Abstract
The proportion of Plasmodium vivax-infected subjects that are carry mature gametocytes, and thus are potentially infectious, remains poorly characterized in endemic settings. Here, we describe a quantitative reverse transcriptase (RT) real-time PCR (qRT-PCR) that targets transcripts of the mature gametocyte-specific pvs25 gene. We found mature gametocytes in 42 of 44 (95.4%) P. vivax infections diagnosed during an ongoing cohort study in northwestern Brazil. SYBR green qRT-PCR was more sensitive than a conventional RT-PCR that targets the same gene. Molecular detection of gametocytes failed, however, when dried bloodspots were used for RNA isolation and complementary DNA synthesis. Estimating the number of pvs25 gene transcripts allowed for examining the potential infectiousness of gametocyte carriers in a quantitative way. We found that most (61.9%) gametocyte carriers were either asymptomatic or had subpatent parasitemias and would have been missed by routine malaria control strategies. However, potentially undiagnosed gametocyte carriers usually had low-density infections and contributed a small fraction (up to 4%) to the overall gametocyte burden in the community. Further studies are required to determine the relative contribution to malaria transmission of long-lasting but low-density gametocytemias in asymptomatic carriers that are left undiagnosed and untreated.
Keywords: Plasmodium vivax, gametocytes, diagnosis, real-time PCR
1. Introduction
Plasmodium vivax, the most widespread human malaria parasite, causes up to 390 million episodes of disease each year in Central and South America, the Middle East, Central, South and Southeast Asia, Oceania and East Africa (Price et al., 2007), where 2.85 billion people are currently at risk of infection (Guerra et al., 2009). Mature gametocytes of P. vivax, in contrast with those of P. falciparum, are found in the bloodstream early in the course of infection, often before symptoms emerge and drug treatment is started. As a consequence, significant transmission of P. vivax may persist even in areas where timely diagnosis and treatment can be achieved (Sattabongkot et al., 2004; Bousema and Drakeley, 2011).
Microscopic examination of Giemsa-stained thick blood smears is poorly sensitive for detecting circulating P. vivax gametocytes. Under ideal conditions, the detection limit of microscopy is around 10–20 parasites per microliter of blood (Gilles, 1993). Plasmodium vivax infections are characterized by low parasitemias, since this parasite finds a limited supply of reticulocytes to infect in the peripheral blood, and only a small proportion of circulating blood stages (around 2%) are gametocytes (McKenzie et al., 2006). Therefore, the proportion of infected subjects that are potentially infectious (i. e., carry mature gametocytes) remains largely unknown in P. vivax- endemic settings.
Over the past decade, molecular methods have been developed to detect RNA transcripts that are specific of mature P. falciparum gametocytes, with a detection limit of 0.02–10 gametocytes per microliter of blood (Babiker et al, 2008). Molecular diagnosis requires the amplification of RNA transcripts of genes expressed exclusively by gametocytes, such as pfs25 and pfg377, with the use of techniques such as reverse transcriptase (RT)-polymerase chain reaction (PCR), quantitative nucleic acid sequence-based amplification (QT-NASBA) and RT loop-mediated isothermal amplification (RT-LAMP) (Babiker et al., 2008). More recently, these methods have been adapted to P. vivax orthologs of gametocyte-expressed genes (Barthi et al., 2006; Beurskens et al., 2009; Chansamut et al., 2012). QT-NASBA detected gametocyte transcripts in nearly two thirds of P. vivax-infected subjects (Beurskens et al., 2009) and the number of copies of mature gametocyte-specific RNA transcripts in blood samples predicted the success of experimental infection of anopheline mosquitoes (Barthi et al., 2006; Chansamut et al., 2012).
Here, we compare techniques for field sample preservation and describe a quantitative RT-PCR for P. vivax gametocyte detection and quantitation. With the use of properly processed samples, we found mature gametocytes in nearly all P. vivax infections diagnosed during an ongoing cohort study in northwestern Brazil, even among asymptomatic carriers of subpatent parasitemias that are missed by conventional microscopy. We discuss the potential implications of these findings for malaria control and elimination strategies.
2. Materials and methods
2.1 Study area and clinical samples
Our study subjects are participants in an ongoing prospective cohort study in the farming settlement of Remansinho, in the state of Amazonas, in the Western Amazon Basin of Brazil, close to the borders with Peru, Bolivia and the Brazilian states of Rondônia and Acre (da Silva-Nunes et al., 2012). The field site is characterized by year-round hypoendemic malaria transmission, with P. vivax prevalence rates varying between 5% and 11%. Plasmodium vivax accounts for more than 90% of all malaria infections diagnosed in this area. All samples used throughout this study were collected from subjects with laboratory-confirmed P. vivax infection under protocols approved by the Institutional Review Board of the University Hospital, University of São Paulo (1025/10), and by the National Research Ethics Committee of the Ministry of Health of Brazil (551/2010).
Three sample sets were analyzed (Table 1). The first set consisted of 44 venous blood samples collected during mass blood surveys between September 2010 and October 2011. Donors were 36 subjects (seven contributed 2 samples and one contributed 3 samples collected on different occasions) aged 5–56 years (median, 26 years) presenting with either symptomatic (n = 24) or asymptomatic (n = 20) single-species infection with P. vivax confirmed by quantitative real-time PCR (qPCR) targeting the a species-specific fragment of the 18S rRNA gene. Parasitemias estimated by qPCR were usually low (geometric mean, 70.9 parasites/µl of blood), ranging between 2 and 7478 parasites/µl of blood; only 21 of these infections were also diagnosed by microscopic examination of Giemsa-stained thick smears. To determine the presence and intensity of 13 malaria-related symptoms (fever, chills, sweating, headache, myalgia, arthralgia, abdominal pain, nausea, vomiting, dizziness, cough, dyspnea, and diarrhea) up to seven days prior to enrollment, we used a structured questionnaire as described (da Silva-Nunes and Ferreira, 2007). Blood samples drawn into ethylenediaminetetraacetic acid (EDTA)-treated vacuum tubes were placed on ice packs until processed in the field laboratory, 4–12 hours later. One 1-ml aliquot of each sample was transferred to cryotubes and frozen at −70°C. After 2–4 weeks, samples were transferred to liquid nitrogen and thawed 1–3 months later for RNA isolation. A 200-µl aliquot of the same samples was kept at −20°C and used for DNA isolation.
