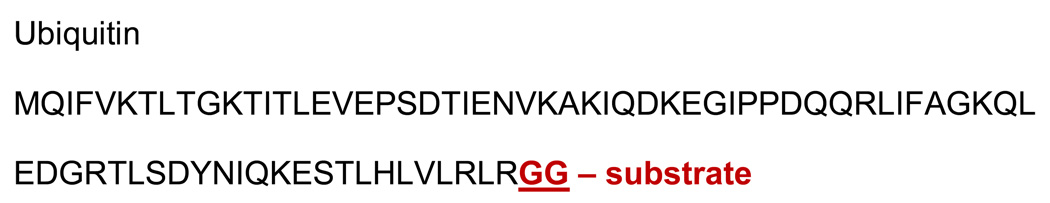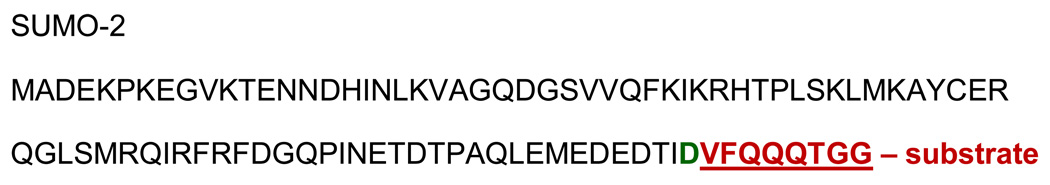Abstract
SUMO (Small-Ubiquitin-like MOdifier) is a post-translational modifier of protein substrates at lysine residues that conjugates to proteins in response to various changes in the cell. As a result of SUMO modification, marked changes in transcription regulation, DNA repair, subcellular localization, and mitosis, among other cellular processes, are known to occur. However, while the identification of ubiquitylation sites by mass spectrometry is aided in part by the presence of a small di-amino acid GlyGly “tag” that remains on lysine residues following tryptic digestion, SUMOylation poses a particular challenge as the absence of a basic residue near to the SUMO C-terminus results in a significant 27 or 32 amino acid sequence branch conjugated to the substrate peptide. MS/MS analyses of these branch peptides generally reveal abundant fragment ions resulting from cleavage of the SUMO tail, but which obscure those needed for characterizing the target peptide sequence. Other approaches for identifying SUMO substrates exist and include overexpression of the SUMO isoforms using an N-terminal histidine tag, as well as site-directed mutagenesis of the C-terminal end of the SUMO sequence. Here, we employ combined enzymatic/chemical approaches which serve to shorten the SUMO tag, and thus help to simplify SUMO spectra, making interpretation of mass spectra and location of the SUMOylation site easier. As described in this report, we demonstrate a method for identifying SUMOylation sites using three commercially available SUMO- modified isoforms, and by employing acid-only and acid/trypsin cleavage strategies. These approaches were carried out using MALDI-TOF and LC/MS instrumentation, along with CID and ETD fragmentation.
Introduction
Background
SUMO is a reversible protein modifier present in all eukaryotes. It is a part of the Ubl (ubiquitin-like) family of proteins, which includes ubiquitin. These proteins are known to modify substrates via an isopeptide bond formed between a lysine residue in the substrate sequence and a C-terminal glycine in the SUMO sequence, which results in a diverse number of cellular processes [1]. SUMO was initially identified in Saccharomyces cerevisiae in a genetic screen for suppressors Mif2, a centromeric protein and was later found attached to Ran GTPase activating protein 1 (RanGap 1), a well-characterized and highly abundant SUMOylated protein in the cell [2]. This ultimately revealed that SUMO could alter the localization of proteins by changing how they interacted with other proteins [2]. There are four SUMO isoforms. SUMO-2 and SUMO-3 have the highest sequence identity (97%), and they have a 41% identity to SUMO-1 [3]. Unlike the other SUMO isoforms, SUMO-4, is not ubiquitously expressed and is only found in immune tissues, kidney cells, and pancreatic islets [4]. As of yet it has not been found to be conjugated to any protein substrates. SUMO conjugation occurs through a process similar to the ubiquitin pathway of conjugation involving three enzymes: an activating enzyme (E1), a conjugating enzyme (E2) and a ligating enzyme (E3). SUMO proteins are first processed by enzymes (deSUMOylating enzymes) that cleave the C-terminus of the modifiers to expose the C-terminal glycine and thus convert SUMO into its mature form [5]. For conjugation to the substrate to occur, the SUMO E1 enzyme interacts with SUMO via a high energy thioester bond, and SUMO is then transferred to the E2 enzyme (Ubc9) via a subsequent thioester bond. This then leads to the activation, transfer, and conjugation of the SUMO to the target substrate [6]. There are a number of E3 ligases involved in SUMOylation. They are not necessary for target conjugation in vitro but have been shown to be necessary for efficient and targeted conjugation in vivo [7]. It has also been found that SUMOs can bind to a consensus sequence found in the target protein: ΨKXD/E, where Ψ is a hydrophobic residue, X is any residue and D/E is an acidic residue [8]. However, not all SUMO conjugation sites are consensus sites and not all consensus sites are SUMOylated. SUMOylation sites can also be found in extended motifs that contain phosphorylated or negatively charged amino acids. These are called phosphorylation-dependent SUMOylation motifs and negatively charged amino acid-dependent SUMOylation motifs, respectively [9]. Like ubiquitin, SUMO has been shown to form chains. SUMO-2 and SUMO-3, which have consensus sites at K11, can form chains both in vitro and in vivo [10]. SUMO-1, however, does not contain a consensus sequence, but has been shown to form chains much less efficiently in vitro and not at all in vivo [10].
SUMO has been found to be involved in a number of processes, including mitosis, DNA repair, transcriptional regulation, and subcellular localization, to name a few. SUMO conjugation is known to result in both transcriptional activation and repression [11]. In DNA repair, for example, SUMO is known to regulate the intra-nuclear trafficking of BLM helicase, a DNA repair protein [12]. In mitosis, primarily SUMO-2/3 isoforms interact with chromatin-associated proteins at centromeres and kineticores, thus indicating major roles in mitosis [13, 14]. SUMOylation is also known to be important for subcellular localization of transcription factors to the nucleus [15]. SUMO has even been implicated in interactions with ubiquitin. Though SUMO is not directly involved in the degradation system, studies by Schimmel and colleagues suggest that modification of certain proteins by SUMO-2/3 leads to the subsequent ubiquitination and degradation of those proteins [16]. Therefore, SUMOylation is very important to overall cell regulation.
While mass spectrometry has been used to identify and characterize ubiquitylated proteins, such analyses have been far more challenging for SUMOylated structures. Tryptic digestion of ubiquitylated proteins cleaves lysine and arginine residues in both the substrate and ubiquitin sequences, leaving (conveniently) a short GlyGly sequence tag attached to the conjugated lysine (SCHEME I). Tandem mass spectrometry treats this simply as a 114 Dalton posttranslational modification in the substrate sequence that is compatible with most data system search engines such as Mascot. In a novel approach reported by Wang et al [17, 18] chemical sulfonation of the N-terminal residues also enables facile, de novo sequencing of the substrate peptide and location of the ubiquitylation site from spectra dominated by y-series ions. In sharp contrast, the sequence residue from SUMO-2 left following tryptic digestion is 32 amino acids in length (SCHEME II). Generally longer than the sequence of the substrate peptide to which it is attached, the SUMO tail is not easily treated as just a larger 3549.48 Dalton PTM, as fragmentation of this tail may dominate the tandem mass spectra, reducing the ability for obtaining an identification using search engines or by de novo approaches. In addition, SUMOylation, like many of PTMs, occurs at low levels at any given time in the cell due in part to the dynamic nature of the conjugation/deconjugation pathways.
SCHEME I.
SCHEME II.
Mutagenesis has also been utilized to enable analysis of SUMOylated structures by mass spectrometry. Knuesel et al. [19], for example, used modified SUMO-1in which the terminal -TGG was mutated to the ubiquitin sequence –RGG. Other studies have focused on the yeast homolog SMTP3, which has an arginine residue proximal to the C-terminus, resulting in a shorter –EQIGG tag [20].
In these studies, we report an approach that utilizes combined chemical and tryptic digestion methods for reducing the size of the SUMO tail and improving characterization of the SUMOylation sites. This approach utilizes a novel microwave-assisted cleavage at aspartic acids that was developed previously [21].
