Abstract
Background
Lynch syndrome is caused by germline mismatch repair (MMR) gene mutations. The PREMM1,2,6 model predicts the likelihood of a MMR gene mutation based on personal and family cancer history.
Objective
To compare strategies using PREMM1,2,6 and tumour testing (microsatellite instability (MSI) and/or immunohistochemistry (IHC) staining) to identify mutation carriers.
Design
Data from population-based or clinic-based patients with colorectal cancers enrolled through the Colon Cancer Family Registry were analysed. Evaluation included MSI, IHC and germline mutation analysis for MLH1, MSH2, MSH6 and PMS2. Personal and family cancer histories were used to calculate PREMM1,2,6 predictions. Discriminative ability to identify carriers from non-carriers using the area under the receiver operating characteristic curve (AUC) was assessed. Predictions were based on logistic regression models for (1) cancer assessment using PREMM1,2,6, (2) MSI, (3) IHC for loss of any MMR protein expression, (4) MSI + IHC, (5) PREMM1,2,6 + MSI, (6) PREMM1,2,6 + IHC, (7) PREMM1,2,6 + IHC + MSI.
Results
Among 1651 subjects, 239 (14%) had mutations (90 MLH1, 125 MSH2, 24 MSH6). PREMM1,2,6 discriminated well with AUC 0.90 (95% CI 0.88 to 0.92). MSI alone, IHC alone, or MSI + IHC each had lower AUCs: 0.77, 0.82 and 0.82, respectively. The added value of IHC + PREMM1,2,6 was slightly greater than PREMM1,2,6 + MSI (AUC 0.94 vs 0.93). Adding MSI to PREMM1,2,6 + IHC did not improve discrimination.
Conclusion
PREMM1,2,6 and IHC showed excellent performance in distinguishing mutation carriers from non-carriers and performed best when combined. MSI may have a greater role in distinguishing Lynch syndrome from other familial colorectal cancer subtypes among cases with high PREMM1,2,6 scores where genetic evaluation does not disclose a MMR mutation.
INTRODUCTION
Lynch syndrome is the most common form of hereditary colorectal cancer (CRC) and accounts for up to 5% of all CRC cases.1 The autosomal dominant syndrome is characterised by CRC diagnosed at an early age and a predilection to the proximal colon, synchronous and metachronous CRCs, accelerated carcinogenesis and an increased risk for numerous extracolonic neoplasms.2
Historically, a number of clinical criteria pertaining to personal and family history of cancer have been employed to identify an individual’s risk for Lynch syndrome and to determine for whom further genetic evaluation is warranted. The original Amsterdam criteria were developed primarily for research purposes to provide consistency among studies but are too stringent for clinical purposes. With the advent of molecular tumour testing, the Bethesda guidelines were established to select those patients with CRC who should undergo microsatellite instability (MSI) analysis, a hallmark feature associated with a defective DNA mismatch repair system. Immunohistochemical (IHC) analysis of tumours for the expression of MMR gene proteins associated with Lynch syndrome is another method to help select individuals who should undergo germline genetic testing and has been advocated as a strategy that should be applied to all newly diagnosed CRC cases.3 Finally, a number of prediction models have recently been developed that rely on a combination of personal and family cancer history to identify patients and families who should undergo genetic evaluation.4–6 Many studies have been conducted to determine an optimal strategy for identifying patients with Lynch syndrome and the majority focused on comparing clinical criteria with molecular diagnostic testing results.4,7–12 However, it is not known how the recently developed prediction models compare with MSI testing and IHC staining in identifying MMR gene mutation carriers.
Our goal was to compare the performance of the PREMM1,2,6 model (http://www.dfci.org/premm) with MSI and IHC tumour testing conducted in patients with CRC with possible Lynch syndrome recruited through the Colon Cancer Family Registry (CCFR). We also evaluated the model’s ability to discriminate gene mutation carriers from non-carriers when MSI and/or IHC are combined in a stepwise manner with the prediction estimates provided by the PREMM1,2,6 model.
METHODS
Study population
We analysed data from 1868 unrelated subjects recruited for the CCFR. The study subjects enrolled in one of six registry centres, including the University of Hawaii (Honolulu, Hawaii, USA), Fred Hutchinson Cancer Research Center (Seattle, Washington, USA), Mayo Clinic (Rochester, Minnesota, USA), University of Southern California Consortium (Los Angeles, California, USA), Cancer Care Ontario (Toronto, Canada) and University of Melbourne (Melbourne, Australia). Families were ascertained through population-based cancer registries (population-based) and high-risk clinics (clinic-based). Some centres recruited all cases of CRC, whereas others oversampled cases with a family history or early age of diagnosis. Standardised procedures were used to collect epidemiological data, blood samples, tumour blocks and pathology reports from patients. Detailed information about the CCFR can be found at http://epi.grants.cancer.gov/CFR/.13 All participants provided informed consent for inclusion in the CCFR through each registry centre which was approved by the institutional review board at each of the institutions.
Classification of personal and family history
Personal and family history data were provided by study participants during an interview. Through standardised procedures implemented by the six registries, family history information was verified by comparing reports from multiple individuals within the same family, when possible. Where available, medical records, death certificates, pathology reports and tumour tissues were also used to confirm reported cancer diagnoses. The six centres used standardised protocols to collect detailed personal/family histories, clinical data and related tumour and blood specimens for analyses, and eligible patients consented to participate in research.
Microsatellite instability testing
MSI was evaluated using a panel of 10 markers (BAT25, BAT26, BAT40, MYCL, D5S346, D17S250, ACTC, D18S55, D10S197, BAT34C4) using standard techniques.13–15 Results were required for at least four markers to determine MSI status. Tumours were deemed MSI-H if instability was seen at ≥30% of markers, MSI-L if >0 and <30% of markers were instable, and MSS if all markers were stable. Results of MSI testing were dichotomised in this study as MSI (includes MSI-H cases) and MSS (includes MSS and MSI-L cases).
Immunohistochemistry testing
IHC testing for MLH1, MSH2, MSH6 and PMS2 proteins was done as previously described.13–15 All subjects with MSI-H or MSI-L tumours had IHC testing regardless of recruitment method. Because of the low frequency of absent protein staining in MSS cases, some CCFR centres did not perform IHC testing on all MSS cases.14 Staining was classified as absent, present, or inconclusive. The IHC results were dichotomised for the purposes of this study as absent versus present/inconclusive.
