Abstract
A DNA nanomechanical device has been developed that enables the positional synthesis of products whose sequences are determined by the state of the device. Consequently, this is a machine that emulates the translational capabilities of the ribosome. The device has been prototyped to make specific DNA sequences. The state of the device is established by the addition of DNA set strands. There is no transcriptional relationship between the set strands and the product strands. The device has potential applications that include designer polymer synthesis, encryption of information and as a variable-input device for DNA-based computation.
We have built a DNA nanomechanical device that mimics the translational capabilities of the ribosome. In response to a DNA signal, it aligns a series of molecules in specific positions; these molecules are then fused together in a specific order. For convenience, we have prototyped this system with DNA, so the products are DNA oligonucleotides of a defined sequence. Thus, in this case, the chemistry of the product is similar to that of the signal molecules, but there is no complementary relationship to the signal sequences. By using DNA molecules to set the states of two DNA PX-JX2 devices (1) independently, we program the synthesis of four different product molecules.
The PX-JX2 device is a sequence-dependent DNA machine whose state is controlled by hybridization topology (1). It can assume two structural states (termed ‘PX’ and ‘JX2’) that differ from each other by a half-turn rotation of one end of the molecule relative to the other end (see Fig. 1A). Two different pairs of ‘set strands’ can bind to the framework of the device, thereby establishing which structural state it adopts. The set strands contain short unpaired segments (‘toeholds’) at one of their ends to facilitate their removal by ‘unset’ strands that bind to the toeholds and then remove the set strands by branch migration (2). In addition to the PX-JX2 device, numerous variants of sequence-dependent control, pioneered in DNA tweezers by Yurke et al. (2), have been reported; these include a DNA actuator (3), a 3-state device (4), and a DNA bipedal walking machine (5). Neither these, nor other ‘shape-shifting’ DNA devices (e.g., ref 6), have been incorporated into a larger context that performs a useful task. By contrast, we have used the structural state of the device reported here for positional control of the products of polymer concatenation.
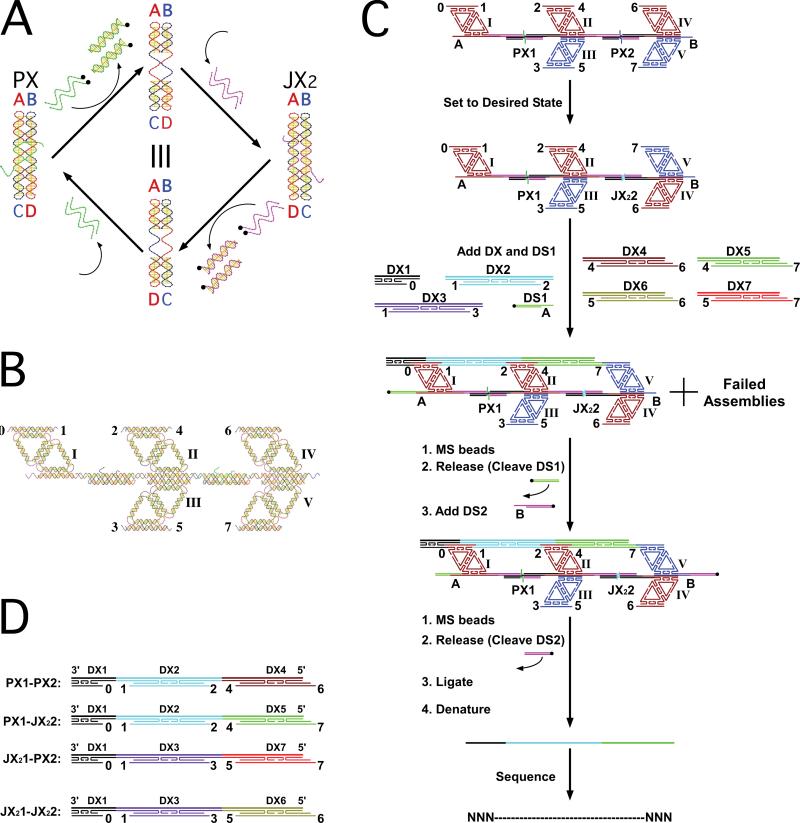
Fig. 1. Schematic Drawings of the System and Its Components.