Microwave-Assisted Acetic Acid and Trypsin Dual Cleavage
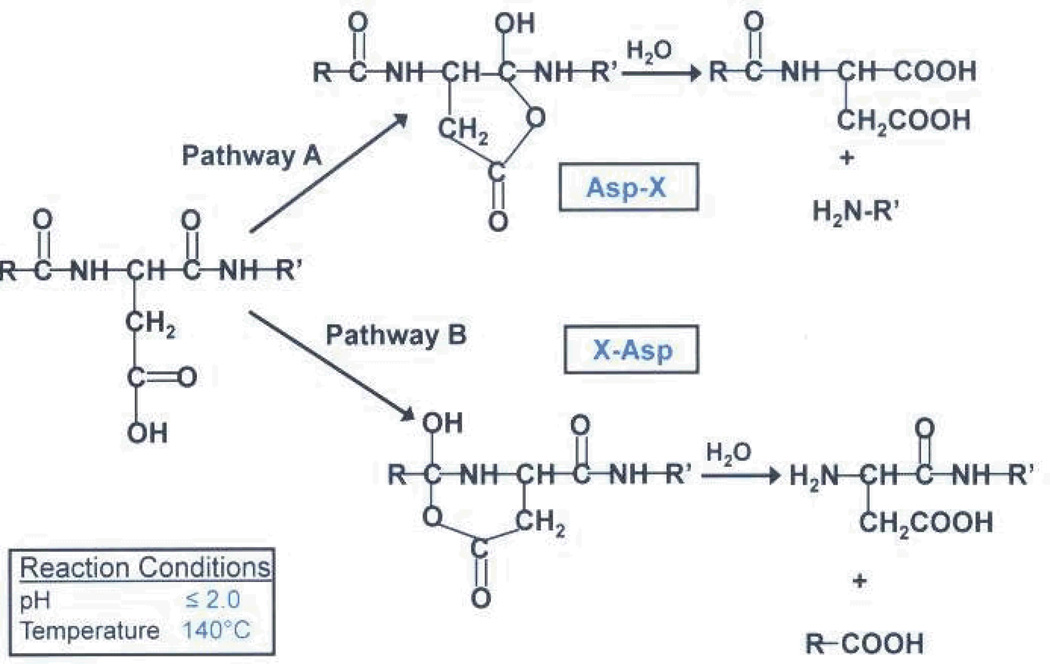
It has been shown that at high temperatures, acid cleaves protein substrates at aspartic acid residues. In 1961, Shultz and colleagues were able to show that aspartic acid residues were particularly susceptible to cleavage by acid hydrolysis by performing a number of experiments on standard proteins. Through timed studies, they were able to show that acid hydrolysis was a first order kinetics reaction [22]. Later, Li and colleagues showed that this cleavage could take place on either side of the residue in a time-dependent manner, as shown in (SCHEME 3) [23]. Using a microwave system, Swatkoski and colleagues were able to analyze the yeast ribosomal proteome, using this cleavage mechanism [21].
SCHEME III.
Enzyme-only hydrolysis approaches have also been useful in targeting aspartic acid residues for proteolytic cleavage. Asp-N, for example, cleaves proteins at the N-terminal side of aspartic acid residues. Zhou and colleagues utilized a trypsin/Asp-N dual enzyme approach, which assisted in revealing a two-step processing mechanism of MIC2/M2AP, an adhesive protein complex produced by the human pathogen, Toxoplasma gondii [24]. In addition, Ni and collegues revealed how Asp-N proteolysis can assist in selectively differentiating between aspartic acid and the post-translational modifier, isoaspartic acid [25]. However, in addition to being more costly, enzymes such as Asp-N usually require several hours for complete hydrolysis to occur, whereas microwave-assisted acid digestion can be accomplished in several minutes in a safer, more controlled environment. In addition, unlike Asp-N digestion, acid digestion allows for cleavage on either side of the aspartic acid residue, resulting in the possibility of generating two species of the same peptide, one containing a C-terminal aspartic acid and one without it. This gives one a higher probability of identifying a particular peptide since more than one species may be present. Therefore, acid hydrolysis provides a useful alternative approach for bottom-up analysis of proteins.
In this study, we apply the acid hydrolysis approach in the presence and absence of trypsin, to standard SUMO modified proteins in order generate a branch peptide containing a shorter SUMOylation tag.
Methods
Commercially available E2-25K substrate, di-SUMO-2 and di-SUMO-3, along with a SUMOylation kit containing SUMO-1, were purchased from Boston Biochem (Boston MA).
The E2-25K substrate was in vitro SUMOylated to SUMO-1 by combining 50mM (5ul) of 10× reaction buffer, 1mM (5ul) of 10× Mg-ATP, 55nM (5ul) of 10× E1 enzyme, 5uM (5ul) of 10× Ubc9, 25uM (5ul) of SUMO-1, and 10uM (10ul) of E2-25K in a 1.5 ml eppendorf tube. The reaction mixture was brought up to 50 ul total volume with the addition of water and allowed to incubate at 37°C for 1 hour. The sample was then speed vacced to dryness and brought up in 20 ul of water. Half of the sample was taken for 1dimensional (1D) gel electrophoresis separation and the other half for cleavage reactions and subsequent mass spectrometric analysis.
Trypsin-only hydrolysis
10 ug of di-SUMO-2 and di-SUMO-3 were brought up separately in 50ul of 50mM NH4HCO3 (pH 7.1) with no sample clean up. E2 25K was conjugated to SUMO-1 as mentioned above, and the reaction mixture was separated by 1D gel electrophoresis and run on a gel. The band corresponding to the modified E2 25K was then digested with trypsin in-gel. 0.4 ug of trypsin (Sigma, St. Louis, MO) was added to the di-SUMO-2 and di-SUMO-3 samples in solution, and all three samples were allowed to incubate for approximately 17.5 hours at 37°C. The samples were then dried and re-suspended in 50% ACN and 50% 0.1% TFA and analyzed by MALDI-TOF.
Acid-only hydrolysis
10ug of di-SUMO-2 and di-SUMO-3 were brought up in 50ul of 12.5 % acetic acid in 1.5 ml eppendorf tubes. The E2 25K substrate was conjugated to SUMO-1 as mentioned above, and the mixture was separated on a 1D gel. The gel band corresponding to the modified E2 25K was excised and re-suspended in 50ul of 12.5 % acetic acid. Each sample was transferred to a reaction vessel, placed in a CEM Corporation (Charlotte NC) Benchmate microwave system and allowed to incubate for 10 minutes at 140°C. Microwave power fluctuated to maintain a constant temperature of 140 °C ± 5 °C. The samples were allowed to cool and were prepared for MALD-TOF and LC/MS analysis.
Dual hydrolysis
Gel slices were digested in-gel with trypsin, and the extracted peptides were speed vacced to dryness and brought up in 50 ul of 12.5% acetic acid. They were then transferred to reaction vessels and placed in the Benchmate system to incubate for 10 minutes at 140°C ± 5 °C. Each sample was then prepared for MALDI-TOF and LC/MS analysis.
MS analysis
Samples subjected to acid/trypsin hydrolysis were analyzed on a Shimadzu (Manchester UK) Axima TOF2 MALDI- tandem time-of-flight mass spectrometer. 1 ul of each digested sample in 50% ACN and 50% 0.1% TFA was spotted on a MALDI plate in a sandwich-style manner with 10 mg/ml of alpha-cyano-4-hydroxycinnamic acid (CHCA) matrix at a total ratio of 2:1 matrix:sample. All MS spectra were acquired in linear mode with an average of 200 profiles per sample and an average power of 70. The observed m/z ratio corresponding to a given branch peptide, as determined by its theoretical mass generated in silico, was isolated and fragmented using post-source decay (PSD) fragmentation in reflectron mode. An average of 500 profiles were acquired at an average power of 90.
