Summary
The four mammalian Argonaute family members are thought to share redundant functions in the microRNA pathway, yet only AGO2 possesses the catalytic “slicer” function required for RNA interference. Whether AGO1, AGO3, or AGO4 possess specialized functions remains unclear. Here we show that AGO4 localizes to spermatocyte nuclei during meiotic prophase I, specifically at sites of asynapsis and the transcriptionally silenced XY sub-domain, the sex body. We generated Ago4 knockout mice and show that Ago4−/− spermatogonia initiate meiosis early, resulting from premature induction of retinoic acid-response genes. During prophase I, the sex body assembles incorrectly in Ago4−/− mice, leading to disrupted meiotic sex chromosome inactivation (MSCI). This is associated with a dramatic loss of microRNAs, >20% of which arise from the X chromosome. Thus, AGO4 regulates meiotic entry and MSCI in mammalian germ cells, implicating small RNA pathways in these processes.
Introduction
Argonaute proteins form two clades within a larger super-family, based on their sequence homology (Cenik and Zamore, 2011; Czech and Hannon, 2011). The PIWI clade includes Mili, Miwi1, and Miwi2 in mice, which are required for retrotransposon silencing in the germ line and normal progression through prophase I in male meiosis (Aravin et al., 2006; Girard et al., 2006; Grivna et al., 2006), in conjunction with the PIWI-interacting RNAs (piRNAs). The second clade are the AGO proteins, of which there are four in mammals (AGO1-4(Steiner and Plasterk, 2006). However, only AGO2 is capable of mediating small RNA-directed endonucleolytic cleavage of mRNA targets, the hallmark of RNA interference (RNAi; Liu et al., 2004; Meister et al., 2004; Song et al., 2003). No individual functions have yet been ascribed to the other mammalian AGOs, but they can all associate with several distinct classes of small RNAs, including microRNAs (miRNAs) and small interfering RNAs (siRNAs).
Endogenous siRNAs and miRNAs are found in the male germ line (Song et al., 2011; Watanabe et al., 2006) and, while some associate with AGO proteins (Kim et al., 2006), their precise roles are unknown. Small RNAs may function in the process of meiotic silencing of unpaired chromosomes, a common feature of prophase I in a number of organisms (Maine, 2010). During meiosis in Neurospora crassa, for example, the presence of unsynapsed DNA triggers the post-transcriptional silencing of any homologous sequences. This process, called Meiotic Silencing of Unpaired DNA (MSUD), requires Ago-protein function and small RNA involvement (Raju et al., 2007; Shiu et al., 2001).
The high expression of mouse Ago4 (and Ago3) in the male mouse germ line (Gonzalez-Gonzalez et al., 2008) indicates that similar meiotic functions for AGO proteins may also exist in mammals. Two distinct levels of meiotic silencing in the male germ line have been described: Meiotic Silencing of Unsynapsed Chromatin (MSUC; Schimenti, 2005) and Meiotic Sex Chromosome Inactivation (MSCI; Fernandez-Capetillo et al., 2003). MSUC monitors complete pairing before permitting progression through prophase I, and protects genome from transposon mobilization by silencing regions that fail to pair because of hemizygous transposon insertions (Turner et al., 2005). MSCI, on the other hand, is unique to the sex chromosomes and results in their heterochromatinization and compartmentalization into a specialized nuclear sub-domain known as the Sex Body (SB; Handel, 2004). This enables the unpaired sex chromatin to bypass meiotic synapsis check points and is essential for meiotic progression in males (Baarends and Grootegoed, 2003). Although neither the mechanism is fully understood, it is notable that the miRNA genes located on the X-chromosome evade MSCI, perhaps indicating a role for small RNAs in silencing (Song et al., 2009).
To examine the role of AGO4 in mammals, we generated an Ago4 null mouse line. Ago4−/− mice are viable, but males have fertility defects including reduced testis size and lower sperm counts. AGO4 localizes to the SB during pachynema of prophase I, and loss of Ago4 perturbs SB morphology leading to an influx of RNA Polymerase II, and an associated failure to silence many sex-linked transcripts, resulting in apoptosis. Interestingly, our analysis also reveals an unexpected role for AGO4 in the onset of meiosis, in that spermatogonia from Ago4−/− males enter prophase I prematurely. These findings reveal a role for nonslicing Argonautes in mammalian germ cell development.
RESULTS AND DISCUSSION
AGO4 localizes to the sex body during prophase I of meiosis
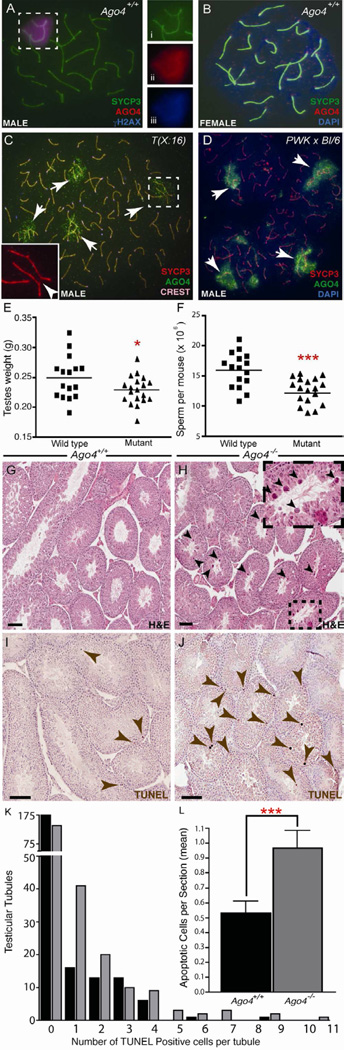
In adult mouse testis, Ago4 mRNA expression is highest at prophase I during pachynema (Gonzalez-Gonzalez et al., 2008), the stage at which homologous autosomes are completely synapsed (paired) and the SB is formed. AGO4 protein localization was assessed on chromosome-spread preparations from prophase I spermatocytes of WT adult male testes (day 70 pp), together with staining of the synaptonemal complex (SC) using antibodies against the lateral element protein, SYCP3 (Figure 1A). AGO4 staining was evident in the nucleus, with the most intense staining at the SB (Figure 1Aii), where it co-localizes with γH2AX (Figure 1Aiii). The AGO4 SB staining pattern was lost when the antibody was pre-incubated with the immunizing peptide, indicating specificity of the signal for AGO4 (Figure S1A) and no AGO4 staining was observed from Ago4-deficient males (Figure S1B,C). Only faint punctate staining was observed in oocytes (Figure 1B), which do not possess an SB.
Figure 1. AGO4 Localization, and phenotypic analysis of Ago4−/− males.
(A–D) AGO4 localization on chromosome spreads from WT and induced asynaptic mouse models. (A) Spermatocyte from a WT mouse stained with anti-SYCP3 (i, green), anti-AGO4 (ii, red), and anti-γH2AX (iii, blue). (B) Pachytene stage oocyte from WT mouse stained with anti-SYCP3 (green) and anti-AGO4 (red) antibodies and DAPI to highlight DNA (blue). (C) Pachytene stage spermatocytes from a T(X:16) male translocation mouse (C) and a PWK × C57BL/6J hybrid mouse (D) which both exhibit significant levels of asynapsis (arrows), stained with anti-SYCP3 (red), CREST autoimmune serum (pink localization on centromeres in panel C only) and anti-AGO4 (green) antibodies. Asynapsis induced by chromosome 16 to X translocation in C is marked by white arrowhead (inset). (E) Testes from Ago4−/− mice (n=19) are significantly reduced in size compared to WT (N=16, *P<0.05, t-test). (F) Caudal epididymal sperm counts shown in millions. Sperm counts from Ago4−/− mice (▲n=19) are reduced when compared to WT (▀ n=16, ***P<0.0005, t-test). (G,H) Testis sections from (G) Ago4+/+ and (H) Ago4−/− mice stained with hematoxylin and eosin. Scale bars, 100 µm. Large multinucleated cells are apparent in Ago4−/− mice (arrows and inset). (I–L) TUNEL staining and quantitation of testis sections from (I) Ago4+/+ and (J) Ago4−/− mice. (K, L) Ago4+/+ (black bars) and Ago4−/− mice (grey bars) expressed per tubule (K) or per section (L) showing significant increase in Ago4−/− mice (n=230, 45.2% increase, ***P<0.0005, t-test).
To explore whether the meiotic localization of AGO4 to the X and Y chromosomes is a consequence of their lack of pairing, we examined the localization of AGO4 in two mouse models displaying varying degrees of induced autosomal asynapsis in spermatocytes: mice carrying a TX(16:X)16H translocation (Figure 1C), or hybrid offspring of two divergent strains (PWKxB6, Figure 1D). In both cases, significant degrees of autosomal asynapsis were evident by the staining pattern for SYCP3 (Figure 1C Arrowhead), and AGO4 co-localized to all these sites (Figure 1C, D arrows). Given that these asynapsed regions of chromatin, are silenced during prophase I (Handel, 2004), these observations suggest a role for AGO4 in meiotic silencing.
Loss of Ago4 results in reduced testis size and epididymal spermatozoa together with increased apoptosis of spermatogenic cells
We generated a floxed Ago4−/− mouse line which, following Cre-driven excision of a LoxP-flanked region, lacks exons 3–17 of the gene (Figure S1D and Supplemental Experimental Procedures). The recombinant allele was backcrossed for six generations from the mixed 129/C57 background onto a pure C57Bl/6J background. Backcrossed homozygous Ago4−/− male and female mice were healthy and viable, and male and female offspring were observed at the expected frequencies (Male: 54%, Female: 46%, n=121). Furthermore, Ago4−/− males exhibit normal mating behavior with no decrease in litter sizes from Ago4+/+ males (Supplemental Table 1). However, testis weights in Ago4−/− adult mice were reduced by 13% compared to Ago4+/+ mice (Figure 1E, Supplementary Table 1). Similarly, epididymal spermatozoa counts were also abnormal in Ago4−/− mice, reduced by 22% compared to WT littermates (Figure 1F, Supplemental Table 1).
Testes from Ago4−/− males undergo increased apoptosis
Histological analysis with H&E staining revealed similar cellular composition of the seminiferous epithelium in Ago4+/+ and Ago4−/− males (Figure 1G–H). In the latter, however, we observed abnormally large, densely staining cells within the seminiferous tubular lumen (Figure 1H). TUNEL staining of testis sections (Figure 1I–J) revealed a significant increase in apoptotic cells within the prophase I layers of the seminiferous epithelium of Ago4−/− mice compared to WT (Figure 1I,J, arrows; Figure 1K,L). These results demonstrate that Ago4−/− mice exhibit an increase in testicular apoptosis, reduced testes weight, and lowered sperm counts, phenotypes previously observed in MSCI-compromised animals (Turner et al., 2004).