MMR mutation data
MMR mutation data analysis was conducted in all probands with abnormal molecular tumour testing, including high or low levels of MSI or loss of normal protein expression on IHC. MMR testing was performed in all probands recruited through any of the clinic-based CCFR registries regardless of molecular tumour testing results. Mutations in MSH2 and MLH1 were detected using a combined approach of denaturing high-pressure liquid chromatography/direct sequencing and multiplex ligation-dependent probe amplification (MLPA). Direct sequencing was used to detect MSH6 mutations in cases with no immunohistochemical staining of MSH6. In addition, large genomic rearrangements were analysed by MLPA analysis. PMS2 mutations were evaluated in patients from four of the CFR centres (Australia, Seattle, Mayo and Ontario) as previously described.12,13 For this analysis, we focus on the gene mutations that are considered to have a clearly deleterious effect based on current evidence, specifically those with (a) changes known or predicted to truncate protein production, including frame shift and nonsense variants, (b) splice site mutations occurring within two base pairs of an intron/exon boundary and (c) missense changes that have been shown to have a deleterious effect.
Derivation of PREMM1,2,6 scores
Detailed clinical information necessary for generating the PREMM1,2,6 score was extracted from the CCFR database for each study participant (proband) and their family members. The following data were used to derive a unique PREMM1,2,6 score for each study participant: (1) proband-specific variables, including gender, occurrence and age of CRC, endometrial and/or other Lynch syndrome-associated cancer diagnoses; extracolonic cancers, including those of the ovary, stomach, kidney, ureter, bile duct, small bowel, brain (glioblastoma multiforme), pancreas, or sebaceous glands; (2) family history related variables, including number of relatives with CRC, endometrial cancer, or other Lynch syndrome-associated cancers; relationship to proband (first- vs second-degree); minimum age at diagnosis of each cancer in the family.
This study was reviewed and approved by the Dana–Farber/Harvard Cancer Center Institutional Review Board. A waiver of consent for study participants was obtained because the analyses were performed on de-identified data and did not require patient contact.
Statistical analyses
Univariate analyses were used to compare personal and family history of cancer, cancer types and ages at diagnosis by gene mutation for probands and relatives. Age was truncated at the lower and upper one centile for ages of CRC and endometrial cancer diagnoses.16 To test for differences among carriers by type of mutated gene, one-way analysis of variance was used. A two-sided p value of <0.05 was considered statistically significant. Analyses were conducted for the entire patient population and then stratified by ascertainment (into population-based and clinic-based).
Seven testing strategies were compared using the area under the receiver operating characteristic curve (AUC). Predictions were based on a series of logistic regression analyses with the presence of any deleterious gene mutation as the outcome variable. The logistic regression models included the log odds of the PREMM1,2,6 risk and the MSI and IHC results as dichotomous variables in various combinations: (1) cancer history assessment using PREMM1,2,6 alone, (2) MSI testing alone (high vs low/stable), (3) IHC testing alone for any loss of MLH1, MSH2, MSH6 and/or PMS2 protein expression (abnormal vs normal/inconclusive), (4) MSI + IHC, (5) PREMM1,2,6 + MSI, (6) PREMM1,2,6 + IHC and (7) PREMM1,2,6 + MSI + IHC. Seven AUCs were generated to distinguish between gene mutation carriers and non-carriers. Only subjects who had undergone clinical genetic testing and had results from tumour molecular testing (IHC and/or MSI) were included in these analyses. Of the 1868 probands enrolled, 217 were excluded because they lacked any results from tumour molecular testing, leaving 1651 subjects available for the analyses. Additional analyses assessed the effect of enrolling site on each strategy’s performance, since each CCFR site had different recruitment criteria, particularly for population-based CRC cases.
To determine the incremental value provided by tumour molecular testing results (IHC and/or MSI) in addition to the estimated probability of mutation carrier status provided by the PREMM1,2,6 model, net reclassification improvement analysis was used. We extended the PREMM1,2,6 model with the inclusion of tumour molecular testing results to determine the additional predictive value when probands were reclassified based on <5%, 5–10% and >10% cut-off points for the risk of carrying a MMR gene mutation. These cut-off points have been previously reported but lack formal support (ie, from cost-effectiveness analyses).4,11 We therefore also considered reclassification over the whole range of possible cut-off values.17
The statistical analysis conducted for this study included SAS statistical software (V.9.1; SAS Institute, Inc) for data management and univariate analysis and SPSS (V.16, SPSS, Inc) and R software (version 2.8.0, R Foundation for Statistical Computing, Vienna, Austria) for the multivariable analysis, validation with reclassification analyses and imputation of missing values. Missing values for molecular tumour testing results were common: 264 subjects had no MSI testing (16%) and IHC was not performed in 75 (4.5%), 59 (3.6%), 203 (12.3%) and 743 (45%) for loss of MLH1, MSH2, MSH6 and/or PMS2 protein expression (abnormal vs normal/inconclusive), respectively. Discarding subjects with incomplete information would have led to inefficient and potentially biased analyses.17 We therefore imputed molecular tumour testing data based on correlations among variables included in the PREMM1,2,6 model, available tumour MSI and IHC results, mutation status and enrolling site. Multiple imputation was used (aregImpute procedure in R) to properly account for missing data uncertainty.18 Five imputed datasets were each analysed to obtain AUC estimates, which were then combined into one overall estimate and variance, incorporating both the within- and between-imputation variability in accordance with Rubin’s rules.18
RESULTS
Data were analysed from 1651 subjects for whom any tumour molecular testing results were available. Of these subjects, 1181 (72%) were population-based cases and 470 (28%) were ascertained through high-risk clinics. In total, 14% (239/1651) of subjects had pathogenic MMR gene mutations with 90 MLH1, 125 MSH2 and 24 MSH6 gene mutation carriers identified (table 1). Sixty-seven per cent (159/239) of all gene mutation carriers were recruited through high-risk clinics.
Table 1.
Characteristics of the study population
| Characteristics | Total cases (N= 1651), N (%) |
Population-based cases (N = 1181), N (%) |
Clinic-based cases (N = 470), N (%) |
|---|---|---|---|
| Age at last follow-up, median (years) | 63 (range 23–93) | 65.0 (range 23–88) | 58 (range 28–93) |
| Sex | |||
| Female | 840 (51.9) | 595 (51.5) | 245 (52.9) |
| Male | 779 (48.1) | 561 (48.5) | 218 (47.1) |
| Missing | 32 (–) | 25 | 7 |
| Clinical criteria | |||
| Amsterdam I | 246 (15.0) | 114 (9.7) | 132 (29.0) |
| Amsterdam II | 306 (18.7) | 136 (11.5) | 170 (37.3) |
| Revised Bethesda guidelines | 1088 (65.9) | 681 (57.7) | 407 (86.6) |
| Pathogenic MMR gene mutations | |||
| Total | 239 (14.5) | 80 (6.8) | 159 (33.8) |
| MLH1 | 90 (5.4) | 27 (2.3) | 63 (13.4) |
| MSH2 | 125 (7.6) | 43 (3.6) | 82 (17.4) |
| MSH6 | 24 (1.5) | 10 (0.9) | 14 (3.0) |
MMR, mismatch repair.
