Abstract
Melanin-concentrating hormone (MCH) is a 19-aa cyclic neuropeptide originally isolated from chum salmon pituitaries. Besides its effects on the aggregation of melanophores in fish several lines of evidence suggest that in mammals MCH functions as a regulator of energy homeostasis. Recently, several groups reported the identification of an orphan G protein-coupled receptor as a receptor for MCH (MCH-1R). We hereby report the identification of a second human MCH receptor termed MCH-2R, which shares about 38% amino acid identity with MCH-1R. MCH-2R displayed high-affinity MCH binding, resulting in inositol phosphate turnover and release of intracellular calcium in mammalian cells. In contrast to MCH-1R, MCH-2R signaling is not sensitive to pertussis toxin and MCH-2R cannot reduce forskolin-stimulated cAMP production, suggesting an exclusive Gαq coupling of the MCH-2R in cell-based systems. Northern blot and in situ hybridization analysis of human and monkey tissue shows that expression of MCH-2R mRNA is restricted to several regions of the brain, including the arcuate nucleus and the ventral medial hypothalamus, areas implicated in regulation of body weight. In addition, the human MCH-2R gene was mapped to the long arm of chromosome 6 at band 6q16.2–16.3, a region reported to be associated with cytogenetic abnormalities of obese patients. The characterization of a second mammalian G protein-coupled receptor for MCH potentially indicates that the control of energy homeostasis in mammals by the MCH neuropeptide system may be more complex than initially anticipated.
The hypothalamic neuropeptide melanin-concentrating hormone (MCH) derived its name from its ability to aggregate melanin-filled granules in melanophores in skin from teleost fish (for review see refs. 1 and 2). Alteration of skin color permits the animal to blend into a background, rendering it less visible to a predator or prey. After the initial isolation of MCH from salmon pituitary (3), determination of the structure for rat and human MCH showed that this neuropeptide is highly conserved during vertebrate evolution (4–6). In addition to the authentic MCH gene on chromosome 12, two other loci on chromosome 5 encoding truncated forms of the MCH gene (variant MCH gene) have been identified in humans (7, 8).
Neurons in the lateral hypothalamus with far-reaching projections throughout the brain are the major site of MCH production in mammals (9). This expression pattern suggested that MCH might participate in the regulation of energy homeostasis. Subsequently, several studies in rodents provided evidence that MCH is an important regulator of food intake and body weight. Qu et al. (10) found that MCH mRNA levels are about 3-fold up-regulated in leptin-deficient ob/ob mice compared with wild-type mice. Fasting also resulted in increased MCH mRNA levels in ob/ob mice and wild-type animals. In addition, injection of MCH into the lateral ventricles in rats led to increased food consumption (10–12). To analyze the physiological role of MCH in mice, Shimada and colleagues (13) generated MCH-deficient mice and found that MCH(−/−) animals are lean and hypophagic and show an increase in metabolic rate. In contrast, overexpression of MCH in transgenic mice leads to obesity and insulin resistance (14). These results suggest that an MCH antagonist might be beneficial for the treatment of obesity. Recently, several groups reported the identification of an orphan G protein-coupled receptor called SLC1/GPR24 as a cognate receptor for MCH (15–19). We now report here the identification and characterization of a second MCH receptor designated MCH-2R.
Materials and Methods
Database Search and Isolation of MCH-2R.
Searching of GenBank databases (20) with the MCH-1R protein sequence (complete coding sequence) using the tblastn search program (21) identified a human genomic DNA sequence (accession no. AQ747249; deposited on 7/19/99, Genome Survey Sequence subset of GenBank) with a significant homology score. The sequence, derived from a human male bacterial artificial chromosome library, encodes a polypeptide sequence with ≈50% protein identity to the MCH-1R in the second through third putative transmembrane domains. The full-length sequence, herein designated MCH-2R, was cloned by a process called RCCA (reduced complexity cDNA analysis) (22, 23). A human fetal brain cDNA library was constructed with a mixture of random and oligo(dT)-primed cDNA and a modified pBlueScript vector (Stratagene). Approximately four million clones were plated at ≈20,000 clones per plate. Plasmid DNA from each plate was prepared and stored in two 96-well plates. Two primers (RRC13: 5′-TGCAGAGAGGCCCCCCAAACACCC-3′ and RPC15: 5′-CTGACATCTATATCTGCAACCTGG-3′) based on the genomic sequence were designed and synthesized. PCRs were carried out by using the two primers to screen superpools of arrayed cDNA libraries. Positive pools were identified. PCR-based race reactions then were carried out by using vector and gene-specific primers as follows: RPC15 + PBS543R (vector primer: 5′-GGGGATGTGCTGCAAGGCGA-3′) or PBS 873F (vector primer: 5′-CCCAGGCTTTACACTTTATGCTTCC-3′), and RPC13 + PBS543R or PBS873F. The products from these reactions were used as templates to carry out secondary PCRs using primers nested within the primary reactions, i.e., RPC55F (inside of RPC15: 5′-TGTGGCTGATTTGGTCCAC-3′) + PBS578R (vector primer: 5′-CCAGGGTTTTCCCAGTCACGAC-3′) or PBS838F (vector primer: 5′-TTGTGTGGAATTGTGAGCGGATAAC-3′) for templates from RPC15-containing primary reactions, and RPC80R (inside of RPC13: 5′-CCTCGGGCCCATTGGTGAATAAG-3′) + PBS578R or PBS838F for templates from RPC13-containing primary reactions. Amplicons were purified and sequenced by using M13 forward and reverse primers and the RPC80R and RPC55F primers. Nucleotide sequences were determined by sequencing both strands using dye terminator cycle sequencing Ready Reactions (Perkin–Elmer) on an ABI 377 instrument.
Putative transmembrane domains were predicted by using the tmpred program on the EMBnet server (http://www.ch.embnet.org/software/TMPRED_form.html).
