Abstract
Dengue diseases have an economic as well as social burden worldwide. In this study, the antiviral activity of protegrin-1 (PG-1, RGGRLCYCRRRFCVCVGR) peptide towards dengue NS2B-NS3pro and viral replication in Rhesus monkey kidney (MK2) cells was investigated. The peptide PG-1 was synthesized by solid-phase peptide synthesis, and disulphide bonds formation followed by peptide purification was confirmed by LC-MS and RPHPLC. Dengue NS2B-NS3pro was produced as a single-chain recombinant protein in E. coli. The NS2B-NS3pro assay was carried out by measuring the florescence emission of catalyzed substrate. Real-time PCR was used to evaluate the inhibition potential of PG-1 towards dengue serotype-2 (DENV-2) replication in MK2 cells. The results showed that PG-1 inhibited dengue NS2B-NS3pro at IC50 of 11.7 μM. The graded concentrations of PG-1 at nontoxic range were able to reduce viral replication significantly (P < 0.001) at 24, 48, and 72 hrs after viral infection. However, the percentage of inhibition was significantly (P < 0.01) higher at 24 hrs compared to 48 and 72 hrs. These data show promising therapeutic potential of PG-1 against dengue infection, hence it warrants further analysis and improvement of the peptide features as a prospective starting point for consideration in designing attractive dengue virus inhibitors.
1. Introduction
Dengue infection causes various clinical symptoms ranging from mild fever to severe hemorrhagic fever and dengue shock syndrome [1–3]. The virus uses host cell ribosomes to translate its genomic RNA to full-length precursor polyprotein. Subsequently, the host cell furin and dengue NS2B-NS3 serine protease (NS2B-NS3pro) cleave viral polyprotein at various regions to produce structural and nonstructural viral proteins [4–6]. The NS3 protein is one of the viral non-structural proteins that possess enzymatic activities. The N-terminal of this protein contains 180 amino acid residues that represent NS3 protease [7, 8], while C-terminal region contains amino acid residues that represent RNA helicase and RNA-stimulated NTPase [9, 10]. The activity of NS2B-NS3pro depends on the interaction with its cofactor NS2B to form a NS2B-NS3pro complex [11]. It has been found that the disruption of NS2B-NS3pro functions inhibits viral replication [12]. Therefore, NS2B-NS3pro is considered as a potential target for the design of antiviral drugs [13].
At present, a legitimate vaccine or treatment to prevent or to cure this disease is unavailable [14]. These facts emphasize the need for a better understanding of the mechanism of viral infection and propagation in the host cell to combat this disease. Recently, computational studies indicated that cationic cyclic peptides have potential inhibition towards dengue NS2B-NS3pro [15, 16]. Protegrin-1 (PG-1, RGGRLCYCRRRFCVCVGR) is an eighteen amino acids cationic cyclic peptide with high content of basic residues and two disulphide bonds. The peptide is originally isolated from porcine white blood cells and considered as an antibiotic agent against a broad range of microorganisms [17, 18]. The formation of two disulphide bonds between cysteine residues endows PG-1 with a stabile β-hairpin secondary structure that is crucial for antimicrobial activity [19, 20]. Therefore, removal of these two disulphide bonds is found to noticeably reduce the antimicrobial activity of PG-1 [21]. The peptide is able to penetrate and disrupt the cell membrane by homodimerization [22, 23]. The mechanism of this activity depends on the secondary structure and the cationic nature of PG-1 molecules which are essential to generate pores in the cell membrane of microorganism [24–27]. In this study, our objective was to examine the efficacy of PG-1 as cationic cyclic peptide to inhibit dengue serine protease and subsequently reduce viral replication in host cells.
2. Methods
2.1. Synthesis of Cyclic PG-1
The linear peptides were prepared by automated peptide synthesis using Symphony parallel synthesizer (Protein Technologies, Tucson, AZ, USA) by standard solid-phase peptide synthesis. The crude peptides were lyophilized before proceeding to folding experiments. The lyophilized peptide was dissolved in 20% DMSO solution in a round bottom flask and stirred on a magnetic stirrer to produce a peptide concentration of 1.1 mM. The progress of the reaction was monitored by HPLC and LCMS. The formation of the first disulphide bond was completed within 24 hrs. The solution was then lyophilized to proceed for second disulfide formation. The lyophilized peptide with the first disulphide bond was dissolved in acetic acid water (4 : 1) so that the peptide concentration was 1.2 mM and iodine (10 equivalents) was added in one portion. The reactions were stirred at 25°C for 60 min and then quenched by diluting with water and extracting with CCl4 to remove excess iodine. The aqueous phase was then lyophilized to give the crude peptide. The identity of the crude peptide was confirmed by LC-MS. Purification of crude cyclised peptide was accomplished by RP-HPLC (Agilent 1200 series). The identity of the purified peptide with 98% purity was confirmed by LC-MS (Shimadzu LC/MS 2020, single quad).