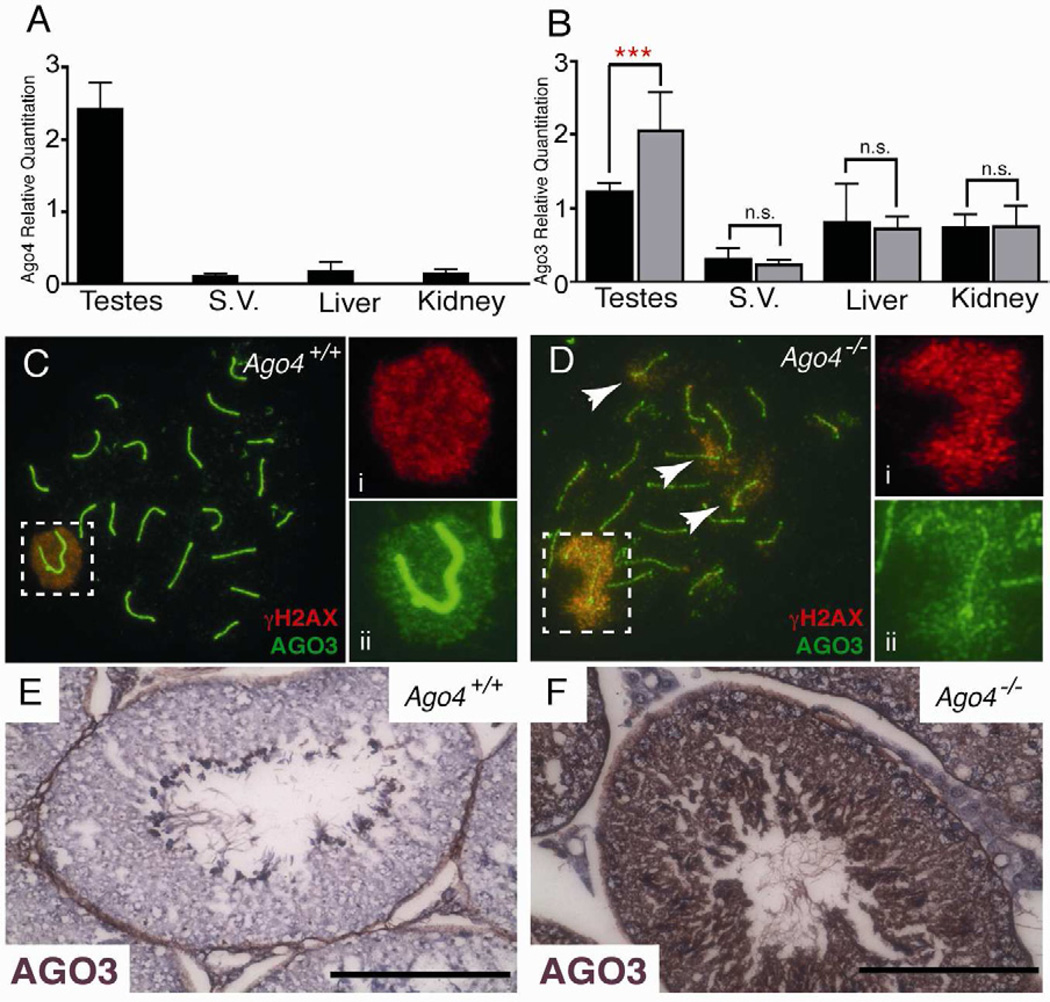
Loss of AGO4 results in increased AGO3 protein
Given the high degree of sequence similarity (Cenik and Zamore, 2011), together with the high expression of Ago3 in testis, we reasoned that overlapping functions might exist between mammalian Argonaute genes. Therefore, we used quantitative reverse transcription PCR (qRT-PCR) to measure the expression of all four AGO family members in testes of Ago4+/+ and Ago4−/− mice. As expected, both Ago3 and Ago4 mRNA are highly expressed in adult testes, and at higher levels than in all other tissues examined (Figure 2A,B). Ago1 was expressed at lower and more uniform levels throughout all of the tissues and Ago2 expression was higher in non-meiotic tissues (Figure S2). Loss of Ago4 resulted in a significant increase in Ago3 expression specifically in testis (Figure 2B, p<0.05, t-test), while Ago1 and Ago2 transcripts were not affected by the loss of AGO4 (Figure S2). Together, these data suggest that Ago3 and Ago4 may function redundantly in the male germline, such that loss of Ago4 results in a compensatory elevation in Ago3 mRNA.
Figure 2. Up-regulation of Ago3 mRNA and protein in the testis of Ago4−/− males.
Quantitative RT-PCR analysis of Ago4 (A) and Ago3 (B) expression in total RNA from whole testes, seminal vesicle, liver and kidney isolated from Ago4+/+ (n=5, black bars) and Ago4−/− (n=5, grey bars) adult littermates (***p<0.001, t-test). (C, D) Spermatocyte spreads from Ago4+/+ (C) and Ago4−/− (D) males stained with anti-γH2AX (red, Ci and Di) and anti-AGO3 (green, Cii and Dii) antibodies and DAPI (blue). “Pseudo-sex-body” regions in panel D denoted by arrows. (E, F) Testis sections from Ago4+/+ (E) and Ago4−/− (F) stained with anti-AGO3 antibody to demonstrate increased AGO3 signal within germ cells of Ago4−/− males.
We next investigated whether AGO3 protein shows similar localization to AGO4 on male prophase I chromosome spreads. Like AGO4, AGO3 localized preferentially to the SB at pachynema in WT spreads (Figure 2Ci). When examining the AGO3 staining pattern on Ago4−/− spreads, however, we found that the chromatin domain occupied by AGO3 signal was dramatically expanded (Figure 2D). Indeed, both AGO3 and γH2AX extended beyond the SB into areas of chromatin associated with the autosomes (Figure 2D, arrows). In mouse mutants that exhibit similar disruption of the SB appearance, these extended γH2AX domains have been termed “pseudo-sex bodies” (Barchi et al., 2005; Bellani et al., 2005; Daniel et al., 2011).
AGO3 localization was also assessed by immunohistochemistry of whole testis sections from Ago4+/+ and Ago4−/− adult males. In WT testis, AGO3 localized to meiotic cells and was absent in the non-meiotic spermatogonial cells (Figure 2E). In Ago4−/− testis, AGO3 signal was increased (Figure 2F), as expected, but still restricted to spermatogenic cells. Thus, the loss of Ago4 results in increased expression of Ago3 in the testis. We conclude that some functions of AGO4 in the male germline may be redundant with AGO3 and, if so, this would imply that the testis weight and sperm count phenotypes observed in Ago4−/− mice result from only a partial defect in these shared functions.
Loss of AGO4 disrupts sex body formation and alters the localization of key sex body components
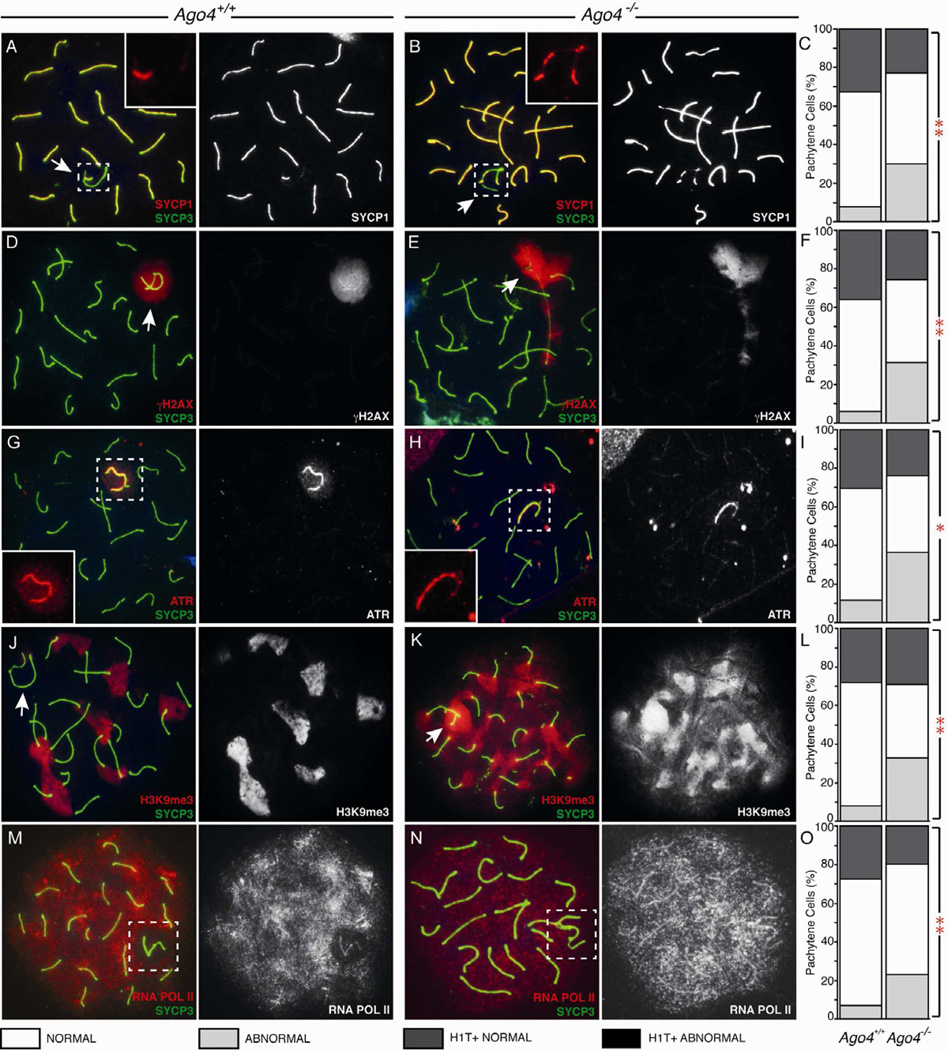
To examine more closely the SB morphology of Ago4−/− males, we investigated the localization of the SC central element protein, SYCP1, together with SYCP3 during prophase I (Figure 3A). In Ago4−/− spreads, SYCP1 localized to unsynapsed regions of the X and Y (Figure 3B) approximately 4-fold more frequently than in the WT (Figure 3C, Ago4+/+ 6.3%. Ago4−/− 22.8% of 300 pachytene spermatocytes, p<0.0005, χ2-test), indicating that loss of AGO4 perturbs SC formation and/or integrity. Importantly, initial events of prophase I progression, including the processing of double strand breaks for recombination, and crossover formation, appeared normal in Ago4−/− males (data not shown).