Table 1.
Methods for field sample processing and storage, RNA isolation and amplification used in this study.
| Type of sample |
No. samples |
Field sample processing and storage | RNA isolation | RT-PCR method |
|---|---|---|---|---|
| Venous blood |
44 | 1 ml blood transferred to cryotubes and kept at −70°C for 2–4 weeks and afterwards in liquid nitrogen (1–3 months) until RNA isolation |
QIAamp Viral RNA kit (Qiagen) |
Conventional and quantitative real time RT-PCR |
| Venous blood |
5 | 125 µl blood spotted onto FTA classic cards, Whatman#3 filter papers, 903 protein saver cards (Whatman) or QIAcard FTA cards (Qiagen) and stored in sealed plastic bags with desiccant at ambient temperature or −20°C (1–2 months) until RNA isolation |
RNA elution with RNA processing buffer (Whatman FTA Protocol BR01) followed by standard TRIzol LS protocol |
Quantitative real time RT-PCR |
| Capillary blood |
36 | 10–30 µl blood spotted onto FTA classic cards (Whatman), stored at ambient temperature (1–3 months) and afterwards at −20°C, in sealed plastic bags with desiccant (up to 8 months) until RNA isolation |
RNA elution with RNA processing buffer (Whatman FTA Protocol BR01) followed by standard TRIzol LS protocol |
Quantitative real time RT-PCR |
The second set consisted of venous blood samples collected from five febrile subjects presenting with single-species P. vivax infection confirmed by both microscopy and qPCR (parasitemia ranging between 859 and 10433 parasites/µl of blood). Up to 4 hours after blood collection, 125-µl aliquots of EDTA-treated whole blood were pipetted onto four types of cards: FTA classic cards, Whatman #3 filter papers, 903 protein saver cards (all from Whatman, Clifton, NJ) and QIAcard FTA cards (Qiagen, Hilden, Germany). Two cards of each type were spotted with each blood sample, air-dried at room temperature and stored in sealed plastic bags with desiccant, for 1–2 months. One card was kept at ambient temperature, while the duplicate was frozen at −20°C until RNA isolation. A 200-l aliquot of each blood sample was kept at −20°C and used for DNA isolation.
The third set consisted of archived capillary blood samples (volume, 10–30 µl) collected by finger prick between March and September 2010 and spotted onto FTA classic cards (Whatman). Donors (n = 36) were patients presenting at the malaria clinic of Remansinho with either symptomatic or asymptomatic infection with single-species P. vivax confirmed by qPCR. Cards had been stored in the field site at ambient temperature for 1–3 months, with no desiccant, before being placed in sealed plastic bags with desiccant and frozen at −20°C until DNA and RNA isolation, up to 8 months later.
2.2 RNA isolation and complementary DNA synthesis
RNA isolation from 200-µl aliquots of venous blood samples was carried out using the QIAmp Viral RNA Mini Kit (Qiagen, Hilden, Germany), following the manufacturers protocol (QIAmp Viral RNA Mini Handbook, third edition, April 2010)). Briefly, clinical samples were incubated with buffer AVL for 10 minutes at room temperature for cell lysis, mixed with ethanol, applied to a spin column, centrifuged and washed with buffers AW1 and AW2. RNA was then eluted from the columns with the AVE buffer and stored at −70°C until complementary DNA (cDNA) synthesis, up to one week later. Since RNA and DNA are isolated in parallel with this procedure, the eluate (4 µl) was treated three times with a RNAse-free DNAse (Fermentas, Burlington, Canada), according to the manufacturers protocol, for removal of residual genomic DNA from RNA preparations to be used as templates for cDNA synthesis.
For RNA isolation from bloodspots on FTA classic cards, Whatman #3 filter papers, 903 protein saver cards and QIAcard FTA cards, we excised 3-mm dried-blood discs with a Harris micropunch. We used the classical TRIzol LS reagent protocol for RNA extraction, essentially as described by the manufacturer (Invitrogen, Carlsbad, CA). Discs were placed in 2-ml reaction tubes, incubated with 400 µl RNA processing buffer (10mM Tris-HCL, pH 8.0; 0.1 mM EDTA, 800 U/ml RNase Out [Invitrogen], 200 µg/ml glycogen and 2 mM dithiothreitol [DTT]) on ice for 15 minutes, with mixing every 5 minutes. The disc was removed and 750 µl of TRIzol LS reagent (Invitrogen) was added. The sample was vortexed for 15 s and incubated at room temperature for 15 min. After adding 200 µl of chloroform, the solution was centrifuged (12,000 × for 15 min at 4°C) and the aqueous phase was transferred to a clean reaction tube. RNA was precipitated with 500 µl isopropyl alcohol on ice for 1 h. The pellet resulting from centrifugation (12,000 × for 45 min at 4°C) was washed with 75% ethanol, air dried, resuspended in 10 µl RNAse-free water and treated with DNAse, as described above, prior to use for cDNA synthesis. Regardless of the source of template RNA, cDNA was synthesized using the single-tube procedure of the Maxima First Strand cDNA synthesis kit for RT-qPCR (Fermentas), according to the manufacturers instructions.
