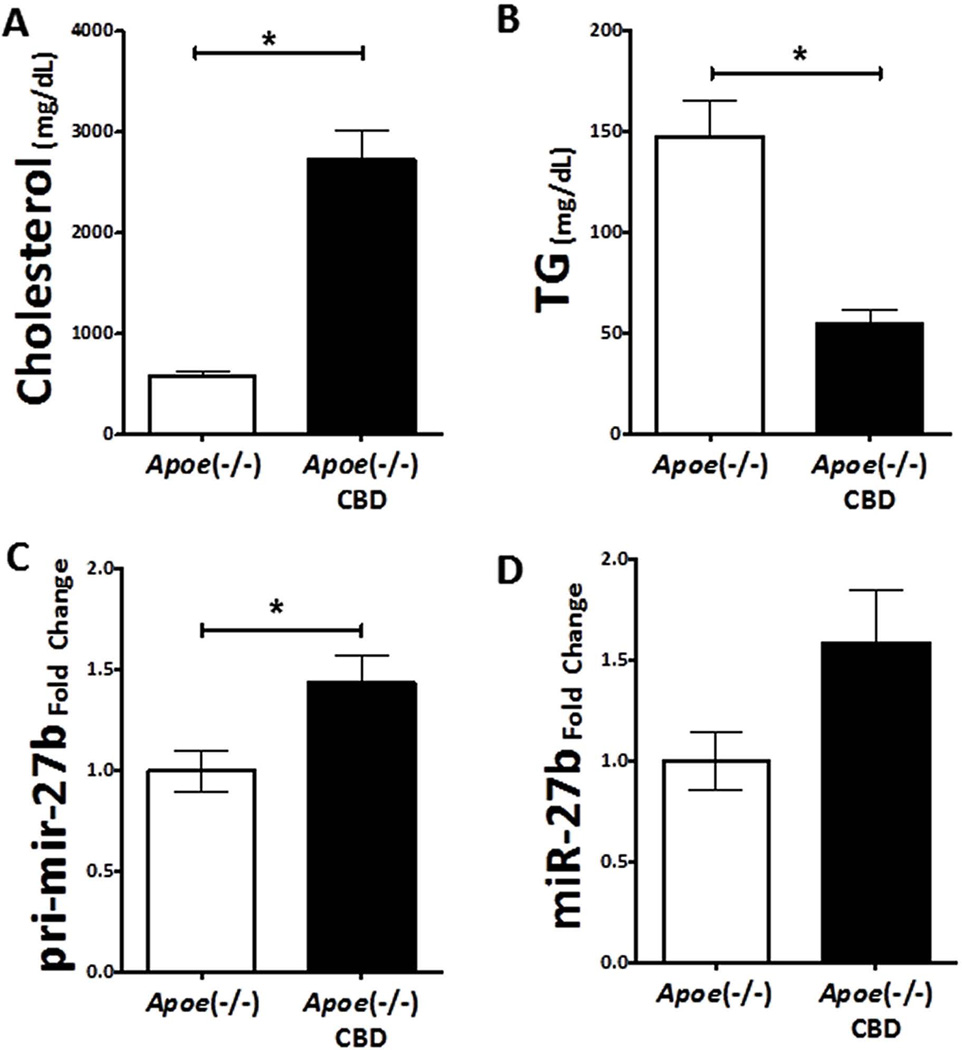
We placed 8-week old Apoe−/− female mice on a high-fat/high-cholesterol diet (21% fat, 7.5% Cocoa butter), which has been shown to induce severe hypercholesterolemia and advanced atherosclerosis(40, 41). To confirm the expected physiologic effects of this diet, we measured plasma total cholesterol and triglyceride levels after four weeks. We observed a significant increase in plasma cholesterol levels (4.6-fold, unpaired t-test P<0.001; Figure 6A) and a significant decrease in plasma triglycerides (~63% loss, unpaired t-test P=0.003; Figure 6B) in the Apoe−/− mice fed the atherogenic diet.
Figure 6. Hepatic miR-27b levels are increased in a mouse model of dyslipidemia and atherosclerosis.
