Abstract
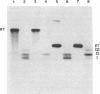
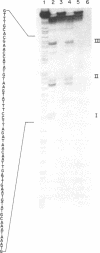
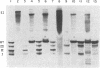
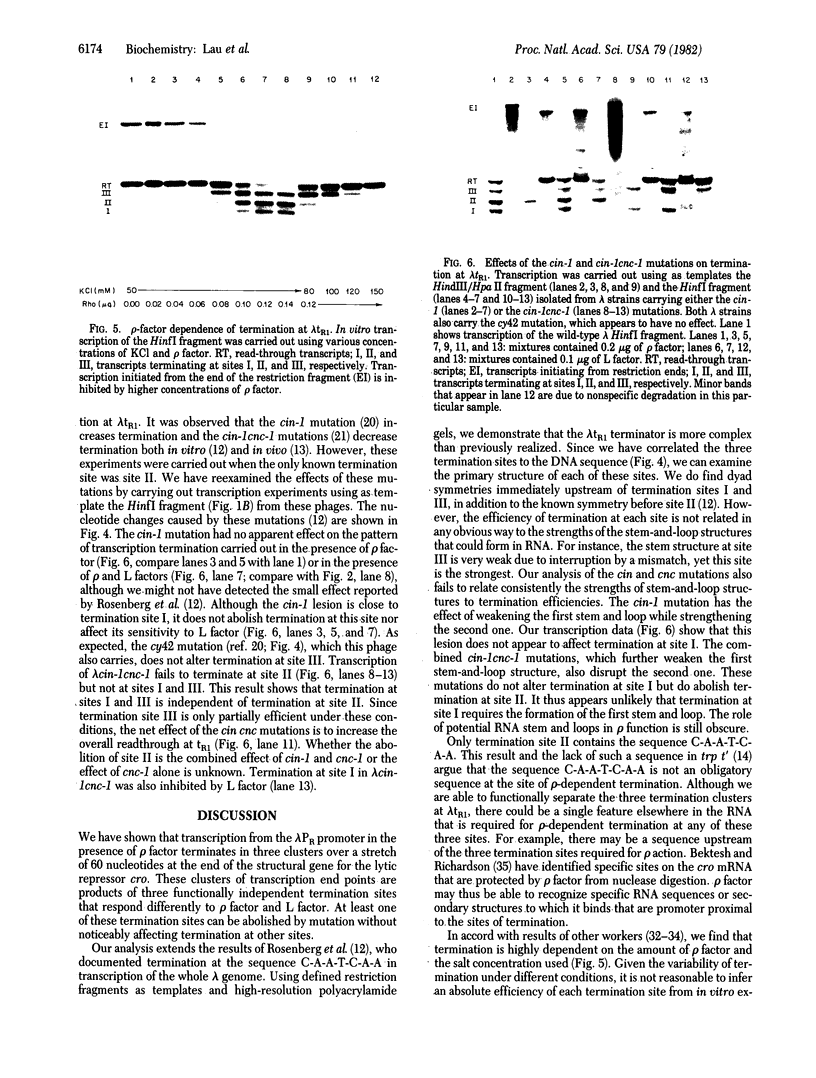
Transcription of the rightward operon of bacteriophage lambda terminates in the presence of rho factor at a region known as tR1, located downstream of the structural gene for the lytic repressor cro. We demonstrate by nuclease Sl mapping that transcription termination at lambda tR1 occurs in vitro over a stretch of 60 nucleotides. End points of transcripts are clustered in three distinct regions, which we refer to as termination sites I, II, and III. Termination at site I is inhibited by L factor, whereas termination at sites II and III is not affected by L factor. The sensitivities of these sites to rho factor are in the order III greater than II greater than I. The cin-lcnc-l mutations abolish termination at site II but not at sites I and III; this result may explain the failure of these mutations to alleviate the phage requirement for N function for growth. Although possible stem-and-loop structures in the RNA transcript immediately upstream of each of the three termination sites can be found, no consistent correlation exists between the strengths of these stem-and-loop structures and the termination efficiencies of their respective sites.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adhya S., Gottesman M. Control of transcription termination. Annu Rev Biochem. 1978;47:967–996. doi: 10.1146/annurev.bi.47.070178.004535. [DOI] [PubMed] [Google Scholar]
- Bektesh S. L., Richardson J. P. A rho-recognition site on phage lambda cro-gene mRNA. Nature. 1980 Jan 3;283(5742):102–104. doi: 10.1038/283102a0. [DOI] [PubMed] [Google Scholar]
- Burgess R. R., Jendrisak J. J. A procedure for the rapid, large-scall purification of Escherichia coli DNA-dependent RNA polymerase involving Polymin P precipitation and DNA-cellulose chromatography. Biochemistry. 1975 Oct 21;14(21):4634–4638. doi: 10.1021/bi00692a011. [DOI] [PubMed] [Google Scholar]
- Butler B., Echols H. Regulation of bacteriophage lambda development by gene N: properties of a mutation that bypasses N control of late protein synthesis. Virology. 1970 Feb;40(2):212–222. doi: 10.1016/0042-6822(70)90396-x. [DOI] [PubMed] [Google Scholar]
- Calva E., Burgess R. R. Characterization of a rho-dependent termination site within the cro gene of bacteriophage lambda. J Biol Chem. 1980 Nov 25;255(22):11017–11022. [PubMed] [Google Scholar]
- Calva E., Rosenvold E. C., Szybalski W., Burgess R. R. Analysis of the in vitro synthesis of 5'-gamma-32P-labeled transcripts from coliphage lambda by gel electrophoresis, RNA-DNA hybridization, and RNase T1 digestion. J Biol Chem. 1980 Nov 25;255(22):11011–11016. [PubMed] [Google Scholar]