2.3 Gametocyte detection by qualitative reverse transcriptase PCR
The conventional, qualitative RT-PCR protocol was standardized with the oligonucleotide primers (forward, AAC GAA GGG CTG GTG CAC CTT T; reverse, AGC AAC CTG CAC TTT GGA TTT CCG) designed by Bharti et al. (2006) to amplify a 267-bp fragment of the Pvs25 gene. The 25-µl PCR reaction mixture contained 9 µl of nuclease-free water, 0.3 µM (0.8 µl) of each primer, 1.5 µl of template cDNA, and 12.5 µl of the GoTaq Green master mix 2 × (Promega, Madison, WI). Thermal cycling, carried out on a GeneAmp PCR System 9700 equipment (Applied Biosystems, Foster City, CA), started with a 2-min denaturation step at 95°C, followed by 35 cycles of 95°C for 30 sec, 55°C for 35 s min and 68°C for 2.5 min, with a final 10-min final extension step at 68°C. Amplicons were visualized after 1.5% agarose gel electrophoresis and staining with 100 µM ethidium bromide. The following negative controls were used: (a) to control for genomic DNA contamination, a RT-minus control (containing all reagents for reverse transcription except for the Maxima enzyme mix [Fermentas]) was run for each RNA sample, using the standard RT-PCR protocol reagents; (b) to control for reagent contamination, no-template controls (containing all reagents for reverse transcription except for the RNA template) were run for every PCR microplate. As a control for cDNA template integrity, for each sample we run a qPCR targeting the 18S rRNA gene of P. vivax (described below, under Laboratory diagnosis of malaria), which is expressed by all parasites blood stages. RNA isolation and cDNA synthesis were repeated whenever amplification of the 18S rRNA control product failed. As a positive control for pvs25 gene amplification, genomic P. vivax DNA templates were run for every PCR microplate.
2.4 Gametocyte detection and quantification by reverse transcriptase real-time PCR
We standardized a SYBR green quantitative real-time RT-PCR (qRT-PCR) to detect and quantify pvs25 gene transcripts using the same oligonucleotide primers described above. The 267-bp amplified gene fragment was cloned into the pGEM-T Easy plasmid vectors (Promega); in every PCR microplate, a standard curve was prepared with 10 tenfold dilutions of the plasmid DNA, starting with 1.4 × 109 plasmid copies/µl. Real-time PCR was carried out in triplicate, on a Mastercycler realplex S real-time thermal cycler (Eppendorf, Hamburg, Germany). Each 15-µL reaction mixture contained 2 µL of sample cDNA, 7.5 µL of 2× Maxima SYBR Green qPCR master mixture (Fermentas) and 0.3 µM of each oligonucleotide primer. Amplification included a template denaturation step at 95° C (2 min) followed by 35 cycles of 30 s at 95°C, 35 s at 55°C and 2.5 minutes at 68°C, with fluorescence acquisition at the end of each extension step and a final extension step of 10 min at 68°C, with a final melting program consisting of 15 seconds at 95°C, 15 s at 60°C, and a stepwise temperature increase of 0.03°C/s until 95°C. Fluorescence acquisition was done at each temperature transition.
2.5 Laboratory diagnosis of malaria
Whenever available, Giemsa-stained thick blood smears had at least 100 fields examined for malaria parasites under 1000 × magnification by two experienced microscopists before being declared negative. Blood samples were further examined for malaria parasites by qPCR. When venous blood samples were available, DNA isolation from 200-µl aliquots, using QIAamp DNA blood kits (Qiagen), was fully automated on a QIAcube equipment (Qiagen) following the manufacturers protocol. DNA isolation from finger- prick blood samples spotted onto FTA classic cards was also carried out on a QIAcube equipment (Qiagen); we used the QIAmp DNA micro kit combined with the QIAcube protocol for the QIAmp DNA investigator kit for optimal DNA yield.
Each 15-µL qPCR mixture contained 2 µL of template DNA (or one washed disk), 7.5 µL of 2× Maxima SYBR Green qPCR master mixture (Fermentas) and 0.5 µM of each primer. We used the forward primer P1 (ACG ATC AGA TAC CGT CGT AAT CTT) and the reverse primer V1 (CAA TCT AAG AAT AAA CTC CGA AGA GAA A), which allow the amplification of a P. vivax-specific 100 kb fragment of the 18S rRNA gene (Kimura et al., 1996). Standard curves were prepared with serial tenfold dilutions of the target sequence, cloned into pGEM-T Easy vectors (Promega, Madison, WI), to allow for species-specific quantitation of parasite loads (number of parasites/µL of blood). We used a Mastercycler realplex S real-time thermal cycler (Eppendorf) for PCR amplification with an initial step at 50°C (2 min), followed by template denaturation at 95°C (10 min) followed by 40 cycles of 15 s at 95°C and 1 minute at 60°C, with fluorescence acquisition at the end of each extension step. Amplification was immediately followed by a melting program consisting of 15 seconds at 95°C, 15 seconds at 60°C, and a stepwise temperature increase of 0.03° C/s until 95°C, with fluorescence acquisition at each temperature transition.
2.6 Data analysis
Variables with overdispersed distribution were summarized as medians or geometric means and interquartile ranges and compared with nonparametric Mann-Whitney U tests. Agreement between tests was assessed using the kappa coefficient. Correlations were assessed using nonparametric Spearman rank correlation tests. Analyses were performed using SPSS 16.0 software (SPSS, Chicago, IL), with statistical significance set at a 5% level.