2.2. Production of NS2B-NS3pro in E. coli
To produce single chain protease NSB2- (G4-T-G4)-NS3, the NS2B fragments were amplified individually by PCR using the primer pairs NS2B-F (5′-ATACTGAGGATCC GCCGATTTGGAACTG-3′) and NS2Blinker-R (ACCTACTAGGTACCTCCTCCACCCAGTGTCTGTTCTTC). The NS2B-NS3 was amplified by NS3linker-F (5′-ATCTATAGGTACCGGCGGTGGAGGTGCTGGAGTATTGTGG-3′) and NS3-R (5′-AGCATAAGCTTAAGCTTCAATTTTCT-3′). The linker sequence was added to NS2Blinker-R and NS3linker-F primers which included the site for KpnI restriction enzyme (all restriction sites are underlined). The PCR product of NS2B fragment was digested with BamHI and KpnI while NS2B fragment was digested with KpnI and HindIII. Purified fragments were cloned into pQE30 plasmid downstream of 6×His tag. The Escherichia coli X-blue strain (Promega, USA) was transformed with pQE30-cNSB-(G4TG4)-NS3 plasmid. The recombinant E. coli was inoculated in Luria-Bertani liquid medium (1% tryptone, 1% NaCl, 0.5% yeast extract, w/v, pH 7.0) supplemented with 100 mg/L ampicillin and cultured overnight at 37°C. In brief, 10 ml of overnight grown culture was added to 1000 mL of medium and incubated with shaking at 37°C until the optical density at 600 nm reached 0.5. Subsequently, isopropylthio-β-D-galactoside (IPTG) was then added to a final concentration of 0.5 mM and the bacteria were cultured for an additional 5 hrs at 37°C in a shaking incubator to induce protein expression. Finally, bacterial cells were harvested by centrifugation at 4000 rpm for 15 min at 4°C.
2.3. Protein Purification
The recombinant NS3pro was produced as soluble proteins and, therefore, the purification had been performed by His GraviTrap Flow precharged Ni Sepharose 6 Fast column (Amersham Biosciences, USA) according to the manufacturer's instructions. In brief, the column was normalized with phosphate buffer (20 mM sodium phosphate buffer and 500 mM NaCl, pH 7.4). The sample was loaded into the column and the column was washed with binding buffer (phosphate buffer containing 20 mM imidazole, pH 7.4). The recombinant protein was eluted with elution buffer (phosphate buffer containing 200 mM imidazole, pH 7.4). Further purification was applied using gel affinity chromatography to achieve more than 95% purity.
2.4. Protease Assay
The bioassay used in this study was modified from the method published by Kiat and coworkers [28]. Reaction mixtures with total volume of 200 μL were prepared. These reaction mixtures consisted of 100 μM fluorogenic peptide substrate (Boc-Gly-Arg-Arg-MCA), 2 μM NS2B-NS3pro complex, with or without PG-1 of varying concentrations, buffered at pH 8.5 with 200 mM Tris-HCl. The PG-1 was initially prepared in Tris-HCl buffer and assayed at five different concentrations. The reaction mixtures without fluorogenic peptide substrate were firstly incubated at 37°Cfor 30 minutes. Subsequently, the substrate was added and the mixture was further incubated at the same temperature for 30 minutes. Triplicates were performed for all measurements and the readings were taken using Tecan Infinite M200 Pro fluorescence spectrophotometer. Substrate cleavage was optimized at the emission at 440 nm upon excitation at 350 nm. The readings were then used for calculating Km values of peptide substrate and IC50 values of peptide inhibitors using nonlinear regression models in GraphPad Prism 5.01 software.
2.5. Maximum Nontoxic Dose Test (MNTD)
The MK2 cell lines were seeded at 1 × 104 cells per well in triplicate at optimal conditions (37°C, 5% CO2 in humidified incubator) in 96 well plates. PG-1 was diluted to serial concentrations 2.5, 12.5, 25, 50, 100 and 200 μM with DMEM media supplemented with 2% FBS. The cell culture was analyzed at 24, 48 and 72 hours using nonradioactive cell proliferation assay (Promega, USA) according to the manufacture protocol.