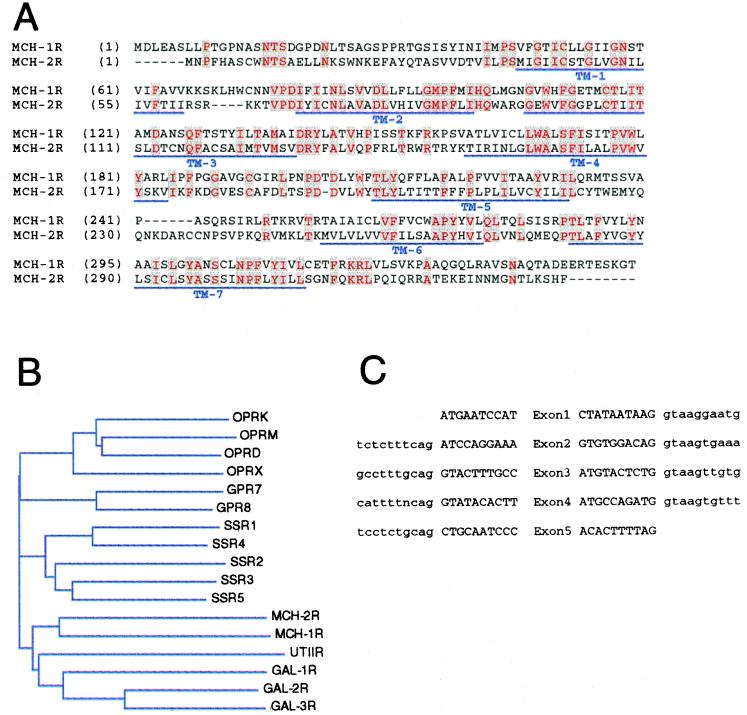
A phylogenetic tree was generated by using the alignx component of the vector nti suite software (InforMax, Bethesda, MD). This program applies the neighbor-joining method of Saitou and Nei (25). Distances between sequences are related to the degree of divergence between sequences. SwissProt and GenBank accession numbers for the other human sequences used in this alignment are: opioid receptors, OPRK P41145 OPRM P35372, OPRD P41143, OPRX P41146, GPR7 P48145, GPR8 P48146; urotensin II receptor, af140631; somatostatin receptors, SSR1 P30872, SSR2 P30874, SSR3 P32745, SSR4 P31391, SSR5 P35745; galanin receptors, GAL1R P47211, GALR2 O43603, GALR3 O60755.
Peptides.
A total of 83 peptides were purchased from Phoenix Pharmaceuticals (St. Joseph, MO), arrayed in a 96-well plate (26), and tested for activation of the human MCH-2R in an aequorin assay.
Radioligand Binding.
Membrane binding assays were performed on transiently transfected COS-7 cells using human MCH-2R in the plasmid vector pCI-neo (Promega) or on a Chinese hamster ovary (CHO) cell line stably expressing the MCH-2R in the plasmid vector pEF1/V5-HisB (Invitrogen). For transient expression, COS-7 cells were cultured in DMEM (GIBCO/BRL) with 10% heat-inactivated FCS. A suspension of 7 × 106 COS-7 cells was transfected with 20 μg of pCI-neo/MCH-2R plasmid by electroporation (27), and cells were harvested after 60–72 h. Membranes were prepared from transient and stable transfectants by hypotonic lysis, frozen in liquid nitrogen, and stored at −80°C as described (28). A scintillation proximity assay (SPA) was developed to measure the specific binding of [125I]Phe13Tyr19-MCH (≈2,000 Ci/mmol; NEN Life Sciences) to MCH-1R- and MCH-2R-containing membranes. SPAs were carried out by using wheat-germ agglutinin-polyvinyltoluene beads (Amersham Pharmacia), in 96-well OptiPlates (Packard). Each well contained 0.25 mg of SPA beads, 1–10 μg of membrane protein, and 200 μl of binding buffer. Binding buffer contained 50 mM Tris (pH 7.4), 8 mM MgCl2, 12% glycerol, 0.1% BSA (Sigma), and protease inhibitors (4 μg/ml leupeptin, 40 μg/ml Bacitracin (Sigma), 5 μg/ml Aprotinin, and 100 μM 4-(2-aminoethyl)benzenesulfonyl fluoride (Roche Molecular Biochemicals). Assays were optimized with respect to membrane preparations: for CHO/MCH-1R membranes, 1 μg of membranes per well yielded a >6× specific binding window and for COS or CHO MCH-2R membranes, 8 μg of membrane protein yielded a window of about 3×. Specific binding is defined as the difference between total binding and nonspecific binding conducted in the presence of 500 nM unlabeled MCH. Beads were coated with membranes for 20 min and dispensed to the 96-wells, various concentrations of test compounds in DMSO were added (final DMSO concentration 1–2%), then 25 nCi of [125I]Phe13Tyr19-MCH was added to the wells. After equilibrating at room temperature for 3 h, the plates were read in a TopCount (Packard). IC50 calculations were performed by using PRISM 3.0 (GraphPad, San Diego).
Functional Analysis of MCH-2R.
The entire coding sequence of human MCH-2R was cloned into the expression plasmids pEF1/V5-HisB or pcDNA-3.1 (Invitrogen), pCI-neo (Promega), and pIRES (CLONTECH). The resultant constructs were transfected into human embryonic kidney cells (HEK293) expressing simian virus 40 large T antigen (HEK293-T) cells or CHO cells by using Effectene (Qiagen, Hilden, Germany) or LipofectAmine PLUS (GIBCO/BRL) according to the manufacturer's instructions. In selected experiments, MCH-2R plasmids were cotransfected with the promiscuous Gα subunit Gα15 (29) to enhance functional responses. HEK293-T or parental HEK293 were maintained in DMEM/F-12 medium (Life Technologies, Rockville, MD) supplemented with 10% FBS, 100 units/ml penicillin-G, and 100 μg/ml streptomycin at 37°C with 5% CO2 in a humidified atmosphere. CHO cells were maintained in Iscove's medium (GIBCO/BRL) and supplemented as above. Stable cell lines were generated in both cell backgrounds after appropriate drug selection. Both FLIPR (fluorometric imaging plate reader, Molecular Devices, Sunnyvale, CA) (26, 30) and aequorin bioluminescence assays (31, 32) were performed to measure agonist-induced calcium mobilization. For the FLIPR, 1 day before assay, MCH-2R/CHO cells were seeded into the black-wall, clear-bottom 96-well plate (Corning 3603) at 5 × 104 cells/100 μl medium/well. Spent medium was discarded, and the cells were incubated with 100 μl/well of the assay buffer (Hanks' balanced salt solution, 0.5% BSA, 20 mM Hepes, 2.5 mM probenecid, pH 7.4) containing 2 μM fluo-4 AM, 0.04% pluronic acid, and 1% FBS in CO2 incubator for 60 min. After washing four times with 100 μl/well of the assay buffer, the cell plate was set into the FLIPR, the assay was started, and fluorescence output was measured. Agonists were dissolved with DMSO and diluted into appropriate concentrations with assay buffer. A total of 50 μl/well of the agonist solution was added. Final DMSO concentration in the reaction was adjusted to 1%. When fluo-4 was used as Ca indicator, the basal fluorescence intensity of dye-loaded cells was 10,000–15,000 and the fluorescence peak on maximal response was 40,000–50,000.
