Abstract
The corticotropin-releasing factor (CRF) family of neuropeptides includes the mammalian peptides CRF, urocortin, and urocortin II, as well as piscine urotensin I and frog sauvagine. The mammalian peptides signal through two G protein-coupled receptor types to modulate endocrine, autonomic, and behavioral responses to stress, as well as a range of peripheral (cardiovascular, gastrointestinal, and immune) activities. The three previously known ligands are differentially distributed anatomically and have distinct specificities for the two major receptor types. Here we describe the characterization of an additional CRF-related peptide, urocortin III, in the human and mouse. In searching the public human genome databases we found a partial expressed sequence tagged (EST) clone with significant sequence identity to mammalian and fish urocortin-related peptides. By using primers based on the human EST sequence, a full-length human clone was isolated from genomic DNA that encodes a protein that includes a predicted putative 38-aa peptide structurally related to other known family members. With a human probe, we then cloned the mouse ortholog from a genomic library. Human and mouse urocortin III share 90% identity in the 38-aa putative mature peptide. In the peptide coding region, both human and mouse urocortin III are 76% identical to pufferfish urocortin-related peptide and more distantly related to urocortin II, CRF, and urocortin from other mammalian species. Mouse urocortin III mRNA expression is found in areas of the brain including the hypothalamus, amygdala, and brainstem, but is not evident in the cerebellum, pituitary, or cerebral cortex; it is also expressed peripherally in small intestine and skin. Urocortin III is selective for type 2 CRF receptors and thus represents another potential endogenous ligand for these receptors.
Corticotropin-releasing factor (CRF), a 41-aa peptide that was first isolated from ovine hypothalamus (1), plays an important role in the regulation of the pituitary-adrenal axis, and in endocrine, autonomic and behavioral responses to stress (2). Until recently, the CRF family of neuropeptides comprised structurally related mammalian and nonmammalian peptides including urocortin (Ucn), a 40-aa peptide originally identified in rat brain (3), fish urotensin I (Uro; ref. 4), and amphibian sauvagine (Svg; ref. 5).
In addition to effects on the pituitary and central nervous system, CRF and related peptides have been shown to modulate cardiovascular and gastrointestinal functions and inflammatory processes in mammals. The actions of CRF-related peptides are mediated via specific binding to G protein-coupled receptors derived from two distinct genes, CRF type 1 (CRF-R1; refs. 6–8) and CRF type 2 (CRF-R2; refs. 9–11) receptors. CRF-R1 and CRF-R2 have distinct pharmacologies and differ in their anatomical distribution (12). CRF and Ucn bind and activate CRF-R1 with similarly high affinities (3). Ucn displays high affinity for CRF-R2, whereas CRF has approximately 10-fold lower affinity for this receptor. We recently identified a second mammalian urocortin II (Ucn II), a 38-aa peptide from mouse brain, that binds CRF-R2 selectively and with high affinity (13). A human sequence closely related to Ucn II (human urocortin related peptide or URP) was also identified, but lacks the typical consensus proteolytic cleavage site associated with C-terminal processing (13). Because we were aware of an additional expressed sequence tag (EST) related to urocortin in the human database, we left open the question of whether human URP may represent the mouse Ucn II ortholog until we could search for additional genes in the mouse. We have now identified both human and mouse genes encoding a deduced 38-aa peptide that represents a new member of the CRF family, which we name urocortin III (Ucn III). Ucn III preferentially binds and activates CRF-R2 and has a discrete central nervous system and peripheral distribution.
Materials and Methods
PCR Amplification and Hybridization Screening.
Nested PCR primers were designed based on the partial human EST sequence, 5′-AAG AGT CCC CAC CGC ACC AAG TTC ACC-3′ and 5′-TCC CTC GAC GTC CCC ACC AAC ATC ATG-3′, and used to screen a Human GenomeWalker Kit (CLONTECH). Nested PCR was performed for 35 cycles at 66°C for 12 min. The amplified fragments were subcloned into pCRIITOPO vector (Invitrogen), sequenced, and found to encode the full-length mature peptide. A 144-bp probe was generated that included the mature peptide region of human Ucn III. This probe was used to screen a mouse genomic library in λFIXII vector (Stratagene) by using standard hybridization techniques (3). By using primers designed from the mouse genomic sequence, a mouse Ucn III cDNA was isolated from whole brain cDNA by PCR. PCR was performed at 55°C for 35 cycles with 2 min extension at 72°C.
Peptide Synthesis.
Human and mouse Ucn III were synthesized manually by using the solid phase approach, and purified by using RP-HPLC as described for mouse Ucn II (13). The peptide was cleaved and deprotected in hydrofluoric acid and then purified on a Vydac (Hesperia, CA) C4 cartridge using 0.1% trifluoroacetic acid (TFA); ref. 14]. Peptides were greater than 95% pure by the criteria of independent HPLC and capillary zone electrophoresis. Mass spectra confirmed the composition of the preparations.
Binding Assays.