(A) Plasma cholesterol levels are significantly increased in apolipoprotein E null (Apoe−/−) mice on a Cocoa butter diet (CBD; black, n=4) compared to Apoe−/− mice on a chow diet (white, n=4). (B) Triglyceride levels are decreased in Apoe−/− mice on a CBD (black, n=4) compared to Apoe−/− mice on a chow diet (white, n=4). (C) Hepatic pri-miR-27b levels are significantly increased in Apoe−/− mice on a CBD (black, n=4) compared to Apoe−/− mice on a chow diet (white, n=4). (D) Hepatic mature miR-27b levels are increased in Apoe−/− mice on a CBD (black, n=4) compared to Apoe−/− mice on a chow diet (white, n=4). P-value computations are based on the unpaired t-test. *, P < 0.05.
After four weeks on the atherogenic diet, levels of both mature miR-27b (1.58-fold, unpaired t-test P = 0.09) and pri-miR-27b (unpaired t-test P = 0.03) were increased in the liver; Figure 6C–D). Based on this finding, we next assessed the hepatic expression of miR-27b target genes. Consistent with the in vitro results, mRNA levels of both Angptl3 (~30% loss) and Gpam (~22% loss) were reduced (Supplementary Figure S4); however, these observations were outside of statistical significance.
DISCUSSION
In this study we provide in silico, in vitro, and in vivo evidence that miR-27b is a strong candidate regulatory hub in lipid metabolism. Based on Monte-Carlo simulations, miR-27b was predicted to target significantly more lipid metabolism-associated genes than expected by chance and more than any other hepatic miRNA. Two of the other miRNAs predicted to be regulatory hubs in lipid metabolism (Figure 1B,C), miR-365 and miR-125, have previously been shown to play roles in either adipocyte differentiation(42) or in cellular lipid uptake(27), respectively, thus validating our approach.
High-throughput small RNA sequencing and real time quantitative PCR analysis revealed that miR-27b is ~3.2-fold up-regulated in the livers of mice on a high-fat diet. miR-27b is encoded with miR-23b and miR-24-1 in the same cistron on mouse chromosome 13. Small RNA sequencing results suggest that both miR-23b and miR-24 are also up-regulated in the liver of wild-type mice after high-fat diet, by ~2.2-fold and ~7.9-fold respectively. However, we did not detect any change in the levels of their primary transcript, which suggests that: (1) post-transcriptional mechanisms are completely responsible for the observed increase in the mature miRNA levels, or (2) there is an increase in transcription of the miR-27b locus but also a concomitant increase in the rate of processing of the primary transcript (i.e. decreased pri-miR-27b stability). In contrast, Apoe−/− mice on an atherogenic diet were found to have increased hepatic levels of both mature miR-27b and pri-miR-27b. While the latter finding is suggestive of enhanced transcription, several other recently reported post-transcriptional mechanisms of miRNA expression control, including pri-miRNA tertiary structure(43) and non-templated nucleotide additions(44), may also be involved and cannot be ruled out.
Microarray and real time quantitative PCR based gene expression analyses in human hepatocytes confirmed robust miR-27b-mediated regulation of key lipid metabolism genes, including PPARG, GPAM, and ANGPTL3. Studies in rodents have revealed that both GPAM and ANGPTL3 regulate lipid metabolism(45, 46). Recent genome-wide association studies in human populations have added to these findings, by identifying genetic polymorphisms in both GPAM and ANGPTL3 that are significantly associated with plasma lipid levels(33).
GPAM is present in a variety of tissues; however, it is most highly expressed in the liver. It is known to catalyze the first committed step in de novo triglyceride synthesis(47), and more recently, has been implicated in regulating cholesterol(33). As such, over-expression of GPAM in mouse liver results in fatty liver, hepatic steatosis, and plasma hyperlipidemia(48). Our data show that hepatic Gpam mRNA levels are reduced in Apoe−/− mice on a four-week atherogenic diet, concomitant with a decrease in plasma triglyceride levels and an increase in hepatic miR-27b expression.
ANGPTL3 is expressed by the liver(49) and secreted into circulation(50), where it suppresses the activity of lipoprotein lipase(51) and endothelial lipase(52), which regulate triglyceride and HDL-cholesterol levels, respectively. Plasma levels of ANGPTL3 correlate with various parameters of lipid/carbohydrate metabolism(53) and atherosclerosis(54), and specific nonsense mutations in ANGPTL3 lead to hypolipidemia(55). While several tissues may contribute to plasma ANGPTL3 levels, our data in this study reveal that hepatic Angptl3 levels are decreased in Apoe−/− mice on a four-week atherogenic diet, concomitant with an increase in hepatic miR-27b expression. It is possible that Gpam and Angptl3 are repressed by miR-27b in the adaptive response to dyslipidemic conditions, in order to mitigate the accumulation of lipids in circulation.