- Christie G. E., Farnham P. J., Platt T. Synthetic sites for transcription termination and a functional comparison with tryptophan operon termination sites in vitro. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4180–4184. doi: 10.1073/pnas.78.7.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court D., Brady C., Rosenberg M., Wulff D. L., Behr M., Mahoney M., Izumi S. U. Control of transcription termination: a rho-dependent termination site in bacteriophage lambda. J Mol Biol. 1980 Apr;138(2):231–254. doi: 10.1016/0022-2836(80)90285-5. [DOI] [PubMed] [Google Scholar]
- Favaloro J., Treisman R., Kamen R. Transcription maps of polyoma virus-specific RNA: analysis by two-dimensional nuclease S1 gel mapping. Methods Enzymol. 1980;65(1):718–749. doi: 10.1016/s0076-6879(80)65070-8. [DOI] [PubMed] [Google Scholar]
- Finger L. R., Richardson J. P. Procedure for purification of Escherichia coli ribonucleic acid synthesis termination protein rho. Biochemistry. 1981 Mar 17;20(6):1640–1645. doi: 10.1021/bi00509a036. [DOI] [PubMed] [Google Scholar]
- Grayhack E. J., Roberts J. W. The phage lambda Q gene product: activity of a transcription antiterminator in vitro. Cell. 1982 Sep;30(2):637–648. doi: 10.1016/0092-8674(82)90260-4. [DOI] [PubMed] [Google Scholar]
- Greenblatt J., Li J., Adhya S., Friedman D. I., Baron L. S., Redfield B., Kung H. F., Weissbach H. L factor that is required for beta-galactosidase synthesis is the nusA gene product involved in transcription termination. Proc Natl Acad Sci U S A. 1980 Apr;77(4):1991–1994. doi: 10.1073/pnas.77.4.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt J., McLimont M., Hanly S. Termination of transcription by nusA gene protein of Escherichia coli. Nature. 1981 Jul 16;292(5820):215–220. doi: 10.1038/292215a0. [DOI] [PubMed] [Google Scholar]
- Greenblatt J. Regulation of transcription termination by the N gene protein of bacteriophage lambda. Cell. 1981 Apr;24(1):8–9. doi: 10.1016/0092-8674(81)90495-5. [DOI] [PubMed] [Google Scholar]
- Herskowitz I., Hagen D. The lysis-lysogeny decision of phage lambda: explicit programming and responsiveness. Annu Rev Genet. 1980;14:399–445. doi: 10.1146/annurev.ge.14.120180.002151. [DOI] [PubMed] [Google Scholar]
- Hopkins N. Bypassing a positive regulator: isolation of a lambda mutant that does not require N product to grow. Virology. 1970 Feb;40(2):223–229. doi: 10.1016/0042-6822(70)90397-1. [DOI] [PubMed] [Google Scholar]
- Kung H., Spears C., Weissbach H. Purification and properties of a soluble factor required for the deoxyribonucleic acid-directed in vitro synthesis of beta-galactosidase. J Biol Chem. 1975 Feb 25;250(4):1556–1562. [PubMed] [Google Scholar]