3. Results and discussion
3.1 Amplification of pvs25 gene transcripts by conventional RT-PCR
We first standardized a conventional, qualitative RT-PCR protocol to detect pvs25 gene transcripts in clinical samples. With this method, 35 of 44 (79.5%) whole-blood samples assayed were positive (Table 2). Parasite cDNA integrity was confirmed in all gametocyte-negative samples by amplification of 18S rRNA gene transcripts from the same templates. Not surprisingly, gametocyte-positive samples had a significantly higher parasite density, as estimated by a qPCR that quantifies both asexual and sexual stages, than negative samples (Mann-Whitney U test, P < 0.0001). The respective geometric mean parasitemias were 131.2 parasites/l for 35 positive samples (range, 2.4–7478 parasites/µl) and 6.5 parasites/µl for 9 negative samples (range, 2.0–15.6 parasites/µl).
Table 2.
Comparison between results obtained with a qualitative, conventional reverse transcriptase (RT)-PCR and a quantitative RT real-time PCR (qRT-PCR), both targeting the pvs25 gene, for gametocyte detection in 44 Plasmodium vivax infections from northwestern Brazil. We observe a fair but significant agreement between both methods (kappa = 0.312, P = 0.004).
| Conventional RT-PCR |
qRT-PCR | ||
|---|---|---|---|
| Negative | Positive | Total | |
| Negative | 2 | 7 | 9 |
| Positive | 0 | 35 | 35 |
| Total | 2 | 42 | 44 |
3.2 Amplification of pvs25 gene transcripts by quantitative real-time RT-PCR
Next, we standardized a simple qRT-PCR to detect and quantify pvs25 gene transcripts in clinical samples. We chose SYBR green dye, a cheap, efficient and flexible fluorescence reporting system, as an alternative to TaqMan probes (Barthi et al., 2006) or molecular beacons (Beurskens et al., 2009). SYBR green qRT-PCR proved to be substantially more sensitive than conventional RT-PCR, being able to detect pvs25 gene transcripts in 42 of 44 (95.4%) whole-blood samples assayed, with a fair but significant agreement between both methods (kappa = 0.312, P = 0.004; Table 2). Both samples with negative results by qRT-PCR had low parasite density (6.3 and 12.8 parasites/µl) estimated by qPCR and were also negative by conventional RT-PCR, but we were able to amplify 18S rRNA gene transcripts from the respective cDNA templates. We thus conclude that nearly all P. vivax-infected subjects in our field site in northwestern Brazil, either symptomatic or not, and regardless of their parasite load, have mature gametocytes circulating in their peripheral blood. Most estimates of P. vivax gametocyte carriage rate currently available for comparison derive from clinical trials that enrolled slide-positive and symptomatic patients (Bousema and Drakeley, 2011). In these selected populations, gametocyte detection rates, by conventional microscopy, in clinical vivax malaria varied widely from 29% to nearly 100% (McKenzie et al., 2006; Nacher et al., 2006; Bousema and Drakeley, 2011).Beurkens et al. (2009) found pvs25 transcripts in 51 of 74 (69%) clinical samples screened with QT-NASBA.
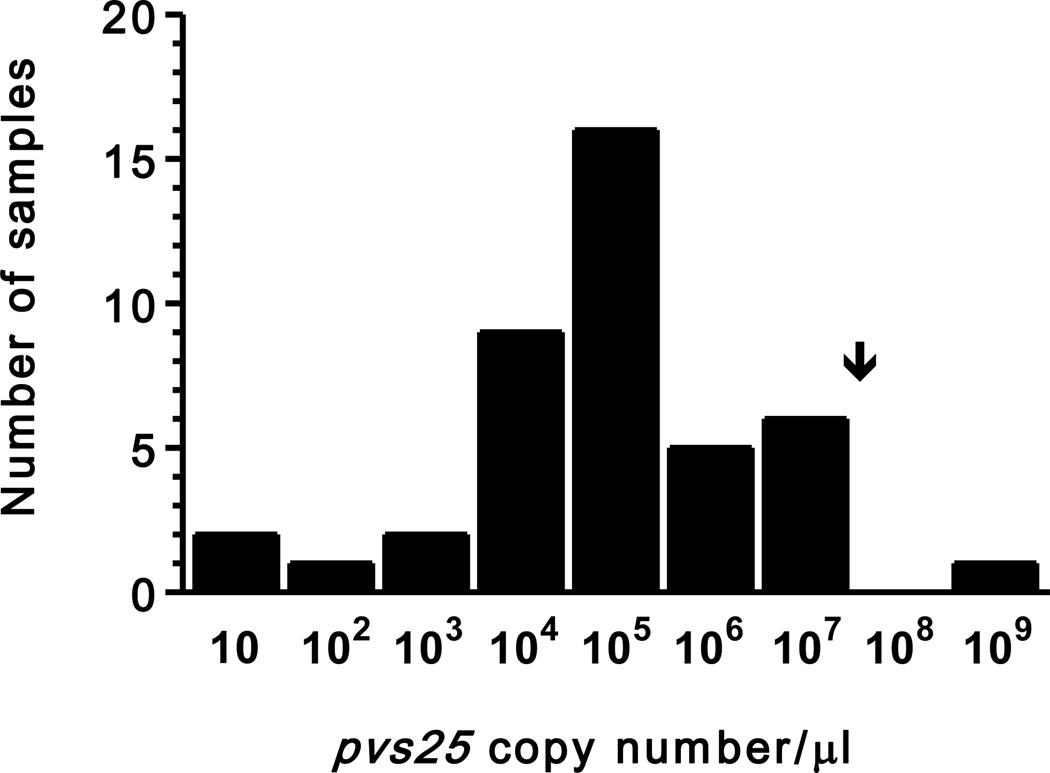
The number of copies of pvs25 gene transcripts detected by qRT-PCR, which correlates positively with (and indirectly estimates) the number of circulating mature gametocytes (Barthi et al., 2006), correlated positively with the overall parasite density estimated by qPCR that targets the 18SrRNA gene and quantifies both asexual and sexual stages (rs= 0.391, P = 0.009). These data are consistent with previous comparisons of gametocyte and asexual parasite densities determined by conventional microscopy (McKenzie et al., 2006). Interestingly, the number of pvs25 gene transcripts in the gametocyte-positive population (n = 42) was clearly overdispersed, with a variance (5.5 × 1015 copies/µl) much greater than the mean (1.2 × 107 copies/µl), ranging between 1.1 and 4.8 × 108 copies/µl of blood. The overdispersed nature of these data is further illustrated in Figure 1, which shows the frequency distribution of pvs25 gene transcript copy number per microliter of blood. The originally overdispersed distribution has been nearly normalized by presenting copy number data on x-axis in a logarithmic scale.