For LC/MS analysis, the dried down samples were brought up in HPLC solvent A (0.1% formic acid in water). Digested samples were loaded directly onto a trap column for 10.5 min at a flow rate of 5uL/min. The peptides were then separated on a reverse phase C18 column packed in-house (12nm pore and S-5um particle gel silica). To elute the peptides, the gradient applied to the C18 column allowed for the percentage of solvent B (0.1% formic acid in 90% acetonitrile) to increase from 10% to 97% over 51 minutes at a flow rate of 300 nL/min. Peptides were analyzed on a Thermo (Sunnyvale CA) LTQ Orbitrap XL mass spectrometer. Precursor ions were measured in the orbitrap at a resolution of 30,000. Analysis was done in data dependent mode where the top 5 most intense ions in each MS spectrum were chosen for MS/MS analysis. Peptides were fragmented using either ETD analysis or CID analysis. For ETD analysis, supplemental activation was allowed, and an activation time of 150 milliseconds was used. In addition, all fragment ions were analyzed in the ion trap. For CID analysis, a normalized collision energy of 35 was used. Fragment ions were analyzed in the Orbitrap for charge state and resolution determination.
Data Analysis/Branch Peptide Identification
All branch peptides were identified manually. Predicted m/z ratios were searched in the raw data, and fragment ions of those precursor m/z ratios in corresponding MS/MS spectra were matched to theoretical fragment ions using available online software (Protein Prospector).
Results and Discussion
MALDI-TOF analyses following trypsin digestion
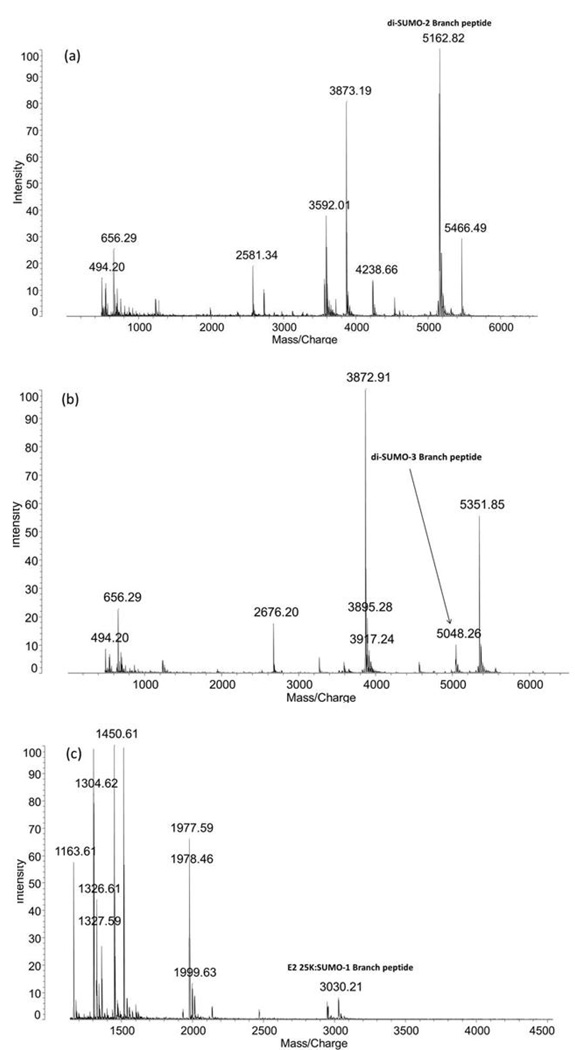
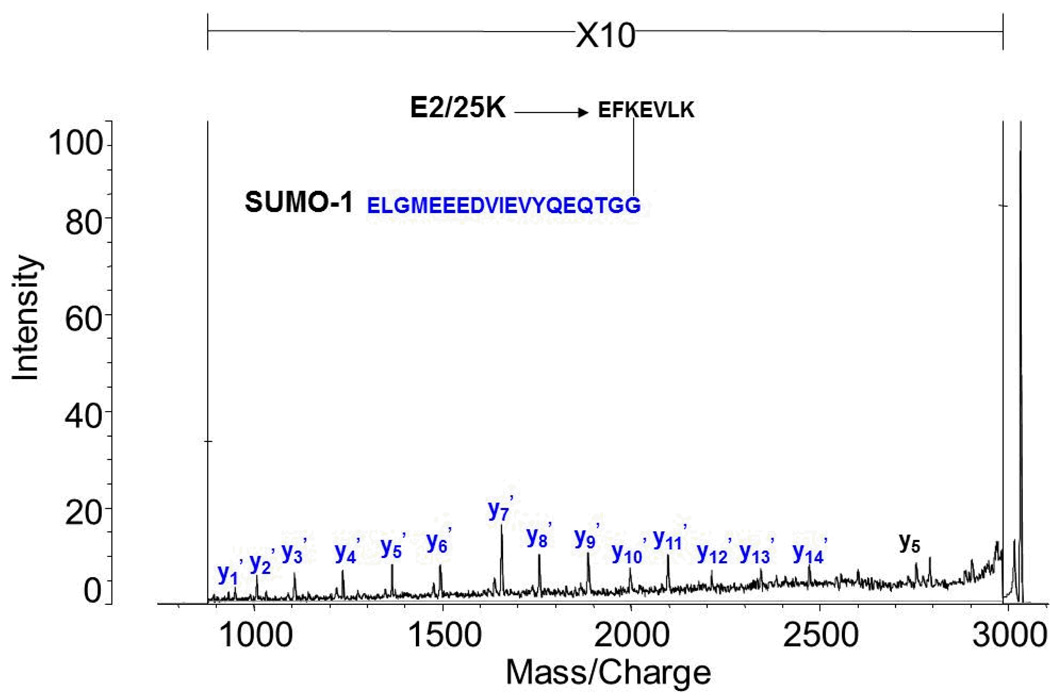
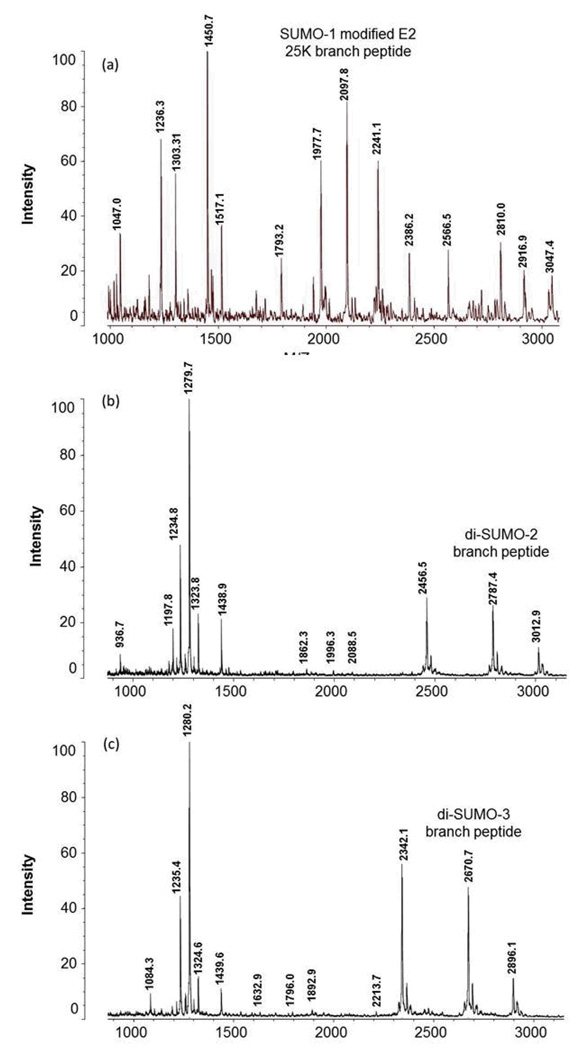
Figure 1 shows the MALDI TOF MS results for the trypsin digests of di-SUMO-2, di-SUMO-3, and SUMO-1 modified E2 25K substrate, respectively. For di-SUMO-2, a branched peptide with an average m/z ratio of 5161.18 was calculated. In the MS spectrum, an m/z of 5162.82 was observed (Figure 1a). This peptide includes the C-terminal sequence from SUMO-2 (3549.48 Daltons) conjugated to K11 on a second molecule of SUMO-2 as its substrate. Since the conjugated lysine is not itself cleaved, this corresponds to the sequence: EGVKTENNDHINLK as the substrate peptide. For di-SUMO-3, an average m/z ratio of 5047.08 was calculated, and an m/z ratio of 5048.26 was observed (Figure 1b). For the SUMO-1 modified E2 25K substrate an m/z ratio of 3028.88 was calculated and a m/z ratio of 3030.21 was observed (Figure 1c). The MALDI MS/MS spectrum of the 3028.88 ion corresponding to the SUMO-1 modified E2 25K branch peptide is shown in Figure 2. It reveals a nearly complete y-ion series from the SUMO-1 tag sequence, but only a single fragmentation from the E2 25K substrate, which would not be sufficient for identification of the peptide in the case of an unknown.