Figure 3. Loss of AGO4 disrupts localization of SB-associated proteins and RNAPII during pachynema.
Pachytene stage spermatocytes with SB (white arrows) from Ago4+/+ (A,D,G,J,M) and Ago4−/− (B,E,H,K,N) mice, stained with anti-SYCP3 (green). For all quantitation graphs (C, F, I, L, O), WT (left bar) and Ago4−/− (right bar). (A–C) SYCP1 in red. Inset panels show magnification of SYCP1 in sex body (n=300 per genotype, **p<0.005 χ2 test). (D–F) γH2AX in red (n=300, **p<0.005 χ2 test). (G–I) ATR in red (n=200, *p<0.05 χ2 test). (J–L) H3K9Me3 in red (n=300, **p<0.005 χ2 test). (M–O) RNAPII staining in red (n=200, **p<0.01 χ2 test).
The mislocalization of SC components in the Ago4−/− SB, together with the importance of SB integrity for meiotic sex chromosome inactivation (MSCI), led us to hypothesize that AGO4 may play a role in transcriptional silencing of the X and Y chromosomes. To test this, we utilized a repertoire of antibodies (γH2AX, TOPBP1, ATR, pCHK2, SUMO1, UbiH2A) against SB components to assess the integrity and functionality of this structure. The histone variant H2AX (Histone 2A family member X) is phosphorylated during prophase I by the ATM and ATR kinases to become γH2AX (Turner et al., 2004). SB-associated H2AX phosphorylation is mediated by ATR during pachynema, a stage at which γH2AX is found almost exclusively in the SB and at any remaining asynapsed regions on autosomes (Figure 3D). However, a large proportion of Ago4−/− pachytene cells (Figure 3E,F, 31% versus 5.5% in WT; n=300, p<0.0005, χ2-test), exhibit expanded γH2AX staining beyond the SB, often associating with synapsed autosomes, or pseudo-sex-bodies. The localization of γH2AX during diplonema is unaffected by loss of AGO4 (data not shown), and in fact, such defects appear to be restricted to early pachytene spermatocytes. To illustrate this, we used an antibody against the testis specific histone variant H1T, which localizes to chromatin from late pachynema (Lin et al., 2000). We found no instances in which the altered pseudo-SB staining of γH2AX, or other SB markers, coincided with the appearance of H1T in late pachytene or diplotene cell populations (Ago4+/+ n=289, Ago4−/− n=241 Figure S3K,L,M). Together, these results suggest that the SB morphology defects occur in early pachynema in Ago4−/− mice. Defective Ago4−/− spermatocytes are likely to be eliminated by the pachytene checkpoint, as evidenced by the increased TUNEL staining.
One explanation for aberrant γH2AX localization in Ago4−/− mice is failure to recruit ATR, which we investigated by staining for both ATR and TopBP1, a regulator of ATR (Kumagai et al., 2006). In WT mice, ATR staining is pronounced on the X and Y chromosome cores and through the SB starting from pachynema (Figure 3G). In Ago4−/− pachytene cells, however, ATR is frequently mislocalized (Figure 3H,I, n=200 cells, p<0.0005, χ2-test). A range of aberrant staining patterns were observed, including a complete loss of signal at the SB, reduced X chromosome core localization, and almost complete loss of Y chromosome core localization. TopBP1 localization is similarly altered by the loss of Ago4 (Figure S3A,B). The loss of ATR at the SB coincides with increased signal on autosome cores, potentially utilizing the limited store of ATR that would normally be required to induce MSCI.
We corroborated ATR mislocalization in Ago4−/− mice using an antibody against the phospho-(Ser/Thr)ATM/ATR substrate motif (Figure S3C) and an antibody specific to phosphorylated CHK2 (Weiss et al., 2002), a substrate of ATM/ATR (Figure S3E). In both cases, ATM/ATR substrate and pCHK2 were absent in pachytene spermatocyte spreads from Ago4−/− mice (Figure S3D and S3F).
SUMO1 and ubiH2A are proteins implicated in meiotic silencing (Brown et al., 2008; Vigodner, 2009). SUMO1 first appears in zygonema localizing to the X and Y cores (not shown) and persists into pachynema (Figure S3G). During pachynema, the ubiH2A histone mark emerges and localizes to the SB (Figure S3I; Baarends et al., 2005). SUMO1 and ubiH2A localization is abolished in a subset of Ago4−/− cells (Figures 3H,J). Together, these studies reveal that a subset of spermatocytes in Ago4−/− males fail to assemble a normal SB structure, leading to the appearance of pseudo-sex bodies and mislocalization of key SB markers.
Ago4−/− mice fail to correctly silence the X and Y chromosomes during meiosis
The defects we observed in SB components prompted us to determine whether silencing of the sex chromosomes during meiosis was normal in the absence of AGO4. The tri-methylation modification of H3K9 is associated with repressed heterochromatinized DNA. During prophase I in WT male mice, H3K9me3 is primarily restricted to autosomal centromeres and the majority of the SB (Kim et al., 2007; Tachibana et al., 2007; Figure 3J). In early pachynema, loss of Ago4 results in diffuse H3K9me3 localization in a significant portion of cells (Figure 3K,L; n=300. P<0.0005, χ2-test). These results suggest that AGO4 contributes to the establishment of chromatin marks associated with silencing, but as meiosis progresses aberrant cells are either eliminated or overcome the defect.
To further assess transcriptional activity of the sex chromosomes, we compared the localization of RNA Polymerase II (RNAP II) in WT and Ago4−/− pachytene spermatocytes. As expected, in Ago4+/+ cells, RNAP II is absent from sites of silencing and heterochromatin (Figure 3M) and is excluded from the SB through pachynema. This exclusion is no longer maintained in a significant fraction of pachytene spermatoctyes from Ago4−/− males (Figure 3N,O, Ago4+/+: 7%, Ago4−/−: 25.5% n=200. p<0.0005, χ2-test), matching the frequency of mislocalization we observed for SB components and further indicating a silencing defect in Ago4−/− spermatocytes.
Loss of Ago4 leads to down-regulation of specific microRNA families
Since Argonaute proteins function with small RNAs, we hypothesized that the phenotypes in Ago4−/− animals reflect alterations in the small RNAs. We therefore cloned and sequenced small RNAs (18–30 nts) from purified pachytene cells isolated from Ago4+/+ and Ago4−/− littermates. We obtained 21.2 and 14.7 million genome-matching reads from the Ago4+/+ and Ago4−/− samples, respectively, a sequencing depth sufficient to robustly profile the small RNAs of pachytene cells.
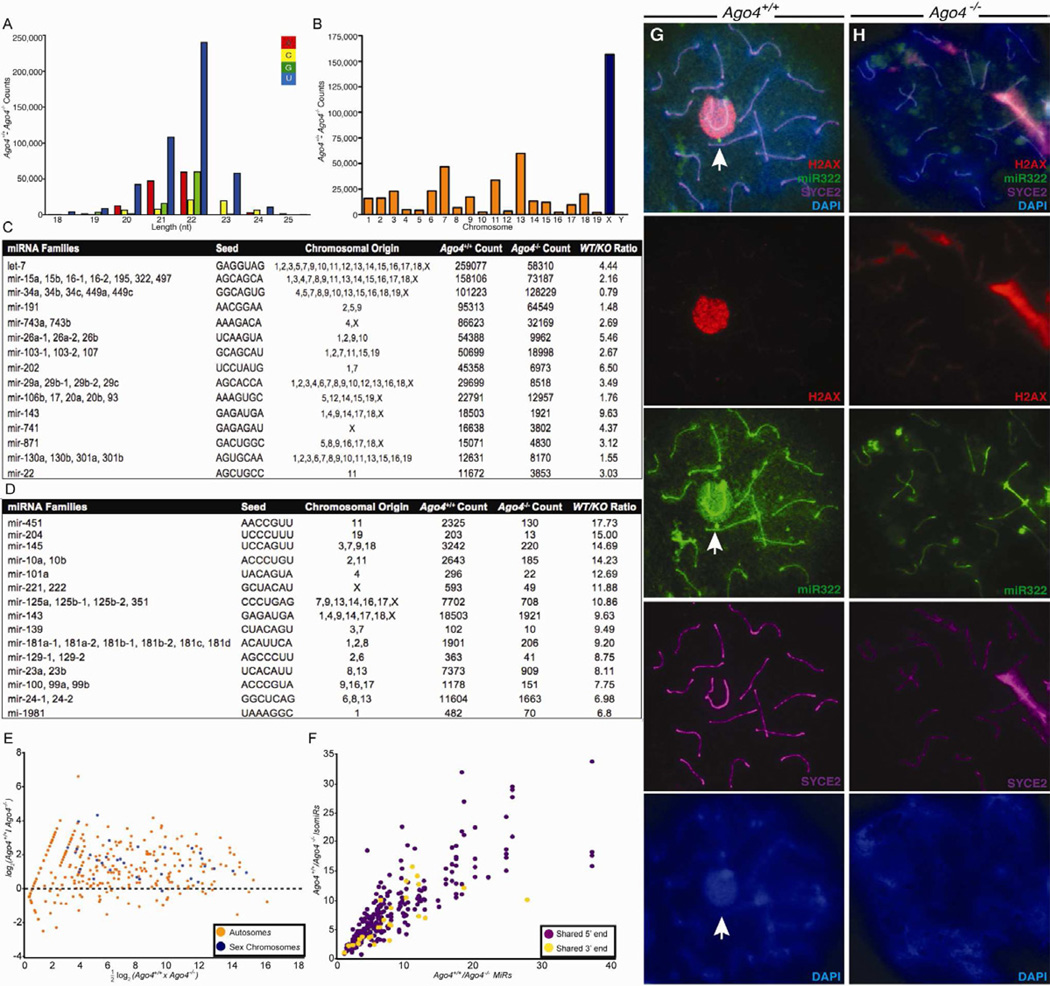
piRNAs are the most abundant small RNAs in pachytene cells (Pillai and Chuma, 2012; Siomi et al., 2011), and loss of Ago4 only minimally perturbed these piRNA populations (Figure S4F,G), as expected. By contrast, loss of Ago4 had a substantial effect on the miRNAs present in pachytene cells. Overall, we observed an approximately 2.3-fold reduction (Figure 4 and S4) in reads matching miRNA hairpins in Ago4−/− pachytene cells (Fig 4A and S4A; 6% Ago4+/+, 2.6%Ago4−/−). However, this reduction was not consistent among different miRNA families, indicating some degree of miRNA specificity for AGO4 (Fig 4C–D,S4C–D, S5). Approximately 20% of the global miRNA down-regulation came from miRNAs expressed from the X chromosome, and interestingly, all miRNAs encoded on the X are significantly less abundant (p<0.01; Wilcoxon-rank sum test) in Ago4−/− cells whereas not all autosomal miRNAs are decreased (Fig 4B–E, S4B–E).