Crude membrane fractions were prepared from Chinese hamster ovary (CHO) cells stably expressing either cloned human CRF-R1 or murine CRF-R2 as described (3). A stably transfected cell line expressing human CRF-R2α was generously provided by Demitri Grigoriadis (Neurocrine Biosciences, San Diego). Test peptides and radio-ligand, [125I-Tyr0,Glu1,Nle17]sauvagine, diluted in assay buffer (20 mM Hepes/2 mM EGTA/0.1% BSA/10% sucrose, pH 7.6), were combined with membrane fractions (10 μg) in 0.22-μm Duropore microtiter plates (Millipore) precoated with 0.1% polyethyleneimine. After 90 min at room temperature, the reaction mixture was filtered and washed twice with assay buffer. The radioligand complex was quantified by gamma-counting. Inhibitory binding constants, Ki's, were determined by using GRAPHPAD software. The affinities of Ucn II and Ucn III for the CRF binding protein (CRF-BP) were estimated on the basis of the displacement of [D-125I-Tyr0]hCRF according to ref. 15.
cAMP Assay.
CHO cells, stably transfected with either human CRF-R1 or murine CRF-R2, were plated into 48-well tissue culture dishes (Costar) and allowed to recover for 24 h. The medium was changed at least 2 h before treatments to DMEM/0.1% FBS. The cells were preincubated with 0.1 mM 3-isobutyl-1-methylxanthine for 30 min and then exposed to peptides for 20 min at 37°C. Rat aortic smooth muscle cells, A7r5, were plated into 48-well culture dishes and allowed to recover for 48 h. Cells were starved in DMEM/0.2% FBS overnight before the experiment. Cells were treated as described above for CHO cells. Rat anterior pituitary cells were established in culture (16) and treated with test peptides for 45 min at 37°C. Intracellular cAMP was extracted from all cells and measured from triplicate wells using a RIA kit (Biomedical Technologies, Stoughton, MA). Potencies were determined by using PRISM GRAPHPAD software.
RNase Protection.
Total RNA was extracted by using Tri Reagent (Molecular Research Center, Cincinnati, OH). Mouse Ucn III and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA levels were measured simultaneously by RNase protection, using mouse GAPDH as an internal loading control. A 528-nt Ucn III antisense riboprobe specific to the mouse Ucn III mRNA was synthesized by using T3 RNA polymerase. A 200-nt antisense riboprobe specific to mouse GAPDH mRNA was synthesized by using T3 RNA polymerase. All riboprobes were synthesized in the presence of [α-32P]UTP [3,000 Ci/mmol (1 Ci = 37 GBq)] and either 20 μM UTP for Ucn III or 200 μM UTP for GAPDH, as described (17). The fragment sizes protected by Ucn III and GAPDH riboprobes are 415 and 135 nucleotides, respectively.
RNase protection analysis was carried out as described (17). RNA samples (50 μg of peripheral tissue or 20–25 μg of brain tissues) were hybridized in 24 μl of deionized formamide plus 6 μl of hybridization buffer containing 107 cpm of Ucn III and 3 × 104 cpm GAPDH antisense riboprobes. After heating to 85°C for 5 min, the samples were hybridized at 42°C for 12 h and subsequently digested by RNase (175 μg/ml RNase A and 500 U/ml RNase T1) at 24°C for 60 min. The samples were resolved on 5% polyacrylamide urea gels. Image analysis was performed by using the PhosphorImager system (Molecular Dynamics) and the IMAGEQUANT 4.0 software package.
In Situ Hybridization.
33P-labeled antisense and sense cRNA probes transcribed from linearized mouse Ucn III cDNA were hybridized to 20 μM coronal brain sections of adult C57BL/6 mice and Sprague–Dawley rats. After hybridization, the slides were dipped in Kodak NTB-2 liquid emulsion and exposed at 4°C for 10 days.
Results
Cloning of Human and Mouse Ucn III.
We screened the human EST database of GenBank by using a pufferfish (Takifugu rubripes) sequence (GenBank assession number AJ251323) related to urocortin (URP) as a probe. A partial human EST was identified that was structurally related to the pufferfish URP sequence and contained the precursor sequence and the first 29 aa of the mature peptide region. To extend sequence information at the C terminus, primers were designed based on the partial human EST sequence to screen a human GenomeWalker Kit (CLONTECH) by PCR. Library pools of human genomic DNA were screened by using gene-specific and anchored primers in a nested PCR strategy. We identified a full-length human gene encoding a protein for a putative peptide, Ucn III, which is related to known CRF family members.
Based on the full-length human Ucn III sequence, a 144-bp probe that spanned the mature peptide region was generated to search for a mouse ortholog. The probe was used to screen a mouse genomic λFIXII library (Stratagene) by low stringency hybridization. A full-length mouse genomic clone was identified and sequenced. To verify expression of the gene, we performed RT-PCR with primers based on the mouse sequence and identified a mouse cDNA from whole brain.