MSI results were available for 1387 cases included in this analysis (table 2). Overall, 155/239 (65%) of gene mutation carriers with CRC had undergone MSI tumour testing. MSI testing had a sensitivity of 94% (146/155 had MSI-H tumours) but specificity was only 66% (810/1232). Among the 1232 non-carriers with available MSI testing, 422 (34%) had MSI-H, 330 (27%) had MSI-L and 480 (39%) had MSS tumours. Of all 1387 tumours cases tested for MSI, 819 were MSS and of these, 40 (5%) did not have IHC testing performed.
Table 2.
MSI and IHC results among probands who had MSI and/or IHC testing
| Molecular tumour characteristics |
Total N (%) |
No mutation N (%) |
MLH1 N (%) |
MSH2 N (%) |
MSH6 N (%) |
|---|---|---|---|---|---|
| Total | 1651 (100) | 1412 (85.5) | 90 (5.5) | 125 (7.6) | 24 (1.5) |
| MSI result | |||||
| Stable/low* | 819 (59.0) | 810 (65.8) | 2 (3.2) | 6 (7.9) | 1 (6.2) |
| High | 568 (41.0) | 422 (34.2) | 61 (96.8) | 70 (92.1) | 15 (93.8) |
| Total | 1387 (100) | 1232 (100) | 63 (100) | 76 (100) | 16 (100) |
| IHC resulty† | |||||
| MLH1 (N=1576) | 366 (23.2) | 288/1349 (21.3) | 78/84‡ (92.8) | 0 (0) | 0 (0) |
| MSH2 (N=1592) | 170 (10.7) | 54/1360 (4.0) | 0 (0) | 116/121§ (95.9) | 0 (0) |
| MSH6 (N=1448) | 165 (11.4) | 53/1234 (4.3) | 0 (0) | 95/108 (87.9) | 17/24 (70.8) |
| PMS2 (N=908) | 257 (28.3) | 196/748 (26.2) | 61/62 (98.4) | 0 (0) | 0 (0) |
MSI-low: total=335, no Mutation=330, MLH1=0, MSH2=5, MSH6=0.
Abnormal IHC results shown (based on abnormal versus normal/inconclusive categories).
Of 90 MLH1 mutation carriers, 84 had IHC testing for MLH1 protein expression.
Of 125 MSH2 mutation carriers, 121 had IHC testing for MSH2 protein expression.
IHC, immunohistochemistry; MSI, microsatellite instability.
MLH1, MSH2, MSH6 and PMS2 IHC testing results were available for 1576, 1592, 1448 and 908 subjects, respectively. Among the 84 subjects with pathogenic MLH1 gene mutations and available IHC results, 78 (93%) had loss of MLH1 protein expression (84% in the population-based group and 97% in the clinic-based group). Among MSH2 gene mutation carriers with available IHC results, 116/121 (96%) had tumours with loss of MSH2 protein expression, with similar results noted in both population- and clinic-based cases. MSH6 gene mutation carriers were less likely to demonstrate loss of MSH6 protein expression in their tumours: 17/24 (71%) in the overall study group with similar IHC results among population- and clinic-based cases. There were a limited number of inconclusive IHC testing results: 33/1576 (2%) for MLH1, 16/1592 (1%) for MSH2 and 47/1448 (3%) for MSH6.
Of the 1651 subjects analysed, 815 (49%) had PREMM1,2,6 scores of >5%, which captured 226 of the 239 (95%) identified gene mutation carriers (table 3). The mean PREMM1,2,6 score for the overall cohort was 14.5% with a mean score of 48% for gene mutation carriers and 9% for subjects without a MMR gene mutation. Compared with cases ascertained through high-risk clinics, population-based cases had a lower mean PREMM1,2,6 score for the overall cohort and among non-carriers at 8.5% and 6%, respectively (table 3). Of the 13 gene mutation carriers with a score of <5%, three were MLH1 gene mutation carriers, six were MSH2 gene mutation carriers and four were MSH6 mutation carriers. There was an almost equal number of gene mutation carriers missed using a 5% cut-off point among both clinic and population-based cases (seven vs six mutation carriers, respectively). The probability of carrying a MMR gene mutation based on varying PREMM1,2,6 cut-off scores, stratified by type of mutated gene is shown in table 1 of the online supplementary data. The positive predictive value of PREMM1,2,6 >5%, MSI and any IHC testing was 28%, 25% and 23%, respectively. The PREMM1,2,6 model discriminated well at distinguishing mutation carriers versus non-carriers with AUC of 0.90 (95% CI 0.88 to 0.92; table 4). The AUCs of MSI alone, IHC alone, or MSI + IHC molecular tumour testing were 0.77, 0.82 and 0.82, respectively. The added value of IHC to PREMM1,2,6 was slightly larger than that for adding MSI: 0.94 vs 0.93. No statistically significant difference was noted for MSI (p=0.33) in the comparison of PREMM1,2,6 + IHC + MSI versus PREMM1,2,6 + IHC (AUC 0.94 vs 0.94). The performance of each strategy was also compared based on method of ascertainment (table 4). The molecular tumour strategy which most improved the discriminative ability of the PREMM model in both population- and clinic-based cases was the addition of IHC tumour testing. There was limited additional value of MSI to the combination of PREMM + IHC. Stratifying for CCFR site of recruitment did not alter the results.
Table 3.