Figure 1.
Frequency distribution of individual gametocyte densities, estimated as the number of pvs25 gene transcripts (copy number/µl of blood) detected by quantitative reverse transcriptase real-time PCR, in 42 Plasmodium vivax-infected subjects from northwestern Brazil. The arrow indicates the arithmetic mean of pvs25 gene transcript copy number (1.2 × 107 copies/µl) in this population. Numbers on x-axis represent the upper limit of each copy number class. Note that by presenting values on x-axis in logarithmic scale we have nearly normalized the originally overdispersed frequency distribution.
3.3 Gametocytes in asymptomatic carriers of subpatent Plasmodium vivax parasitemias
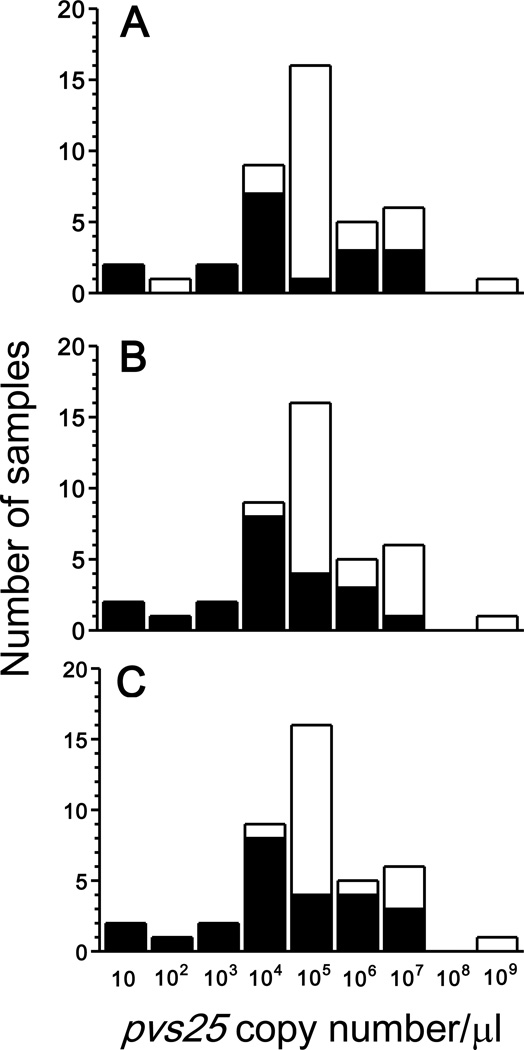
Asymptomatic infections, which are missed by routine active and passive case detection (ACD and PCD, respectively), have been hypothesized to represent a major source of infective gametocytes across the Amazon Basin of Brazil (Alves et al., 2005; da Silva-Nunes et al., 2012). ACD implies periodic visits to households, with collection of thick blood smears from every person having had fever since the last visit, while PCD targets febrile subjects visiting malaria diagnosis outposts who have a blood sample tested positive for malaria parasites. Because both ACD and PCD target only symptomatic infections (defined as “fever cases” in the classical literature [e. g., MacDonald, 1957]), asymptomatic carriers remain undetected and untreated (Coura et al., 2006). We thus examined the relative contribution of asymptomatic carriers of low-density parasitemias, who would have been missed by ACD and PCD based on conventional microscopy, to the overall P. vivax gametocyte burden (estimated by summing pvs25 gene transcript numbers) in the 42 gametocyte-positive infections diagnosed by qRT-PCR. In the first hypothetical scenario, only symptomatic subjects are screened, but a highly sensitive diagnostic method is used for parasite detection. Twenty of 42 (47.6%) gametocyte carriers found by qRT-PCR (corresponding to the black bar segments in Figure 2A) would have been missed by ACD and PCD simply because they have no malaria-related symptoms. Interestingly, some subjects in this area may remain asymptomatic despite their relatively high parasitemias and gametocyte densities (i. e., high numbers of copies of pvs25 gene transcripts), most likely due to acquired clinical immunity (da Silva-Nunes et al., 2012). In fact, we have found no clear-cut association between overall parasite burden (number of circulating asexual and sexual blood stages) and frequency and intensity of malaria-related symptoms in this population (Amanda B. Gozze and MUF, unpublished results), suggesting that the parasite density threshold associated with symptoms varies widely according to the hosts' age and cumulative exposure to malaria. However, most potentially undiagnosed asymptomatic infections have low gametocyte densities; as a consequence, symptom-less gametocyte carriers overall contribute a very small (around 0.04%) fraction of the total gametocyte burden in the host population.
Figure 2.