For the aequorin assay, activation of the MCH-2R expressed in the aequorin stable reporter cell line HEK293-AEQ17(33) was performed by using a Luminoskan RT luminometer (Labsystems, Gaithersburg, MD) controlled by custom software (31). HEK293-AEQ17 cells (8 × 105 cells plated in a T75 flask) stably expressing MCH-2R were incubated with the essential chromophore coelenterazine cp (10 μM; Molecular Probes) under reducing conditions (300 μM reduced glutathione in ECB buffer: 140 mM NaCl/20 mM KCl/20 mM Hepes-NaOH, pH 7.4/5 mM glucose/1 mM MgCl2/1 mM CaCl2/0.1 mg/ml BSA) to charge the apo-aequorin. The cells were harvested, washed once in ECB medium, and resuspended to 500,000 cells/ml. One hundred microliters of cell suspension (corresponding to 5 × 104 cells) then was injected into a 96-well test plate, and the integrated light emission was recorded over 30 s in 0.5-s units. Twenty microliters of lysis buffer (0.1% final Triton X-100 concentration) then was injected, and the integrated light emission was recorded over 10 s in 0.5-s units. The “fractional response” values for each well were calculated by taking the ratio of the integrated response to the initial challenge to the total integrated luminescence including the Triton X-100 lysis response.
For measuring pertussis toxin (PTX) sensitivity of intracellular calcium mobilization, stable HEK293-AEQ17 cells expressing human MCH-1R or MCH-2R were treated for 4 h with 0, 150 ng/ml, 500 ng/ml, or 5,000 ng/ml PTX (Sigma) in the growth medium before the aequorin assay.
For analysis of inositol phosphate turnover, HEK293-AEQ17 cells stably expressing human MCH-1R or MCH-2R were plated in 12-well dishes (2.75 × 105 cells/well) and incubated with 4.8 μCi 3H-myo inositol (TRK 912, Amersham Pharmacia) overnight at 37°C. After stimulation with MCH for 1 h at 37°C the reaction was stopped by replacing the medium with 800 μl of 10 mM formic acid. Cell extracts were collected after a 15-min incubation period at 4°C and neutralized with 100 μl of 50 mM ammonium hydroxide. Subsequently, the inositol phosphate fraction was isolated by anion exchange chromatography as described by Berridge et al. (34).
Expression Analysis.
Northern blot analysis.
Human multitissue Northern blots were purchased from CLONTECH. The entire coding sequence of human MCH-2R was radio-labeled by using α32P-dCTP (ReadyPrime kit, Amersham Pharmacia). Hybridizations were carried out in ExpressHyb buffer (CLONTECH) at 68°C for 4 h. The blots then were washed at high stringency (20 min at 68°C twice in 2× SSC, 0.05% SDS followed by 30 min final wash at 68°C, in 0.1%× SSC, 0.1% SDS) and exposed to x-ray film at −70°C for 3 days.
In situ hybridization analysis.
33P- and biotin-labeled riboprobes were generated by in vitro transcription (Roche Molecular Biochemicals) from a 1,086-bp MCH-1R cDNA or a 1,022-bp MCH-2R cDNA. Eight-micrometer thick sections of fresh frozen monkey brains (Merck Research Laboratories) were immersion-fixed in 4% paraformaldehyde for 45 min, and in situ hybridization was carried out as described (35). Detection of biotin probes was done by using the TSA Indirect Kit or TSA Direct Kit (NEN Life Sciences) according to manufacturer's instructions. For 33P-labeled probes, autoradiography was carried out with Kodak BioMax MR film at room temperature for 1 week. Sections were examined microscopically, and images were digitally acquired, pseudocolored, and quantified by using an M5 MCID (Imaging Research, St. Catherine's, ON, Canada) or Metamorph imaging program (Universal Imaging, West Chester, PA).
Genomic Structure and Chromosomal Localization.
Isolation of genomic clones containing the human MCH-2R gene.
A human P1 artificial chromosome (PAC) genomic library was obtained from Genome Systems (St. Louis) with individual PAC clones spotted in duplicate in an array. The filters were hybridized at 32°C with a radio-labeled (random prime labeled; Promega) human MCH-2R probe. Posthybridization washing stringency was at 65°C in 0.1× SSC, 0.1% SDS for 1 h. Three individual clones hybridized strongly under these conditions and were subjected to further analysis. PAC plasmid DNA was isolated from 30-ml overnight cultures grown in LB/25 μg/ml kanamycin, by an alkaline lysis protocol as per the manufacturer's instructions (Genome Systems) and dissolved in 10 mM Tris⋅HCl, pH 8.3/1 mM EDTA). Two of the clones (60p05 and 214d23) were chosen for subsequent characterization. PAC DNA was purified according to the supplier's protocol (Qiagen Large-Construct Kit) and used directly for sequencing.
Fluorescence in situ hybridization.