Probability of a MMR gene mutation according to PREMM1,2,6, by ascertainment
| Characteristics | Total N (%) |
No mutation N (%) |
With mutation N (%) |
|---|---|---|---|
| Overall | |||
| PREMM1,2,6 (N=1651) | |||
| <5% | 836 (50.6) | 823 (58.3) | 13 (5.4) |
| 5–9% | 331 (20.1) | 306 (21.7) | 25 (10.5) |
| 10–19% | 174 (10.5) | 151 (10.7) | 23 (9.6) |
| 20–29% | 68 (4.1) | 44 (3.1) | 24 (10.0) |
| 30–39% | 53 (3.2) | 28 (2.0) | 25 (10.5) |
| ≥40% | 189 (11.5) | 60 (4.2) | 129 (54.0) |
| Total | 1651 (100.0) | 1412 (100.0) | 239 (100.0) |
| Mean score (±SD) | 14.4 (±22.4) | 8.60 (±13.7) | 48.4 (±31.7) |
| Population-based | |||
| PREMM1,2,6 (N=1181) | |||
| <5% | 738 (62.5) | 731 (66.4) | 7 (8.8) |
| 5–9% | 239 (20.2) | 225 (20.4) | 14 (17.5) |
| 10–19% | 99 (8.4) | 92 (8.4) | 7 (8.7) |
| 20–29% | 31 (2.6) | 20 (1.8) | 11 (13.7) |
| 30–39% | 18 (1.5) | 10 (0.9) | 8 (10) |
| ≥40% | 56 (4.8) | 23 (2.1) | 33 (41.3) |
| Total | 1181 (100.0) | 1101 (100.0) | 80 (100.0) |
| Mean score (±SD) | 8.5 (±22.4) | 6.3 (±10.3) | 38.7 (±30.3) |
| Clinic-based | |||
| PREMM1,2,6 (N=470) | |||
| <5% | 98 (20.8) | 92 (29.6) | 6 (3.8) |
| 5–9% | 92 (19.6) | 81 (26.0) | 11 (6.9) |
| 10–19% | 75 (16.0) | 59 (19.0) | 16 (10.0) |
| 20–29% | 37 (7.9) | 24 (7.7) | 13 (8.2) |
| 30–39% | 35 (7.4) | 18 (5.8) | 17 (10.7) |
| ≥40% | 133 (28.3) | 37 (11.9) | 96 (60.4) |
| Total | 470 (100.0) | 311 (100.0) | 159 (100.0) |
| Mean score (±SD) | 29.2 (±29.8) | 16.8 (±19.9) | 53.3 (±31.4) |
MMR, mismatch repair.
Table 4.
Discriminative performance of MMR prediction strategies, stratified by ascertainment
| Strategy | Total cohort (N=1651), c-statistic |
Population-based (N=1181), c-statistic |
Clinic-based (N=470), c-statistic |
|---|---|---|---|
| PREMM1,2,6 | 0.90 (0.88 to 0.92) | 0.84 (0.81 to 0.88) | 0.88 (0.84 to 0.93) |
| MSI | 0.77 (0.74 to 0.79) | 0.82 (0.78 to 0.86) | 0.79 (0.76 to 0.82) |
| IHC | 0.82 (0.80 to 0.84) | 0.88 (0.85 to 0.90) | 0.79 (0.76 to 0.82) |
| MSI + IHC | 0.82 (0.80 to 0.84) | 0.90 (0.87 to 0.92) | 0.80 (0.78 to 0.83) |
| PREMM1,2,6 + MSI | 0.93 (0.92 to 0.95) | 0.92 (0.89 to 0.94) | 0.93 (0.89 to 0.96) |
| PREMM1,2,6 + IHC | 0.94 (0.92 to 0.96) | 0.94 (0.92 to 0.96) | 0.92 (0.88 to 0.96) |
| PREMM1,2,6 + MSI + IHC | 0.94 (0.93 to 0.96) | 0.94 (0.92 to 0.97) | 0.94 (0.89 to 0.96) |
IHC, immunohistochemistry; MSI, microsatellite instability.
The addition of IHC to PREMM1,2,6 reclassified 364 subjects with moderate or high probability as non-carriers, whereas PREMM1,2,6 alone classified 161 subjects as non-carriers that the extended PREMM1,2,6 + IHC model would have reclassified as moderate or high probability of carrying a MMR gene mutation. Overall, extending the prediction model with IHC increased the sensitivity of PREMM in identifying MMR gene mutation carriers by 9.2% and the specificity by 10%. The increase in sensitivity and specificity by adding MSI to predictions provided by PREMM1,2,6 + IHC was minor: + 0.4% and + 0.2%, respectively. Reclassification plots confirmed that MSI had limited additional value once IHC results were added to PREMM1,2,6.
Conversely, when using IHC alone as the initial strategy, the overall sensitivity among the 229 mutation carriers with available IHC results was 92%. When adding PREMM1,2,6 prediction to IHC testing, the sensitivity improved: 97.8% for IHC + PREMM1,2,6>5%, 96.1% for IHC + PREMM1,2,6>10% and 95.2% for IHC + PREMM1,2,6>15% and at higher cutoff points the sensitivity approached that of IHC alone. The sensitivity of IHC for MLH1, MSH2 and MSH6 protein loss individually was determined at various cut-off scores for PREMM1,2,6. A score of ≥15% improved sensitivity for all three IHC testing approaches: 92.9% to 94.3% for MLH1 loss in IHC, 95.9% to 98% for MSH2 loss in IHC and 70.8% to 77.8% for MSH6 loss in IHC. The specificity for IHC with the addition of PREMM1,2,6 prediction at a cut-off score of <5% also improved from 78.7% to 92.5% for MLH1, 96% to 97.4% for MSH2 and 95.7% to 97% for MSH6 gene mutations.
To determine whether the proband’s age at CRC diagnosis affects the ability of PREMM1,2,6, MSI and IHC in discriminating between mutation carriers and non-carriers, we examined each individual strategy stratified by age. The age categories assessed were for initial CRC diagnoses made by age 50, 60 and 70 years. The overall performance of IHC testing decreased with every 10-year increase in the age of CRC diagnosis: AUC of 0.85 for probands diagnosed by 50 years, 0.84 for CRC diagnosed by 60 years and 0.82 for CRC diagnoses by 70 years; the specificity of IHC testing with loss of MLH1 protein expression decreased from 90.4% to 87.1% to 82.6% for CRC cases diagnosed by 50, 60 and 70 years, respectively, while sensitivity increased from 93% to 94%. In contrast, the performance of PREMM1,2,6 increased slightly with every 10-year increase in age at time of CRC diagnosis: AUC of 0.87 to 0.88 to 0.90 for CRC diagnoses made by 50, 60 and 70 years, respectively. For MSI, the AUC was 0.83 for CRC cases diagnosed by ages 50 and 60 years but decreased to 0.81 for CRC cases diagnosed by 70 years.
DISCUSSION
There is active debate about the optimal and standard approach to screening for MMR deficiency in patients diagnosed with CRC. Although a variety of strategies have been advocated, including universal IHC on all CRCs,3 MSI testing based on clinical guidelines,19 comprehensive family history assessment for all newly diagnosed patients with CRC as well as healthy individuals,20,21 there is no standard approach and great variability exists in care practices in the USA and around the world. We compared a variety of strategies and found that all approaches can reasonably distinguish mutation carriers from non-carriers, each with its advantages and disadvantages.