2.6. Treatment of DENV-2-Infected Cells with PG-1 Peptide
The MK2 cell lines were grown in a 24-well tissue culture plate (1 × 105 cells/well), incubated 24 hrs under optimal conditions (37°C and 5% CO2). DENV-2 was added to the wells (MOI of 2) followed by incubation for 1 hr with gentle shaking every 10 min for optimal virus to cell contact. The virus supernatant was removed, and the cells were washed twice with fresh serum free DMEM media to remove residual virus. New complete DMEM media containing 2.5, 7.5 and 12.5 μM of PG-1 were added and the cultures were incubated for 24, 48 and 72 hrs. Afterwards, cellular supernatants were collected and stored at −80°C for viral quantification by real-time PCR.
2.7. Real-Time PCR
For quantification of DENV-2 copies, the standard curve was generated by 10-fold serial dilution of known copies of DENV-2 RNA. Viral RNA was extracted from culture supernatant using QIAmp viral RNA minikit (QIAGEN, Germany) according to the manufacturer's instructions. A fragment located at the 5′UTR region of the virus genome was used to generate the primers. One-step RT-PCR using SyBr Green Master Kit (Qiagen, Germany) was used to conduct absolute quantification using ABI7300 machine from Applied Biosystems (Foster City, CA). The PCR programme included 1 cycle of 50°C for 2 min, 1 cycle of 95°C for 10 min, and 40 cycles of 95°C for 15 sec and 60°C for 1 min. Dissociation curve analysis was added to the end of each run. Results were analyzed using Sequence Detection Software Version 1.3 (Applied Biosystems, Foster City, CA, USA).
2.8. Statistical Analysis
All the assays were done in triplicates and the statistical analyses were performed using GraphPad Prism version 5.01 (GraphPad Software, San Diego, CA). P values <0.05 were considered significant. Error bars are expressed as ± SD.
3. Results and Discussion
The recombinant NS2B-NS3pro was produced as a soluble protein in E. coli and purified by nickel column (Figure 1(a)). Further purification was applied using gel affinity chromatography to achieve more than 95% of enzyme purity (Figure 1(b)). The activity of purified enzyme had been assessed at 37°C by catalyzing the fluorogenic peptide substrate t-Butyloxycarbonylglycyl-L-arginyl-L-arginyl-L-4-methylcoumaryl-7-amide (Boc-Gly-Arg-Arg-MCA). The PG-1 peptide was added to the protease reaction at different concentrations and the inhibition profile was plotted as shown in Figure 2.
Figure 1.
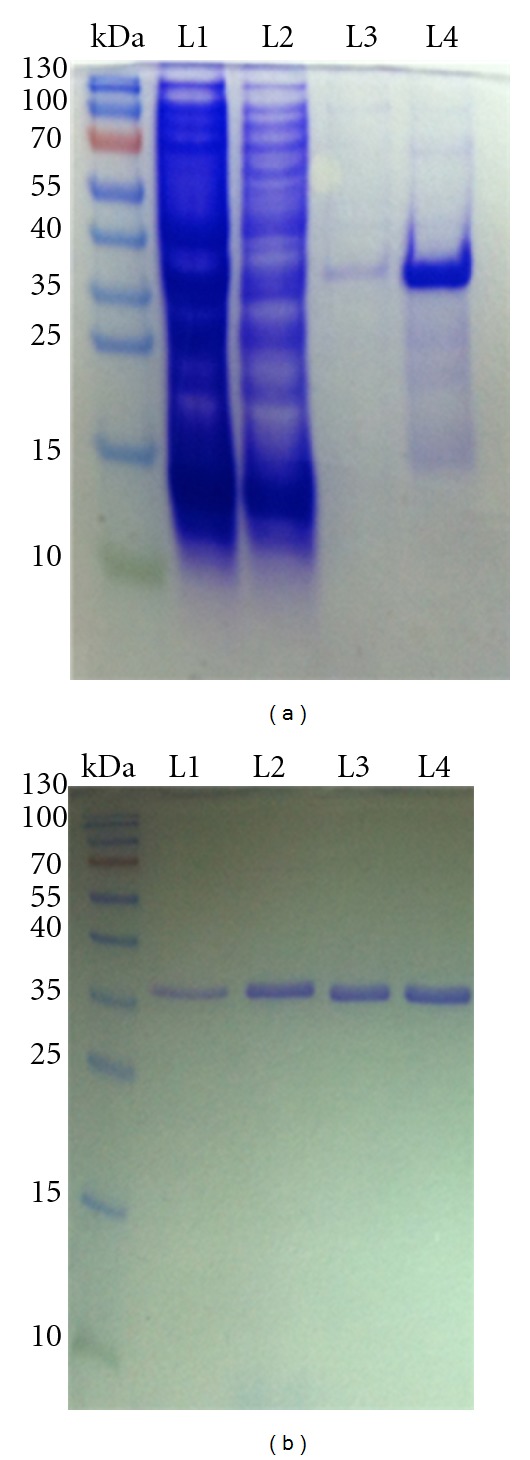
Production of single chain protease NSB2- (G4-T-G4)-NS3pro in E. coli. (a) The recombinant NS2B-NS3pro was produced as soluble proteins and purified by nickel column. L1: cell lysate, L2: washing flow through, L3: elution 1, and L4: elution 2. (b) Further purification was applied using gel affinity chromatography (95% of enzyme purity) L1 to L4: Elution 1 to 4. The activity of purified enzyme was assessed by catalyzing the fluorogenic peptide substrate (Boc-Gly-Arg-Arg-MCA).