- Küpper H., Sekiya T., Rosenberg M., Egan J., Landy A. A rho-dependent termination site in the gene coding for tyrosine tRNA su3 of Escherichia coli. Nature. 1978 Mar 30;272(5652):423–428. doi: 10.1038/272423a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe P. A., Hager D. A., Burgess R. R. Purification and properties of the sigma subunit of Escherichia coli DNA-dependent RNA polymerase. Biochemistry. 1979 Apr 3;18(7):1344–1352. doi: 10.1021/bi00574a034. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McDermit M., Pierce M., Staley D., Shimaji M., Shaw R., Wulff D. L. Mutations masking the lambda cin-1 mutation. Genetics. 1976 Mar 25;82(3):417–422. doi: 10.1093/genetics/82.3.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt T. Termination of transcription and its regulation in the tryptophan operon of E. coli. Cell. 1981 Apr;24(1):10–23. doi: 10.1016/0092-8674(81)90496-7. [DOI] [PubMed] [Google Scholar]
- Roberts J. W. Termination factor for RNA synthesis. Nature. 1969 Dec 20;224(5225):1168–1174. doi: 10.1038/2241168a0. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D., Shimatake H., Brady C., Wulff D. L. The relationship between function and DNA sequence in an intercistronic regulatory region in phage lambda. Nature. 1978 Mar 30;272(5652):414–423. doi: 10.1038/272414a0. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Weissman S., deCrombrugghe B. Termination of transcription in bacteriophage lambda. Heterogeneous, 3'-terminal oligo-adenylate additions and the effects of rho factor. J Biol Chem. 1975 Jun 25;250(12):4755–4764. [PubMed] [Google Scholar]
- Rossi J., Egan J., Hudson L., Landy A. The tyrT locus: termination and processing of a complex transcript. Cell. 1981 Nov;26(3 Pt 1):305–314. doi: 10.1016/0092-8674(81)90199-9. [DOI] [PubMed] [Google Scholar]
- Schwarz E., Scherer G., Hobom G., Kössel H. Nucleotide sequence of cro, cII and part of the O gene in phage lambda DNA. Nature. 1978 Mar 30;272(5652):410–414. doi: 10.1038/272410a0. [DOI] [PubMed] [Google Scholar]
- Treisman R., Kamen R. Structure of polyoma virus late nuclear RNA. J Mol Biol. 1981 May 25;148(3):273–301. doi: 10.1016/0022-2836(81)90539-8. [DOI] [PubMed] [Google Scholar]
- Wu A. M., Christie G. E., Platt T. Tandem termination sites in the tryptophan operon of Escherichia coli. Proc Natl Acad Sci U S A. 1981 May;78(5):2913–2917. doi: 10.1073/pnas.78.5.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R. Nucleotide sequence analysis of DNA. I. Partial sequence of the cohesive ends of bacteriophage lambda and 186 DNA. J Mol Biol. 1970 Aug;51(3):501–521. doi: 10.1016/0022-2836(70)90004-5. [DOI] [PubMed] [Google Scholar]
- Wulff D. L. Lambda cin-1, a new mutation which enhances lysogenization by bacteriophage lambda, and the genetic structure of the lambda cy region. Genetics. 1976 Mar 25;82(3):401–416. doi: 10.1093/genetics/82.3.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky C. Attenuation in the control of expression of bacterial operons. Nature. 1981 Feb 26;289(5800):751–758. doi: 10.1038/289751a0. [DOI] [PubMed] [Google Scholar]