The best approach, and one that is feasible for the vast majority of clinical practices, is the combination of personal and family history assessment using the PREMM1,2,6 model and IHC for the MLH1, MSH2, MSH6 and PMS2 proteins. The added value of IHC to PREMM1,2,6 was greater than that for PREMM1,2,6 + MSI testing. In addition, no incremental gain in discriminating mutation carriers from non-carriers was noted when adding MSI to the combined testing strategy of PREMM1,2,6 + IHC.
Personal and family history assessment have always been the cornerstone for the diagnosis of Lynch syndrome and a variety of clinical criteria exist to aid the clinician, including the Amsterdam criteria and the Bethesda guidelines,19,22,23 each with their respective sensitivities and specificities.24 However, recent studies have consistently shown that Lynch syndrome prediction models outperform the existing clinical criteria,4,11,25 and it is increasingly being suggested that the models should replace the existing clinical criteria as prescreening tools for identifying at-risk individuals for possible germline mutation testing.25–27 We elected to evaluate PREMM1,2,6 because we did not have all the data required to compute predictions using the other models, but our findings are probably generalisable to the other models as well. A comprehensive family history obtained through PREMM1,2,6 did a better job at distinguishing mutation carriers from non-carriers in the entire cohort than either MSI or IHC alone (AUC of 0.90 vs 0.77 and 0.82, respectively); when stratified by ascertainment it appears that all three modalities are equivalent in population-based cases, but the model performs better in the higher-risk clinic-based cases.
Unfortunately, computing risk predictions for all newly diagnosed CRCs may not be an achievable goal as family history assessments are known to be suboptimal in clinical practice,28,29 and therefore universal molecular analysis has also been advocated. MSI and IHC staining, either separately or in combination, are important intermediate instruments of screening for Lynch syndrome among patients with a personal diagnosis of CRC.30 Nevertheless, both techniques for molecular testing have some advantages and some limitations. Recent studies show a high interobserver variability among pathologists in interpreting cases with indefinite IHC staining.31,32 To ensure the highest accuracy attainable by molecular tumour testing, pathologists suggest that MMR protein immunostaining should be restricted to experienced gastrointestinal pathologists in specialised settings, which potentially limits the feasibility and/or accuracy of universal IHC on all newly diagnosed CRCs. Also, tumours from MSH6 gene mutation carriers may not display the MSI phenotype despite an inactive DNA MMR system and there are pathogenic missense mutations that do not completely abrogate protein expression, yielding false-negative results by IHC testing.33 In this study, 28% (7/24) of carriers with pathogenic MSH6 gene mutations had tumours which displayed normal protein expression on IHC staining. As expected, IHC tumour testing was more informative for MLH1 and MSH2 gene mutation carriers: 93% of MLH1 carriers had correlating loss of MLH1 protein expression and 96% of MSH2 carriers had loss of MSH2 protein expression. However, MSS or MSI-L tumours were noted in 3% and 8% of MLH1 and MSH2 gene mutation carriers, respectively. In addition, the ability of MSI and IHC testing to discern between gene mutation carriers and non-carriers also decreased among CRC cases diagnosed after the age of 60 years, probably because somatic hypermethylation of MLH1 contributes more to a microsatellite unstable tumour than a germline MMR gene mutation.
The combination strategy of PREMM1,2,6 and IHC offers several advantages. In high-risk families, where a high prediction score (≥15%) is obtained, further germline testing should be considered even if MSI and IHC are normal, owing to the possibility of false-negative results. Conversely, when IHC testing reveals aberrant MLH1 tumour staining but the personal and family history evaluation through PREMM1,2,6 yields a low score, this case probably represents a sporadic case of CRC due to hypermethylation of the MLH1 gene promoter and may not warrant germline analysis; alternative approaches to confirm this via promoter methylation studies or BRAF mutation analysis can be pursued first. In our study, 21% of tumours tested for MLH1 protein expression showed deficiency but no associated MLH1 germline mutation; the addition of a <5% PREMM1,2,6 score improved the specificity of MLH1 IHC testing from 79% to 93%.
What about the role of MSI? We found that MSI testing performed less well than either PREMM1,2,6 estimates or IHC alone and the combination of PREMM1,2,6 and IHC did not improve the distinction between mutation carriers and noncarriers in the overall cohort. However, MSI may in fact be helpful in the counselling of patients who are at high risk (PREMM1,2,6 ≥15%), but are not found to carry a mutation. In these cases, MSI analysis would help in categorising the family as an indeterminate negative if there is evidence of MSI (and should still be followed up as a Lynch syndrome family) versus a ‘familial colorectal cancer type X’ family if the tumour is microsatellite stable, where surveillance guidelines are not so arduous and extracolonic surveillance may not be necessary.34 In addition, MSI testing is increasingly being performed to help with treatment decision-making and prognosis of CRC and ultimately both MSI analysis and IHC may be deemed necessary for different purposes.
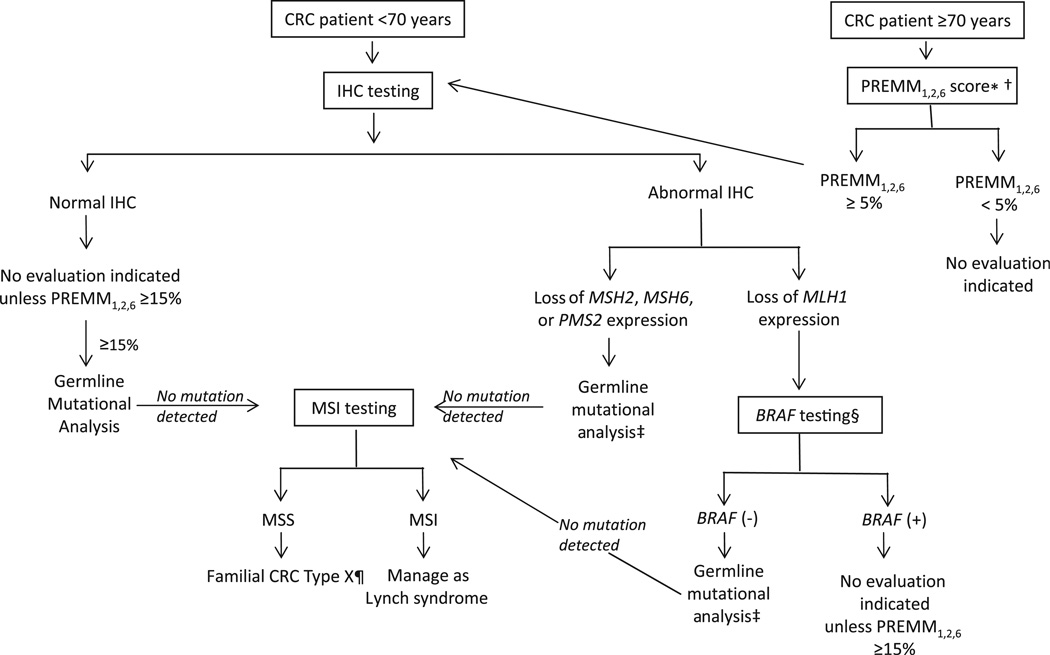
As there are benefits and limitations to each approach when used individually, we propose an algorithm for the systematic evaluation for Lynch syndrome in patients diagnosed with CRC based on the results of this study, which combines PREMM1,2,6 prediction, IHC and MSI (figure 1). Since our data demonstrate that specificity of IHC decreases significantly after age 70, presumably owing to MLH1 promoter hypermethylation, and the AUC of PREMM1,2,6 predictions remain high, we propose that evaluations are initially stratified by age of CRC diagnosis at ≥70 or <70 years. For those patients with CRC diagnosed at age <70 years, IHC testing is an acceptable first step. When tumours display loss of MLH1 protein expression on IHC testing, BRAF testing can be pursued unless there is a strong family cancer history (PREMM1,2,6≥15%) when direct germline mutational analysis should still be performed. Family cancer history is important in evaluating cases diagnosed at ages ≥70 years since those with PREMM1,2,6 scores <5% will be unlikely to have a germline mutation. This approach limits screening all CRC cases for Lynch syndrome through reflex IHC testing, but expands significantly the age cut-off point of 50 years proposed in the Bethesda guidelines. MSI testing should be reserved for those cases with (a) abnormal IHC tumour testing results but where no germline MMR gene mutation is detected or (b) normal IHC testing but high PREMM1,2,6 score of ≥15%.