Figure 2.
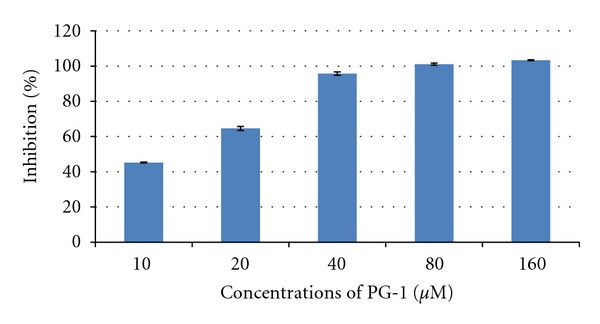
Dengue protease inhibition assay. The reaction mixtures consisted of fluorogenic peptide substrate and NS2B-NS3pro, with or without PG-1 of varying concentrations. The PG-1 was initially prepared in Tris-HCl buffer and assayed at five different concentrations, 10–160 μM. The best inhibition with low concentration of PG1 was observed at 40 μM with 95.7% inhibition.
It was observed that the inhibition potential increased with PG-1 concentration and the highest inhibition (95.7%) with low concentrations of PG-1 was at 40 μM. The kinetic assay study indicated that PG-1 competitively inhibited NS2B-NS3pro activity (alpha value 3.14) with Ki value 5.85 μM (Table 1). Intriguingly, the maximum enzyme velocity decreased threefold when PG-1 concentration was 20 μM (Figure 3). Previous studies have shown that various types of natural and chemical compounds were able to inhibit dengue NS2B-NS3pro activity. For example, some of plant natural compounds, such as Chalcones, have shown good inhibition potential against the dengue protease (Ki value 21–25 μM) [28]. The synthesized peptidic α-keto-amide compound inhibited dengue NS2B-NS3pro with Ki value of 47 μM [29]. Most recent study indicated that the retrotripeptides with an arylcyanoacrylamide group as N-terminal cap exhibited high inhibition potential against dengue protease with Ki value of 4.6 μM [30]. Most recent study showed that the inhibition potential of some chemical compounds towards NS2B-NS3pro measured by IC50 was 15.4, 20.4, and 27.0 μM [31].
Table 1.
The parameters of PG-1 Inhibition potential towards NS2B-NS3pro.
| Parameter* | Values | Std. Error |
|---|---|---|
| V max | 0.28 μmol/min | ±0.01 |
| Alpha | 3.14 | ±0.81 |
| K i | 5.85 μM | ±1.82 |
| IC50 | 11.70 μM | ±2.23 |
| K m | 109.10 μM | ±7.50 |
*Vmax: maximum enzyme velocity (μmol/min). Km: Michaelis-Menten constant (μM). Ki: inhibition constant (μM). Alpha: constant that determines mechanism. If alpha = 1, this is the same as noncompetitive. If alpha is very high, then the model approaches a competitive model. If alpha is very low (but greater than zero), the model approaches an uncompetitive model. All parameters were calculated by GraphPad Prism 5.0 software.
Figure 3.