Proportion of gametocyte carriers that would be undiagnosed (represented as black bar segments) in 42 Plasmodium vivax-infected subjects from northwestern Brazil, under different hypothetical scenarios for malaria diagnosis, according to gametocyte densities, estimated as the number of pvs25 gene transcripts (copy number/µl of blood) detected by quantitative reverse transcriptase real-time PCR. In A, only symptomatic subjects are screened for malaria parasites, but a highly sensitive diagnostic method is used for parasite detection. As a consequence, even high-density infections may be missed if parasite carriers remain asymptomatic. In B, only slide-positive infections, regardless of the presence of symptoms, are detected. As a consequence, low-density (or subpatent) infections are missed even when parasite carriers are symptomatic. In C, only symptomatic subjects are screened for malaria parasites by conventional microscopy. As a consequence, all microscopy-negative infections (regardless of the presence of symptoms) and asymptomatic infections (regardless of parasite density) are missed. Note that most of the potentially undiagnosed infections have low gametocyte densities (i. e., low numbers of pvs25 gene transcripts). Values on x-axis are shown in logarithmic scale; numbers represent the upper limit of each copy number class.
In the second scenario, only slide-positive infections, regardless of the presence of symptoms, are detected. This corresponds to the strategy known as aggressive active case detection (AACD; Macauley, 2005), which consists of population-wide screenings for malaria parasites using conventional microscopy, irrespective of any clinical symptoms. We show that conventional microscopy would miss 23 of 42 (54.8%) gametocyte carriers detected by qRT-PCR, but most slide-negative infections have low gametocyte densities (Figure 2B). Therefore, gametocyte carriers missed by conventional microscopy contribute a small (around 1.7%) fraction of the overall gametocyte burden.
In the last scenario, only symptomatic subjects are screened for malaria parasites by conventional microscopy. This is exactly what routine ACD and PCD do in Brazil (Oliveira-Ferreira et al., 2010) and other endemic countries with well-structured national malaria control programs. This strategy would miss a substantial proportion (26 of 44, 61.9%) of gametocyte carriers, who contribute 4.0% of the total gametocyte burden in the host population (Figure 2C).
We therefore show that routine ACD and PCD miss a sizeable proportion of P. vivax gametocyte carriers. For example, more than two thirds of malaria infections in farming settlements in the Amazon are slide-negative (being detected only by PCR) and asymptomatic (da Silva et al., 2010). However, since most undiagnosed and untreated infections have very low gametocyte densities, their relative contribution to total gametocyte burden and thus malaria transmission might be considered almost negligible. In fact, the number of circulating P. vivax gametocytes correlates positively with the success rate of experimental infections of both South American (Barthi et al., 2006) and Southeast Asian (Chansamut et al., 2012) anopheline mosquitoes, with blood donors carrying low-density gametocytemias being less infectious to these vectors. Nevertheless, the correlation between gametocyte density and mosquito infection rates is by no means linear and submicroscopic infections may still be important for malaria transmission (Alves et al., 2005). In addition, untreated asymptomatic infections allow for gametocyte carriage and potential infectiousness over extended periods of time, while P. vivax gametocytes in treated patients have a median clearance time of 2 hours following standard chloroquine-primaquine chemotherapy (Pukrittayakamee et al., 2008). As a consequence, the relative contribution of asymptomatic and/or submicroscopic gametocyte carriage to P. vivax transmission in communities may be severely underestimated if one does not consider the duration of the infectious period, which in turn can be measured in well-designed cohort studies with frequent blood sampling.
3.4 Failed attempts to amplify pvs25 gene transcripts from dried bloodspots
The widespread use of molecular methods for gametocyte detection as a public health tool is severely constrained by the need to freeze field-collected blood samples at −70°C or in liquid nitrogen until RNA isolation and cDNA synthesis. A previous report showed that P. falciparum culture material spotted onto 903 protein saver cards and stored at 25°C or 37°C for up to 3 months remained suitable for amplifying pfs25 transcripts by nested RT-PCR (Mlambo et al., 2008). This method for sample storage has been used for active detection of P. falciparum gametocyte carriers in Zambia (Stresman et al, 2010). No similar study has been carried out with P. vivax gametocytes from patient-derived blood samples. We thus tested whether our qRT-PCR can detect pvs25 gene transcripts from cDNA templates prepared from dried bloodspots.
From blood samples spotted on four different types of cards (FTA classic cards, Whatman #3 filter papers, 903 protein saver cards, and QIAcard FTA cards; Table 1), kept at ambient temperature or frozen at −20°C for 1–2 months prior to further processing, we were able to obtain parasite cDNA templates and amplify transcripts of the 18S rRNA gene of P. vivax. However, pvs25 gene transcript amplification failed in all 40 cDNA templates tested (five whole-blood samples spotted on duplicates of four different cards and maintained at different temperatures). Similarly, we amplified 18S rRNA gene transcripts from most (28 of 36; 77.8%) archived bloodspots collected on FTA classic cards and stored in the field at ambient temperature, but none of these samples yielded pvs25 gene transcripts. We thus were able to amplify only highly abundant P. vivax-specific transcripts, such as those of the 18S rRNA gene (Kamau et al., 2011), from dried blood spots.
Mlambo et al. (2008) were able to detect pfs25 gene transcripts by nested RT-PCR, which may be more sensitive that our qRT-PCR, by using a commercial kit (RNAeasy Mini-kit, Qiagen) for RNA extraction from in vitro culture material spotted onto 903 protein saver cards. Moreover, dried blood spots stored under different temperatures have been successfully used as a source of RNA for quantification of HIV-1 viral load. A recent report from India, for example, showed a sensitivity of 95% for HIV-1 detection from dried blood spots on 903 protein saver cards kept at room temperature for 3–6 months (Vidya et al., 2012). However, a weak correlation was found between viral RNA measurements from dried blood spots and plasma samples from patients with low (<2000 copies/mL) viral loads (Vidya et al., 2012). These findings suggest that further improvements in RNA isolation and pvs25 gene amplification strategies may render our qRT-PCR suitable for gametocyte transcript amplification from dried blood spots, although the ability of accurately quantifying low-density gametocytemias from these samples remains to be determined.