Figure 1.
Proposed algorithm for systematic evaluation for Lynch syndrome in patients with colorectal cancer. *PREMM1,2,6 score can be calculated at website http://www.dfci.org/premm; †other models (MMRpro, MMRpredict) may also be used with their own specified cut-off scores; ‡gene-specific germline mutational analysis. §BRAF testing: (+)= mutation present,(−)= mutation absent/wild-type; ¶Surveillance recommendations based on personal and family history. CRC, colorectal cancer; IHC, immunohistochemistry; MSI, microsatellite instability; MSS, microsatellite stable. Reprinted with permission (Kastrinos F, Syngal S: Cancer J 17: 405–415, 2011).
Our study has some limitations. We elected to impute missing molecular tumour testing data using a multiple imputation method in order to minimise a biased analysis that would be encountered when restricting the analysis to include only those subjects with complete clinical, MSI and IHC data. Based on correlations among variables in the PREMM1,2,6 model, available MSI and IHC results, mutation carrier status and participating recruitment site, five imputed datasets were created and each analysed to obtain AUC estimates and thereafter combined into one overall estimate and variance by incorporating both the within- and between-imputation variability.18 We believe that this approach in missing data imputation offers a more accurate assessment and comparison of the different risk assessment strategies. Another potential limitation may involve the variability by which each participating centre in the CCFR defines population-based cases. Differing eligibility criteria for population-based cases exist between sites where probands may have been selected based on their age at first CRC diagnosis. However, we do not believe that this affects our results as performance of the PREMM1,2,6 model was as robust and slightly better among cases recruited through high-risk clinics. Additionally, performance of the PREMM1,2,6 model, together with the other risk assessment models and the existing clinical criteria, require good quality family data, where history limited only to the proband (ie, personal CRC history at age <50 years) is not sufficient. The CCFR registries may represent an idealised version of family history accuracy and the performance of the models would be less reliable if providers input erroneous or incomplete data.
Lastly, we did not take into account the costs related to each testing strategy (individually or in combination) or assess the additional costs incurred when evaluation based on one approach requires subsequent testing, including gene mutation analyses. A recent cost-effectiveness study based on a Markov model evaluating approaches for diagnosing Lynch syndrome among patients with CRC found that any strategy (ie, use of risk model estimates, molecular tumour testing, direct genetic testing or different combinations of the above) was very sensitive to the number of relatives who would undergo gene-specific testing after an identified mutation was found in the affected proband.35 The cost-effective strategy of IHC with BRAF testing was preferred, particularly when the implementation rate of clinical criteria or prediction model estimates decreased. The use of both MSI and IHC testing was the least cost-effective strategy and supports our findings in a large patient cohort that the performance of this approach led to little (if any) incremental gain in identifying MMR gene mutation carriers. While this study was not designed to determine a preferred approach among unaffected people at-risk for Lynch syndrome, recent data also support direct genetic testing with a prediction score of ≥5% when tumour tissue is unavailable or cannot be obtained from an affected relative.21
In summary, our study demonstrates that there are several excellent options available, including personal and family history assessment using PREMM1,2,6 and molecular tumour diagnostic testing, to distinguish between MMR gene mutation carriers and non-carriers. Based on our study and others, it is time that all clinical centres adopted a systematic strategy that can be implemented as a standard of care for patients with newly diagnosed CRC based on the specifics of their own institutions so that individuals with Lynch syndrome and their families are identified and offered an opportunity to prevent future cancers through appropriate screening and risk-reducing options.
Supplementary Material
Significance of this study.
What is already known about this subject?
-
►
Lynch syndrome is the most common inherited colorectal cancer (CRC) syndrome and is caused by germline mutations in the DNA mismatch repair (MMR) system.
-
►
There is active debate about the standard approach to screening for MMR deficiency in patients diagnosed with CRC, which may include use of clinical criteria, molecular tumour testing and risk estimation provided by prediction models.
-
►
The performance characteristics of prediction models compared with molecular tumour testing involving microsatellite instability (MSI) and immunohistochemistry (IHC) testing are not known.
-
►
The PREMM1,2,6 model is a validated, web-based clinical prediction model that offers personal and family cancer history risk assessment to identify patients at risk for Lynch syndrome.
What are the new findings?
-
►
The best approach in identifying MMR gene mutation carriers involves the combination of PREMM1,2,6 assessment and IHC tumour testing, as determined by this large international cohort of patients with CRC undergoing genetic testing.
-
►
The added value of IHC to PREMM1,2,6 prediction was greater than that for PREMM1,2,6 and MSI testing.
-
►
PREMM1,2,6 prediction estimates are especially informative in CRC cases owing to somatic hypermethylation of the MLH1 gene, and among MSH6 gene mutation carriers with pathogenic missense mutations which may yield false-negative IHC results.
How might the findings impact clinical practice in the foreseeable future?
-
►
Combining molecular tumour testing results with risk prediction estimates using the PREMM1,2,6 model may better aid healthcare providers in standardising the approach to identifying Lynch syndrome among patients with CRC.