Both human and mouse Ucn III genes contain two potential initiation sites for translation. The nucleotide sequence of the human gene encodes a protein deduced to be either 161 or 159 aa, with preference for the first methionine (ref. 18; Fig. 1A). Similarly, the mouse gene encodes a protein deduced to be either a 164- or 162-aa precursor also with preference for translation beginning at the first methionine (Fig. 1B). The C-terminal sequence of both human and mouse Ucn III peptides contains a pair of basic residues, RK for the human and KK for the mouse, immediately preceded by a glycine, presumed to be involved in C-terminal amidation. From among several options, we tentatively selected a cleavage site that would generate the same length peptide (38 aa) in both mouse and human, but the size of the mature Ucn III has yet to be established. The predicted mature peptide regions of the human and mouse Ucn III peptides are shown in the boxed regions (Fig. 1 A and B). Because the mature peptide regions of human and mouse Ucn III differ only by 4-aa residues, we consider them likely to be orthologous.
Figure 1.
(A) Predicted amino acid sequence encoding human Ucn III and (B) mouse Ucn III. Amino acids are numbered starting with the initiating methionine. The putative mature peptide coding region is in the boxed area. (C) Alignment of putative mature peptide regions of human and mouse Ucn III with pufferfish urocortins, human and mouse Ucn II, human and ovine CRF, pufferfish urotensin (Uro), frog sauvagine, and human and mouse Ucn. Residues identical to human Ucn III sequence are boxed. Alignment was made by using the CLUSTAL method of MEGALIGN in dnastar. Z, pyroglutamic acid; ■, amidation site (putative for human Ucn II). (D) The phylogenetic tree groups human and mouse Ucn III with the pufferfish urocortins and human and mouse Ucn II. The more distantly related group is comprised of ovine and human CRF, human and mouse Ucn, pufferfish Uro, and frog sauvagine. The scale beneath the tree measures sequence distances. The phylogenetic tree was generated by DNASTAR.
In Fig. 1C, the putative 38-aa mature peptide region of human Ucn III sequence is compared with that of other family members. Human and mouse Ucn III share 40% identity to mouse Ucn II and they share 37% identity to human URP. They are more distantly related to Ucn and CRF. Human and mouse Ucn III share 21% and 18% identity with human and mouse Ucn, respectively. Human and mouse Ucn III share 32% and 26% identity to human/rat CRF. The mammalian urocortins II and III appear to be a separate but related evolutionary branch of the CRF-family with closer ties to two pufferfish urocortins than to mammalian Ucn or CRF (Fig. 1D). At the amino acid level, the pufferfish URP (GenBank accession no. AJ251323) is most closely related (76% identity) to both human and mouse Ucn III, and more distantly related to human URP (42%) and mouse Ucn II (37%). A second pufferfish urocortin sequence (GenBank accession no. AL175143) is also more closely related to Ucn III (53%) than to any of the other mammalian CRF family members. A third pufferfish peptide (GenBank accession no. AL218869) is most similar to the fish urotensin I with highest amino acid identity to flounder urotensin I (63%).
Receptor and CRF-BP Binding and Cellular Activation.
Receptor binding.
Comparisons of the binding affinities and potencies for stimulating cAMP accumulation in cells stably expressing the human CRF-R1, rat CRF-R2α, and mouse CRF-R2β are shown in Table 1. Ucn is significantly more potent than either Ucn II or Ucn III in binding to CRF-R1. Both Ucn II and Ucn III selectively bind both splice variants of CRF-R2, compared with CRF-R1. Human Ucn III displays lower affinity for either type 2 receptor than Ucn or Ucn II. Mouse Ucn III displays considerably higher affinity for murine CRF-R2 receptors than does human Ucn III. Both Ucn II and Ucn III display slightly higher affinities for CRF-R2β compared with CRF-R2α. The potency advantage of mouse Ucn III over human Ucn III is also observed for binding to human CRF-R2α (data not shown).
Table 1.
Binding properties and functional activities of CRF-family ligands
| Peptide | Binding* hCRF-R1 (Ki, nM) | Binding* rCRF-R2α (Ki, nM) | Binding* mCRF-R2β (Ki, nM) | cAMP† hCRF-R1 (EC50, nM) | cAMP† rCRF-R2α (EC50, nM) | cAMP† mCRF-R2β (EC50, nM) |
|---|---|---|---|---|---|---|
| Ucn (rat) | 0.32 (0.14–0.77) | 2.2 (0.91–5.4) | 0.62 (0.14–2.8) | 0.15 (0.03–0.64) | 0.063 (0.014–0.28) | 0.087 (0.017–0.43) |
| Ucn II (human) | >100 | 1.7 (0.73–4.1) | 0.50 (0.22–1.16) | >100 | 0.26 (0.11–0.61) | 0.42 (0.16–1.1) |
| Ucn II (mouse) | >100 | 2.1 (0.78–5.4) | 0.66 (0.13–3.3) | >100 | 0.14 (0.04–0.43) | 0.05 (0.02–0.12) |
| Ucn III (human) | >100 | 21.7 (8.2–57) | 13.5 (9.2–19.7) | >100 | 0.16 (0.09–0.28) | 0.12 (0.06–0.20) |
| Ucn III (mouse) | >100 | 5.0 (4.0–6.3) | 1.8 (0.77–4.1) | >100 | 0.073 (0.052–0.10) | 0.081 (0.08–0.80) |
The values were determined from three to six independent experiments using stably transfected Chinese hamster ovary cells (†) or their membranes (*) for each test peptide. EC50 and Ki values were determined by using prism software. Their log10 values were averaged (γ). The average EC50 or Ki was taken to be 10γ. The standard deviation of the log10 values was calculated (δ). The ranges given were taken to be [(10γ)10δ or 10γ/10δ]. Ucn, urocortin.