PAC clone 60p05 was used by Incyte Genomics (St. Louis) for fluorescence in situ hybridization according to standard procedures. In brief, purified DNA was labeled with digoxigenin dUTP and hybridized to metaphase chromosomes derived from phytohemagglutinin-stimulated peripheral blood lymphocytes. Specific hybridization signals were detected by fluorescinated antidigoxigenin antibodies followed by counterstaining with 4′,6-diamidino-2-phenylindole. After an initial experiment suggested labeling of the long arm of chromosome 6 on the basis of size, morphology, and banding pattern a second experiment was conducted in which a marker for the 6p12 region was cohybridized with PAC clone 60p05. The distance of the hybridization signal from the centromere and telomere was used to assign the chromosomal location of PAC clone 60p05 to band 6q16.2–16.3.
Radiation hybrid mapping.
Chromosomal mapping of the MCH-2R and the SIM1 gene was performed by using GENEBRIDGE 4 panel, consisting of 93 radiation hybrid clones (Research Genetics, Huntsville, AL). Primer pairs used were: MCH-2R exon 4, 5′-GTATACACTTTATTTGACGATAACAAC-3′ (2R588+), 5′-CCTTATTCTGTTGATACATCTCCC-3′ (2R697−), resulting in a 109-bp fragment; SIM1 exon 1, 5′-AGAAGAAAGGGGGAACAAGACACA-3′ (SIME1+), 5′-CTGGGAACACCACTCTCATTTTGA-3′ (SIME1−), for a 195- bp fragment; SIM1 exon 11, 5′-ACATCATGTGAG CCTGTTTCAAATA3′ (SIME11+), 5′-CATAGTAAATGCTGGTAATGGGGTAT-3′ (SIME11−) for a 952-bp fragment. PCR was performed with AmpliTaq Gold (Perkin–Elmer) using the following cycling parameters: 94°C for 9 min, 94°C 2 min, 62°C 30 s, 72°C for 1 min, 32 cycles, 72°C 7 min. Results were submitted to the Whitehead Institute Genome server (http://www.genome.wi.mit.edu/cgi-bin/contig/rhmapper.pl) and confirmed by Quantum Somatic Cell Hybrid PCRable Panel (Quantum, Durham, NC) and Stanford G3 Radiation Hybrid Panel (Research Genetics).
Animals.
Animals used in this study were kept in accord with rules and guidelines of the Merck Research Laboratories Institutional Animal Care and Use Committee and the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals.
Results and Discussion
Identification of MCH-2R.
Searching the GenBank database by using the complete human MCH-1R amino acid sequence as query, we identified a segment of human genomic sequence derived from a bacterial artificial chromosome clone (AQ747249), which displayed a 70-aa region of about 50% amino acid identity to the MCH-1R sequence between the TM2 and TM3 regions. Primers complementary to this region were able to detect expression of the corresponding RNA transcript in a human fetal brain cDNA library with PCR. Subsequently, a 1,023-bp cDNA clone was isolated and its nucleotide sequence was determined. Translation of the ORF contained within the cDNA clone predicts a 340-aa protein that shares an overall homology of about 38% with MCH-1R at the amino acid level (Fig. 1A). Its homology to members of the somatostatin, galanin, and opioid receptor gene family is between 32% and 35% amino acid identity. Phylogenetic analysis placed the clone most closely related to MCH-1R in this family of the neuropeptide receptors (Fig. 1B), and it was designated MCH-2R.
Figure 1.
(A) Amino acid sequence comparison of the human MCH-1R and MCH-2R. Identical residues are colored in red. Predicted transmembrane domains are indicated with a blue line. (B) Phylogenetic tree of the MCH neuropeptide receptor family including opioid, somatostatin, galanin, urotensin 2, and orphan receptors (GPR7, GPR8). (C) Exon/intron boarders of the human MCH-2R gene. Exon sequence is indicated by capital letters; intron sequence is noted in lowercase letters.
In Vitro Characterization of MCH-2R.
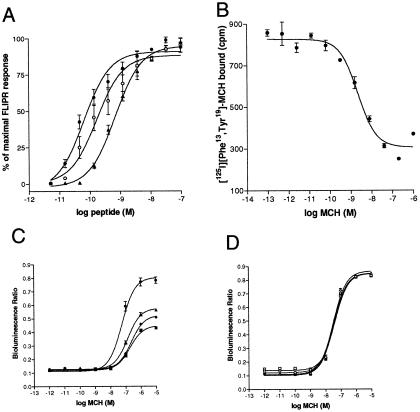
We identified that MCH-2R is highly sensitive to MCH by stable expression of MCH-2R in a CHO cell line and monitoring mobilization of intracellular calcium with FLIPR (fluorometric imaging plate reader) technology (Fig. 2A), thus confirming that MCH-2R is a functional receptor for the MCH. Similar to MCH, the modified peptide Phe13Tyr19-MCH (36) and salmon MCH are able to activate MCH-2R, albeit with a reduced potency (Table 1).
Figure 2.
(A) Agonist-induced calcium mobilization in a CHO cell line stably expressing human MCH-2R as detected by a fluorometric imaging plate reader (FLIPR) assay. MCH (●), Phe13Tyr19-MCH (○), salmon MCH (▴). (B) Membrane binding assay using a scintillation proximity assay measuring specific binding of [125I]Phe13Tyr19-MCH to MCH-2R containing membranes. (C and D) PTX sensitivity of intracellular calcium mobilization mediated by MCH-1R (C) or MCH-2R (D) in HEK293-AEQ17 cell lines. Stable transfected HEK293-AEQ17 cell lines were treated with 0 ng/ml (circles), 150 ng/ml (triangles), 500 ng/ml (diamonds), or 5,000 ng/ml (rectangles) PTX before the aequorin assay.
Table 1.