Figure 4. Loss of Ago4 results in loss of microRNAs originating from chromosome X and wide-scale alterations in the expression of most microRNA families.
(A) The difference between microRNAs cloned from pachytene cells of WT and Ago4−/− males, distributed by microRNA length and base identity of the most 5′ nucleotide. (B) For microRNAs with a single chromosomal origin, the difference between WT and mutant counts of these microRNAs is shown by chromosome (autosomes in orange, X chromosome in blue). (C, D) MicroRNA families, grouped by common ‘seed’ (nucleotides 2–7), with the highest expression in WT pachytene cells (C) and those showing the largest decrease in expression in the Ago4−/− (D). (E) MA plot showing the intensity of expression (x-axis) versus change in expression (y-axis) for WT and mutant microRNAs reveals a preferential decrease in microRNA levels in the mutant for microRNAs originating from the sex chromosomes. (F) Comparison of the ratio of WT to mutant microRNA counts between each miRNA and other variant species derived from the same hairpin. (G, H) miRNA fluorescence in situ hybridization for miR-322 using pachytene stage spermatocytes from Ago4+/+ (G) and Ago4−/− (H) mice stained with anti-γH2AX (red), anti-SYCE2 (purple) and DAPI (blue).
We considered two explanations for the alterations in the miRNA profile of Ago4−/− cells. First, AGO proteins might associate with miRNAs at different efficacies, thus, in the absence of AGO4, the sequenced miRNAs would represent the propensities of AGO1, AGO2 and AGO3 to form complexes with specific miRNAs. Second, the altered profile might be a downstream consequence of altered transcriptional regulation of miRNA genes. To distinguish between these possibilities, we compared the change in abundance for each mature miRNA sequence to each variant miRNA sequence originating from the same precursor. The normal processing of miRNA precursors (Ruby et al., 2006; Ruby et al., 2007) results in the production of a mature miRNA species together with rarer shorter or longer variant miRNAs. Such variations can alter propensities for association with AGO proteins (Okamura et al., 2009; Wang et al., 2011) while alterations in the transcription of miRNA genes would be reflected equally by all variant products (isomiRs) of a particular miRNA. We found that changes in the abundance of pairs of miRNAs and variant miRNA species were significantly correlated (Fig 4F; R=0.68; P≤10≥45; Figure S5, Supplemental Table 2), a result most consistent with transcriptional differences for miRNA genes in Ago4−/− cells. Interestingly, when comparing miRs and isomiRs with shared 5′ ends and shared 3′ ends, those with shared 5′ ends were somewhat (P=0.09) more correlated (Figure 4F; R values of 0.70, red) than those with shared 3′ ends (R=0.41, blue). These results are consistent with loss of Ago4 resulting in both changes in miRNA transcription, together with different propensities for certain miRNA species to form complexes with different AGO proteins. Importantly, loss of AGO4 results in a significant reduction in specific microRNAs, and despite increased RNAP II in the SB, a significant fraction of these arise from the X chromosome.
To correlate changes in small RNA species identified by small RNA cloning with changes in their localization, we examined the localization of microRNAs originating from the X chromosome. For example, miR-322 is known to escape MSCI (Song et al., 2009), and was shown in our small RNA cloning to be dramatically down-regulated in the absence of AGO4. We find miR-322 localizes to the SB in WT spermatocytes (Figure 4G), while its localization is decreased in those Ago4−/− pachytene spermatocytes that display aberrant SB morphology (Figure 4H). Furthermore, we confirm the SB localization of miR-24 as demonstrated previously (Marcon et al., 2008), and observed no signal from miR-206, consistent with the small RNA cloning data (Figure S4H,I). Together, these studies demonstrate that certain miRNAs are dependent on AGO4 and suggest an accumulation of some miRNA species in the SB during pachynema.
Expression profiling of pachytene spermatocytes reveals loss of silencing of sex-linked genes
To better understand alterations in gene expression that result from loss of Ago4, we used RNAseq to profile the transcriptome of Ago4−/− and WT cell populations. Our profiling was performed on whole testis extracts at day 3–4 and day 8–9 pp, both comprised solely of somatic cells and spermatogonia, together with isolated meiotic cell fractions from adult testis at leptonema/zygonema and at pachynema. RNA from adult kidney and seminal vesicles served as controls for each genotype.
Contrary to our expectation, we found no evidence for a global failure in sex chromosome silencing in Ago4−/− pachytene cells (Figure 5A). However, numerous Y-linked transcripts did show upregulated expression in the absence of Ago4, including Zfy2 and Ube1y1 (Figure 5B,C). Zfy2 is one of the “suicide” genes (along with Zfy1) whose expression in the absence of MSCI results in apoptosis (Royo et al., 2010). Zfy2 expression was elevated approximately 2-fold in pachytenes from Ago4−/− males (p<0.03). Importantly, the increased transcription seen for certain sex-linked genes in the absence of AGO4 is likely contributed only by those pachytene cells showing SB aberrations, such that the overall extent of the loss of silencing is likely lost in the heterogeneous population of aberrant and normal pachytene cells obtained for RNAseq analysis, suggesting that the effect of AGO4 loss on MSCI is under-estimated by our RNAseq analysis.
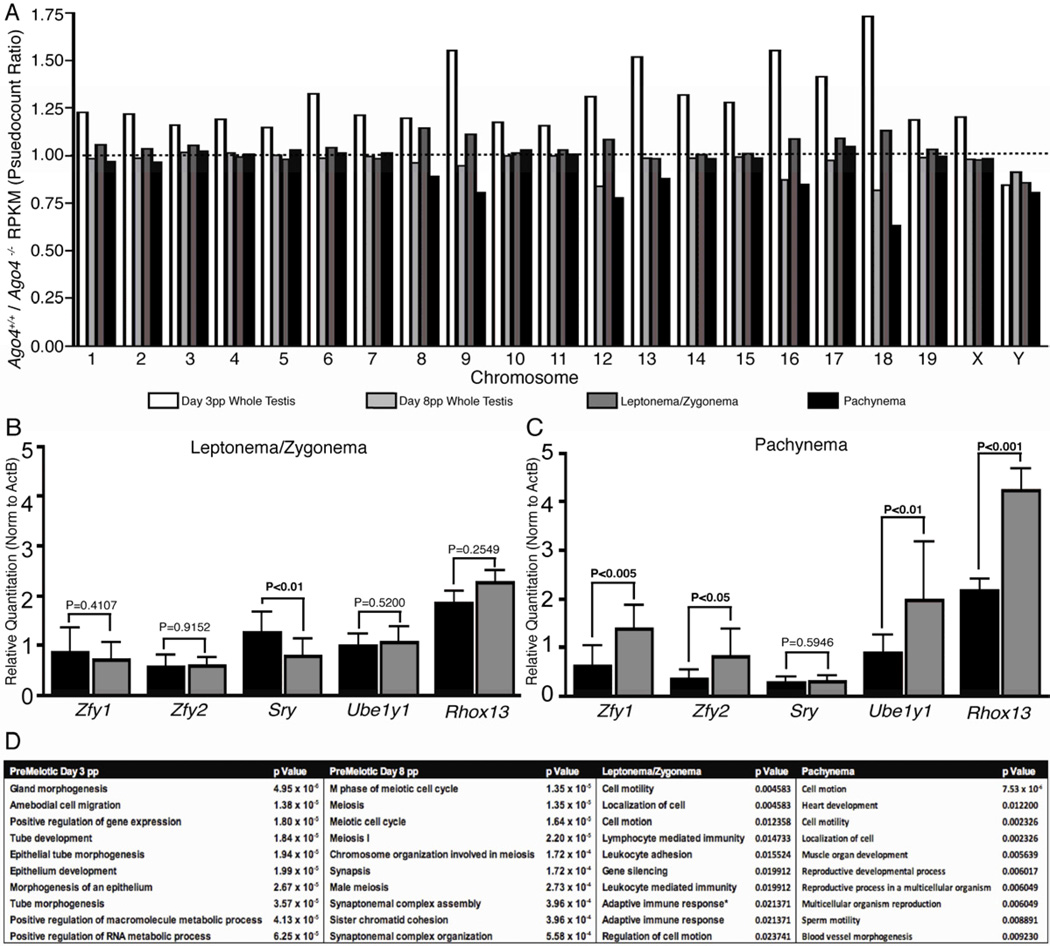
Figure 5. RNAseq profiling of purified spermatocytes from different stages of prophase I, and from pre-meiotic testes.
(A) Differential expression between WT and Ago4−/− transcript levels was determined by RNAseq and depicted by ratios of WT to Ago4−/− mRNA levels by chromosome. (B,C) Quantitative RT-PCR analysis of XY-linked gene expression in total RNA from isolated (B) leptotene/zygotene and (C) pachytene spermatocytes from Ago4+/+ (n=3, black bars) and Ago4−/− (n=3, grey bars) adult littermates (significant values in bold). (D) Gene ontology analysis of transcripts showing significant differential expression (Q<0.05) in mRNA levels; the most significant groups of transcripts differentially expressed are shown for each cell type.
Many aspects MSCI initiation and maintenance remain elusive. Establishment of MSCI requires H2AX phosphorylation by ATR in the SB. Thus, during meiotic prophase I, ATR and γH2AX accumulate on sex chromatin in spermatocytes (Fernandez-Capetillo et al., 2003; Turner et al., 2004; Turner et al., 2005). The prevailing model proposes that to maintain MSCI, the limited pool of ATR functions to prevent expression of toxic Y-linked genes (Royo et al., 2010). Thus, ATR must be recruited to the SB by first being depleted from the autosomes following synapsis. In various SC or recombination repair mouse mutants, therefore, ATR persists on the autosomes due to the more generalized MSUC mechanism, resulting in up-regulation of gene expression from the sex chromosomes including Zfy1/Zfy2. It is the expression of these genes that results in meiotic arrest, rather than any effects of recombination failure per se (Royo et al., 2010). Importantly, RNAseq analysis of RNA from the leptotene/zygotene and pachytene stages did not show any differences in expression of Atr or Brca1 between WT and mutant (Supplemental Table 3). These genes are essential for phosphorylation of H2AX during MSCI (Turner et al., 2004), suggesting that the derepression of XY-linked genes is not due to failure to phosphorylate H2AX.