Figure 1.
MALDI TOF mass spectra of the peptides resulting from tryptic digestion of (a) di-SUMO-2, (b) di-SUMO-3 and (c) SUMO-1 modified E2 25K. The branch peptide in each case is indicated.
Figure 2.
MALDI TOF MS/MS spectrum of the MH+ molecular ions from the branch peptides resulting from tryptic digestion of SUMO-1 modified E2 25K.
Acid/Trypsin Hydrolysis-MALDI-TOF analysis
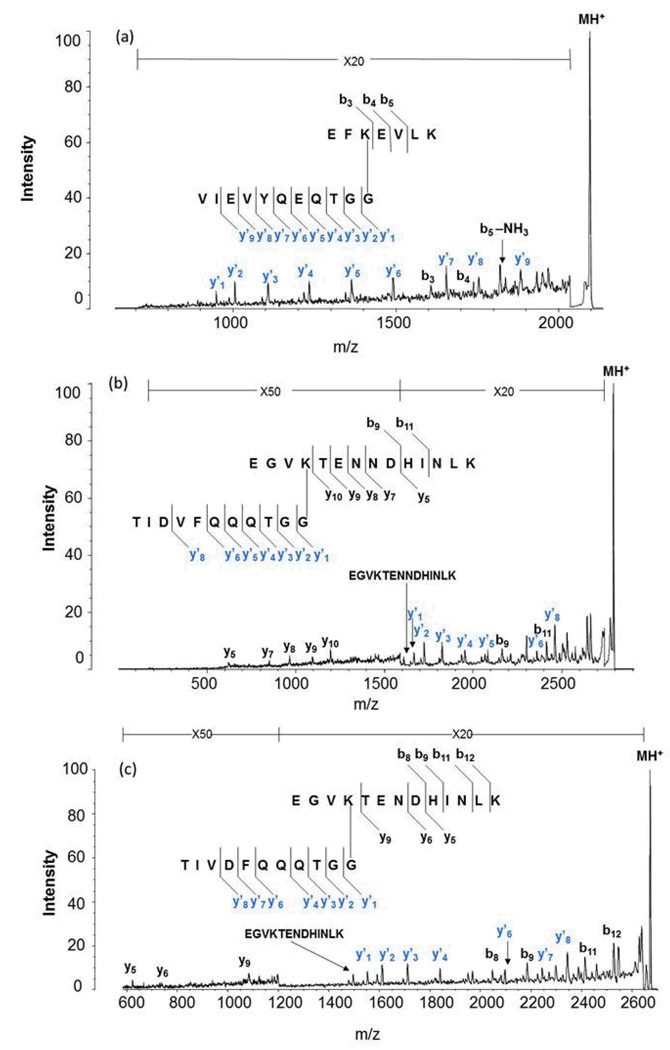
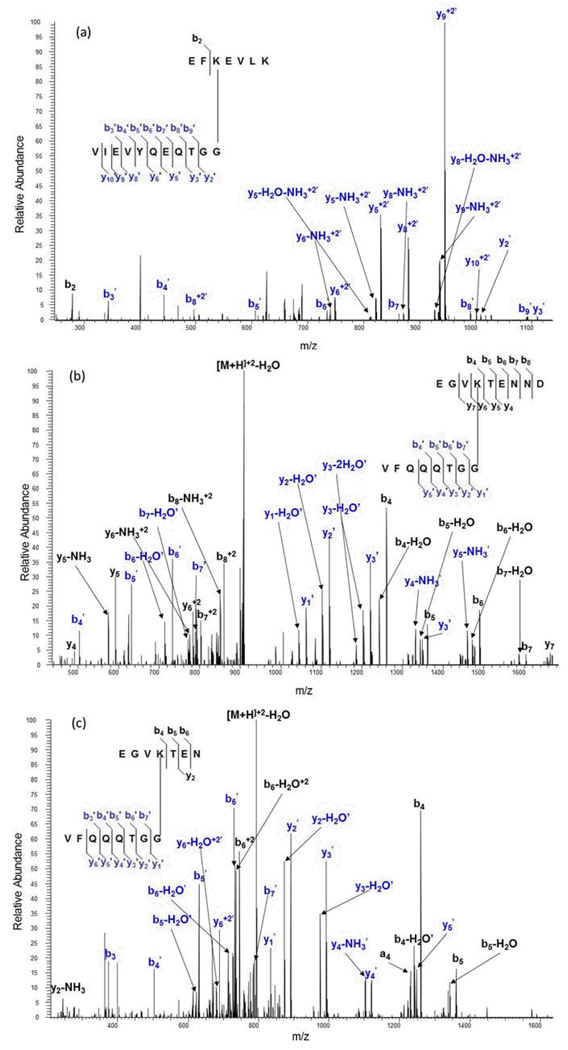
Microwave-assisted acid hydrolysis cleaves at the aspartic acid residues of the SUMO moiety that is conjugated to proteins, resulting in a considerably shorter branched sequence than observed using trypsin. Scheme 4 shows the resulting 11 amino acid sequence for SUMO-1 which will contribute a mass of 1203.54 Daltons to the substrate peptide. Scheme 5 shows the 8 amino acid sequence from SUMO-2/3 weighing 846.99 Daltons. We therefore obtained MALDI MS spectra for the SUMO dimers following combined acid hydrolysis and trypsin digestion. Based on the high specificity of acid and trypsin for Asp and Lys respectively, we were able to make predictions about the branch peptide species for each SUMO isoform that we expected to see in the MALDI-TOF MS spectrum. We know that E2 25K, a ubiquitin conjugating enzyme, is modified at K14. For SUMO-1 modified E2 25K, we expected a precursor mass of 2097.39. This corresponds to the E2 25K sequence, EFKEVLK, conjugated to a SUMO-1 tag having a molecular weight of 1203.54 Da (accounting for water loss), where acid has cleaved at Asp 86, resulting in an eleven-amino acid tag (VIEVYQEQTGG). Figure 3a shows the MALD-TOF MS spectrum identifying the predicted mass of the SUMO-1 E2 25K branch peptide. Figure 4a then shows the MS/MS spectrum of the SUMO-1 modified E2 25K branch peptide. Much like the trypsin-only analysis of this branch peptide species, we were able to obtain an almost complete y-ion series for the SUMO tag; however, we were able to obtain slightly better fragmentation of the substrate sequence compared to trypsin-only analysis.
SCHEME IV.
SCHEME V.
Figure 3.
MALDI TOF mass spectra of the peptides resulting from combined aspartic acid cleavage/trypsin digestion of (a) SUMO-1 modified E2 25K, (b) di-SUMO-2 and (c) di-SUMO-3. The branch peptide in each case is indicated.
Figure 4.
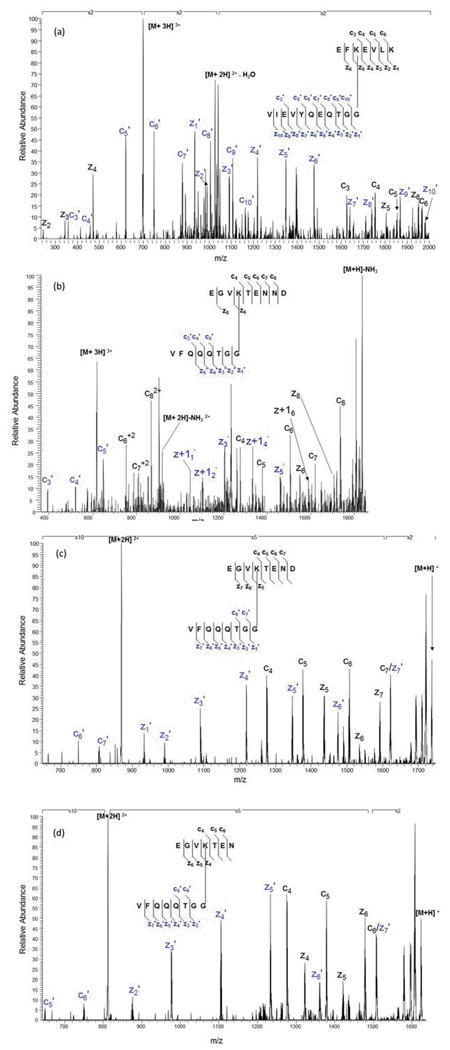
MALDI TOF MS/MS spectra of the MH+ molecular ions from the branch peptides resulting from combined aspartic acid cleavage/trypsin digestion of the SUMO-conjugated proteins: (a) MS/MS spectrum of the m/z 2097.8 ion from SUMO-1 modified E2 25K, (b) MS/MS spectrum of the m/z 2787.4 ion from di-SUMO-2 and (c) MS/MS spectrum of the m/z 2670.7 ion from di-SUMO-3. Fragment ions resulting from cleavage on the SUMO moiety are indicated with a prime.