Activation of adenylate cyclase in receptor-transfected cells.
Intracellular cAMP accumulation in cells stably transfected with either CRF-R1 or CRF-R2 was used as a measure of receptor activation. Both Ucn II and Ucn III have very low potencies to activate CRF-R1 (>100 nM), contrasting sharply with that of Ucn, whose EC50 is ≈0.15 nM. Indeed, Ucn III shows no activation of CRF-R1 even at very high doses (1 μM). The potencies of Ucn II and III in activating CRF-R2α and CRF-R2β are approximately equal and nearly equivalent to that of Ucn. Thus, in the cAMP stimulation assay both Ucn II and III show selectivity for CRF-R2 over CRF-R1, but no preference with respect to CRF-R2α and CRF-R2β. Further, the relative potencies of murine and human Ucn II and Ucn III to functionally activate CRF-R2 overlap despite the lower affinity of human Ucn III for binding to CRF-R2.
Activation of adenylate cyclase in cells expressing endogenous receptors.
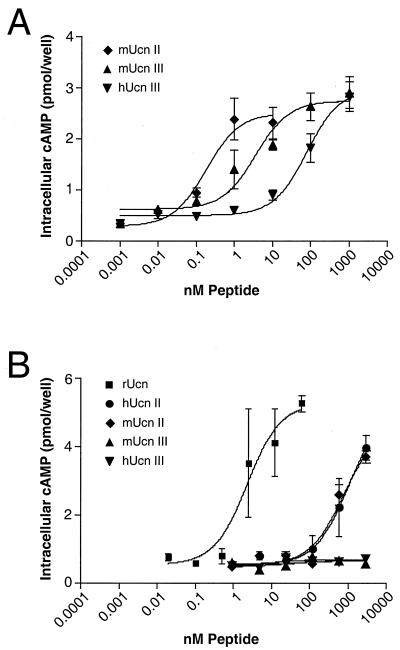
The abilities of Ucn II and Ucn III to activate adenylate cyclase in cells expressing endogenous CRF-R1 (cultured primary anterior pituitary cells; ref. 3) or CRF-R2β (A7r5 cells; ref. 19) are shown in Fig. 2. Ucn II and Ucn III are able to activate endogenous CRF-R2β at sub- or low nanomolar concentrations of ligand (Fig. 2A). Expectedly, Ucn II exhibits low potency to increase cAMP in cultured pituitary cells expressing CRF-R1, whereas Ucn III is inactive in this assay even at concentrations >1 μM (Fig. 2B).
Figure 2.
Effects of Ucn-related peptides on cAMP accumulation in a CRF-R2β-expressing cell line (A) and primary rat anterior pituitary cells (B). (A) A7r5 rat aortic smooth muscle cells. EC50: mUcn II, 0.18 nM; mUcn III, 3.7 nM; hUcn III, 80.9 nM. (B) Primary rat anterior pituitary cells were established in culture and were stimulated with various peptides for 45 min. EC50: rUcn, 2.3 nM; hUcn II, 1 μM; mUcn II, 0.75 μM (EC50 for hUcn II and mUcn II estimated by using the plateau of rUcn).
Binding to CRF binding protein.
As opposed to CRF and urocortin, which have subnanomolar affinities for CRF-BP, neither Ucn II nor Ucn III exhibit appreciable affinity for this protein (data not shown).
Ucn III mRNA Expression.
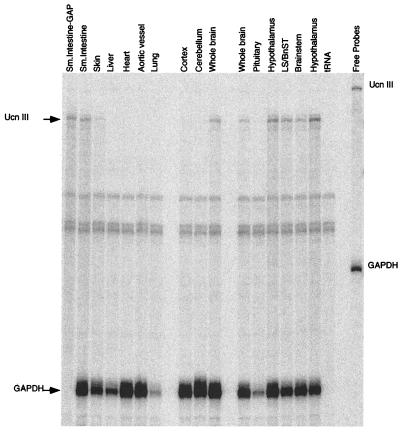
RNase protection assays were performed to determine the tissue distribution of mouse Ucn III mRNA. Total RNA from tissue encompassing several regions of the central nervous system and peripheral tissues was hybridized with a 528-bp cRNA probe that spanned the mature peptide region and gave a protected fragment of 415 bp. In the central nervous system, sites of mRNA expression include the hypothalamus, brainstem, and lateral septum (LS)/bed nucleus of stria terminalis (BnST) (Fig. 3). The pituitary, cerebellum, and cerebral cortex showed no detectable mRNA expression. In the periphery, Ucn III mRNA is expressed in small intestine and skin, with no detectable expression in heart, aortic vessel, liver, or lung (Fig. 3).