Acknowledgements
We would like to acknowledge the following Colon Cancer Family Registries that provided data for the analysis: Australasian Colorectal Cancer Family Registry (U01 CA097735), the University of Southern California (USC) Familial Colorectal Neoplasia Collaborative Group (U01 CA074799), Mayo Clinic Cooperative Family Registry for Colon Cancer Studies (U01 CA074800), Ontario Registry for Studies of Familial Colorectal Cancer (U01 CA074783), Seattle Colorectal Cancer Family Registry (U01 CA074794) and University of Hawaii Colorectal Cancer Family Registry (U01 CA074806).
Funding This work was supported by the National Cancer Institute (K07 CA151769-02 to FK; R01 CA132829 and K24 CA113433 to SS), National Institutes of Health under RFA # CA95-011 and through cooperative agreements with members of the Colon Cancer Family Registry and PIs. The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centres in the CFRs, nor does mention of trade names, commercial products, or organisations imply endorsement by the US Government or the CFR. The study sponsors/funders had no role in the study design; in the collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit the article for publication.
Footnotes
Contributors All authors have substantially contributed to (1) the study conception and design, acquisition of data or analysis and interpretation of data; (2) drafting of the article or revising it critically for important intellectual content; and (3) final approval of the version published.
Competing interests All authors have completed the unified competing interest form at http://www.icmje.org/coi_disclosure.pdf (available on request from the corresponding author) and declare that (1) FK, JB, SG, GC, LL, NL, ST, SS have support from the National Institutes of Health/National Cancer Institute for the submitted work and ES, RM, RH, JH, PN have no support from any organisation for the submitted work; (2) ES, JB, RM, RH, GC, JH, LL, NL, PN, ST have no financial relationships with any organisations that might have an interest in the submitted work in the previous 3 years; FK has been paid for consultancy outside the submitted work by Marinabio, Inc. SS has been paid for consultancy outside the submitted work by Archimedes Inc., Quest Diagnostics Inc., Interquest Inc. and has stock/stock options outside the submitted work through Marinabio Inc.; SG has been paid for consultancy outside the submitted work by Roche; (3) all authors have no other relationships or activities that might appear to have influenced the submitted work.
Patient consent This article does not contain any personal medical information about any identifiable living individuals. All participants provided informed consent for inclusion in the Colon Cancer Family Registry through each registry centre: University of Hawaii (Honolulu, Hawaii), Fred Hutchinson Cancer Research Center (Seattle, Washington), Mayo Clinic (Rochester, Minnesota), University of Southern California Consortium (Los Angeles, California), Cancer Care Ontario (Toronto, Canada) and University of Melbourne (Melbourne, Australia). This study was reviewed and approved by the Dana-Farber/Harvard Cancer Center Institutional Review Board. A waiver of consent for study participants was obtained because the analyses were performed on de-identified data and did not require patient contact.
Ethics approval Ethics approval was provided by the institutional review board at all institutions in the Colon Cancer Family Registry; the Dana-Farber/Harvard Cancer Center Institutional Review Board.
Provenance and peer review Not commissioned; externally peer reviewed.
REFERENCES
- 1.Lynch HT, de la Chapelle A. Genetic susceptibility to non-polyposis colorectal cancer. J Med Genet. 1999;36:801–818. [PMC free article] [PubMed] [Google Scholar]
- 2.Lynch HT, Boland CR, Gong G, et al. Phenotypic and genotypic heterogeneity in the Lynch syndrome: diagnostic, surveillance and management implications. Eur J Hum Genet. 2006;14:390–402. doi: 10.1038/sj.ejhg.5201584. [DOI] [PubMed] [Google Scholar]
- 3.Hampel H, Frankel WL, Martin E, et al. Feasibility of screening for Lynch syndrome among patients with colorectal cancer. J Clin Oncol. 2008;26:5783–5788. doi: 10.1200/JCO.2008.17.5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balmaña J, Stockwell DH, Steyerberg EW, et al. Prediction of MLH1 and MSH2 Mutations in Lynch syndrome. JAMA. 2006;296:1469–1478. doi: 10.1001/jama.296.12.1469. [DOI] [PubMed] [Google Scholar]
- 5.Barnetson RA, Tenesa A, Farrington SM, et al. Identification and survival of carriers of mutations in DNA mismatch repair genes in colon cancers. N Engl J Med. 2006;354:2751–2763. doi: 10.1056/NEJMoa053493. [DOI] [PubMed] [Google Scholar]
- 6.Chen S, Wang W, Lee S, et al. Prediction of germline mutations and cancer risk in the Lynch syndrome. JAMA. 2006;296:1479–1487. doi: 10.1001/jama.296.12.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reyes CM, Allen BA, Terdiman JP, et al. Comparison of selection strategies for genetic testing of patients with hereditary nonpolyposis colorectal carcinoma: effectiveness and cost-effectiveness. Cancer. 2002;95:1848–1856. doi: 10.1002/cncr.10910. [DOI] [PubMed] [Google Scholar]
- 8.Piñol V, Castells A, Andreu M, et al. Accuracy of revised Bethesda guidelines, microsatellite instability and immunohistochemistry for the identification of patients with hereditary nonpolyposis colorectal cancer. JAMA. 2005;293:1986–1994. doi: 10.1001/jama.293.16.1986. [DOI] [PubMed] [Google Scholar]
- 9.Rodriguez-Moranta F, Castells A, Andreu M, et al. Gastrointestinal Oncology Group of the Spanish Gastroenterological Association. Clinical performance of original and revised Bethesda guidelines for the identification of MSH2/MLH1 gene carriers in patients with newly diagnosed colorectal cancer: proposal of a new and simpler set of recommendations. Am J Gastroenterol. 2006;101:1104–1111. doi: 10.1111/j.1572-0241.2006.00522.x. [DOI] [PubMed] [Google Scholar]
- 10.Julié C, Trésallet C, Brouquet A, et al. Identification in daily practice of patients with Lynch syndrome (hereditary nonpolyposis colorectal cancer): revised Bethesda guidelines-based approach versus molecular screening. Am J Gastroenterol. 2008;103:2825–2835. doi: 10.1111/j.1572-0241.2008.02084.x. quiz 2836. [DOI] [PubMed] [Google Scholar]