Because SUMO-2 and SUMO-3 are known to form chains both in vivo and in vitro, SUMO chains were used to test the validity of the acid/trypsin digest strategy. It is known that chain formation occurs primarily at K11 in the SUMO2/3 sequences [REF?]. This particular site is also a SUMO consensus site, having the sequence VKTE (see Figures 4b and 4c). In the case of SUMO dimers, one SUMO acts as the substrate, while the other one acts as the modifier, which obviates the need to perform an in-vitro SUMOylation reaction. For the di-SUMO-2 standard, based upon possible cleavage on either side of an aspartic acid residue, we predicted branch peptides where the substrate moiety has a sequence of EGVKTENN or EGVKTENND conjugated to the modifier with the sequence VFQQQTGG, with m/z 1735.82 and 1850.85, respectively. These were not observed. Rather, there was a missed cleavage at the Asp 16 in the substrate peptide, and a missed cleavage at Asp 85 in the SUMO branch. The branched peptide shown in Figure 3b agrees with the calculated mass at 2787.03. This ion was then isolated and fragmented, to confirm its identity (Figure 4b). For this branch peptide species, we were able to obtain better fragment coverage from both the substrate and the SUMO-2 tag compared to the SUMO-1 modified E2 25K branch peptide.
SUMO-2 and SUMO-3 sequences are nearly identical with an overall sequence identity of 97%. As modifiers, they have identical tag sequences after enzymatic and/or acid digestion. As a result, there is no distinguishing between the two of them. However, SUMO-3 differs from SUMO-2 by a single asparagine residue that falls in the substrate peptide sequence containing the consensus lysine, causing their branch peptide masses to differ. For di-SUMO-3, the branched peptides, assuming no missed cleavages, would have ions with m/z ratios of 1621.78 for the species without the C-terminal Asp and 1736.80 for the species with the C-terminal Asp. While not observed, we again found a branched peptide with a missed cleavage in both the substrate and the SUMO tag sequence (Figure 3c) matching a calculated m/z of 2671.33 We were then able to confirm the identity of this species based on MS/MS analysis (Figure 4c). The fragmentation coverage for the di-SUMO-3 branch peptide was very similar to that of the di-SUMO-2 branch peptide, which is expected since the branch peptides are nearly identical.
Acid/Trypsin Digestion-LC/MS analysis (CID)
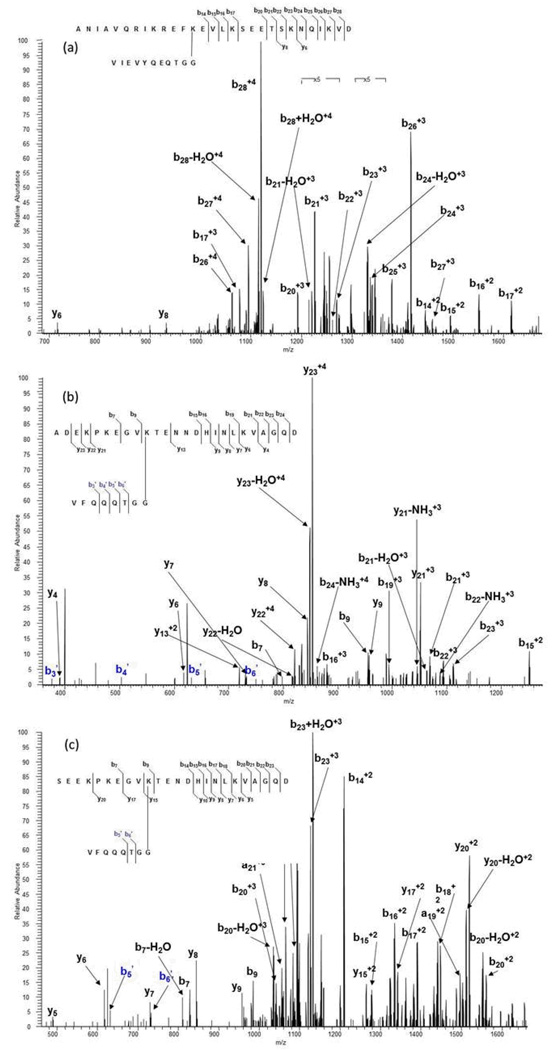
On-line LCMS analysis using electrospray ionization on an LTQ Orbitrap mass spectrometer allows us to separate peptides online before MS analysis and distinguish any structural isoforms, to possibly improve fragmentation from multiply-charged ions, to utilize both CID and electron transfer dissociation (ETD), and to take advantage of high mass resolution and accuracy. We looked first at the CID mass spectra of the most abundant multiply-charged molecular ion for the branched peptides from each of the three dimers. Figure 5a shows the CID MS/MS fragmentation spectrum of the triply-charged (+3) molecular ion at m/z 699.71 from the SUMO-1 modified E2 25K branch peptide, corresponding to a molecular mass of 2096.39 Daltons. As expected, we were able to obtain almost complete fragmentation coverage of the SUMO tag sequence; however, the fragmentation coverage of the substrate sequence was in this case worse than the MALDI-TOF fragmentation of the same species, fragmenting at only one peptide bond in the sequence.
Figure 5.
CID MS/MS spectra from an electrospray ionization LTQ/Orbitrap mass spectrometer obtained for the molecular ions from the branch peptides resulting from combined aspartic acid cleavage/trypsin digestion of the SUMO-conjugated proteins: (a) MS/MS spectrum of the +3 ion at m/z 699.71from SUMO-1 modified E2 25K, (b) MS/MS spectrum of the +2 ion at m/z 925.93 from di-SUMO-2 and (c) MS/MS spectrum of the +2 ion at m/z 811.84 from di-SUMO-3. Fragment ions resulting from cleavage on the SUMO moiety are indicated with a prime.
For the di-SUMO standards, on-line LC/MS separations enabled us to locate and identify the branched peptides resulting from no missed cleavages. The MS/MS spectrum of the doubly-charged molecular ion (m/z 925.93) from the completely cleaved species containing the C-terminal Asp for di-SUMO-2 is shown in Figure 5b. We were able to obtain a robust spectrum, yielding nearly complete coverage from both the substrate and the SUMO tag sequence. For the di-SUMO-3 standard, a +2 species lacking the C-terminal Asp was found with an m/z ratio of 811.84. It yielded similar results to di-SUMO-2, though the substrate sequence coverage was not as complete (Figure 5c).
Acid/Trypsin Digestion-LC/MS analysis (ETD)
For the SUMO-1 modified E2 25K standard, we found the fully cleaved branched peptide with a charge state of +4 and an m/z ratio of 899.95. Figure 6a shows the ETD MS/MS spectrum of the peptide. For this standard, we obtained almost complete fragmentation coverage, missing only four ions in the entire c- and z-ion series from both moieties of the branch peptide.
Figure 6.
ETD MS/MS spectra from an electrospray ionization LTQ/Orbitrap mass spectrometer obtained for the molecular ions from the branch peptides resulting from combined aspartic acid cleavage/trypsin digestion of the SUMO-conjugated proteins: (a) MS/MS spectrum of the +3 ion at m/z 699.71 from SUMO-1 modified E2 25K, (b) MS/MS spectrum of the +3 ion at m/z 617.62 from di-SUMO-2, (c) MS/MS spectrum of the +2 ion at m/z 869.39 from di-SUMO-3, and (d) MS/MS spectrum of the +2 ion at m/z 812.38 from di-SUMO-3. Fragment ions resulting from cleavage on the SUMO moiety are indicated with a prime.