Figure 3.
Expression of mouse Ucn III mRNA in brain and peripheral tissues. A representative image of RNase protection assay of Ucn III mRNA. Total RNA isolated from each tissue listed was hybridized with the mouse Ucn III antisense probe and mouse GAPDH. The protected fragments were resolved on a 6% polyacrylamide urea gel. BnST, bed nucleus of stria terminalis.
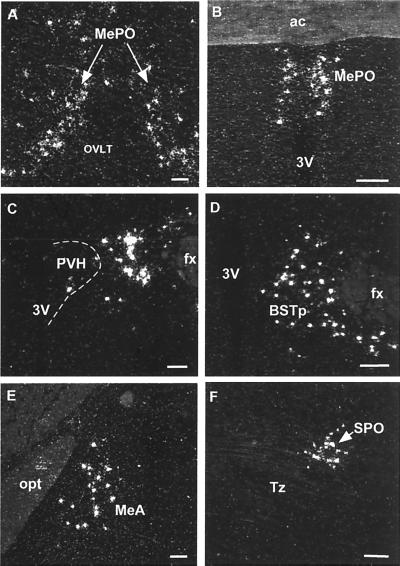
To reveal a more detailed pattern of expression of Ucn III in the brain, we carried out in situ hybridization on both rat and mouse brain sections by using antisense and sense cRNA probes generated from a 528-bp mouse Ucn III cDNA template. A positive Ucn III mRNA signal was observed only in sections hybridized with the antisense probe. The distribution of Ucn III mRNA expression was found to be limited mainly to a few discrete regions of the ventral forebrain (Fig. 4). One was in the median preoptic nucleus, where a continuous band of positively labeled cells comprised an inverted Y-shaped midline grouping. Neither of the circumventricular cell groups with which the median preoptic nucleus is intimately associated (the vascular organ of the lamina terminalis and the subfornical organ) contained positive hybridization signals, and the only additional sites of Ucn III mRNA expression at this level were over scattered cells in medial and lateral preoptic areas. A second major locus of Ucn III mRNA expression appeared as a longitudinally organized cluster of labeled cells associated with (and essentially encircling) the columns of the fornix throughout the rostral hypothalamus. This cluster includes cells situated within the posterior part of the bed nucleus of the stria terminalis, anterior and lateral hypothalamic areas, and the ill-defined region just lateral to (but seldom within) the paraventricular nucleus of the hypothalamus. The caudal extension of this grouping occupied an analagous position, mainly dorsal and lateral to the rostral aspects of the dorsomedial hypothalamic nucleus. Apart from this “rostral perifornical” group, scattered positively hybridized neurons were seen reliably in the ventral part of the anterior periventricular nucleus and the retrochiasmatic area. The third major site of Ucn III mRNA expression was over a subset of cells in the anterodorsal part of the medial amygdaloid nucleus. In the brainstem, the only reliable site of Ucn III expression was localized discretely to an auditory-related cell group, the superior paraolivary nucleus (Fig. 4).
Figure 4.
Hybridization histochemical localization of Ucn III mRNA in the rat brain. Positive hybridizing signal was most prominent in three regions of the ventral forebrain. These included the median preoptic nucleus (A and B), the rostral perifornical area that encompasses areas just lateral to the paraventricular nucleus (C), the posterior part of the bed nucleus of stria terminalis (D), and the medial amygdaloid nucleus (E). In the brainstem, positive hybridization signals were detected mainly in the superior paraolivary nucleus (F). 3V, third ventricle; ac, anterior commissure; BSTp, posterior part of the bed nucleus of stria terminalis; fx, fornix; MeA, medial nucleus of amygdala; MePO, median preoptic nucleus; OVLT, vascular organ of the lamina terminalis; opt, optic tract; PVH, paraventricular nucleus of hypothalamus; SPO, superior paraolivary nucleus; Tz, nucleus of the trapezoid body. (Scale bars, 50 μm.)
Discussion
We identifed Ucn III by sequence homology screening of the human GenBank database. Analyses of the peptide or nucleotide sequences of the complete proteins or putative mature peptide regions of Ucn III and Ucn II suggest that they represent a separate branch of the CRF family more closely related to one another than they are to other members (Fig. 1D). With 76% identity within the putative mature peptide domain, Ucn III and pufferfish URP are likely to be orthologous. It is likely that the human URP gene, which is most related to mouse Ucn II, is the human Ucn II ortholog. We propose that the human URP gene be renamed the human urocortin II gene. The fact that the human Ucn II precursor lacks consensus proteolytic processing residues at the putative C-terminal region (although the potential amide-donating glycine is present) and has multiple potential N-terminal cleavage sites raises the possibility that the product of the human Ucn II prohormone may be an extended peptide or even a cyclic protein that is larger than the other members of the family. The orthologous relationship between murine and human Ucn II notwithstanding, the chemical nature and functions of Ucn II in the human remain at issue.