(A) The PX state of the device is shown at the left, with green set strands. These are removed by biotinylated (black dots) unset strands to leave a naked frame (top). Adding the purple set strands puts the device in the JX2 state. The bottom of the cycle shows restoration of the PX state. The two states differ by a half-turn rotation, as highlighted by the letters ‘A’, ‘B’, ‘C’ and ‘D’ flanking the helices. (B) Five diamond motifs (6) are labeled by Roman numerals, and the sticky ends are labeled by Arabic numerals. The diamonds are connected to form double-diamond wings via a PX linkage, to give the wings dyad symmetry (14). There is an ‘initiator’ diamond motif at the left (I), while two double diamond motifs are at the center and right. The initiator diamond and the double diamonds are connected by PX-JX2 devices, so the relative orientations of the sticky ends can be varied. (C) The initial assembly of the device is shown at the top, similar to the structure in (B). Diamonds III and IV have been drawn with different colors for clarity. ‘A’ and ‘B’ indicate sticky ends used in purification. The next step entails setting the state of the device. The DX molecules are all added to the solution, along with an initiator DX1 (black) and a biotinylated double strand, DS1, complementary to A. Following left-side purification by streptavidin magnetic beads, and release by cleaving DS1, right-side purification is achieved similarly using DS2 and adding magnetic beads again. The purified complex is then ligated, and the continuous strand is denatured, purified, and sequenced. (D) Using the same color coding as (C), the four ligated DX molecules are shown. The continuous strands across the top are the final products that are sequenced.
We have incorporated two PX-JX2 devices in succession, thereby controlling the relative orientations of a diamond-shaped motif (7) and a pair of double-diamond-shaped wings Fig. 1B. The Arabic numerals in that drawing label individual sticky ends; these sticky ends are available to bind DNA double crossover (DX) molecules (8) that contain a continuous DNA strand extending from one end to the other. This continuous strand ultimately will be a component of the product. The rest of the DX molecule plays a role analogous to that of tRNA in translation: It serves as an adaptor between the strand that it carries and the device. The set strands of the device contain the signal or ‘message’ that configures the sticky ends to bind one of a pair of DX molecules in each of the two gaps.
Figure 1C illustrates the flow-chart for the experiments performed here. First, the device is constructed with both PX-JX2 machines in the PX state. Unset strands, followed by specific set strands, are then added to the device, setting its state. The complete set of DX molecules is added to the solution, and the correct ones (cyan and green in Fig. 1C) bind in the correct sites between the diamond structures, as dictated by the sticky ends. In addition, an ‘initiator’ DX (black in Fig. 1C) is bound on the top of the leftmost double diamond. Following ligation, the DX molecules are dissociated, the target strand is isolated, and its sequence is determined. Its target length is longer than any other strand in the system, so it is easy to isolate from failure products and fortuitous molecules that bind to the opposite (bottom) side of the device. Fig. 1D shows the way that the selection of set strands directs the synthesis of different products, in the same way that different mRNA molecules direct the synthesis of different polypeptide chains. Experimental methods are described in (9).