- 11.Balmaña J, Balaguer F, Castellví-Bel S, et al. Comparison of predictive models, clinical criteria and molecular tumour screening for the identification of patients with Lynch syndrome in a population-based cohort of colorectal cancer patients. J Med Genet. 2008;45:557–563. doi: 10.1136/jmg.2008.059311. [DOI] [PubMed] [Google Scholar]
- 12.Mueller J, Gazzoli I, Bandipalliam P, et al. Comprehensive molecular analysis of mismatch repair gene defects in suspected Lynch syndrome (hereditary nonpolyposis colorectal cancer) cases. Cancer Res. 2009;69:7053–7061. doi: 10.1158/0008-5472.CAN-09-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newcomb PA, Baron J, Cotterchio M, et al. Colon Cancer Family Registry. Colon Cancer Family Registry: an international resource for studies of the genetic epidemiology of colon cancer. Cancer Epidemiol Biomarkers Prev. 2007;16:2331–2343. doi: 10.1158/1055-9965.EPI-07-0648. [DOI] [PubMed] [Google Scholar]
- 14.Poynter JN, Siegmund KD, Weisenberger DJ, et al. Colon Cancer Family Registry Investigators. Molecular characterization of MSI-H colorectal cancer by MLHI promoter methylation, immunohistochemistry and mismatch repair germline mutation screening. Cancer Epidemiol Biomarkers Prev. 2008;17:3208–3215. doi: 10.1158/1055-9965.EPI-08-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cicek MS, Lindor NM, Gallinger S, et al. Quality assessment and correlation of microsatellite instability and immunohistochemical markers among population- and clinic-based colorectal tumors: results from the Colon Cancer Family Registry. J Mol Diagn. 2011;13:271–281. doi: 10.1016/j.jmoldx.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steyerberg EW, Balmaña J, Stockwell DH, et al. Data reduction for prediction: a case study on robust coding of age and family history for the risk of having a genetic mutation. Stat Med. 2007;26:5545–5556. doi: 10.1002/sim.3119. [DOI] [PubMed] [Google Scholar]
- 17.Steyerberg EW, Vickers AJ, Cook NR, et al. Assessing the performance of prediction models: a framework for traditional and novel measures. Epidemiology. 2010;21:128–138. doi: 10.1097/EDE.0b013e3181c30fb2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marshall A, Altman DG, Holder RL, et al. Combining estimates of interest in prognostic modelling studies after multiple imputation: current practice and guidelines. BMC Med Res Methodol. 2009;9:57–64. doi: 10.1186/1471-2288-9-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodriquez-Bigas MA, Boland CR, Hamilton SR, et al. An NCI workshop on HNPCC: meeting highlights and Bethesda guidelines. J Natl Cancer Inst. 1997;89:1758–1762. doi: 10.1093/jnci/89.23.1758. [DOI] [PubMed] [Google Scholar]
- 20.Kastrinos F, Steyerberg EW, Mercado R, et al. The PREMM1,2,6 model predicts risk of MLH1, MSH2 and MSH6 germline mutations based on cancer history. Gastroenterology. 2011;140:73–81. doi: 10.1053/j.gastro.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dinh TA, Rosner BI, Atwood JC, et al. Health benefits and cost-effectiveness of primary genetic screening for Lynch syndrome in the general population. Cancer Prev Res (Phila) 2011;4:9–22. doi: 10.1158/1940-6207.CAPR-10-0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vasen HF, Watson P, Mecklin JP, et al. New clinical criteria for hereditary nonpolyposis colorectal cancer proposed by the International Collaborative group on HNPCC. Gastroenterology. 1999;116:1453–1456. doi: 10.1016/s0016-5085(99)70510-x. [DOI] [PubMed] [Google Scholar]
- 23.Umar A, Boland CR, Terdiman JP, et al. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Syngal S, Fox EA, Eng C, et al. Sensitivity and specificity of clinical criteria for hereditary non-polyposis colorectal cancer associated mutations in MSH2 and MLH1. J Med Genet. 2000;37:641–645. doi: 10.1136/jmg.37.9.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Green RC, Parfrey PS, Woods MO, et al. Prediction of Lynch syndrome in consecutive patients with colorectal cancer. J Natl Cancer Inst. 2009;101:331–340. doi: 10.1093/jnci/djn499. [DOI] [PubMed] [Google Scholar]
- 26.Monzon JG, Cremin C, Armstrong L, et al. Validation of predictive models for germline mutations in DNA mismatch repair genes in colorectal cancer. Int J Cancer. 2010;126:930–939. doi: 10.1002/ijc.24808. [DOI] [PubMed] [Google Scholar]
- 27.Boland CR. Moshe Shike. Report from the Jerusalem Workshop on Lynch Syndrome- Hereditary Nonpolyposis Colorectal Cancer. Gastroenterology. 2010;138:2197–2201. doi: 10.1053/j.gastro.2010.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hampel H, de la Chapelle A. The search for unaffected individuals with Lynch syndrome: do the ends justify the means? Cancer Prev Res. 2011;4:1–5. doi: 10.1158/1940-6207.CAPR-10-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grover S, Stoffel EM, Bussone L, et al. Physician assessment of family cancer history and referral for genetic evaluation in colorectal cancer patients. Clin Gastroenterol Hepatol. 2004;2:813–819. doi: 10.1016/s1542-3565(04)00352-0. [DOI] [PubMed] [Google Scholar]
- 30.Lynch HT, Lynch PM. Molecular screening for the Lynch syndrome-better than family history? N Engl J Med. 2005;352:1920–1922. doi: 10.1056/NEJMe058058. [DOI] [PubMed] [Google Scholar]
- 31.Overbeek LI, Ligtenber MJ, Willems RW, et al. Interpretation of immunohistochemistry for mismatch repair proteins is only reliable in a specialized setting. Am J Surg Pathol. 2008;32:1246–1251. doi: 10.1097/pas.0b013e31816401bb. [DOI] [PubMed] [Google Scholar]
- 32.Klarskov L, Ladelund S, Holck S, et al. Interobserver variability in the evaluation of mismatch repair protein immunostaining. Hum Pathol. 2010;41:1387–1396. doi: 10.1016/j.humpath.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 33.Lynch HT, Boland CR, Rodriguez-Bigas MA, et al. Who should be sent for genetic testing in hereditary colorectal cancer syndromes? J Clin Oncol. 2007;25:3534–3542. doi: 10.1200/JCO.2006.10.3119. [DOI] [PubMed] [Google Scholar]
- 34.Lindor NM, Rabe K, Petersen GM, et al. Lower cancer incidence in Amsterdam-I criteria families without mismatch repair deficiency: familial colorectal cancer type X. JAMA. 2005;293:1979–1985. doi: 10.1001/jama.293.16.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ladabaum U, Wang G, Terdiman J, et al. Strategies to identify the Lynch syndrome among patients with colorectal cancer:a cost-effectiveness analysis. Ann Intern Med. 2011;155:69–79. doi: 10.7326/0003-4819-155-2-201107190-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.