Both mouse and human Ucn III are highly selective for the type 2 CRF receptors and, like Ucn II, exhibit low affinities for type 1 receptors and minimal abilities to induce cAMP production in cells expressing either endogenous (anterior pituitary corticotropes; Fig. 2B) or transfected (Table 1) CRF-R1. Human Ucn III has lower affinity for the type 2 receptors than do either mouse Ucn III or human or mouse Ucn II. Only four residues differ between human and mouse Ucn III, providing structure/function insight regarding the requirements for high affinity CRF-R2 binding. Human Ucn III is also less potent on human CRF-R2α (data not shown), indicating that the affinity differences between mouse and human Ucn III are not related to the species source of the receptor. However, despite its relatively low binding affinity for CRF-R2, human Ucn III is functionally quite potent (EC50 < 1 nM) to promote cAMP production by cells expressing this receptor. Therefore, both human and mouse Ucn III exhibit sufficient potency to serve as native ligands for CRF-R2. Neither of the CRF-R2 selective ligands, Ucn III or Ucn II, are bound by CRF-BP with high affinity. By contrast, the two ligands with high affinity for CRF-R1 also have high affinity for CRF-BP.
From the RNase protection analyses, it is feasible that Ucn III could gain access to receptors derived from CRF-R2 in both the brain and periphery. In the periphery, Ucn III mRNA is found in the small intestine and skin, although the cell types in which Ucn III mRNA is expressed remain to be determined. In the GI tract, CRF-R2 has been shown to be involved in modulating gut motility (20).
In the brain, Ucn III mRNA is found in discrete subcortical regions where its distribution is distinct from that of CRF (21), Ucn (22), and Ucn II (13). Although identification of the contexts in which Ucn III may operate must await the results of detailed immunohistochemical and functional analyses, some initial insights may be gleaned from its major sites of expression in the median preoptic nucleus, what we term here the rostral perifornical region, and the medial nucleus of the amygdala. By virtue of receiving inputs from both circumventricular structures of the lamina terminalis and the brainstem, the median preoptic nucleus is considered a key site for the integration of neural and humoral signals from the viscera, related to fluid and cardiovascular homeostasis (23, 24). Among the major targets of its projections are multiple relevant neurosecretory and preautonomic populations of the paraventricular and/or supraoptic nuclei of the hypothalamus (25), at least some of which are known to express CRF-R2 (26). It is decidedly more difficult to assign potential functions to the rostral perifornical group of Ucn III-expressing cells, because this cluster spans several cytoarchitectonically defined cell groups, and is distinct from the perifornical hypothalamic nucleus recognized by some authors (27). It may be pointed out, however, that the perifornical region has been identified as a sensitive site of action for several neuroactive agents in stimulating ingestive behavior (28–30), and for excitatory amino acids in eliciting cardiovascular responses (31). Aspects of the perifornical region have been identified as projecting to such major sites of CRF-R2 expression as the lateral septal and ventromedial hypothalamic nuclei (32–34), and the sources of local inhibitory (GABAergic) projections to stress-related neuroendocrine and autonomic effectors in the paraventricular nucleus includes a rostral perifornical component (35) whose distribution is very similar to that of Ucn III-expressing neurons. With regard to the third major site of Ucn III expression identified in our material, anatomical and functional studies have indicated that medial nucleus of the amygdala projects extensively to the hypothalamus (including the ventromedial nucleus) and other limbic forebrain structures (36) and is involved in modulation of behaviors (37, 38) and neuroendocrine function (39, 40) related particularly to reproduction and stress. Overall, the central sites of Ucn III expression described here are consistent with potential roles for this peptide in modulating stress-related autonomic, neuroendocrine, and behavioral function, perhaps including some previously thought to be a province of other members of this peptide family.
The distributions of the three murine urocortins exhibit some potential for interacting with type 2 CRF receptors, and each exhibits high affinity for these receptors. By contrast, CRF itself is highly selective for CRF-R1 and has lower affinity for CRF-R2. Under the assumption that the nomenclature for the three urocortin-related peptides becomes accepted, it would be reasonable to consider CRF-R2 to be a urocortin receptor both in function and in name. The “CRF system” now includes ligands with selectivity for each receptor type as well as the bivalent ligand, urocortin. Careful anatomic and physiologic studies will be required for the understanding of the roles of the various components of this complex and important regulatory network.
Acknowledgments
We gratefully acknowledge the contributions of K. Anderson, D. Kirby, W. Low, R. Kaiser, and A. Craig. The human and rat CRF-R2α receptor was generously provided by Demitri Grigoriadis (Neurocrine Biosciences, San Diego). This work was supported by National Institutes of Health Grant DK-26741, The Adler Foundation, The Kleberg Foundation, The Foundation for Medical Research, and The Foundation for Research. W.W.V. and P.E.S. are Senior Investigators for the Foundation for Research and the Foundation for Medical Research, respectively. T.M.R. is supported by a National Research Service Award (DK-10135).