Comparison of half-maximal concentrations for binding at and functional activation of human MCH-1R and MCH-2R by MCH, Phe13Tyr19-MCH, and salmon MCH
| Ligand | Binding IC50, nM ± SEM (n)
|
Function EC50, nM ± SEM (n)
|
||
|---|---|---|---|---|
| MCH-1R | MCH-2R | MCH-1R | MCH-2R | |
| MCH | 0.3 ± 0.1 (10) | 1.5 ± 0.9 (8) | 3.9 ± 1.2 (3) | 0.1 ± 0.1 (3) |
| Phe13Tyr19-MCH | 0.3 ± 0.1 (3) | 0.8 ± 0.1 (3) | 8.5 ± 0.9 (3) | 0.2 ± 0.1 (3) |
| Salmon MCH | 0.2 ± 0.1 (5) | 436.7 ± 143.8 (3) | 35.0 ± 14.8 (3) | 0.8 ± 0.1 (3) |
Membrane binding was assessed by competition of [125I]Phe13Tyr19-MCH in a scintillation proximity assay. Functional activation was measured by mobilization of intracellular calcium detected in a fluorometric imaging plate reader assay. Values shown are nanomolar concentrations and averaged from at least three independent experiments.
To confirm that MCH is a high-affinity ligand for the MCH-2R we performed radioligand binding assays using membranes from cells transiently or stably expressing MCH-2R. MCH as well as Phe13Tyr19-MCH or salmon MCH displaced iodinated Phe13Tyr19-MCH radioligand (36) with low and subnanomolar half-maximal inhibition (IC50) concentrations (Table 1).
For a more detailed analysis we generated a HEK293-AEQ17 cell line that stably expressed MCH-2R. In this cell line mobilization of intracellular calcium can be detected by an aequorin assay based on bioluminescence of jellyfish aequorin upon calcium binding (33). MCH elicited a robust and dose-dependent response in this cell line with an effective half-maximal concentration (EC50) of 31 nM. This response is similar to the signal obtained with MCH at the MCH-1R in this cellular background, applying similar assay conditions (Fig. 2 C and D). To confirm specificity of the MCH-mediated MCH-2R activation, we evaluated a variety of other neuropeptides for their ability to activate the MCH-2R (see Materials and Methods), and we show that none of the other peptides tested activated the MCH-2R. This analysis included the peptides NGE and NEI, which are predicted to be cleaved from the same peptide precursor that encodes MCH.
In addition to signaling through intracellular calcium, it was shown by several laboratories that MCH-1R can inhibit forskolin-induced cAMP production (15–18). We were unable to detect a similar effect with MCH-2R, and we conclude that MCH-2R is not coupled to the Gαi/o pathway in these cells (data not shown). If MCH-2R is not able to activate Gαi/o proteins an interaction with Gαq is most likely. Therefore we examined a possible role of MCH-2R activation in the inositol phospholipid turnover. Receptor activation of Gαq-coupled receptors leads to an increase in phospholipid turnover by cleavage of inositol-3-phosphate (IP3) from IP3-acylated lipids in the cell membrane. MCH potently stimulated IP3 turnover through the MCH-2R. MCH-2R displayed an EC50 of 2.7 ± 0.6 nM (n = 3) whereas MCH-1R showed an EC50 of about 88.7 ± 16 nM (n = 4) in these stably expressing HEK293-AEQ17 cell lines.
To extend this observation, we analyzed the PTX sensitivity of calcium mobilization evoked by MCH-1R and MCH-2R in HEK293 cells. PTX can ADP-ribosylate the α subunit of Gi proteins and thus inhibit signaling mediated through Gi proteins but has no effect on signaling mediated by Gq (37). Using various PTX concentrations we found that intracellular calcium mobilization through MCH-2R is not affected by PTX, whereas MCH-1R signaling can be reduced in a dose-dependent manner to about 50% (Fig. 2 C and D).
Our result of MCH-1R signaling confirms results by Hawes and colleagues (38), which reported incomplete PTX inhibition of calcium mobilization (40% inhibition) and inositol phosphate production (60% inhibition) in a CHO cell line. Both results are in contrast to Lembo et al. (17) who found complete PTX inhibition of MCH-1R signaling. This difference might be attributed to different expression levels of the MCH-1R in the cell lines used for the analysis. Interestingly, Tadayyon and colleagues (39) recently reported a PTX-insensitive MCH response in insulin producing cells such as CRI-G1 and RINm5F.
Expression Analysis of MCH-2R.
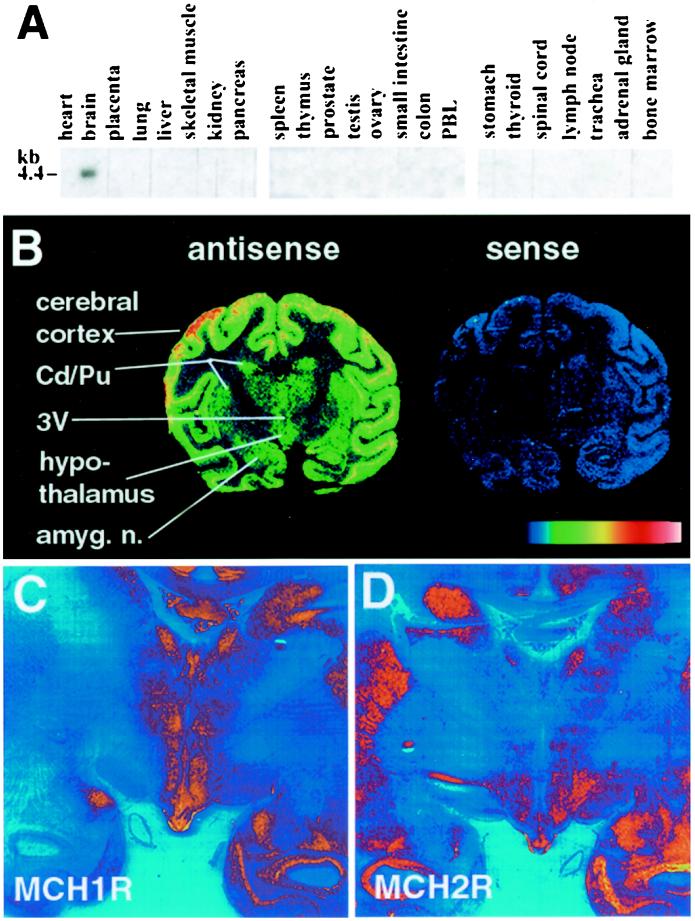
Northern blot analysis of multiple human tissues using radio-labeled MCH-2R cDNA as a probe detected a 4.4-kb mRNA transcript for MCH-2R specifically expressed in brain (Fig. 3A). To increase spatial resolution we continued our expression analysis by in situ hybridization of rhesus monkey brain tissue using radio-labeled human MCH-2R riboprobes (Fig. 3B). The antisense probe demonstrated high levels of MCH-2R mRNA expression in cerebral cortex, hippocampus, and hypothalamus, with lower levels of expression in the caudate nucleus, putamen, and thalamus. Sense control probe showed no signal. To compare expression of the two MCH receptors in the hypothalamus, adjacent coronal sections of African green monkey brain were hybridized with riboprobes complementary to one or the other MCH receptor (Fig. 3 C and D). In the dorsomedial hypothalamus, there was strong MCH-1R expression but little or no MCH-2R expression. Conversely, in the anterior and lateral hypothalamic areas, MCH-2R expression was abundant, whereas MCH-1R signal was barely detectable. The ventromedial hypothalamic nuclei displayed abundant signal for both MCH-1R and MCH-2R.