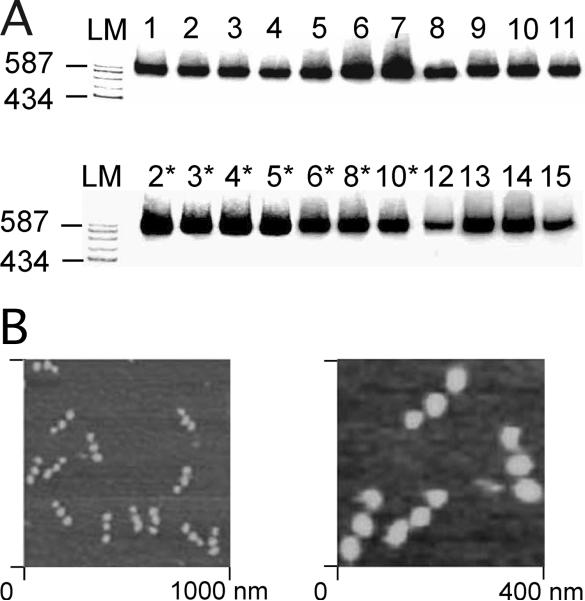
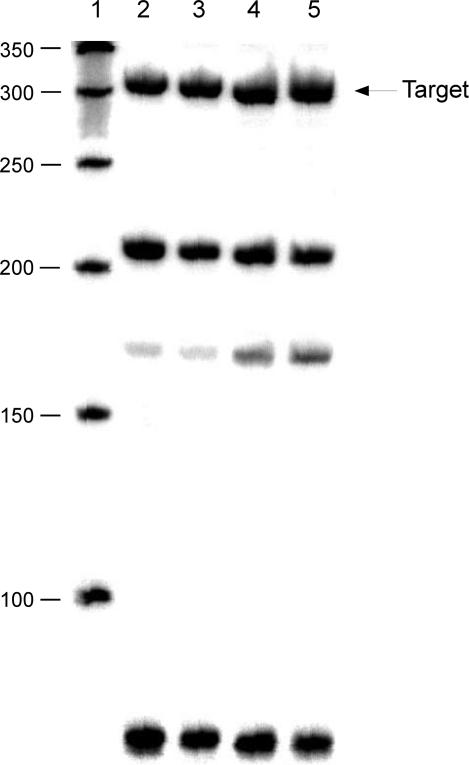
Figure 2A shows a non-denaturing gel in which each of the 22 strands of a wing component is labeled individually. The uniform nature of the gel demonstrates that the complex contains all of the strands, and that no strand partially denatures from it. Atomic force microscopy images (Fig. 2B) show purification of the device in the absence of DX molecules. Each molecule contains two large components, corresponding to the double diamond wings, and a smaller one on the end, corresponding to the single diamond that supports the initiator DX. The product strands from the four different combinations are shown in a denaturing gel in Figure 3. Numerous ligation failures are evident, but the target molecules containing 307 nucleotides are well represented on the gels. In each case the target molecule contains the expected sequence. The designed sequences of the device components and the set strands, the experimental sequences of the products and their sequencing traces (Figs. S4-S7) are available in the supporting online material (9).
Fig. 2.
(A) This is a 3.5% non-denaturing gel of the complex, composed of all 22 strands. LM indicates linear markers. Each of the strands (numbering in (9, Figs. S1-S3)) is labeled separately with radioactive phosphate. The number above each lane shows the labeled strand. In each case, the strand is incorporated cleanly into the complex, with no doubling and no breakdown products evident. (B) The AFM images were prepared as described previously (1). The DX molecules were omitted for clarity in the image. The left image is a large field, and the right image is a zoom. Note that the single- and double-diamond nature of the complex is evident from the small-large-large nature of the sample molecules. It is clear that the purification protocol used is quite successful at eliminating failure products from the sample.
Fig 3.
This is a 6% denaturing gel autoradiogram illustrating the products of the ligation. The first lane contains a 50 nucleotide ladder. Lanes 2-5 contain, respectively, the products of setting the device states to PX-PX, PX-JX2, JX2-PX, and JX2-JX2. The black strand (Fig. 1D) is labeled in each case. The target band is prominent in all lanes, although failure products are evident. The band between 150 and 200 may be an erroneous product; it represents 0.3%, 0.2%, 1.0% and 1.2%, of the material in lanes 2-5, respectively.