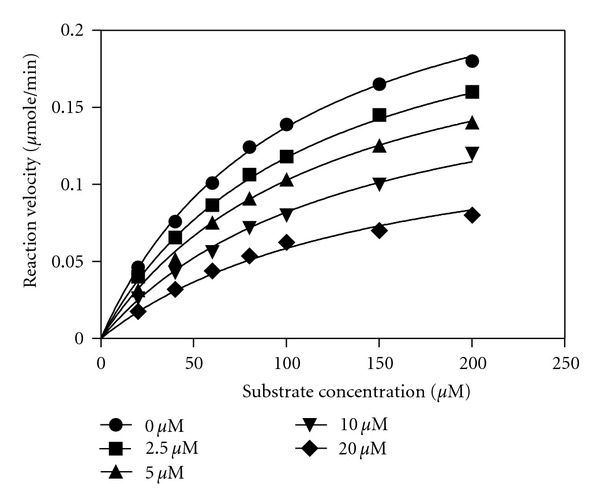
Kinetic assay plot for the inhibition of the NS2B-NS3pro from dengue virus by PG-1 peptide. This assay was carried out using increasing concentration of inhibitor while all other conditions were kept constant. The data was analysed by Michaelis-Menten model under nonlinear regression curve fit in GraphPad Prism 5 software. The peptide concentrations were 0, 2.5, 5, 10, and 20 μM. The maximum enzyme velocity decreased from approximately threefold when PG-1 concentration was 20 μM.
Cytotoxicity and compound stability can be considered as major limitations of the practical application of protease inhibitors. In this study, to test PG-1 toxicity, MK2 cell lines were incubated with increasing concentrations of PG-1 for 24, 48, and 72 hrs. The PG-1 peptide showed toxic effect against MK2 cell lines at concentrations greater than 12.5 μM (Figure 4). Other studies indicated that PG-1 was also toxic to 293A cell lines (human embryonic kidney cells) at 50 μg/ml (25 μM) [32] and more than 50% of human red blood cells lyses were observed at 80 μg/ml (40 μM) [33]. Therefore, three concentrations at nontoxic range were used to test PG-1 stability and ability to reduce dengue viral replication in MK2 cell lines. The results showed that the viral copy number significantly (P < 0.001) reduced with increasing concentrations of PG-1 (Figure 5(a)). Furthermore, the highest inhibition percentage was observed when the PG-1 concentration was 12.5 μM at 24, 48 and 72 hrs (Figure 5(b)). However, the low concentrations exhibited less inhibition percentage at 48 and 72 hrs as compared with 24 hrs, indicating that the PG-1 stability declined with longer incubation in culture media (Figures 5(a) and 5(b)). Similarly, it has been showed that at low doses (4 μg/ml), PG-1 has significant in vitro antimicrobial activity with low in vivo toxicity (up to 8 mg/kg i.v. mouse injection) [34]. This may be accounted by its short half-life in vivo as its level in mice plasma that was injected with 4 mg/ml i.v. declined rapidly to 28 μg/ml after 5 min [34]. Although PG-1 showed significant inhibition profile towards dengue virus in this study and to human immunodeficiency virus 1 (HIV1) in another study [35], the peptide instability should be considered as a major concern. The results in this study may give a clear picture that would then help in engineering new sequence of peptides to retain the antiviral activity against dengue while increasing its stability and eliminating the toxic characteristics of PG-1.
Figure 4.
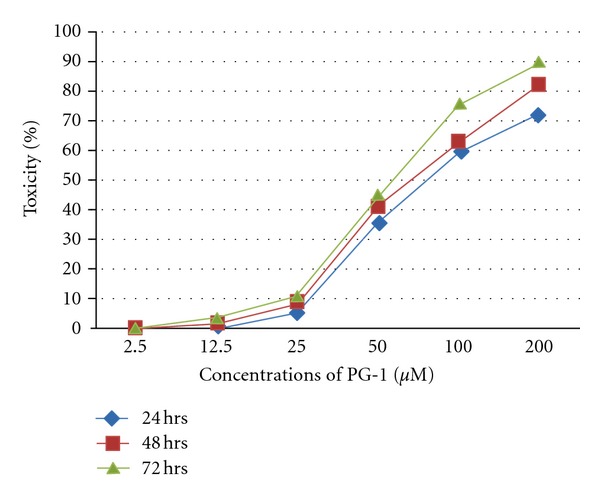
Maximum nontoxic dose (MNTD) of PG-1.The MK2 cell lines were seeded at 1 × 104 cells per well in triplicate at optimal conditions (37°C, 5% CO2 in humidified incubator) in 96 well plates. PG-1 was diluted to serial concentrations 2.5, 12.5, 25, 50, 100, and 200 μM with DMEM media supplemented with 2% FBS. The cell culture was analyzed at 24, 48, and 72 hrs using Non-Radioactive Cell Proliferation assay (Promega, USA) according to the manufacture protocol. The toxic dose of MK2 cells was identified to be greater than 12.5 μM.
Figure 5.