For di-SUMO-2, we identified the fully cleaved branch peptide species containing the C-terminal Asp, much like we did for our CID analysis. This species was found as a +3 ion with an m/z ratio of 617.62. As shown in the ETD MS/MS spectrum in Figure 6b, fragmentation coverage for this branch peptide was composed primarily of c-ions for the substrate moiety and primarily z-ions for the modifier moiety. For di-SUMO-3, we found two species of the branch peptide, one containing the C-terminal Asp residue and one lacking this residue, both as doubly-charged molecular ions appearing at m/z 869.39 and 812.38, respectively. MS/MS spectra shown in Figures 6c and 6d reveal that they have nearly identical fragmentation patterns, where we able to obtain an almost complete z-ion series for the modifier moiety and a mixture of c- and z-ions for the substrate moiety.
Acid-Digestion-LC/MS analysis (CID)
We also employed acid-only hydrolysis to generate SUMO branched peptides. Because of the proximity of the Asp residue to the C-terminal branch point, we expect to generate an identical SUMO tag to the one generated from the acid/trypsin hydrolysis, thereby generating the same shift in the mass of the substrate. However, the substrate mass would be larger. We first employed CID analysis.
In the SUMO-1 modified E2 25K protein the first Asp residue occurs in the amino acid sequence of the substrate at position 30. Because SUMO conjugation occurs at K14, the branch peptide after acid cleavage contains the entire 30-amino acid sequence, a much larger peptide compared to that from the acid/trypsin hydrolysis of the protein having a molecular weight of 4603.47 Da. We observed its molecular ion species with a +4 charge state and an m/z ratio of 1152.39. Figure 7a shows the MS/MS spectrum of the SUMO-1 modified E2 25K branch peptide generated from acid hydrolysis. In this case, fragmentation occurred entirely in the substrate sequence. In addition, most of the fragmentation of the substrate occurred at the C-terminal end and consisted primarily of b-ions. This again confirms that peptide length has an effect on the fragmentation coverage of the branch peptide as a whole. SUMO-2 contains an Asp residue at position 3 and one at position 16 within its N-terminal region. Because we know that SUMO-2 is modified at K11 in this region, we expect to generate a completely cleaved SUMO-2 substrate that is twelve or thirteen amino acids long, which is not much larger than the nine-amino acid acid/trypsin substrate species. This species was not observed; rather we found a species that contained two missed cleavages in the substrate sequence of the branch peptide, resulting in a twenty-five amino acid SUMO-2 substrate peptide conjugated to a completely cleaved SUMO-2 modifier peptide. This species was found as a +4 ion with an m/z of 899.95. The MS/MS spectrum is found in Figure 7b. Fragmentation of di-SUMO-2 resulted in roughly equal b-ions and y-ions along the substrate backbone. We were able to obtain a small amount of fragmentation of the SUMO-2 tag, though the relative intensity of these fragment ions was low. This, and the MS/MS spectrum of the SUMO-1 modified E2 25K branched peptide suggest that for large substrate sequences, CID fragmentation of the SUMO tag is not as reliable.
Figure 7.
CID MS/MS spectra from an electrospray ionization LTQ/Orbitrap mass spectrometer obtained for the molecular ions from the branch peptides resulting from aspartic acid-only digestion of SUMO-conjugated proteins: (a) MS/MS spectrum of the +3 ion at m/z 1152.39 from SUMO-1 modified E2 25K, (b) MS/MS spectrum of the +4 ion at m/z 899.95 from di-SUMO-2 and (c) MS/MS spectrum of the +3 ion at m/z 1171.59 from di-SUMO-3. Fragment ions resulting from cleavage on the SUMO moiety are indicated with a prime.
The di-SUMO-3 branch peptide analysis yielded similar results. We identified a species containing one missed cleavage in the substrate sequence, generating a branch peptide species with a +3 charge and an m/z ratio of 1171.59. The length of the substrate moiety was 24 amino acids long and contained the C-terminal Asp. CID fragmentation generated the bulk of fragments at the C-terminal end of the substrate moiety, as well as two fragments from the SUMO tag using CID analysis. Figure 7c shows the MS/MS spectrum of the di-SUMO-3 branch peptide.
Acid-Digestion-LC/MS analysis (ETD)
Additionally, we analyzed the acid cleaved samples using ETD fragmentation. Because this fragmentation method has been shown to be optimal for larger peptides, it was of interest to investigate whether these larger branch peptides generated from an acid-only approach would benefit from ETD fragmentation. Data-dependent analysis did not result in an MS/MS spectrum for any branch peptide species from the SUMO-1 modified E2 25K standard, but we did identify di-SUMO-2 and di-SUMO-3 branch peptides.
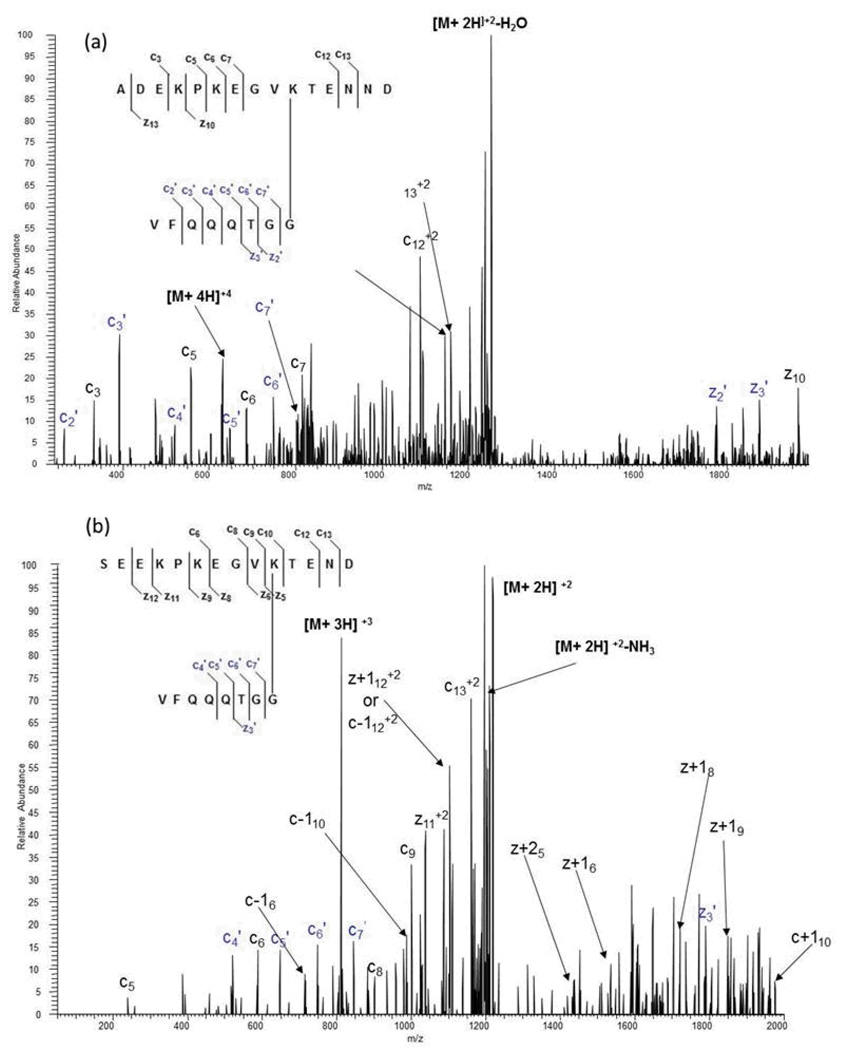
Figure 8a shows the ETD MS/MS spectrum of a di-SUMO-2 branch peptide generated from an acid-only digest. The molecular ion at m/z 630.80 corresponds to the +4 ion for a branched peptide with one missed cleavage at Asp 3, resulting in a 15 amino acid substrate species containing the C-terminal Asp. As expected, ETD fragmentation generated fragment ions from the substrate moiety of the branch peptide, and these ions were primarily c-ions. More notably, ETD also generated abundant ions from the SUMO tag. All but one of the fragments from the c-ion series is represented, along with two ions from the z-ion series.
Figure 8.
ETD MS/MS spectra from an electrospray ionization LTQ/Orbitrap mass spectrometer obtained for the molecular ions from the branch peptides resulting from aspartic acid-only digestion of SUMO-conjugated proteins: (a) MS/MS spectrum of the +4 ion at m/z 630.80 from di-SUMO-2 and (b) MS/MS spectrum of the +3 ion at m/z 812.72 from di-SUMO-3. Fragment ions resulting from cleavage on the SUMO moiety are indicated with a prime.