We have produced a device that translates a DNA signal into an unrelated sequence. The connection between the signals and the products (the ‘genetic code’ for this system) has been established arbitrarily, so that there is no transcriptional relationship between them. It is evident that this simple device prototypes an arbitrary, but general, encryption method (10). In addition, this type of device has been suggested as the basis for a finite-state machine with variable input, whose output can be used for universal computation (11). From a chemical standpoint, we expect to be able to couple this system with a recent method that adds reactive groups to the backbone residues of nucleotides; that method enables the attachment of arbitrary polymers to the DNA (12). Adding a reactive group to the continuous chain at a few accessible sites (e.g., once per helical turn) would be independent of steric effects. Such groups could be used in this system to scaffold the construction of diverse and unprecedented polymers of well-defined size and composition. This device lacks the translocational capability of the ribosome, so that product length is similar to device size; nevertheless, translocation is well within the scope of DNA nanotechnology (13) suggesting that it can be incorporated in future versions of the device.
Supplementary Material
Acknowledgments
We would like to thank Dr. Ruojie Sha for valuable discussions. This research has been supported by grants from the Office of Naval Research, the National Institute of General Medical Sciences, the National Science Foundation and Nanoscience Technologies, Inc.
Footnotes
A DNA-nanomechanical device enables positional synthesis of polymers using an arbitrary translation code.
REFERENCES
- 1.Yan H, Zhang X, Shen Z, Seeman NC. Nature. 2002;415:62. doi: 10.1038/415062a. [DOI] [PubMed] [Google Scholar]
- 2.Yurke B, Turberfield AJ, Mills AP, Jr., Simmel FC, Neumann JL. Nature. 2000;406:605. doi: 10.1038/35020524. [DOI] [PubMed] [Google Scholar]
- 3.Simmel FC, Yurke B. Phys. Rev. E. 2001;63:041913. doi: 10.1103/PhysRevE.63.041913. [DOI] [PubMed] [Google Scholar]
- 4.Simmel FC, Yurke B. Appl. Phys. Lett. 2002;80:883. [Google Scholar]
- 5.Sherman WB, Seeman NC. NanoLett. 2004;4:1203. [Google Scholar]
- 6.Mao C, Sun W, Shen Z, Seeman NC. Nature. 1999;397:144. doi: 10.1038/16437. [DOI] [PubMed] [Google Scholar]
- 7.Yan H, Seeman NC. J. Supramol. Chem. 2001;1:229. [Google Scholar]
- 8.Fu T-J, Seeman NC. Biochem. 1993;32:3211. doi: 10.1021/bi00064a003. [DOI] [PubMed] [Google Scholar]
- 9.Strand numbering, sequences and materials and methods are available on Science Online.
- 10.Landweber LF, Lipton RJ, Rabin MO. In: Proceedings of the 3rd DIMACS Meeting on DNA-Based Computers, University of Pennsylvania, June 23-25, 1997. Rubin H, Wood D, editors. Am. Math. Soc.; Providence, RI: 1997. p. 161. [Google Scholar]
- 11.Jonoska N, Liao S, Seeman NC. Aspects of Molecular Computing, Lecture Notes in Computer Science 2340. Springer-Verlag; Berlin: 2004. p. 219. [Google Scholar]
- 12.Zhu L, Lukeman PS, Canary J, Seeman NC. J. Am. Chem. Soc. 2003;125:10178. doi: 10.1021/ja035186r. [DOI] [PubMed] [Google Scholar]
- 13.Feng L, Park SH, Reif JH, Yan H. Angew. Chem. Int. Ed. 2003;42:4342. doi: 10.1002/anie.200351818. [DOI] [PubMed] [Google Scholar]
- 14.Shen Z, Yan H, Wang T, Seeman NC. J. Am. Chem. Soc. 2004;126:1666–1674. doi: 10.1021/ja038381e. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.