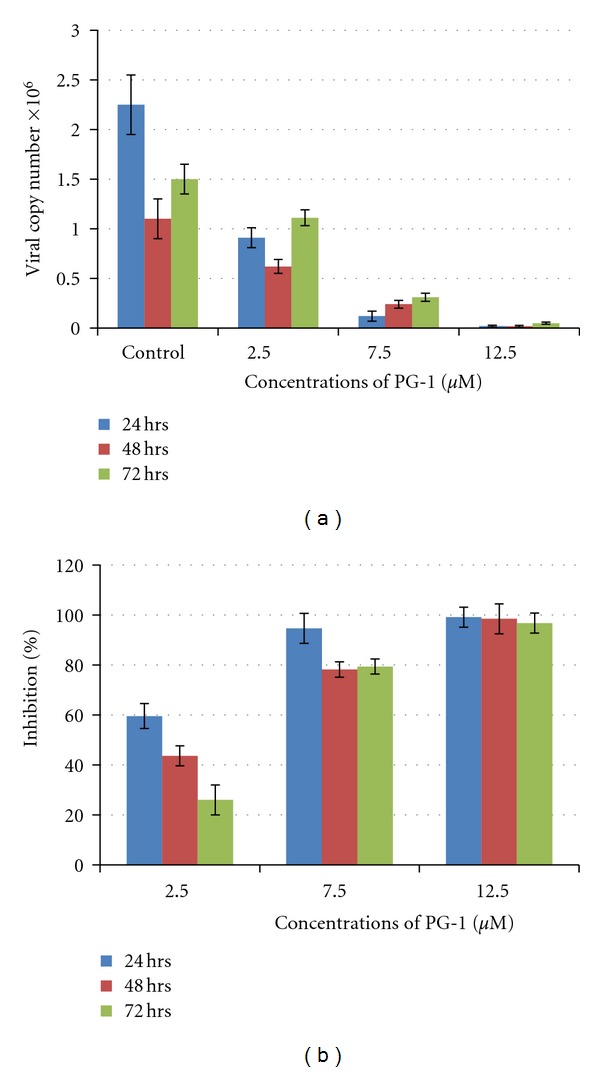
RNA quantification and the percentage of inhibition after applying different concentrations of PG-1 on MK2 cell lines infected with dengue virus. The standard curve was generated by 10-fold serial dilution of known copies of dengue RNA. Viral RNA was extracted from cellular supernatant and the fragment located at the 5′UTR region of the virus genome was amplified using one-step RT-PCR. Significant reduction (P < 0.001) in viral copy number was observed with increasing PG-1 concentration (a). The highest inhibition percentage was observed with 12.5 μM at 24, 48, and 72 hrs (b). PG-1 significantly (P < 0.01) inhibited DENV-2 replication in a dosedependent manner with the greatest inhibition effects at 24 hrs (two-way ANOVA with Bonferroni posttest, mean ± SD).
Conflict of Interests
The authors have declared that no competing interests exist.
Acknowledgment
This project was funded by the University of Malaya and Ministry of Science, Technology and Innovation (IPharm Grant 53-02-03-1049).
Abbreviations
- PG-1:
Protegrin-1 (RGGRLCYCRRRFCVCVGR)
- NS2B:
NS2B cofactor amino acids sequence 49–95 in DEN2 NS2B and 1394–1440 in DENV-2 polyprotein
- NS3:
NS3 protease amino acids sequence 1–185 in NS3 protease and 1476–1660 in DENV-2 polyprotein
- DENV-2:
Dengue virus serotype 2
- NS2B-NS3pro:
NS2B fused to NS3 via 9 amino acids (G4-T-G4)
- MCA:
fluorogenic peptide substrate (Boc-Gly-Arg-Arg-AMC)
- MK2 cells:
Rhesus monkey kidney cell lines.