Figure 3.
(A) Northern blot analysis of human tissues using a radio-labeled human MCH-2R probe. (B) In situ hybridization using radio-labeled human antisense or sense MCH-2R probes on coronal brain sections from rhesus monkey. (C and D) Comparison of the expression of MCH-1R and MCH-2R by in situ hybridization using radio-labeled antisense probes for human MCH-1R or MCH-2R on adjacent coronal brain sections from African green monkey (magnification: <0.5×).
Chromosomal Localization of MCH-2R.
To determine the chromosomal localization of the human MCH-2R gene we isolated two overlapping human PAC genomic clones (60p05, 214d23) that contain the MCH-2R gene. To confirm the presence of the MCH-2R gene we directly sequenced parts of these clones by using primers complementary to the MCH-2R cDNA. This analysis revealed that the coding region of the MCH-2R is distributed over five exons (Fig. 1C). One of the two genomic clones (60p05) contained the first two exons whereas the other (214d23) contained all five exons of the coding region of MCH-2R. Subsequently, PAC clone 60p05 was used for fluorescence in situ hybridization and was localized to the long arm of chromosome 6 to the band 6q16.2–16.3. We confirmed this result by radiation hybrid analyses. A PCR fragment specific for exon 4 was located about 6.83 centiRays distal to the marker WI-6516.
During our analysis of the human MCH-2R gene searches in the high-throughput genomic subdivision of GenBank revealed an additional PAC clone assigned to chromosome 6 (accession no. AC027643; Nt_023456), which was successively sequenced and currently (February 9, 2001) contains a 158-kb sequence that encompasses the MCH-2R gene.
There are several reports of obese patients with cytogenetic alterations on the long arm of chromosome 6 (40–44). A recent report by Holder et al. (40) described an obese girl who has a balanced translocation in this region on chromosome 6. The authors show that a cytogenetic alteration disrupts the SIM1 gene, the human homolog of the Drosophila single-minded gene which also is located at 6q16.2. To determine the distance of the two genes relative to each other we repeated the radiation hybrid mapping with primers specific for exon 1 and exon 11 of the SIM1 gene and found that both fragments were placed 7.26 centiRays distal to marker WI-6516. Considering the effective resolution of the GENEBRIDGE 4 panel (45), the MCH-2R gene and the SIM1 gene should be located within 1 Mb of each other. Fine mapping of the MCH-2R locus has been initiated and will provide more detailed information relating to the physical proximity of the SIM1 and MCH-2R genes. In addition to the results by Holder et al., a total of four other obese patients with cytogenetic alterations in this region have been reported (41–44). As noted by Gilhuis and colleagues (44), all four patients have in common some clinical features including obesity, hypotonia, and developmental delays resembling Prader–Willi syndrome (PWS), the most common pathological obesity syndrome at pediatric clinics (46, 47). However, their behavior, facial features, and additional neurological abnormalities as well as a lack of cytogenetic changes or imprinting mutations at chromosome 15, which are the hallmarks for PWS diagnosis, clearly distinguishes this PWS-like phenotype from PWS patients.
A more detailed analysis of the MCH-2R locus in PWS-like patients who appear to be normal at chromosome 15 may provide insight in the role of the MCH-2R gene in the pathology of PWS-like symptoms including obesity.
In addition to the link of this genomic locus to patients with obesity this region has been implicated in neuropsychiatric disorders such as autism (48) and schizophrenia (24, 49–51).
In conclusion we present here the discovery of human MCH-2R as a second G protein-coupled receptor for MCH. Considering the wide range of possible physiological activities attributed to MCH and the selective expression of MCH-2R in human brain, especially in the hypothalamus, it will be of interest to define the role of this receptor in human physiology.
Abbreviations
- MCH
melanin-concentrating hormone
- MCH-1R and MCH-2R
MCH1 and 2 receptors
- CHO
Chinese hamster ovary
- HEK
human embryonic kidney
- PTX
pertussis toxin
- PAC
P1 artificial chromosome
- PWS
Prader–Willi syndrome
Note Added in Proof.
During the review of this manuscript two other research groups reported the identification and cloning of the same MCH receptor (MCH2, STL) (52, 53).
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY029596).