3.5 Conclusions
We describe methods for RNA isolation and SYBR green qRT-PCR quantitation of mature P. vivax gametocytes from whole-blood samples. We show that qRT-PCR is more sensitive than a conventional RT-PCR that targets the same transcripts. In addition, qRT-PCR is faster and has a reduced risk of carry-over contamination, since amplicon handling (e. g., agarose gel electrophoresis) is not required. Molecular detection of gametocytes failed, however, when dried bloodspots were used for RNA isolation and cDNA synthesis.
Estimating the number of transcripts that are specific for mature gametocytes allows for examining the potential infectiousness of gametocyte carriers in a quantitative way. To illustrate this point, we found that nearly all asymptomatic carriers of submicroscopic P. vivax infections from an endemic setting in Brazil have circulating mature gametocytes, but usually at very low densities, contributing a small fraction to the overall gametocyte burden in the community. However, carriers of low gametocyte densities can still infect mosquitoes (Barthi et al., 2006; Chansamut et al., 2012) and, if left untreated, may potentially remain infectious over several months. We conclude that further community-based studies are required to determine the relative contribution of long-lasting but low-density gametocyte carriage to P. vivax transmission in areas approaching malaria elimination, where most high-density infections result in clinical episodes of malaria that are diagnosed by ACD or PCD and promptly treated.
Highlights.
Mature gametocyte are detected by molecular methods in nearly all incident Plasmodium vivax infections in a cohort study in the Amazon
Most gametocyte carriers in this endemic setting are either asymptomatic or have subpatent parasitemias and would be missed by routine malaria control strategies
Further studies are required to determine the relative contribution to malaria transmission of long-lasting but low-density gametocyte carriage
Acknowledgments
We thank the patients from Remansinho for their enthusiastic cooperation, and Amanda B. Gozze, Vanessa C. Nicolete, Raquel M. Gonçalves, Pablo S. Fontoura, Mônica da Silva-Nunes, Carlos E. Cavasini, and Rosely S. Malafronte for their support during field work, and Maria José Menezes and Gerhard Wunderlich (University of São Paulo) for their laboratory support and advice. This research was supported by research grants from the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health of USA (RO1 AI 075416 to MUF and U19 AI089681 to Joseph M. Vinetz) and the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP, 2009/52729-9 to MUF), Brazil. NFL received a research training scholarship from FAPESP and is currently supported by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Brazil. MUF receives a senior researcher scholarship from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) of Brazil. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations
- AACD
aggressive active case detection
- ACD
active case detection
- cDNA
complementary DNA
- CAPES
Coordenaçã o de Aperfeiçoamento de Pessoal de Nível Superior, Brazil
- CNPq
Conselho Nacional de Desenvolvimento Científico e Tecnológico, Brazil
- DTT
dithiothreitol
- EDTA
ethylenediaminetetraacetic acid
- FAPESP
Fundação de Amparo àPesquisa do Estado de São Paulo, Brazil
- LAMP
loop-mediated isothermal amplification
- NIAID
National Institute of Allergy and Infectious Diseases
- NIH
National Institutes of Health, United States
- PCD
passive case detection
- PCR
polymerase chain reaction
- qPCR
quantitative real-time PCR
- QT-NASBA
quantitative nucleic acid sequence-based amplification
- rRT-PCR
quantitative reverse transcriptase real-time PCR
- RT
reverse transcriptase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alves FP, Gil LH, Marrelli MT, Ribolla PE, Camargo EP, da Silva LH. Asymptomatic carriers of Plasmodium spp as infection source for malaria vector mosquitoes in the Brazilian Amazon. J. Med. Entomol. 2005;42:777–779. doi: 10.1093/jmedent/42.5.777. [DOI] [PubMed] [Google Scholar]
- Babiker HA, Schneider P, Reece SE. Gametocytes: insights gained during a decade of molecular monitoring. Trends Parasitol. 2008;24:525–530. doi: 10.1016/j.pt.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beurskens M, Mens P, Schallig H, Syafruddin D, Asih PB, Hermsen R, Sauerwein R. Quantitative determination of Plasmodium vivax gametocytes by real-time quantitative nucleic acid sequencebased amplification in clinical samples. Am. J. Trop. Med. Hyg. 2009;81:366–369. [PubMed] [Google Scholar]
- Bharti AR, Chuquiyauri R, Brouwer KC, Stancil J, Lin J, Llanos- Cuentas A, Vinetz JM. Experimental infection of the neotropical malaria vector Anopheles darlingi by human patient-derived Plasmodium vivax in the Peruvian Amazon. Am. J. Trop. Med. Hyg. 2006;75:610–616. [PMC free article] [PubMed] [Google Scholar]
- Bousema T, Drakeley C. Epidemiology and infectivity of Plasmodium falciparum and Plasmodium vivax gametocytes in relation to malaria control and elimination. Clin. Microbiol. Rev. 2011;24:377–410. doi: 10.1128/CMR.00051-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chansamut N, Buates S, Takhampunya R, Udomsangpetch R, Bantuchai S, Sattabongkot J. Correlation of Pfg377 ortholog gene expression of Plasmodium vivax and mosquito infection. Trop. Med. Int. Health. 2012 doi: 10.1111/j.1365-3156.2011.02940.x. in press. [DOI] [PubMed] [Google Scholar]