References
- 1.Rigau-Pérez JG, Clark GG, Gubler DJ, Reiter P, Sanders EJ, Vorndam AV. Dengue and dengue haemorrhagic fever. The Lancet. 1998;352(9132):971–977. doi: 10.1016/s0140-6736(97)12483-7. [DOI] [PubMed] [Google Scholar]
- 2.Monath TP. Dengue: the risk to developed and developing countries. Proceedings of the National Academy of Sciences of the United States of America. 1994;91(7):2395–2400. doi: 10.1073/pnas.91.7.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gubler DJ, Clark GG. Dengue/dengue hemorrhagic fever: the emergence of a global health problem. Emerging Infectious Diseases. 1995;1(2):55–57. doi: 10.3201/eid0102.952004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin C, Amberg SM, Chambers TJ, Rice CM. Cleavage at a novel site in the NS4A region by the yellow fever virus NS2B-3 proteinase is a prerequisite for processing at the downstream 4A/4B signalase site. Journal of Virology. 1993;67(4):2327–2335. doi: 10.1128/jvi.67.4.2327-2335.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arias CF, Preugschat F, Strass JH. Dengue 2 virus NS2B and NS3 form a stable complex that can cleave NS3 within the helicase domain. Virology. 1993;193(2):888–899. doi: 10.1006/viro.1993.1198. [DOI] [PubMed] [Google Scholar]
- 6.Lobigs M. Flavivirus premembrane protein cleavage and spike heterodimer secretion require the function of the viral proteinase NS3. Proceedings of the National Academy of Sciences of the United States of America. 1993;90(13):6218–6222. doi: 10.1073/pnas.90.13.6218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chambers TJ, Hahn CS, Galler R, Rice CM. Flavivirus genome organization, expression, and replication. Annual Review of Microbiology. 1990;44:649–688. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- 8.Falgout B, Pethel M, Zhang YM, Lai CJ. Both nonstructural proteins NS2B and NS3 are required for the proteolytic processing of dengue virus nonstructural proteins. Journal of Virology. 1991;65(5):2467–2475. doi: 10.1128/jvi.65.5.2467-2475.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gorbalenya AE, Koonin EV, Donchenko AP, Blinov VM. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Research. 1989;17(12):4713–4730. doi: 10.1093/nar/17.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wengler G, Wengler G. The NS3 nonstructural protein of flaviviruses contains an RNA triphosphatase activity. Virology. 1993;197(1):265–273. doi: 10.1006/viro.1993.1587. [DOI] [PubMed] [Google Scholar]
- 11.Yusof R, Clum S, Wetzel M, Murthy HMK, Padmanabhan R. Purified NS2B/NS3 serine protease of dengue virus type 2 exhibits cofactor NS2B dependence for cleavage of substrates with dibasic amino acids in vitro. The Journal of Biological Chemistry. 2000;275(14):9963–9969. doi: 10.1074/jbc.275.14.9963. [DOI] [PubMed] [Google Scholar]
- 12.Geiss BJ, Stahla H, Hannah AM, Gari HH, Keenan SM. Focus on flaviviruses: current and future drug targets. Future Medicinal Chemistry. 2009;1(2):327–344. doi: 10.4155/fmc.09.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tomlinson SM, Malmstrom RD, Watowich SJ. New approaches to structure-based discovery of Dengue protease inhibitors. Infectious Disorders—Drug Targets. 2009;9(3):327–343. doi: 10.2174/1871526510909030327. [DOI] [PubMed] [Google Scholar]
- 14.Martina BEE, Koraka P, Osterhaus ADME. Dengue virus pathogenesis: an integrated view. Clinical Microbiology Reviews. 2009;22(4):564–581. doi: 10.1128/CMR.00035-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tambunan US, Alamudi S. Designing cyclic peptide inhibitor of dengue virus NS3-NS2B protease by using molecular docking approach. Bioinformation. 2010;5:250–254. doi: 10.6026/97320630005250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tambunan USF, Apriyanti N, Parikesit AA, Chua W, Wuryani K. Computational design of disulfide cyclic peptide as potential inhibitor of complex NS2B-NS3 dengue virus protease. African Journal of Biotechnology. 2011;10(57):12281–12290. [Google Scholar]
- 17.Kokryakov VN, Harwig SSL, Panyutich EA, et al. Protegrins: leukocyte antimicrobial peptides that combine features of corticostatic defensins and tachyplesins. FEBS Letters. 1993;327(2):231–236. doi: 10.1016/0014-5793(93)80175-t. [DOI] [PubMed] [Google Scholar]
- 18.Miyasaki KT, Lehrer RI. β-sheet antibiotic peptides as potential dental therapeutics. International Journal of Antimicrobial Agents. 1998;9(4):269–280. doi: 10.1016/s0924-8579(98)00006-5. [DOI] [PubMed] [Google Scholar]