The derepression of certain XY-linked transcripts was examined further by performing qRT-PCR on RNA from isolated leptotene/zygotene and pachytene spermatocytes (Figure 5B,C, respectively). Three out of four Y-linked genes examined (Zfy1, Zfy2, Ube1y1) and one X-linked gene (Rhox13) showed significant increases in expression in Ago4−/− spermatocytes compared to WT specifically at pachynema (Figure 5C), indicating that many XY-linked genes escape MSCI in the absence of AGO4.
Given our model for AGO4 involvement in MSCI, it was somewhat surprising to see only a modest loss of silencing in the SB, associated with modest persistence of ATR on the X, but not the Y, chromosome, and only a slight increase in ATR localization on the autosomes. This could reflect partial redundancy with AGO3, which is up-regulated in Ago4−/− spermatocytes. Alternatively, this partial loss of MSCI could reflect the fact that only ~30% of cells observed in early pachynema have abnormal SB morphologies, which are likely eradicated by apoptosis, thus those that survive are functionally intact with respect to MSCI processes. This possibility is supported by the finding that all abnormal SBs are observed in cells that are H1T-negative, indicative of early pachynema, while only normal SB morphologies are observed in cells that are H1T-positive, indicative of late pachynema.
Loss of Ago4 drives premature entry into meiosis
While comparing gene expression profiles between Ago4−/− and WT littermate males, gene ontology (GO) analysis of RNAseq data revealed a dramatic up-regulation of transcripts encoding proteins involved in spermatogonial proliferation in Ago4−/− testes on day 3–4 pp relative to WT, together with a distinct induction of genes regulating meiotic entry within day 8–9 pp Ago4−/− testes (Figure 5D). This suggests that loss of AGO4 facilitates spermatogonial differentiation and proliferation leading to premature meiotic initiation. In WT mice, meiotic initiation is brought about by the production of Retinoic acid (RA) via the action of the RALDH2 enzyme (the product of the Aldh1a2 gene) in Sertoli (and other) cells of the postnatal testis. The oxidation of RA by the P450 enzyme, CYP26B1, prevents RA-induced meiotic entry in fetal testes at a time when fetal oocytes initiate meiosis because they lack this enzyme (Hogarth and Griswold, 2010). In postnatal males, at around day 5–6 pp, RA binds to the retinoic acid receptor G (RARG) on the surface of Type A spermatogonia (Gely-Pernot et al., 2012) and induces a cascade of genes, including Stra8, Esco2, and Uba6 (Hogarth et al., 2011), while negative regulators of meiotic onset act to repress Stra8 function and promote spermatogonial maintenance. These negative regulators of meiotic induction include Nanos2 and Fgf9 (Suzuki and Saga, 2008).
Genes involved in spermatogonial stem cell maintenance and proliferation were up-regulated in RNA samples from day 3–4 pp Ago4−/− testes (Figure 6A). These included Sox2 (Figure 6A) and Sox9 (not shown), members of the Sox family of transcription factors that are involved in the reactivation of stem cell pluripotency (Yabuta et al., 2006). Similarly, the RNA binding protein, Lin28b, was up-regulated along with its downstream target, Prdm1, while let7 was down-regulated, in Ago4−/− testes. Under the influence of RA, LIN28b is required to repress the somatic mesodermal program by sequestering the let-7 miRNA family, which in turn allows PRDM1 to act as an inducer of germ cell lineages (Bowles and Koopman, 2010). Thus, the day 3–4 pp GO analysis of RNA from whole Ago4−/− testes revealed a switch from spermatogonial stem cell maintenance to one of spermatogonial proliferation and/or differentiation.
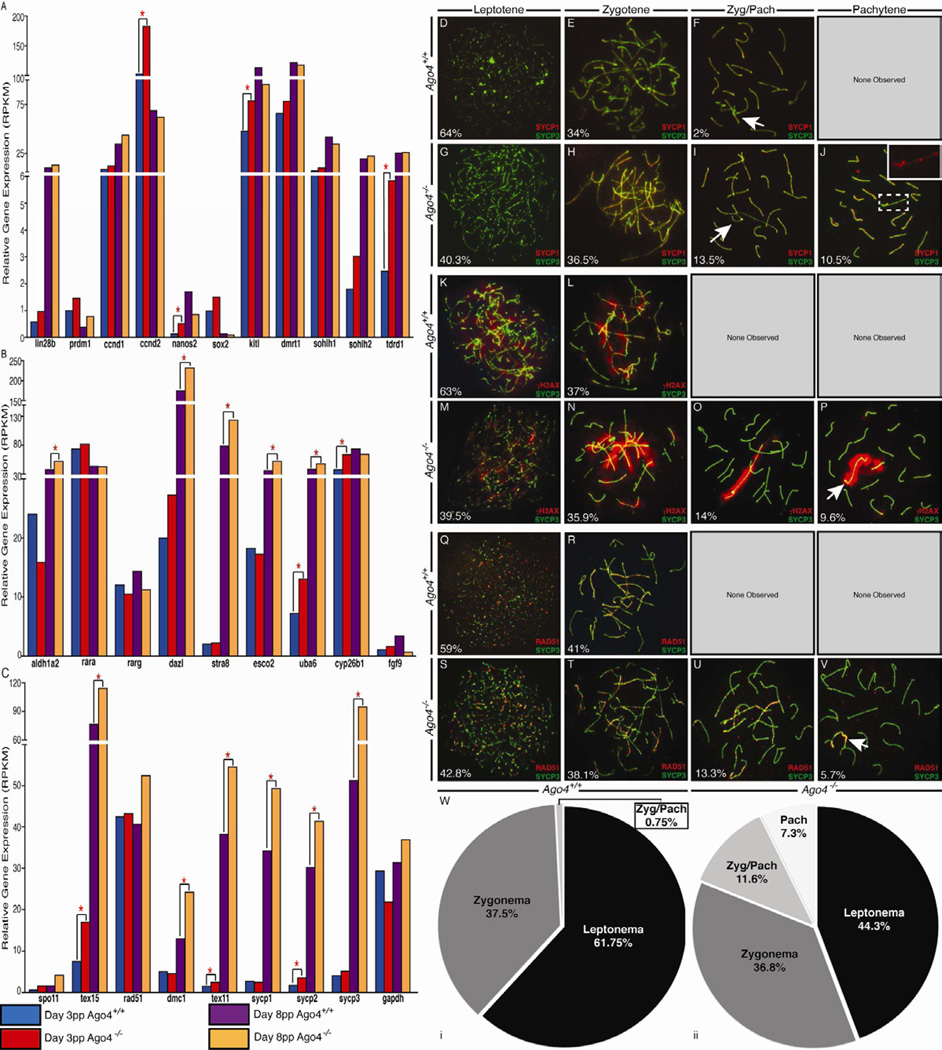
Figure 6. Loss of AGO4 induces early entry into meiosis in postnatal testes.
(A–C) Quantification of transcripts from RNAseq performed on pre-meiotic testis from Ago4+/+ and Ago4−/− males at day 3–4 pp (blue (Ago4+/+) and red (Ago4−/−) bars) and day 8–9 pp (purple (Ago4+/+) and orange (Ago4−/−) bars. GAPDH is shown as a control. Genes with a significant degree of differential expression between Ago4+/+ and Ago4−/− males (Q<0.05) are indicated with an asterisk (*). Genes in panel A are associated with spermatogonial proliferation and/or differentiation. Genes in panel B are involved in the initiation of meiosis, via RA. Genes involved in early prophase I progression are in panel C. (D–V) Chromosome immunofluorescence from day 8–9 pp testes from Ago4+/+ (D,E,F,K,L,Q,R,Wi) and Ago4−/− (G,H,I,J,M,N,O,P,S,T,U,V,Wii) littermates stained with anti-SYCP3 (green), together with either anti-SYCP1 (D–J), anti-γH2AX (K–P), or anti-RAD51 (Q–V), all in red. Progression through prophase I is depicted by horizontal panels from left to right. If no image is given for a particular stage of prophase I, then that stage was not observed in the staining for this genotype. % cells found at each stage is shown. (Wi, Wii) Quantification of prophase I stages for Ago4+/+ and Ago4−/−, respectively, n=400. χ2 analysis revealed significant differences between the two populations (p<0.0001***).
At day 8–9 pp in Ago4−/− testes, Cyp26b1, Nanos2 and Fgf9 levels are all reduced while Aldh1a2, Rarg and Stra8 levels are greatly enhanced compared to Ago4+/+ males. Similarly, Esco2 and Uba6 show premature increases in expression at day 8–9 pp in the Ago4−/− testes (Figure 6B). These alterations in the induction of RA pathway suggest that the absence of AGO4 results in premature induction of meiotic genes approximately 5 days earlier than in the WT.
Early meiotic initiation in young Ago4−/− male pups was supported by the onset of expression of genes known to be essential and specific to meiotic prophase I, which usually occurs around day 9 pp in the male mouse (Goetz et al., 1984). These include the genes encoding SPO11, the topoisomerase that generates double strand breaks (DSB) that initiate meiotic recombination; the RecA homologs, DMC1 and RAD51, which process DSBs to facilitate homology searching and recombination (reviewed by Handel and Schimenti, 2010); TEX15, which facilitates loading of the RecA homologs (Yang et al., 2008); and the synaptonemal complex components, SYCP1 and SYCP3 (Figure 6C).