References
- 1.Baker B I. Int Rev Cytol. 1991;126:1–47. doi: 10.1016/s0074-7696(08)60681-6. [DOI] [PubMed] [Google Scholar]
- 2.Nahon J L. Crit Rev Neurobiol. 1994;8:221–262. [PubMed] [Google Scholar]
- 3.Kawauchi H, Kawazoe I, Tsubokawa M, Kishida M, Baker B I. Nature (London) 1983;305:321–323. doi: 10.1038/305321a0. [DOI] [PubMed] [Google Scholar]
- 4.Presse F, Nahon J L, Fischer W H, Vale W. Mol Endocrinol. 1990;4:632–637. doi: 10.1210/mend-4-4-632. [DOI] [PubMed] [Google Scholar]
- 5.Vaughan J M, Fischer W H, Hoeger C, Rivier J, Vale W. Endocrinology. 1989;125:1660–1665. doi: 10.1210/endo-125-3-1660. [DOI] [PubMed] [Google Scholar]
- 6.Zamir N, Skofitsch G, Bannon M J, Jacobowitz D M. Proc Natl Acad Sci USA. 1986;83:1528–1531. doi: 10.1073/pnas.83.5.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breton C, Schorpp M, Nahon J L. Brain Res Mol Brain Res. 1993;18:297–310. doi: 10.1016/0169-328x(93)90093-5. [DOI] [PubMed] [Google Scholar]
- 8.Viale A, Ortola C, Richard F, Vernier P, Presse F, Schilling S, Dutrillaux B, Nahon J L. Mol Biol Evol. 1998;15:196–214. doi: 10.1093/oxfordjournals.molbev.a025915. [DOI] [PubMed] [Google Scholar]
- 9.Bittencourt J C, Presse F, Arias C, Peto C, Vaughan J, Nahon J L, Vale W, Sawchenko P E. J Comp Neurol. 1992;319:218–245. doi: 10.1002/cne.903190204. [DOI] [PubMed] [Google Scholar]
- 10.Qu D, Ludwig D S, Gammeltoft S, Piper M, Pelleymounter M A, Cullen M J, Mathes W F, Przypek R, Kanarek R, Maratos-Flier E. Nature (London) 1996;380:243–247. doi: 10.1038/380243a0. [DOI] [PubMed] [Google Scholar]
- 11.Ludwig D S, Mountjoy K G, Tatro J B, Gillette J A, Frederich R C, Flier J S, Maratos-Flier E. Am J Physiol. 1998;274:E627–E633. doi: 10.1152/ajpendo.1998.274.4.E627. [DOI] [PubMed] [Google Scholar]
- 12.Rossi M, Choi S J, O'Shea D, Miyoshi T, Ghatei M A, Bloom S R. Endocrinology. 1997;138:351–355. doi: 10.1210/endo.138.1.4887. [DOI] [PubMed] [Google Scholar]
- 13.Shimada M, Tritos N A, Lowell B B, Flier J S, Maratos-Flier E. Nature (London) 1998;396:670–674. doi: 10.1038/25341. [DOI] [PubMed] [Google Scholar]
- 14.Ludwig D S, Tritos N A, Mastaitis J W, Kulkarni R, Kokkotou E, Elmquist J, Lowell B, Flier J S, Maratos-Flier E. J Clin Invest. 2001;107:379–386. doi: 10.1172/JCI10660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chambers J, Ames R S, Bergsma D, Muir A, Fitzgerald L R, Hervieu G, Dytko G M, Foley J J, Martin J, Liu W S, et al. Nature (London) 1999;400:261–265. doi: 10.1038/22313. [DOI] [PubMed] [Google Scholar]
- 16.Saito Y, Nothacker H P, Wang Z, Lin S H, Leslie F, Civelli O. Nature (London) 1999;400:265–269. doi: 10.1038/22321. [DOI] [PubMed] [Google Scholar]
- 17.Lembo P M, Grazzini E, Cao J, Hubatsch D A, Pelletier M, Hoffert C, St-Onge S, Pou C, Labrecque J, Groblewski T, et al. Nat Cell Biol. 1999;1:267–271. doi: 10.1038/12978. [DOI] [PubMed] [Google Scholar]
- 18.Shimomura Y, Mori M, Sugo T, Ishibashi Y, Abe M, Kurokawa T, Onda H, Nishimura O, Sumino Y, Fujino M. Biochem Biophys Res Commun. 1999;261:622–626. doi: 10.1006/bbrc.1999.1104. [DOI] [PubMed] [Google Scholar]
- 19.Bachner D, Kreienkamp H, Weise C, Buck F, Richter D. FEBS Lett. 1999;457:522–524. doi: 10.1016/s0014-5793(99)01092-3. [DOI] [PubMed] [Google Scholar]
- 20.Benson D A, Boguski M S, Lipman D J, Ostell J, Ouellette B F. Nucleic Acids Res. 1998;26:1–7. doi: 10.1093/nar/26.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 22.McDonald T, Wang R, Bailey W, Xie G, Chen F, Caskey C T, Liu Q. Biochem Biophys Res Commun. 1998;247:266–270. doi: 10.1006/bbrc.1998.8774. [DOI] [PubMed] [Google Scholar]
- 23.Liu Q, Bai C, Chen F, Wang R, MacDonald T, Gu M, Zhang Q, Morsy M A, Caskey C T. Gene. 1998;207:1–7. doi: 10.1016/s0378-1119(97)00596-9. [DOI] [PubMed] [Google Scholar]
- 24.Martinez M, Goldin L R, Cao Q, Zhang J, Sanders A R, Nancarrow D J, Taylor J M, Levinson D F, Kirby A, Crowe R R, et al. Am J Med Genet. 1999;88:337–343. [PubMed] [Google Scholar]
- 25.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 26.Howard A D, Wang R, Pong S S, Mellin T N, Strack A, Guan X M, Zeng Z, Williams D L, Jr, Feighner S D, Nunes C N, et al. Nature (London) 2000;406:70–74. doi: 10.1038/35017610. [DOI] [PubMed] [Google Scholar]
- 27.Strader C, Sigal I, Register R, Candelore M, Rands E, Dixon R. Proc Natl Acad Sci USA. 1987;84:4384–4388. doi: 10.1073/pnas.84.13.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacNeil D J, Occi J L, Hey P J, Strader C D, Graziano M P. Biochem Biophys Res Commun. 1994;198:328–334. doi: 10.1006/bbrc.1994.1046. [DOI] [PubMed] [Google Scholar]