Abbreviations
- CRF
corticotropin-releasing factor
- CRF-R1
CRF receptor type 1
- CRF-R2
CRF receptor type 2
- CRF-BP
CRF binding protein
- Ucn
urocortin
- CHO
Chinese hamster ovary
- EST
expressed sequence tag
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- URP
human urocortin related peptide
Note Added in Proof.
Since submission of this manuscript, Hsu and Hsueh [(2001) Nat. Med. 7, 605–611] have described putative precursor peptides for two new CRF/urocortin family members identified in the human genome, named stresscopin and stresscopin-related peptide, which are identical to human urocortin III, reported here, and human urocortin II, published previously (13). Both groups agree that it is uncertain whether and how the human urocortin II precursor may be processed to yield a functional peptide. However, the presence of canonical proteolytic cleavage sites in murine urocortin II (13) and murine urocortin III (present work) strongly support the existence of both peptides in the mouse.
Footnotes
References
- 1.Vale W, Spiess J, Rivier C, Rivier J. Science. 1981;213:1394–1397. doi: 10.1126/science.6267699. [DOI] [PubMed] [Google Scholar]
- 2.Koob G F, Heinrichs S C. Brain Res. 1999;848:141–152. doi: 10.1016/s0006-8993(99)01991-5. [DOI] [PubMed] [Google Scholar]
- 3.Vaughan J M, Donaldson C, Bittencourt J, Perrin M H, Lewis K, Sutton S, Chan R, Turnbull A, Lovejoy D, Rivier C, et al. Nature (London) 1995;378:287–292. doi: 10.1038/378287a0. [DOI] [PubMed] [Google Scholar]
- 4.Lederis K, Vale W W, Rivier J E, MacCannell K L, McMaster D, Kobayashi Y, Suess U, Lawrence J. Proc West Pharmacol Soc. 1982;25:223–227. [PubMed] [Google Scholar]
- 5.Montecucchi P C, Anastasi A, de Castiglione R, Erspamer V. Int J Pept Protein Res. 1980;16:191–199. [PubMed] [Google Scholar]
- 6.Chen R, Lewis K A, Perrin M H, Vale W W. Proc Natl Acad Sci USA. 1993;90:8967–8971. doi: 10.1073/pnas.90.19.8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vita N, Laurent P, Lefort S, Chalon P, Lelias J-M, Kaghad M, Le Fur G, Caput D, Ferrara P. FEBS Lett. 1993;335:1–5. doi: 10.1016/0014-5793(93)80427-v. [DOI] [PubMed] [Google Scholar]
- 8.Chang C P, Pearse R V, O'Connell S, Rosenfeld M G. Neuron. 1993;11:1187–1195. doi: 10.1016/0896-6273(93)90230-o. [DOI] [PubMed] [Google Scholar]
- 9.Perrin M H, Donaldson C J, Chen R, Lewis K A, Vale W W. Endocrinology. 1993;133:3058–3061. doi: 10.1210/endo.133.6.8243338. [DOI] [PubMed] [Google Scholar]
- 10.Kishimoto T, Pearse R V, Lin C R, Rosenfeld M G. Proc Natl Acad Sci USA. 1995;92:1108–1112. doi: 10.1073/pnas.92.4.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lovenberg T W, Liaw C W, Grigoriadis D E, Clevenger W, Chalmers D T, DeSouza E B, Oltersdort T. Proc Natl Acad Sci USA. 1995;92:836–840. doi: 10.1073/pnas.92.3.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Pett K, Viau V, Bittencourt J C, Chan R K W, Li H-Y, Arias C, Prins G S, Perrin M, Vale W, Sawchenko P E. J Comp Neurol. 2000;428:191–212. doi: 10.1002/1096-9861(20001211)428:2<191::aid-cne1>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 13.Reyes T M, Lewis K, Perrin M H, Kunitake K S, Vaughan J, Arias C A, Hogenesch J B, Gulyas J, Rivier J, Vale W W, et al. Proc Natl Acad Sci USA. 2001;98:2843–2848. doi: 10.1073/pnas.051626398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller C, Rivier J. Biopolymers. 1996;40:265–317. doi: 10.1002/(SICI)1097-0282(1996)40:3%3C265::AID-BIP2%3E3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 15.Sutton S W, Behan D P, Lahrichi S, Kaiser R, Corrigan A, Lowry P, Potter E, Perrin M, Rivier J, Vale W W. Endocrinology. 1995;136:1097–1102. doi: 10.1210/endo.136.3.7867564. [DOI] [PubMed] [Google Scholar]