For the di-SUMO-3 standard, we found the completely cleaved species with an m/z ratio of 812.72 (+3). Figure 8b shows an MS/MS spectrum of the branch peptide generated from the acid-only approach. Much like the di-SUMO-2 standard, we were able to generate ions from both the substrate moiety and the modifier moiety of the branch peptide, though modifier fragmentation generated less fragment ions.
Summary and Conclusions
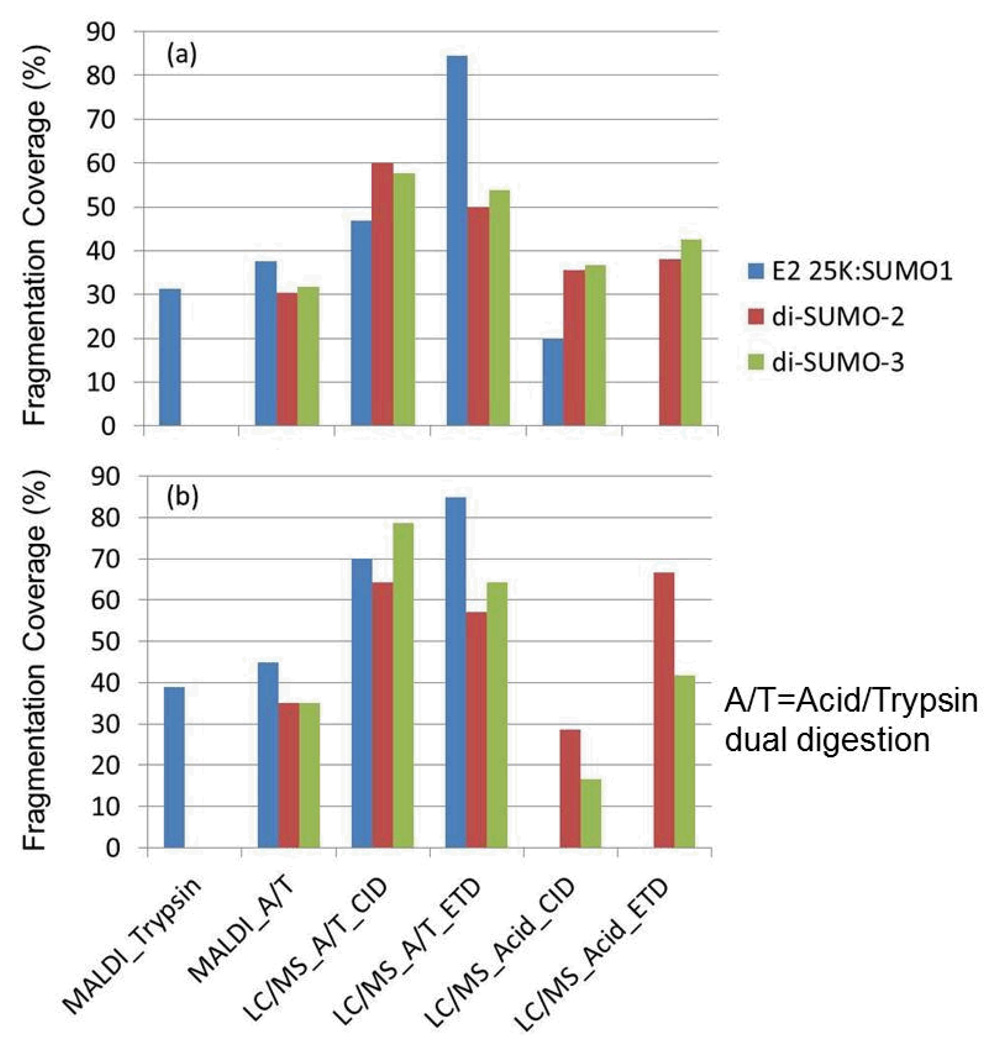
In this study, we used commercially available human SUMO isoform standards to validate the usefulness of acid hydrolysis for identifying SUMOylated branch peptides. Utilizing microwave assisted cleavage at aspartic acid with and without trypsin, we show that we can generate abundant fragmentation, sufficient to identify the SUMO branch peptides. Figure 9 compares the percent fragmentation coverage for the substrate and the SUMO branch for each of the methods described. The acid/trypsin approach, whether using CID or ETD fragmentation, provided the most fragmentation for the substrate moiety and the modifier moiety.
Figure 9.
Summary of the fragment ion coverage for (a) the substrate moiety and (b) the SUMO tail for the ionization, digestion and fragmentation methods utilized in this report.
We did, however, observe missed cleavages for some of the branch peptides that were analyzed. The prevalence of these missed cleavages is a result of limiting digestion times. At longer digestion times, we would expect to see a decrease in the number of overall missed cleavages. In addition, as it relates to the acid hydrolysis event, digestion time is the main factor in determining the type of species that we observe. Because species generation is time dependent, at shorter times, we expect the species with the Asp residue on the C-terminus to be more prevalent, while at longer times, the species without the C-terminus should be more prevalent (see Scheme III). As seen in the results, digestion times of 10 minutes or less appear to favor the former overall, suggesting that longer times are required for generating the alternative species. However, missed cleavages in either the substrate or modifier sequences of the branch peptides did not hinder our ability, or make it more difficult, to successfully identify the branch peptides.
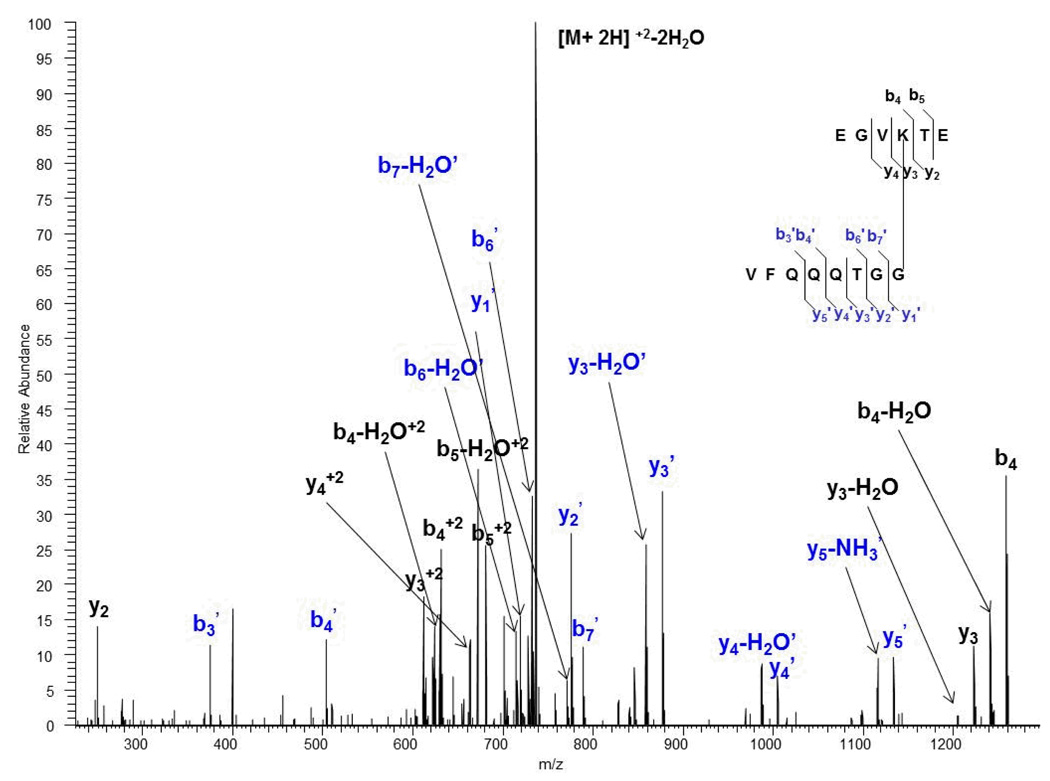
The advantage of the enzymatic/chemical shortening of the SUMO tag lies in the fact that identification of the branch peptides should be much easier due to the de-convolution of the MS/MS spectra which makes interpretation easier, a feat that is more challenging with the large SUMO tag generated after a trypsin-only digest. In addition, because acid hydrolysis gives us the ability to generate two species of branch peptides in a time-dependent manner, our approach serves as an inherent indication of reproducibility. We were able to show based on acid/trypsin hydrolysis of the di-SUMO-3 standard, for example, that we can identify the species with the C-terminal Asp and the one without it (See figures 6 (c) and (d)). Consequently, identification of both species, in a sense, reinforces the reproducibility of generating and identifying the branch peptide as a whole. A more interesting finding which adds to this concept is the deamination product of the SUMO-2/3 standards that we inadvertently discovered. SUMO-2 contains two Asn residues at positions 14 and 15, while SUMO-3 contains one Asn residue at position 14. Based on work done analyzing these samples using reconstructed ion chromatograms (data not shown), we identified species that appeared to be cleaved at the Asn positions. For example, a species of the di-SUMO-2 and di-SUMO-3 branch peptides corresponding to a substrate with the sequence EGVKTE (missing two Asn residues at the C-terminal end) conjugated to the SUMO-2 tag sequence VFQQQTGG was identified as a +2 ion with a precursor mass of 754.37. This corresponded to a mass accuracy of 0.80 ppm, and we were able to successfully match theoretical fragment ions with observed product ions in the MS/MS spectrum. Figure 10 (ADDED FIGURE) shows the MS/MS spectrum of the di-SUMO-2 branch peptide cleaved at positions 14 and 15 in the substrate sequence. Upon closer evaluation, we hypothesize that at low pH, deamination of Asn to Asp is favored [26], thus resulting in cleavage at these positions. The acidic character of the acetic acid provides an ideal environment for this process to take place. As a result, deamination provides another species that, along with acid cleavage, reinforces the reproducibility of the approach and that can easily be accounted for using available search engines. Consequently, the efficiency and wealth of information generated using this approach can possibly provide advantages for identifying SUMOylated peptides in complex mixtures and biologically relevant samples.