To confirm the early onset of prophase I at the cytological level, chromosome preps were examined from pups at day 5–6 pp and day 8–9 pp. At day 5–6 pp, no cells were found to be in prophase I in Ago4+/+ males, whereas a large number of cells from Ago4−/− males were found to be in pre-leptonema (not shown). This difference was even more dramatic by day 8–9 pp, when large numbers of spermatocytes from Ago4−/− males were found to have entered, and progressed through, the early stages of prophase I. Progression through prophase I was demonstrated by staining with antibodies against SYCP3, together with SYCP1 (Figure 6D–J), or with γH2AX (Figure 6K–P), and with RAD51 (Figure 6Q–V). In all cases, the major alteration to the spermatocyte pool in Ago4−/− males appeared to be a decrease in total leptotene cells (collective counts shown in figure 6W, n=400, p<0.005 t-test), and a concomitant increase in zygonema/pachynema intermediate cells (“Zyg/Pach” Figure 6W, n=400, p<0.005 t-test). The emergence of pachytene cells in Ago4−/− males was approximately 5–6 days earlier than in WT males. Interestingly, all spermatocytes observed to reach the pachytene stage in the young mutant pups (Figure 6J,P) displayed defects identical to the altered staining patterns described earlier for spermatocytes from adult males. This observation suggests that meiotic progression in the adult can be partially rescued, perhaps by AGO3, while the initial waves of meiotic prophase I in early postnatal life are more wholly dependent on AGO4.
Together, our observations demonstrate early entry into meiosis in Ago4−/− males caused by premature induction of RA response genes that trigger meiotic initiation. These studies are in line with a recent report which demonstrates that the C. elegans argonautes ALG-1 and ALG-2 are required for germ cell proliferation and entry into meiosis (Bukhari et al., 2012). In the worm, ALG-1 and ALG-2 localize to distal tip cells (DTC), specialized somatic cells in the gonad that regulate the switch from mitosis to meiosis (Kimble and Crittenden, 2007) suggesting a cell non-autonomous miRNA mechanism for regulating meiotic entry.
CONCLUSIONS
The studies described herein demonstrate a role for AGO4 in the nucleus of male germ cells. Our data support a model whereby AGO4-associated small RNAs directly participate in MSCI (Figure 7). Consistent with the discovery of nuclear RNAi components in human cells (Ahlenstiel et al., 2011; Robb et al., 2005), our study reveals distinct localization patterns for both AGO3 and AGO4 within the nuclei of mouse spermatogenic cells, along with certain miRNAs. These observations support the idea that AGO4 may recruit small RNAs to the SB, or vice versa. Importantly, we show that in the absence of AGO4, disproportionate numbers of down-regulated miRNAs originate from the X chromosome, and that miRNA localization in the SB is markedly reduced. This loss of miRNAs specifically from X and Y is contrary to expectation given the increased localization of RNAP II in the SB, but is in line with the fact that X-linked miRNAs have been found to escape repression during MSCI of prophase I (Song et al., 2009).
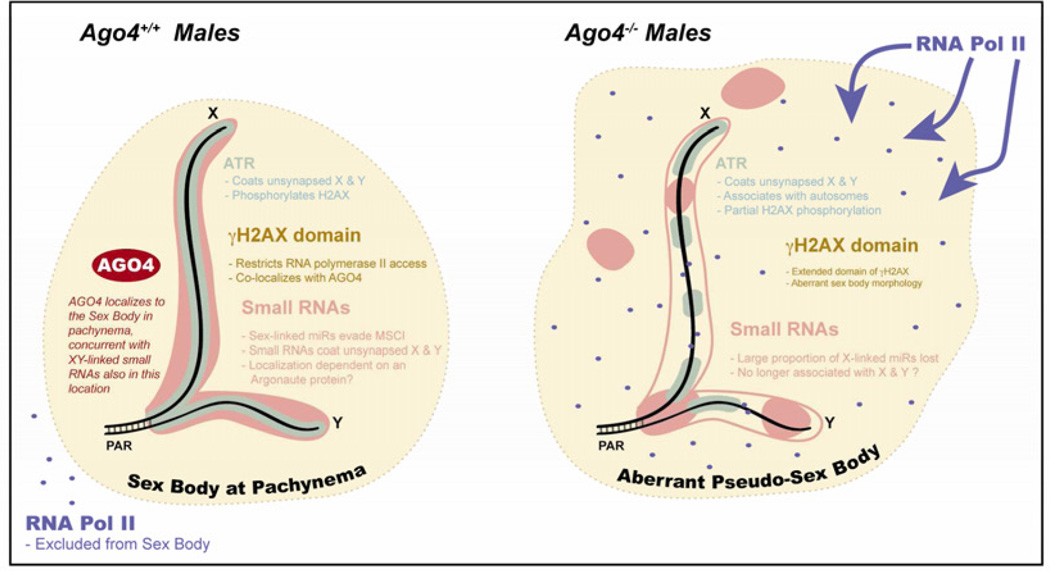
Figure 7. Model for the role of AGO4 in meiotic silencing.
In WT males at pachynema (left side), the SB domain is defined by localization of γH2AX (yellow background), whose phosphorylation is mediated by the ATR kinase, which coats the unsynapsed lengths of the X and Y chromosomes (green). Sex chromosomes are also coated with small RNAs (pink) at this time. We propose that the presence of these RNA species may depend on AGO4, and the sum effect is the exclusion of RNA polymerase II from the SB and silencing of the sex chromosomes by MSCI. In the absence of AGO4 (right side), ATR localization is drastically reduced along abnormal SB morphology. Sex-linked small RNAs are drastically reduced and no longer reside in the SB, leading to RNAP II infiltration and failed silencing. These effects are distinct from those involving RA signaling and meiotic initiation described in Figure 6.
Given the localization of AGO4 to asynapsed regions and the SB in pachytnema, together with its co-localization with mediators of MSCI (ATR and γH2AX), and the dependence of these mediators on AGO4, the results presented herein indicate a role for AGO4 in the initiation and/or maintenance of transcriptional silencing during prophase I. We hypothesize that this transcriptional regulation is regulated by AGO4 in a manner reminiscent of the RITS complex seen in S. pombe and C. elegans (van Wolfswinkel and Ketting, 2010). While small RNAs have been postulated to be involved in MSCI (Burgoyne et al., 2009), our data provide direct evidence for AGO4 in these events (Figure 7).
The role of AGO4 in meiotic initiation appears to originate in at least two cell types: the RA producing Sertoli cells and the spermatogonia. However, AGO4 expression appears to be confined to the germ lineages, leading us to postulate an indirect effect on RA production by the somatic cells of the testis. In this scenario, loss of AGO4 results in loss of a signal from the germ cells to the Sertoli cells that would usually repress RA production. The action of AGO4 in the germ cells themselves may be directed towards RA signal transduction and/or towards downstream mediators. Indeed, treatment of fetal testes with trichostatin A, an inhibitor of class I/II histone deacetylases, promotes premature activation of Stra8 expression and early meiotic entry (Wang and Tilly, 2010), suggesting epigenetic regulation of these events. Moreover, there is evidence supporting the role of RNA binding proteins and small RNAs in self-renewal of spermatogonial stem cells (Niu et al., 2011), as well as in the response of spermatogonia to RA signaling (Griswold et al., 2012; Ro et al., 2007). For example, DAZL, an RNAbinding protein expressed in germ cells, which is up-regulated on day 8–9 pp in the absence of AGO4, facilitates germ cell responses to RA to promote meiotic initiation (Gill et al., 2011; Lin et al., 2008; Lin and Page, 2005). In contrast, two miRNA clusters have been implicated during spermatogonial differentiation in mice, miR-17–92 and miR-106b-25 (Tong et al., 2011), and both are down-regulated in Ago4−/− testes.
Taken together, our studies demonstrate important roles for AGO4 in meiotic induction and progression in male mice. Of particular note is the observation that AGO4 is found in the nucleus of prophase I cells, suggesting the existence of a RITS-like complex at the level of heterochromatin in meiotic cells. Finally, it is important to note that the two roles for AGO4 in mammalian germ cells that we describe appear to be distinct, since treatment of vitamin A deficient mice with RA does not lead to MSCI defects or meiotic disruption (van Pelt and de Rooij, 1990). Indeed, it is plausible that the early meiotic entry and the MSCI events require different roles for AGO4 in the context of their nuclear and cytoplasmic activities. Continued studies are aimed at elucidating the mechanisms of AGO4 involvement in these mammalian germ cell events.
EXPERIMENTAL PROCEDURES
Generation of Ago4 mutant mice
An Ago4 targeting construct, containing a 2.5-kbp 5′ arm from intron 2 of Ago4, a LoxP site, 25.4 kb of Ago4 from exons 3–17, an Frt-flanked Pgk-neo cassette, a second LoxP site and a 1.6-kbp 3′ arm within intron 17 of Ago4 (Figure S1D–F), was used to generate chimeric mice using standard mouse mutagenesis techniques. Following FlpE and CRE-mediated excision of the Frt-flanked and LoxP-flanked cassettes, respectively, the resulting Ago4−/− allele (lacking exons 3–17) was bred to homozygosity onto a C57BL/6 background.
Testes weights, Sperm counts and Histology
Whole testes were removed from Ago4+/+ and Ago4−/− littermates and weighed, and epididymal sperm counts assessed (Edelmann et al., 1996). For histological analysis, testes were fixed in Bouins (for H&E staining) or in 10% formalin (for all other staining and for TUNEL) overnight at 4°C. Paraffin embedded tissues were sectioned at 5 µm before dewaxing and histological staining.
Chromosome spreading and immunofluorescent staining
Prophase I chromosome spreads and antibody staining were prepared as previously described, (Holloway et al., 2008; Holloway et al., 2011; Kolas et al., 2005; Lipkin et al., 2002) using antibodies from various sources (see supplemental Experimental Procedures).
RT-PCR and Q-PCR
Total RNA was extracted from whole tissue or isolated spermatocytes using TRIZOL reagent (Invitrogen). Reverse transcription was performed using Superscript III (Invitrogen). For real-time PCR analysis, TaqMan probes (Applied Biosystems, Carlsbad CA, USA) were used on the 7500 Real Time PCR System (Applied Biosystems). Raw data was analyzed using the 7500 System Sequence Detection Software (Applied Biosystems). Probesets are provided in supplemental Experimental Procedures.
Isolation of mouse spermatogenic cells
Testes from adult Ago4+/+ and Ago4 −/− adult mice (day 70–80 pp) were removed, weighed and decapsulated prior to enrichment of specific spermatogenic cells types using the STA-PUT method based on separation by cell diameter/density at unit gravity (Bellve, 1993). Purity of resulting fractions was determined by microscopy based on cell diameter and morphology. Pachytene cells were at approximately 95% purification with potential contamination from spermatocytes of slightly earlier or later developmental timing.
mRNA transcript sequencing and analysis
mRNA transcripts were isolated and prepared for sequencing using Illumina’s TRUseq kit. The completed library was sequenced on an Illumina HiSeq 2000 and aligned to the genome using TopHat and Cuffdiff programs (Trapnell et al., 2009) to determine differential expression between WT and mutant samples. A q-value, which represents a false discovery rate-corrected p-value (Klipper-Aurbach et al., 1995), <.05 was chosen as the cutoff for determining whether differential gene expression was significant. For ontology analysis, genes with significant differential expression were analyzed using DAVID (Huang da et al., 2009a, b).