- 16.Vale W, Vaughan J, Yamamoto G, Bruhn T, Douglas C, Dalton D, Rivier C, Rivier J. In: Methods in Enzymology: Neuroendocrine Peptides. Conn P M, editor. New York: Academic; 1983. pp. 565–577. [DOI] [PubMed] [Google Scholar]
- 17.Bilezikjian L M, Corrigan A Z, Blount A L, Vale W W. Endocrinology. 1996;137:4277–4284. doi: 10.1210/endo.137.10.8828487. [DOI] [PubMed] [Google Scholar]
- 18.Pedersen A G, Nielsen H. Proc Int Conf Intell Syst Mol Biol. 1997;5:226–233. [PubMed] [Google Scholar]
- 19.Kageyama K, Gaudriault G E, Bradbury M J, Vale W W. Endocrinology. 2000;141:2285–2293. doi: 10.1210/endo.141.7.7572. [DOI] [PubMed] [Google Scholar]
- 20.Nozu T, Martínez V, Rivier J, Taché Y. Am J Physiol. 1999;39:G867–G874. doi: 10.1152/ajpgi.1999.276.4.G867. [DOI] [PubMed] [Google Scholar]
- 21.Swanson L W, Sawchenko P E, Rivier J, Vale W W. Neuroendocrinology. 1983;36:165–186. doi: 10.1159/000123454. [DOI] [PubMed] [Google Scholar]
- 22.Bittencourt J C, Vaughan J, Arias C, Rissman R A, Vale W W, Sawchenko P E. J Comp Neurol. 1999;415:285–312. [PubMed] [Google Scholar]
- 23.Johnson A K, Loewy A D. In: Central Regulation of Autonomic Functions. Loewy A D, Spyer K M, editors. New York: Oxford Univ. Press; 1990. pp. 246–267. [Google Scholar]
- 24.Saper C B, Levisohn D. Brain Res. 1983;288:21–31. doi: 10.1016/0006-8993(83)90078-1. [DOI] [PubMed] [Google Scholar]
- 25.Sawchenko P E, Swanson L W. J Comp Neurol. 1983;218:121–144. doi: 10.1002/cne.902180202. [DOI] [PubMed] [Google Scholar]
- 26.Chalmers D T, Lovenberg T W, De Souza E B. J Neurosci. 1995;15:6340–6350. doi: 10.1523/JNEUROSCI.15-10-06340.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paxinos G, Watson C. The Rat Brain: In Stereotaxic Coordinates. San Diego: Academic; 1986. [Google Scholar]
- 28.Gillard E R, Khan A M, Grewal R S, Mouradi B, Wolfsohn S D, Stanley B G. J Neurosci. 1998;18:2646–2652. doi: 10.1523/JNEUROSCI.18-07-02646.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stanley B G, Magdalin W, Seirafi A, Thomas W J, Leibowitz S F. Brain Res. 1993;604:304–317. doi: 10.1016/0006-8993(93)90382-w. [DOI] [PubMed] [Google Scholar]
- 30.Leibowitz S F, Rossakis C. Brain Res. 1979;172:101–113. doi: 10.1016/0006-8993(79)90898-9. [DOI] [PubMed] [Google Scholar]
- 31.Allen G V, Cechetto D F. J Comp Neurol. 1993;330:421–438. doi: 10.1002/cne.903300310. [DOI] [PubMed] [Google Scholar]
- 32.Kita H, Oomura Y. Brain Res Bull. 1982;8:53–62. doi: 10.1016/0361-9230(82)90027-2. [DOI] [PubMed] [Google Scholar]
- 33.Onteniente B, Menetrey D, Arai R, Calas A. Cell Tissue Res. 1989;256:585–592. doi: 10.1007/BF00225608. [DOI] [PubMed] [Google Scholar]
- 34.Szeidemann Z, Shanabrough M, Leranth C. J Comp Neurol. 1995;358:573–583. doi: 10.1002/cne.903580410. [DOI] [PubMed] [Google Scholar]
- 35.Roland B L, Sawchenko P E. J Comp Neurol. 1993;332:123–143. doi: 10.1002/cne.903320109. [DOI] [PubMed] [Google Scholar]
- 36.Canteras N S, Simerly R B, Swanson L W. J Comp Neurol. 1995;360:213–245. doi: 10.1002/cne.903600203. [DOI] [PubMed] [Google Scholar]
- 37.Newman S W. Ann NY Acad Sci. 1999;877:242–257. doi: 10.1111/j.1749-6632.1999.tb09271.x. [DOI] [PubMed] [Google Scholar]
- 38.Rajendren G, Moss R L. Brain Res. 1993;617:81–86. doi: 10.1016/0006-8993(93)90616-u. [DOI] [PubMed] [Google Scholar]
- 39.Feldman S, Conforti N, Saphier D. Neuroscience. 1990;37:775–779. doi: 10.1016/0306-4522(90)90107-f. [DOI] [PubMed] [Google Scholar]
- 40.Dayas C V, Buller K M, Day T A. Eur J Neurosci. 1999;11:2312–2322. doi: 10.1046/j.1460-9568.1999.00645.x. [DOI] [PubMed] [Google Scholar]