Figure 10.
Orbitrap CID MS/MS acid/trypsin spectrum of the +2 ion at m/z 754.37 of di-SUMO-2 branch peptide cleaved by acid at both Asn positions, suggesting deamidation prior to digestion.
Acknowledgments
This work was supported in part by a grant U54 RR020839 (Boeke, J.) from the National Institutes of Health and instrumentation grant NIH S10 RR023025 (Cotter, R). The work was carried out at the Middle Atlantic Mass Spectrometry Laboratory.
Literature Cited
- 1.Johnson ES. Protein modification by SUMO. Annual Review of Biochemistry. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 2.Melchior F, Geiss-Friedlander R. Concepts in sumoylation: a decade on. Nature Reviews Molecular Cell Biology. 2007;8(12):947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 3.Sixma TK, et al. Noncovalent interaction between Ubc9 and SUMO promotes SUMO chain formation. Embo Journal. 2007;26(11):2797–2807. doi: 10.1038/sj.emboj.7601711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo D, et al. Proteomic analysis of SUMO4 substrates in HEK293 cells under serum starvation-induced stress. Biochem Biophys Res Commun. 2005;337(4):1308–1318. doi: 10.1016/j.bbrc.2005.09.191. [DOI] [PubMed] [Google Scholar]
- 5.Sarge KD, Park-Sarge OK. Detection of proteins sumoylated in vivo and in vitro. Methods Mol Biol. 2009;590:265–277. doi: 10.1007/978-1-60327-378-7_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miura K, Hasegawa PM. Sumoylation and other ubiquitin-like post-translational modifications in plants. Trends Cell Biol. 2010;20(4):223–232. doi: 10.1016/j.tcb.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 7.Verger A, Perdomo J, Crossley M. Modification with SUMO. A role in transcriptional regulation. EMBO Rep. 2003;4(2):137–142. doi: 10.1038/sj.embor.embor738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu L, et al. Protein sumoylation sites prediction based on two-stage feature selection. Mol Divers. 2010;14(1):81–86. doi: 10.1007/s11030-009-9149-5. [DOI] [PubMed] [Google Scholar]
- 9.Sistonen L, et al. Novel Proteomics Strategy Brings Insight into the Prevalence of SUMO-2 Target Sites. Molecular & Cellular Proteomics. 2009;8(6):1382–1390. doi: 10.1074/mcp.M800551-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ulrich HD. The Fast-Growing Business of SUMO Chains. Molecular Cell. 2008;32(3):301–305. doi: 10.1016/j.molcel.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 11.Hay RT, Girdwood DWH, Tatham MH. SUMO and transcriptional regulation. Seminars in Cell & Developmental Biology. 2004;15(2):201–210. doi: 10.1016/j.semcdb.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 12.Ellis NA, et al. Intra-nuclear trafficking of the BLM helicase to DNA damage-induced foci is regulated by SUMO modification. Human Molecular Genetics. 2005;14(10):1351–1365. doi: 10.1093/hmg/ddi145. [DOI] [PubMed] [Google Scholar]
- 13.Ryu H, et al. PIASy-dependent SUMOylation regulates DNA topoisomerase II alpha activity. Journal of Cell Biology. 2010;191(4):783–794. doi: 10.1083/jcb.201004033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Azuma Y, et al. PIASy mediates SUMO-2 conjugation of Topoisomerase-II on mitotic chromosomes. Embo Journal. 2005;24(12):2172–2182. doi: 10.1038/sj.emboj.7600700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larsson O, et al. SUMOylation Mediates the Nuclear Translocation and Signaling of the IGF-1 Receptor. Science Signaling. 2010;3(108) doi: 10.1126/scisignal.2000628. [DOI] [PubMed] [Google Scholar]
- 16.Vertegaal ACO, et al. The Ubiquitin-Proteasome System Is a Key Component of the SUMO-2/3 Cycle. Molecular & Cellular Proteomics. 2008;7(11):2107–2122. doi: 10.1074/mcp.M800025-MCP200. [DOI] [PubMed] [Google Scholar]
- 17.Wang DX, Cotter RJ. Approach for determining protein ubiquitination sites by MALDI-TOF mass spectrometry. Analytical Chemistry. 2005;77(5):1458–1466. doi: 10.1021/ac048834d. [DOI] [PubMed] [Google Scholar]
- 18.Wang DX, et al. Direct identification of ubiquitination sites on ubiquitin-conjugated CHIP using MALDI mass spectrometry. Journal of Proteome Research. 2005;4(5):1554–1560. doi: 10.1021/pr050104e. [DOI] [PubMed] [Google Scholar]
- 19.Knuesel M, et al. A method of mapping protein sumoylation sites by mass spectrometry using a modified small ubiquitin-like modifier 1 (SUMO-1) and a computational program. Molecular & Cellular Proteomics. 2005;4(10):1626–1636. doi: 10.1074/mcp.T500011-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Denison C, et al. A proteomic strategy for gaining insights into protein sumoylation in yeast. Molecular & Cellular Proteomics. 2005;4(3):246–254. doi: 10.1074/mcp.M400154-MCP200. [DOI] [PubMed] [Google Scholar]
- 21.Fenselau C, et al. Evaluation of microwave-accelerated residue-specific acid cleavage for proteomic applications. Journal of Proteome Research. 2008;7(2):579–586. doi: 10.1021/pr070502c. [DOI] [PubMed] [Google Scholar]
- 22.Schultz J, Allison H, Grice M. Specificity of the cleavage of proteins by dilute acid. I. Release of aspartic acid from insulin, ribonuclease, and glucagon. Biochemistry. 1962;1:694–698. doi: 10.1021/bi00910a024. [DOI] [PubMed] [Google Scholar]
- 23.Li AQ, et al. Chemical cleavage at aspartyl residues for protein identification. Analytical Chemistry. 2001;73(22):5395–5402. doi: 10.1021/ac010619z. [DOI] [PubMed] [Google Scholar]
- 24.Zhou XW, et al. Proteomic analysis of cleavage events reveals a dynamic two-step mechanism for proteolysis of a key parasite adhesive complex. Molecular & Cellular Proteomics. 2004;3(6):565–576. doi: 10.1074/mcp.M300123-MCP200. [DOI] [PubMed] [Google Scholar]
- 25.Ni W, et al. Analysis of isoaspartic Acid by selective proteolysis with Asp-N and electron transfer dissociation mass spectrometry. Analytical Chemistry. 2010;82(17):7485–7491. doi: 10.1021/ac101806e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peters B, Trout BL. Asparagine deamidation: pH-dependent mechanism from density functional theory. Biochemistry. 2006;45(16):5384–5392. doi: 10.1021/bi052438n. [DOI] [PubMed] [Google Scholar]