Small RNA sequencing and Analysis
Small RNA sequencing was performed as described previously (Grimson et al., 2008). Sequences were filtered to remove reads containing undefined bases (‘N’) and reads under 14 nt in length. Sequences were aligned to the genome using Bowtie (version 0.12.07, (Langmead et al., 2009)). MicroRNAs were determined by alignment of sequences to mouse miRNA hairpins (miRBase Release 18, (Griffiths-Jones, 2004; Griffiths-Jones et al., 2006; Griffiths-Jones et al., 2008)).
miRNA Fluorescent In Situ Hybridization and Immunofluorescence
Slides were prepared and stained as described above with modifications to retain RNA (see supplemental Experimental Procedures). Following immunofluorescence with primary antibodies as described previously (Holloway et al., 2010; Kolas et al., 2005), and using an anti-SYCE2 antibody (from Howard Cooke, Edinburgh, Scotland), Fluorescein conjugated Locked Nucleic Acid (LNA) probes (Exiqon, Denmark) were used to probe for mature miRNA localization. Probes were first denatured by incubation at 90°C for 4 minutes and hybridizations were carried out at the appropriate incubation temperature overnight. Slides were then washed, mounted with antifade (Prolong Gold, Molecular Probes) and visualized.
Supplementary Material
Highlights.
AGO4 localizes to mammalian germ cell nuclei, particularly sites of unpaired DNA
AGO4 functions in mammalian germ cells to regulate entry into meiosis in males
AGO4 is a nuclear non-slicing Argonaute involved in X-linked microRNA production
AGO4 also contributes to meiotic sex chromosome inactivation (MSCI)
ACKNOWLEDGEMENTS
This work was supported by the Vertebrate Genomics Center at Cornell University (VERGE postdoctoral award to R.J.H. and predoctoral award to S.H.) and by funding from the NIH to P.E.C. (HD041012). A.M. was supported by NIH training grants in Genetics and Development (T32GM00761), and in Reproductive Genomics (T32HD052471); S.H. was supported by an NIH training grant in Biochemistry, Molecular and Cellular Biology. The authors acknowledge with gratitude the generosity of Dr. Mary Ann Handel (The Jackson Laboratory) for providing the anti-H1T antibody, Dr. Raimundo Freire (Tenerife, Spain) for providing the anti-TOPBP1 antibody, Drs. Ian Adams and Howard Cooke (University of Edinburgh, Scotland) for providing the anti-SYCE2 antibody, and Dr. George Enders (University of Kansas Medical Center) for providing the GCNA1 hybridoma supernatant. We are grateful to Ian Welsh for technical advice, and to Ewelina Bolcun-Filas, Jen Grenier, Kim Holloway, and Mark Roberson for reading this manuscript and for helpful suggestions. We also thank Kim Holloway for extensive guidance in preparing figures and editorial assistance. We thank Peter L. Borst for assistance in mouse care.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ahlenstiel CL, Lim HG, Cooper DA, Ishida T, Kelleher AD, Suzuki K. Direct evidence of nuclear Argonaute distribution during transcriptional silencing links the actin cytoskeleton to nuclear RNAi machinery in human cells. Nucleic Acids Research. 2011 doi: 10.1093/nar/gkr891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- Baarends WM, Grootegoed A. Chromatin dynamics in the male meiotic prophase. Cytogenet Genome Res. 2003;103:225–234. doi: 10.1159/000076808. [DOI] [PubMed] [Google Scholar]
- Baarends WM, Wassenaar E, van der Laan R, Hoogerbrugge J, Sleddens-Linkels E, Hoeijmakers JH, de Boer P, Grootegoed JA. Silencing of unpaired chromatin and histone H2A ubiquitination in mammalian meiosis. Molecular and cellular biology. 2005;25:1041–1053. doi: 10.1128/MCB.25.3.1041-1053.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barchi M, Mahadevaiah S, Di Giacomo M, Baudat F, de Rooij DG, Burgoyne PS, Jasin M, Keeney S. Surveillance of different recombination defects in mouse spermatocytes yields distinct responses despite elimination at an identical developmental stage. Mol Cell Biol. 2005;25:7203–7215. doi: 10.1128/MCB.25.16.7203-7215.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellani MA, Romanienko PJ, Cairatti DA, Camerini-Otero RD. SPO11 is required for sex-body formation, and Spo11 heterozygosity rescues the prophase arrest of Atm−/− spermatocytes. J Cell Sci. 2005;118:3233–3245. doi: 10.1242/jcs.02466. [DOI] [PubMed] [Google Scholar]
- Bellve AR. Purification, culture, and fractionation of spermatogenic cells. Methods Enzymol. 1993;225:84–113. doi: 10.1016/0076-6879(93)25009-q. [DOI] [PubMed] [Google Scholar]
- Bowles J, Koopman P. Sex determination in mammalian germ cells: extrinsic versus intrinsic factors. Reproduction. 2010;139:943–958. doi: 10.1530/REP-10-0075. [DOI] [PubMed] [Google Scholar]
- Brown PW, Hwang K, Schlegel PN, Morris PL. Small ubiquitin-related modifier (SUMO)-1, SUMO-2/3 and SUMOylation are involved with centromeric heterochromatin of chromosomes 9 and 1 and proteins of the synaptonemal complex during meiosis in men. Human Reproduction. 2008;23:2850–2857. doi: 10.1093/humrep/den300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukhari SI, Vasquez-Rifo A, Gagne D, Paquet ER, Zetka M, Robert C, Masson JY, Simard MJ. The microRNA pathway controls germ cell proliferation and differentiation in C. elegans. Cell research. 2012;22:1034–1045. doi: 10.1038/cr.2012.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgoyne PS, Mahadevaiah SK, Turner JM. The consequences of asynapsis for mammalian meiosis. Nat Rev Genet. 2009;10:207–216. doi: 10.1038/nrg2505. [DOI] [PubMed] [Google Scholar]
- Cenik ES, Zamore PD. Argonaute proteins. Current biology : CB. 2011;21:R446–R449. doi: 10.1016/j.cub.2011.05.020. [DOI] [PubMed] [Google Scholar]
- Czech B, Hannon GJ. Small RNA sorting: matchmaking for Argonautes. Nature reviews Genetics. 2011;12:19–31. doi: 10.1038/nrg2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel K, Lange J, Hached K, Fu J, Anastassiadis K, Roig I, Cooke HJ, Stewart AF, Wassmann K, Jasin M, et al. Meiotic homologue alignment and its quality surveillance are controlled by mouse HORMAD1. Nature Cell Biology. 2011;13:599–610. doi: 10.1038/ncb2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann W, Cohen PE, Kane M, Lau K, Morrow B, Bennett S, Umar A, Kunkel T, Cattoretti G, Chaganti R, et al. Meiotic pachytene arrest in MLH1-deficient mice. Cell. 1996;85:1125–1134. doi: 10.1016/s0092-8674(00)81312-4. [DOI] [PubMed] [Google Scholar]
- Fernandez-Capetillo O, Mahadevaiah SK, Celeste A, Romanienko PJ, Camerini-Otero RD, Bonner WM, Manova K, Burgoyne P, Nussenzweig A. H2AX is required for chromatin remodeling and inactivation of sex chromosomes in male mouse meiosis. Dev Cell. 2003;4:497–508. doi: 10.1016/s1534-5807(03)00093-5. [DOI] [PubMed] [Google Scholar]
- Gely-Pernot A, Raverdeau M, Celebi C, Dennefeld C, Feret B, Klopfenstein M, Yoshida S, Ghyselinck NB, Mark M. Spermatogonia differentiation requires retinoic acid receptor gamma. Endocrinology. 2012;153:438–449. doi: 10.1210/en.2011-1102. [DOI] [PubMed] [Google Scholar]
- Gill ME, Hu YC, Lin Y, Page DC. Licensing of gametogenesis, dependent on RNA binding protein DAZL, as a gateway to sexual differentiation of fetal germ cells. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:7443–7448. doi: 10.1073/pnas.1104501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- Goetz P, Chandley AC, Speed RM. Morphological and temporal sequence of meiotic prophase development at puberty in the male mouse. Journal of Cell Science. 1984;65:249–263. doi: 10.1242/jcs.65.1.249. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Gonzalez E, Lopez-Casas PP, del Mazo J. The expression patterns of genes involved in the RNAi pathways are tissue-dependent and differ in the germ and somatic cells of mouse testis. Biochim Biophys Acta. 2008;1779:306–311. doi: 10.1016/j.bbagrm.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S. The microRNA Registry. Nucleic Acids Research. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Research. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Research. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, King N, Degnan BM, Rokhsar DS, Bartel DP. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008;455:1193–1197. doi: 10.1038/nature07415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griswold MD, Hogarth CA, Bowles J, Koopman P. Initiating meiosis: the case for retinoic Acid. Biology of Reproduction. 2012;86:35. doi: 10.1095/biolreprod.111.096610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivna ST, Beyret E, Wang Z, Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006;20:1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handel M. The XY body: a specialized meiotic chromatin domain. Exp Cell Res. 2004;296:57–63. doi: 10.1016/j.yexcr.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Handel MA, Schimenti JC. Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat Rev Genet. 2010;11:124–136. doi: 10.1038/nrg2723. [DOI] [PubMed] [Google Scholar]
- Hogarth CA, Griswold MD. The key role of vitamin A in spermatogenesis. The Journal of clinical investigation. 2010;120:956–962. doi: 10.1172/JCI41303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogarth CA, Mitchell D, Evanoff R, Small C, Griswold M. Identification and expression of potential regulators of the mammalian mitotic-to-meiotic transition. Biology of Reproduction. 2011;84:34–42. doi: 10.1095/biolreprod.110.086215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway JK, Booth J, Edelmann W, McGowan CH, Cohen PE. MUS81 generates a subset of MLH1–MLH3-independent crossovers in mammalian meiosis. PLoS Genet. 2008;4 doi: 10.1371/journal.pgen.1000186. e1000186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway JK, Mohan S, Balmus G, Sun X, Modzelewski A, Borst PL, Freire R, Weiss RS, Cohen PE. Mammalian BTBD12 (SLX4) protects against genomic instability during mammalian spermatogenesis. PLoS Genetics. 2011 doi: 10.1371/journal.pgen.1002094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway JK, Morelli MA, Borst PL, Cohen PE. Mammalian BLM helicase is critical for integrating multiple pathways of meiotic recombination. J Cell Biol. 2010;188:779–789. doi: 10.1083/jcb.200909048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Kim DH, Villeneuve LM, Morris KV, Rossi JJ. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat Struct Mol Biol. 2006;13:793–797. doi: 10.1038/nsmb1142. [DOI] [PubMed] [Google Scholar]