Further detailed in vivo experimentation is required to determine the extent to which miR-27b targeting of GPAM and ANGPTL3 is required for controlling plasma lipid levels, and whether modulation of endogenous miR-27b levels could serve as an effective therapeutic strategy for lipid-related disorders.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the intramural programs of the National Human Genome Research Institute (NHGRI) and the National Heart, Lung, and Blood Institute (NHLBI) supported this study, a K22 grant HL113039-01 (K.C.V) from the NHLBI and an R00 grant DK091318-01 (P.S.) from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK). The authors thank Yanqin Yang, Ph.D. for help with bioinformatics, Alonzo Jalan for animal studies, Maureen Sampson for plasma lipid analysis, the NHLBI DNA Sequencing Core Facility (Jun Zhu, Ph.D.), the NHLBI Genomics Core Facility (Nalini Raghavachari, Ph.D.), and the NHGRI Microarray Core Facility (Abdel Elkahloun and Bhavesh Borate).
REFERENCES
- 1.Goldstein JL, DeBose-Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell. 2006;124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 2.Brown MS, Goldstein JL. Multivalent feedback regulation of HMG CoA reductase, a control mechanism coordinating isoprenoid synthesis and cell growth. J Lipid Res. 1980;21:505–517. [PubMed] [Google Scholar]
- 3.Osborne TF. Transcriptional control mechanisms in the regulation of cholesterol balance. Crit Rev Eukaryot Gene Expr. 1995;5:317–335. doi: 10.1615/critreveukargeneexpr.v5.i3-4.50. [DOI] [PubMed] [Google Scholar]
- 4.LaRosa JC, Hunninghake D, Bush D, Criqui MH, Getz GS, Gotto AM, Jr, Grundy SM, et al. The cholesterol facts. A summary of the evidence relating dietary fats, serum cholesterol, and coronary heart disease. A joint statement by the American Heart Association and the National Heart, Lung, and Blood Institute. The Task Force on Cholesterol Issues, American Heart Association. Circulation. 1990;81:1721–1733. doi: 10.1161/01.cir.81.5.1721. [DOI] [PubMed] [Google Scholar]
- 5.Espenshade PJ. SREBPs: sterol-regulated transcription factors. J Cell Sci. 2006;119:973–976. doi: 10.1242/jcs.02866. [DOI] [PubMed] [Google Scholar]
- 6.Mourelatos Z. Small RNAs: The seeds of silence. Nature. 2008;455:44–45. doi: 10.1038/455044a. [DOI] [PubMed] [Google Scholar]
- 7.Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr Opin Cell Biol. 2009;21:452–460. doi: 10.1016/j.ceb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 8.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sethupathy P, Collins FS. MicroRNA target site polymorphisms and human disease. Trends Genet. 2008;24:489–497. doi: 10.1016/j.tig.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 10.Couzin J. MicroRNAs make big impression in disease after disease. Science. 2008;319:1782–1784. doi: 10.1126/science.319.5871.1782. [DOI] [PubMed] [Google Scholar]
- 11.Mishra PJ, Merlino G. MicroRNA reexpression as differentiation therapy in cancer. J Clin Invest. 2009;119:2119–2123. doi: 10.1172/JCI40107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Rooij E, Marshall WS, Olson EN. Toward microRNA-based therapeutics for heart disease: the sense in antisense. Circ Res. 2008;103:919–928. doi: 10.1161/CIRCRESAHA.108.183426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moore KJ, Rayner KJ, Suarez Y, Fernandez-Hernando C. microRNAs and cholesterol metabolism. Trends Endocrinol Metab. 2010;21:699–706. doi: 10.1016/j.tem.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with 'antagomirs'. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 15.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 16.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rayner KJ, Suarez Y, Davalos A, Parathath S, Fitzgerald ML, Tamehiro N, Fisher EA, et al. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328:1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marquart TJ, Allen RM, Ory DS, Baldan A. miR-33 links SREBP-2 induction to repression of sterol transporters. Proc Natl Acad Sci U S A. 2010;107:12228–12232. doi: 10.1073/pnas.1005191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE, Naar AM. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. 2010;328:1566–1569. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davalos A, Goedeke L, Smibert P, Ramirez CM, Warrier NP, Andreo U, Cirera-Salinas D, et al. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc Natl Acad Sci U S A. 2011;108:9232–9237. doi: 10.1073/pnas.1102281108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vickers KC, Remaley AT. MicroRNAs in atherosclerosis and lipoprotein metabolism. Curr Opin Endocrinol Diabetes Obes. 2010;17:150–155. doi: 10.1097/MED.0b013e32833727a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu H, Sun S, Tu K, Gao Y, Xie B, Krainer AR, Zhu J. A splicing-independent function of SF2/ASF in microRNA processing. Mol Cell. 2010;38:67–77. doi: 10.1016/j.molcel.2010.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, Johnston WK, et al. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev. 2010;24:992–1009. doi: 10.1101/gad.1884710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moore KJ, Rayner KJ, Suarez Y, Fernandez-Hernando C. The Role of MicroRNAs in Cholesterol Efflux and Hepatic Lipid Metabolism. Annu Rev Nutr. 2011;31:49–63. doi: 10.1146/annurev-nutr-081810-160756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J, Yang Y, Yang T, Liu Y, Li A, Fu S, Wu M, et al. microRNA-22, downregulated in hepatocellular carcinoma and correlated with prognosis, suppresses cell proliferation and tumourigenicity. Br J Cancer. 2010;103:1215–1220. doi: 10.1038/sj.bjc.6605895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen T, Huang Z, Wang L, Wang Y, Wu F, Meng S, Wang C. MicroRNA-125a-5p partly regulates the inflammatory response, lipid uptake, and ORP9 expression in oxLDL-stimulated monocyte/macrophages. Cardiovasc Res. 2009;83:131–139. doi: 10.1093/cvr/cvp121. [DOI] [PubMed] [Google Scholar]
- 28.Hand NJ, Master ZR, Eauclaire SF, Weinblatt DE, Matthews RP, Friedman JR. The microRNA-30 family is required for vertebrate hepatobiliary development. Gastroenterology. 2009;136:1081–1090. doi: 10.1053/j.gastro.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roderburg C, Urban GW, Bettermann K, Vucur M, Zimmermann H, Schmidt S, Janssen J, et al. Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology. 2011;53:209–218. doi: 10.1002/hep.23922. [DOI] [PubMed] [Google Scholar]
- 30.Stone N, Pangilinan F, Molloy AM, Shane B, Scott JM, Ueland PM, Mills JL, et al. Bioinformatic and Genetic Association Analysis of MicroRNA Target Sites in One-Carbon Metabolism Genes. PLoS One. 2011;6:e21851. doi: 10.1371/journal.pone.0021851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horton JD, Shah NA, Warrington JA, Anderson NN, Park SW, Brown MS, Goldstein JL. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci U S A. 2003;100:12027–12032. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bartz F, Kern L, Erz D, Zhu M, Gilbert D, Meinhof T, Wirkner U, et al. Identification of cholesterol-regulating genes by targeted RNAi screening. Cell Metab. 2009;10:63–75. doi: 10.1016/j.cmet.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 33.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Dongen S, Abreu-Goodger C, Enright AJ. Detecting microRNA binding and siRNA off-target effects from expression data. Nat Methods. 2008;5:1023–1025. doi: 10.1038/nmeth.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shin C, Nam JW, Farh KK, Chiang HR, Shkumatava A, Bartel DP. Expanding the microRNA targeting code: functional sites with centered pairing. Mol Cell. 2010;38:789–802. doi: 10.1016/j.molcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gu S, Jin L, Zhang F, Sarnow P, Kay MA. Biological basis for restriction of microRNA targets to the 3' untranslated region in mammalian mRNAs. Nat Struct Mol Biol. 2009;16:144–150. doi: 10.1038/nsmb.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Ree JH, van den Broek WJ, Dahlmans VE, Groot PH, Vidgeon-Hart M, Frants RR, Wieringa B, et al. Diet-induced hypercholesterolemia and atherosclerosis in heterozygous apolipoprotein E-deficient mice. Atherosclerosis. 1994;111:25–37. doi: 10.1016/0021-9150(94)90188-0. [DOI] [PubMed] [Google Scholar]