- Coura JR, Suarez-Mutis M, Ladeia-Andrade S. A new challenge for malaria control in Brazil: asymptomatic Plasmodium infection - a review. Mem. Inst. Oswaldo Cruz. 2006;101:229–237. doi: 10.1590/s0074-02762006000300001. [DOI] [PubMed] [Google Scholar]
- da Silva-Nunes M, Ferreira MU. Clinical spectrum of uncomplicated malaria in semi-immune Amazonians: beyond the "symptomatic" vs "asymptomatic" dichotomy. Mem Inst Oswaldo Cruz. 2007;102:341–347. doi: 10.1590/s0074-02762007005000051. [DOI] [PubMed] [Google Scholar]
- da Silva NS, da Silva-Nunes M, Malafronte RS, Menezes MJ, D'Arcadia RR, Komatsu NT, Scopel KK, Braga EM, Cavasini CE, Cordeiro JA, Ferreira MU. Epidemiology and control of frontier malaria in Brazil: lessons from community-based studies in rural Amazonia. Trans. R. Soc. Trop. Med. Hyg. 2010;104:343–350. doi: 10.1016/j.trstmh.2009.12.010. [DOI] [PubMed] [Google Scholar]
- da Silva-Nunes M, Moreno M, Conn JE, Gamboa D, Abeles S, Vinetz JM, Ferreira MU. Amazonian malaria: Asymptomatic human reservoirs, diagnostic challenges, environmentally driven changes in mosquito vector populations, and the mandate for sustainable control strategies. Acta Trop. 2012;121:281–291. doi: 10.1016/j.actatropica.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilles HM. Diagnostic methods in malaria. In: Gilles HM, Warrell DA, editors. Bruce Chwatt's Essential Malariology. London: Edward Arnold; 1993. pp. 78–95. [Google Scholar]
- Guerra CA, Howes RE, Patil AP, Gething PW, Van Boeckel TP, Temperley WH, Kabaria CW, Tatem AJ, Manh BH, Elyazar IR, Baird JK, Snow RW, Hay SI. The international limits and population at risk of Plasmodium vivax transmission in 2009. PLoS Negl. Trop. Dis. 2010;4:e774. doi: 10.1371/journal.pntd.0000774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamau E, Tolbert LS, Kortepeter L, Pratt M, Nyakoe N, Muringo L, Ogutu B, Waitumbi JN, Ockenhouse CF. Development of a highly sensitive genus-specific quantitative reverse transcriptase realtime PCR assay for detection and quantitation of Plasmodium by amplifying RNA and DNA of the 18S rRNA genes. J. Clin. Microbiol. 2011;49:2946–2953. doi: 10.1128/JCM.00276-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M, Kaneko O, Liu Q, Zhou M, Kawamoto F, Wataya Y, Otani S, Yamaguchi Y, Tanabe K. Identification of the four species of human malaria parasites by nested PCR that targets variant sequences in the small subunit rRNA gene. Parasitol. Int. 1997;46:91–99. [Google Scholar]
- Macauley C. Aggressive active case detection: a malaria control strategy based on the Brazilian model. Soc. Sci. Med. 2005;60:563–573. doi: 10.1016/j.socscimed.2004.05.025. [DOI] [PubMed] [Google Scholar]
- MacDonald G. The Epidemiology and Control of Malaria. Oxford: Oxford University Press; 1957. [Google Scholar]
- McKenzie FE, Wongsrichanalai C, Magill AJ, Forney JR, Permpanich B, Lucas C, Erhart LM, O'Meara WP, Smith DL, Sirichaisinthop J, Gasser RA., Jr Gametocytemia in Plasmodium vivax and Plasmodium falciparum infections. J. Parasitol. 2006;92:1281–1285. doi: 10.1645/GE-911R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlambo G, Vasquez Y, LeBlanc R, Sullivan D, Kumar N. Short report: a filter paper method for the detection of Plasmodium falciparum gametocytes by reverse transcription-polymerase chain reaction. Am. J. Trop. Med. Hyg. 2008;78:114–116. [PubMed] [Google Scholar]
- Nacher M, Silachamroon U, Singhasivanon P, Wilairatana P, Phumratanaprapin W, Fontanet A, Looareesuwan S. Risk factors for Plasmodium vivax gametocyte carriage in Thailand. Am. J. Trop. Med. Hyg. 2004;71:693–695. [PubMed] [Google Scholar]
- Oliveira-Ferreira J, Lacerda MV, Brasil P, Ladislau JL, Tauil PL, Daniel-Ribeiro CT. Malaria in Brazil: an overview. Malar. J. 2010;9:115. doi: 10.1186/1475-2875-9-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price RN, Tjitra E, Guerra CA, Yeung S, White NJ, Anstey NM. Vivax malaria: neglected and not benign. Am. J. Trop. Med. Hyg. 2007;77(6 Suppl):79–87. [PMC free article] [PubMed] [Google Scholar]
- Pukrittayakamee S, Imwong M, Singhasivanon P, Stepniewska K, Day NJ, White NJ. Effects of different antimalarial drugs on gametocyte carriage in P. vivax malaria. Am. J. Trop. Med. Hyg. 2008;79:378–384. [PubMed] [Google Scholar]
- Sattabongkot J, Tsuboi T, Zollner GE, Sirichaisinthop J, Cui L. Plasmodium vivaxtransmission: chances for control? Trends Parasitol. 2004;20:192–198. doi: 10.1016/j.pt.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Schneider P, Bousema JT, Gouagna LC, Otieno S, van de Vegte- Bolmer M, Omar SA, Sauerwein RW. Submicroscopic Plasmodium falciparum gametocyte densities frequently result in mosquito infection. Am. J. Trop. Med. Hyg. 2007;76:470–474. [PubMed] [Google Scholar]
- Vidya M, Saravanan S, Rifkin S, Solomon SS, Waldrop G, Mayer KH, Solomon S, Balakrishnan P. Dried blood spots versus plasma for the quantification of HIV-1 RNA usind a real-time PCR, m2000rt assay. J. Virol. Methods. 2012;181:177–181. doi: 10.1016/j.jviromet.2012.02.006. [DOI] [PubMed] [Google Scholar]