- Kim S, Namekawa SH, Niswander LM, Ward JO, Lee JT, Bardwell VJ, Zarkower D. A mammal-specific Doublesex homolog associates with male sex chromatin and is required for male meiosis. PLoS Genet. 2007;3:e62. doi: 10.1371/journal.pgen.0030062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J, Crittenden SL. Controls of germline stem cells, entry into meiosis, and the sperm/oocyte decision in Caenorhabditis elegans. Annual review of cell and developmental biology. 2007;23:405–433. doi: 10.1146/annurev.cellbio.23.090506.123326. [DOI] [PubMed] [Google Scholar]
- Klipper-Aurbach Y, Wasserman M, Braunspiegel-Weintrob N, Borstein D, Peleg S, Assa S, Karp M, Benjamini Y, Hochberg Y, Laron Z. Mathematical formulae for the prediction of the residual beta cell function during the first two years of disease in children and adolescents with insulin-dependent diabetes mellitus. Med Hypotheses. 1995;45:486–490. doi: 10.1016/0306-9877(95)90228-7. [DOI] [PubMed] [Google Scholar]
- Kolas NK, Svetlanov A, Lenzi ML, Macaluso FP, Lipkin SM, Liskay RM, Greally J, Edelmann W, Cohen PE. Localization of MMR proteins on meiotic chromosomes in mice indicates distinct functions during prophase I. J Cell Biol. 2005;171:447–458. doi: 10.1083/jcb.200506170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Sirotkin A, Skoultchi AI. Normal spermatogenesis in mice lacking the testis-specific linker histone H1t. Mol Cell Biol. 2000;20:2122–2128. doi: 10.1128/mcb.20.6.2122-2128.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Gill ME, Koubova J, Page DC. Germ cell-intrinsic and - extrinsic factors govern meiotic initiation in mouse embryos. Science. 2008;322:1685–1687. doi: 10.1126/science.1166340. [DOI] [PubMed] [Google Scholar]
- Lin Y, Page DC. Dazl deficiency leads to embryonic arrest of germ cell development in XY C57BL/6 mice. Developmental Biology. 2005;288:309–316. doi: 10.1016/j.ydbio.2005.06.032. [DOI] [PubMed] [Google Scholar]
- Lipkin SM, Moens PB, Wang V, Lenzi M, Shanmugarajah D, Gilgeous A, Thomas J, Cheng J, Touchman JW, Green ED, et al. Meiotic arrest and aneuploidy in MLH3-deficient mice. Nat Genet. 2002;31:385–390. doi: 10.1038/ng931. [DOI] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Maine EM. Meiotic silencing in Caenorhabditis elegans. Int Rev Cell Mol Biol. 2010;282:91–134. doi: 10.1016/S1937-6448(10)82002-7. [DOI] [PubMed] [Google Scholar]
- Marcon E, Babak T, Chua G, Hughes T, Moens PB. miRNA and piRNA localization in the male mammalian meiotic nucleus. Chromosome Res. 2008 doi: 10.1007/s10577-007-1190-6. [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Niu Z, Goodyear SM, Rao S, Wu X, Tobias JW, Avarbock MR, Brinster RL. MicroRNA-21 regulates the self-renewal of mouse spermatogonial stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:12740–12745. doi: 10.1073/pnas.1109987108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Liu N, Lai EC. Distinct mechanisms for microRNA strand selection by Drosophila Argonautes. Molecular Cell. 2009;36:431–444. doi: 10.1016/j.molcel.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai RS, Chuma S. piRNAs and their involvement in male germline development in mice. Dev Growth Differ. 2012 doi: 10.1111/j.1440-169X.2011.01320.x. [DOI] [PubMed] [Google Scholar]
- Raju NB, Metzenberg RL, Shiu PK. Neurospora spore killers Sk-2 and Sk-3 suppress meiotic silencing by unpaired DNA. Genetics. 2007;176:43–52. doi: 10.1534/genetics.106.069161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ro S, Park C, Sanders KM, McCarrey JR, Yan W. Cloning and expression profiling of testis-expressed microRNAs. Dev Biol. 2007;311:592–602. doi: 10.1016/j.ydbio.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb GB, Brown KM, Khurana J, Rana TM. Specific and potent RNAi in the nucleus of human cells. Nature structural & molecular biology. 2005;12:133–137. doi: 10.1038/nsmb886. [DOI] [PubMed] [Google Scholar]
- Royo H, Polikiewicz G, Mahadevaiah SK, Prosser H, Mitchell M, Bradley A, de Rooij DG, Burgoyne PS, Turner JM. Evidence that meiotic sex chromosome inactivation is essential for male fertility. Current biology : CB. 2010;20:2117–2123. doi: 10.1016/j.cub.2010.11.010. [DOI] [PubMed] [Google Scholar]
- Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome research. 2007;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimenti J. Synapsis or silence. Nat Genet. 2005;37:11–13. doi: 10.1038/ng0105-11. [DOI] [PubMed] [Google Scholar]
- Shiu PK, Raju NB, Zickler D, Metzenberg RL. Meiotic silencing by unpaired DNA. Cell. 2001;107:905–916. doi: 10.1016/s0092-8674(01)00609-2. [DOI] [PubMed] [Google Scholar]
- Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nature reviews Molecular cell biology. 2011;12:246–258. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- Song JJ, Liu J, Tolia NH, Schneiderman J, Smith SK, Martienssen RA, Hannon GJ, Joshua-Tor L. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat Struct Biol. 2003;10:1026–1032. doi: 10.1038/nsb1016. [DOI] [PubMed] [Google Scholar]
- Song R, Hennig GW, Wu Q, Jose C, Zheng H, Yan W. Male germ cells express abundant endogenous siRNAs. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:13159–13164. doi: 10.1073/pnas.1108567108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song R, Ro S, Michaels JD, Park C, McCarrey JR, Yan W. Many X-linked microRNAs escape meiotic sex chromosome inactivation. Nature genetics. 2009;41:488–493. doi: 10.1038/ng.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner FA, Plasterk RH. Knocking out the Argonautes. Cell. 2006;127:667–668. doi: 10.1016/j.cell.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Saga Y. Nanos2 suppresses meiosis and promotes male germ cell differentiation. Genes & Development. 2008;22:430–435. doi: 10.1101/gad.1612708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Nozaki M, Takeda N, Shinkai Y. Functional dynamics of H3K9 methylation during meiotic prophase progression. The EMBO journal. 2007;26:3346–3359. doi: 10.1038/sj.emboj.7601767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong MH, Mitchell D, Evanoff R, Griswold MD. Expression of Mirlet7 family microRNAs in response to retinoic acid-induced spermatogonial differentiation in mice. Biology of Reproduction. 2011;85:189–197. doi: 10.1095/biolreprod.110.089458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JM. Meiotic sex chromosome inactivation. Development. 2007;134:1823–1831. doi: 10.1242/dev.000018. [DOI] [PubMed] [Google Scholar]
- Turner JM, Aprelikova O, Xu X, Wang R, Kim S, Chandramouli GV, Barrett JC, Burgoyne PS, Deng CX. BRCA1, histone H2AX phosphorylation, and male meiotic sex chromosome inactivation. Curr Biol. 2004;14:2135–2142. doi: 10.1016/j.cub.2004.11.032. [DOI] [PubMed] [Google Scholar]
- Turner JM, Mahadevaiah SK, Fernandez-Capetillo O, Nussenzweig A, Xu X, Deng CX, Burgoyne PS. Silencing of unsynapsed meiotic chromosomes in the mouse. Nat Genet. 2005;37:41–47. doi: 10.1038/ng1484. [DOI] [PubMed] [Google Scholar]
- van Pelt AM, de Rooij DG. Synchronization of the seminiferous epithelium after vitamin A replacement in vitamin A-deficient mice. Biology of Reproduction. 1990;43:363–367. doi: 10.1095/biolreprod43.3.363. [DOI] [PubMed] [Google Scholar]
- van Wolfswinkel JC, Ketting RF. The role of small non-coding RNAs in genome stability and chromatin organization. Journal of Cell Science. 2010;123:1825–1839. doi: 10.1242/jcs.061713. [DOI] [PubMed] [Google Scholar]
- Vigodner M. Sumoylation precedes accumulation of phosphorylated H2AX on sex chromosomes during their meiotic inactivation. Chromosome research : an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology. 2009;17:37–45. doi: 10.1007/s10577-008-9006-x. [DOI] [PubMed] [Google Scholar]
- Wang H, Zhang X, Liu J, Kiba T, Woo J, Ojo T, Hafner M, Tuschl T, Chua NH, Wang XJ. Deep sequencing of small RNAs specifically associated with Arabidopsis AGO1 and AGO4 uncovers new AGO functions. The Plant journal : for cell and molecular biology. 2011 doi: 10.1111/j.1365-313X.2011.04594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Tilly JL. Epigenetic status determines germ cell meiotic commitment in embryonic and postnatal mammalian gonads. Cell Cycle. 2010;9:339–349. doi: 10.4161/cc.9.2.10447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Takeda A, Tsukiyama T, Mise K, Okuno T, Sasaki H, Minami N, Imai H. Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes & development. 2006;20:1732–1743. doi: 10.1101/gad.1425706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss RS, Matsuoka S, Elledge SJ, Leder P. Hus1 acts upstream of chk1 in a mammalian DNA damage response pathway. Current biology : CB. 2002;12:73–77. doi: 10.1016/s0960-9822(01)00626-1. [DOI] [PubMed] [Google Scholar]
- Yabuta Y, Kurimoto K, Ohinata Y, Seki Y, Saitou M. Gene expression dynamics during germline specification in mice identified by quantitative single-cell gene expression profiling. Biology of Reproduction. 2006;75:705–716. doi: 10.1095/biolreprod.106.053686. [DOI] [PubMed] [Google Scholar]
- Yang F, Eckardt S, Leu NA, McLaughlin KJ, Wang PJ. Mouse TEX15 is essential for DNA double-strand break repair and chromosomal synapsis during male meiosis. The Journal of Cell Biology. 2008;180:673–679. doi: 10.1083/jcb.200709057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.