- 41.Getz GS, Reardon CA. Diet and murine atherosclerosis. Arterioscler Thromb Vasc Biol. 2006;26:242–249. doi: 10.1161/01.ATV.0000201071.49029.17. [DOI] [PubMed] [Google Scholar]
- 42.Sun L, Xie H, Mori MA, Alexander R, Yuan B, Hattangadi SM, Liu Q, et al. Mir193b-365 is essential for brown fat differentiation. Nat Cell Biol. 2011 doi: 10.1038/ncb2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chaulk SG, Thede GL, Kent OA, Xu Z, Gesner E, Veldhoen RA, Khanna SK, et al. Role of pri-miRNA tertiary structure in miR-17~92 miRNA biogenesis. RNA Biol. 2011;8 doi: 10.4161/rna.8.6.17410. [DOI] [PubMed] [Google Scholar]
- 44.Kim YK, Heo I, Kim VN. Modifications of small RNAs and their associated proteins. Cell. 2010;143:703–709. doi: 10.1016/j.cell.2010.11.018. [DOI] [PubMed] [Google Scholar]
- 45.Koishi R, Ando Y, Ono M, Shimamura M, Yasumo H, Fujiwara T, Horikoshi H, et al. Angptl3 regulates lipid metabolism in mice. Nat Genet. 2002;30:151–157. doi: 10.1038/ng814. [DOI] [PubMed] [Google Scholar]
- 46.Coleman RA, Lee DP. Enzymes of triacylglycerol synthesis and their regulation. Prog Lipid Res. 2004;43:134–176. doi: 10.1016/s0163-7827(03)00051-1. [DOI] [PubMed] [Google Scholar]
- 47.Gonzalez-Baro MR, Lewin TM, Coleman RA. Regulation of Triglyceride Metabolism. II. Function of mitochondrial GPAT1 in the regulation of triacylglycerol biosynthesis and insulin action. Am J Physiol Gastrointest Liver Physiol. 2007;292:G1195–G1199. doi: 10.1152/ajpgi.00553.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Linden D, William-Olsson L, Ahnmark A, Ekroos K, Hallberg C, Sjogren HP, Becker B, et al. Liver-directed overexpression of mitochondrial glycerol-3-phosphate acyltransferase results in hepatic steatosis, increased triacylglycerol secretion and reduced fatty acid oxidation. FASEB J. 2006;20:434–443. doi: 10.1096/fj.05-4568com. [DOI] [PubMed] [Google Scholar]
- 49.Conklin D, Gilbertson D, Taft DW, Maurer MF, Whitmore TE, Smith DL, Walker KM, et al. Identification of a mammalian angiopoietin-related protein expressed specifically in liver. Genomics. 1999;62:477–482. doi: 10.1006/geno.1999.6041. [DOI] [PubMed] [Google Scholar]
- 50.Clark HF, Gurney AL, Abaya E, Baker K, Baldwin D, Brush J, Chen J, et al. The secreted protein discovery initiative (SPDI), a large-scale effort to identify novel human secreted and transmembrane proteins: a bioinformatics assessment. Genome Res. 2003;13:2265–2270. doi: 10.1101/gr.1293003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shimizugawa T, Ono M, Shimamura M, Yoshida K, Ando Y, Koishi R, Ueda K, et al. ANGPTL3 decreases very low density lipoprotein triglyceride clearance by inhibition of lipoprotein lipase. J Biol Chem. 2002;277:33742–33748. doi: 10.1074/jbc.M203215200. [DOI] [PubMed] [Google Scholar]
- 52.Shimamura M, Matsuda M, Yasumo H, Okazaki M, Fujimoto K, Kono K, Shimizugawa T, et al. Angiopoietin-like protein3 regulates plasma HDL cholesterol through suppression of endothelial lipase. Arterioscler Thromb Vasc Biol. 2007;27:366–372. doi: 10.1161/01.ATV.0000252827.51626.89. [DOI] [PubMed] [Google Scholar]
- 53.Robciuc MR, Tahvanainen E, Jauhiainen M, Ehnholm C. Quantitation of serum angiopoietin-like proteins 3 and 4 in a Finnish population sample. J Lipid Res. 2010;51:824–831. doi: 10.1194/jlr.M002618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hatsuda S, Shoji T, Shinohara K, Kimoto E, Mori K, Fukumoto S, Koyama H, et al. Association between plasma angiopoietin-like protein 3 and arterial wall thickness in healthy subjects. J Vasc Res. 2007;44:61–66. doi: 10.1159/000098153. [DOI] [PubMed] [Google Scholar]
- 55.Musunuru K, Pirruccello JP, Do R, Peloso GM, Guiducci C, Sougnez C, Garimella KV, et al. Exome sequencing, ANGPTL3 mutations, and familial combined hypolipidemia. N Engl J Med. 2010;363:2220–2227. doi: 10.1056/NEJMoa1002926. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.