- 19.Fahrner RL, Dieckmann T, Harwig SSL, Lehrer RI, Eisenberg D, Feigon J. Solution structure of protegrin-1, a broad-spectrum antimicrobial peptide from porcine leukocytes. Chemistry and Biology. 1996;3(7):543–550. doi: 10.1016/s1074-5521(96)90145-3. [DOI] [PubMed] [Google Scholar]
- 20.Lai JR, Huck BR, Weisblum B, Gellman SH. Design of non-cysteine-containing antimicrobial β-hairpins: structure-activity relationship studies with linear protegrin-1 analogues. Biochemistry. 2002;41(42):12835–12842. doi: 10.1021/bi026127d. [DOI] [PubMed] [Google Scholar]
- 21.Harwig SSL. Intramolecular disulfide bonds enhance the antimicrobial and lytic activities of protegrins at physiological sodium chloride concentrations. European Journal of Biochemistry. 1996;240(2):352–357. doi: 10.1111/j.1432-1033.1996.0352h.x. [DOI] [PubMed] [Google Scholar]
- 22.Panchal RG, Smart ML, Bowser DN, Williams DA, Petrou S. Pore-forming proteins and their application in biotechnology. Current Pharmaceutical Biotechnology. 2002;3(2):99–115. doi: 10.2174/1389201023378418. [DOI] [PubMed] [Google Scholar]
- 23.Sokolov Y, Mirzabekov T, Martin DW, Lehrer RI, Kagan BL. Membrane channel formation by antimicrobial protegrins. Biochimica et Biophysica Acta. 1999;1420(1-2):23–29. doi: 10.1016/s0005-2736(99)00086-3. [DOI] [PubMed] [Google Scholar]
- 24.Drin G, Temsamani J. Translocation of protegrin I through phospholipid membranes: role of peptide folding. Biochimica et Biophysica Acta. 2002;1559(2):160–170. doi: 10.1016/s0005-2736(01)00447-3. [DOI] [PubMed] [Google Scholar]
- 25.Gidalevitz D, Ishitsuka Y, Muresan AS, et al. Interaction of antimicrobial peptide protegrin with biomembranes. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(11):6302–6307. doi: 10.1073/pnas.0934731100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sitaram N, Nagaraj R. Interaction of antimicrobial peptides with biological and model membranes: structural and charge requirements for activity. Biochimica et Biophysica Acta. 1999;1462(1-2):29–54. doi: 10.1016/s0005-2736(99)00199-6. [DOI] [PubMed] [Google Scholar]
- 27.Hancock REW, Lehrer R. Cationic peptides: a new source of antibiotics. Trends in Biotechnology. 1998;16(2):82–88. doi: 10.1016/s0167-7799(97)01156-6. [DOI] [PubMed] [Google Scholar]
- 28.Kiat TS, Pippen R, Yusof R, Ibrahim H, Khalid N, Rahman NA. Inhibitory activity of cyclohexenyl chalcone derivatives and flavonoids of fingerroot, Boesenbergia rotunda (L.), towards dengue-2 virus NS3 protease. Bioorganic and Medicinal Chemistry Letters. 2006;16(12):3337–3340. doi: 10.1016/j.bmcl.2005.12.075. [DOI] [PubMed] [Google Scholar]
- 29.Yin Z, Patel SJ, Wang WL, et al. Peptide inhibitors of dengue virus NS3 protease. Part 2: SAR study of tetrapeptide aldehyde inhibitors. Bioorganic and Medicinal Chemistry Letters. 2006;16(1):40–43. doi: 10.1016/j.bmcl.2005.09.049. [DOI] [PubMed] [Google Scholar]
- 30.Nitsche C, Behnam MAM, Steuer C, Klein CD. Retro peptide-hybrids as selective inhibitors of the Dengue virus NS2B-NS3 protease. Antiviral Research. 2012;94(1):72–79. doi: 10.1016/j.antiviral.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 31.Yang CC, Hsieh YC, Lee SJ, et al. Novel dengue virus-specific NS2B/NS3 protease inhibitor, BP2109, discovered by a high-throughput screening assay. Antimicrobial Agents and Chemotherapy. 2011;55(1):229–238. doi: 10.1128/AAC.00855-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steinstraesser L, Tippler B, Mertens J, et al. Inhibition of early steps in the lentiviral replication cycle by cathelicidin host defense peptides. Retrovirology. 2005;2, article 2 doi: 10.1186/1742-4690-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langham AA, Khandelia H, Kaznessis YN. How can a β-sheet peptide be both a potent antimicrobial and harmfully toxic? Molecular dynamics simulations of protegrin-1 in micelles. Peptide Science. 2006;84(2):219–231. doi: 10.1002/bip.20397. [DOI] [PubMed] [Google Scholar]
- 34.Steinberg DA, Hurst MA, Fujii CA, et al. Protegrin-1: a broad-spectrum, rapidly microbicidal peptide with in vivo activity. Antimicrobial Agents and Chemotherapy. 1997;41(8):1738–1742. doi: 10.1128/aac.41.8.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tamamura H, Murakami T, Horiuchi S, et al. Synthesis of protegrin-related peptides and their antibacterial and anti-human immunodeficiency virus activity. Chemical and Pharmaceutical Bulletin. 1995;43(5):853–858. doi: 10.1248/cpb.43.853. [DOI] [PubMed] [Google Scholar]