- 29.Offermanns S, Simon M I. J Biol Chem. 1995;270:15175–15180. doi: 10.1074/jbc.270.25.15175. [DOI] [PubMed] [Google Scholar]
- 30.Liu Q, Pong S S, Zeng Z, Zhang Q, Howard A D, Williams D L, Jr, Davidoff M, Wang R, Austin C P, McDonald T P, et al. Biochem Biophys Res Commun. 1999;266:174–178. doi: 10.1006/bbrc.1999.1796. [DOI] [PubMed] [Google Scholar]
- 31.Ungrin M D, Singh L M, Stocco R, Sas D E, Abramovitz M. Anal Biochem. 1999;272:34–42. doi: 10.1006/abio.1999.4145. [DOI] [PubMed] [Google Scholar]
- 32.Feighner S D, Tan C P, McKee K K, Palyha O C, Hreniuk D L, Pong S S, Austin C P, Figueroa D, MacNeil D, Cascieri M A, et al. Science. 1999;284:2184–2188. doi: 10.1126/science.284.5423.2184. [DOI] [PubMed] [Google Scholar]
- 33.Button D, Brownstein M. Cell Calcium. 1993;14:663–671. doi: 10.1016/0143-4160(93)90091-j. [DOI] [PubMed] [Google Scholar]
- 34.Berridge M J, Dawson R M, Downes C P, Heslop J P, Irvine R F. Biochem J. 1983;212:473–482. doi: 10.1042/bj2120473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petrukhin K, Koisti M J, Bakall B, Li W, Xie G, Marknell T, Sandgren O, Forsman K, Holmgren G, Andreasson S, et al. Nat Genet. 1998;19:241–247. doi: 10.1038/915. [DOI] [PubMed] [Google Scholar]
- 36.Drozdz R, Siegrist W, Baker B I, Chluba-de Tapia J, Eberle A N. FEBS Lett. 1995;359:199–202. doi: 10.1016/0014-5793(95)00043-9. [DOI] [PubMed] [Google Scholar]
- 37.Fields T A, Casey P J. Biochem J. 1997;321:561–571. doi: 10.1042/bj3210561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hawes B E, Kil E, Green B, O'Neill K, Fried S, Graziano M P. Endocrinology. 2000;141:4524–4532. doi: 10.1210/endo.141.12.7833. [DOI] [PubMed] [Google Scholar]
- 39.Tadayyon M, Welters H J, Haynes A C, Cluderay J E, Hervieu G. Biochem Biophys Res Commun. 2000;275:709–712. doi: 10.1006/bbrc.2000.3357. [DOI] [PubMed] [Google Scholar]
- 40.Holder J L, Jr, Butte N F, Zinn A R. Hum Mol Genet. 2000;9:101–108. doi: 10.1093/hmg/9.1.101. [DOI] [PubMed] [Google Scholar]
- 41.Villa A, Urioste M, Bofarull J M, Martinez-Frias M L. Am J Med Genet. 1995;55:379–383. doi: 10.1002/ajmg.1320550326. [DOI] [PubMed] [Google Scholar]
- 42.Stein C K, Stred S E, Thomson L L, Smith F C, Hoo J J. Clin Genet. 1996;49:306–310. doi: 10.1111/j.1399-0004.1996.tb03794.x. [DOI] [PubMed] [Google Scholar]
- 43.Turleau C, Demay G, Cabanis M O, Lenoir G, de Grouchy J. Clin Genet. 1988;34:38–42. doi: 10.1111/j.1399-0004.1988.tb02613.x. [DOI] [PubMed] [Google Scholar]
- 44.Gilhuis H J, van Ravenswaaij C M, Hamel B J, Gabreels F J. Eur J Paediatr Neurol. 2000;4:39–43. doi: 10.1053/ejpn.1999.0259. [DOI] [PubMed] [Google Scholar]
- 45.Walter M A, Spillett D J, Thomas P, Weissenbach J, Goodfellow P N. Nat Genet. 1994;7:22–28. doi: 10.1038/ng0594-22. [DOI] [PubMed] [Google Scholar]
- 46.Bray G A. Clin Cornerstone. 1999;2:1–15. doi: 10.1016/s1098-3597(99)90001-7. [DOI] [PubMed] [Google Scholar]
- 47.Ledbetter D H, Riccardi V M, Airhart S D, Strobel R J, Keenan B S, Crawford J D. N Engl J Med. 1981;304:325–329. doi: 10.1056/NEJM198102053040604. [DOI] [PubMed] [Google Scholar]
- 48.Philippe A, Martinez M, Guilloud-Bataille M, Gillberg C, Rastam M, Sponheim E, Coleman M, Zappella M, Aschauer H, Van Maldergem L, et al. Hum Mol Genet. 1999;8:805–812. doi: 10.1093/hmg/8.5.805. [DOI] [PubMed] [Google Scholar]
- 49.Cao Q, Martinez M, Zhang J, Sanders A R, Badner J A, Cravchik A, Markey C J, Beshah E, Guroff J J, Maxwell M E, et al. Genomics. 1997;43:1–8. doi: 10.1006/geno.1997.4815. [DOI] [PubMed] [Google Scholar]
- 50.Kaufmann C A, Suarez B, Malaspina D, Pepple J, Svrakic D, Markel P D, Meyer J, Zambuto C T, Schmitt K, Matise T C, et al. Am J Med Genet. 1998;81:282–289. [PubMed] [Google Scholar]
- 51.Levinson D F, Holmans P, Straub R E, Owen M J, Wildenauer D B, Gejman P V, Pulver A E, Laurent C, Kendler K S, Walsh D, et al. Am J Hum Genet. 2000;67:652–663. doi: 10.1086/303041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mori M, Harada M, Terao Y, Sugo T, Watanabe T, Shimomura Y, Abe M, Shintani Y, Onda H, Nishimura O, et al. Biochem Biophys Res Commun. 2001;283:1013–1018. doi: 10.1006/bbrc.2001.4893. [DOI] [PubMed] [Google Scholar]
- 53.Hill, J., Duckworth, M., Murdock, P., Rennie, G., Sabido-David, C., Ames, R. S., Szekeres, P., Wilson, S., Bergsma, D. J., Gloger, I. S., et al. (2001) J. Biol. Chem., in press. [DOI] [PubMed]