Abstract
Natural product-derived bengamides possess potent antiproliferative activity and target human methionine aminopeptidases (MetAPs) for their cellular effects. Several derivatives were designed, synthesized, and evaluated as MetAP inhibitors. Here, we present four new X-ray structures of human MetAP1 in complex with the inhibitors. Together with the previous structures of bengamide derivatives with human MetAP2 and tubercular MtMetAP1c, analysis of the interactions of these inhibitors at the active site provides structural basis for further modification of these bengamide inhibitors for improved potency and selectivity as anticancer and antibacterial therapeutics.
Bengamides are natural products that were isolated from marine sponge.1 Bengamides A and B (1 and 2 in Fig. 1) showed antiproliferative activity with nanomolar potency against cancer cell lines,2, 3 and bengamides arrest mammalian cells at the G1 and G2/M phases of the cell cycle.2, 4 The cellular targets of bengamides were identified as two human methionine aminopeptidases (HsMetAP1 and HsMetAP2) by a proteomic approach.5 Bengamides inhibit the two human MetAP isozymes with similar potencies.5
Figure 1.
Chemical structures of natural bengamides (1 and 2) and their synthetic derivatives reported in literature (3 and 4) and designed and synthesized in this laboratory (5-9).
MetAP is ubiquitous and carries out N-terminal methionine excision from majority of newly synthesized proteins.6 The importance of this cotranslational modification is underscored by the observed lethality when the single MetAP gene is deleted in bacteria, such as Escherichia coli 7 and Salmonella typhimurium.8 Therefore, MetAP is a potential target for developing novel antibacterial drugs.9 Eukaryotic cells usually have two MetAPs, type 1 and type 2. Deletion of either of the two MetAP genes in Saccharomyces cerevisiae rendered a slow growth phenotype, and lethality was observed when both genes were deleted.10 Mammalian MetAP1 and MetAP2 play important roles in cell proliferation and angiogenesis,11, 12 and using a conditional MetAP2 knockout mouse, the gene disruption resulted in an embryonic gastrulation defect and endothelial cell growth arrest.13 Fumagillin is another natural product that inhibits angiogenesis and suppresses tumor growth,14 and it targets MetAP2 enzyme specifically.11, 15 Clinical trials have been carried out for anticancer therapy, using the synthetic bengamide derivative LAF389 (3)16 and a fumagillin derivative.17, 18
The unique bound conformation of bengamides at the MetAP active site was first revealed by the X-ray structure of the human enzyme HsMetAP2 in complex with a bengamide derivative LAF153 (4) (pdb code 1QZY, Fig. 2A).5 In the dimetalated structure, the triol moiety of 4 coordinates with the two Co(II) ions to form two octahedral geometries, which is reminiscent of the binding of a bestatin-derived transition state inhibitor.19 The spatial arrangement of three hydroxyl groups may uniquely satisfy the coordination requirement and possibly confer the high affinity. The t-butylalkene substituent occupies the site often seen for the terminal methionine in a peptide substrate, and on the other side of the triol moiety, a caprolactam ring beyond the amide bond interacts with residues towards the opening of the active site pocket. This unique binding mode of bengamides has guided our design and synthesis of several bengamide derivatives (5-9), and some of them displayed potent inhibition of the Mycobacterium tuberculosis MetAPs (MtMetAP1a and MtMetAP1c) and modest antitubercular activity.20
Figure 2.
Coordination of bengamide derivatives with the metal ions at the dinuclear catalytic site. A. Inhibitor 4 with two Co(II) ions in HsMetAP2. B. Inhibitors 5, 7-9 with two Mn(II) ions in HsMetAP1. Inhibitors and protein residues are shown as sticks, and metal ions are shown as spheres. Coordination between the metal ions and the heteroatoms of the inhibitors or protein residues is shown as dashed lines. Only metal coordinating protein residues are shown. For coloring carbon atoms, 4 is yellow, 5 is green, 7 is orange, 8 is magenta, 9 is cyan, and protein residues are grey. For coloring non-carbon atoms, oxygen is red, and nitrogen is blue. For coloring the metal ions, Co(II) is green, and Mn(II) is orange.
Human HsMetAP1 regulates cell cycle and is a promising target for the discovery and development of new anticancer agents.12 However, although bengamides inhibit HsMetAP1 potently,5 there is currently no structural information for binding of bengamides. In fact, there are only seven X-ray structures of HsMetAP1 found in the PDB databank, and only four structures with a ligand bound at the active site.12, 21, 22 Here, we report four new X-ray structures of HsMetAP1 in complex with four different bengamide derivatives (5, 7-9), and the binding mode of bengamides in these structures is significantly different from those in the previous HsMetAP1 structures. Recently, we reported five X-ray structures of the tubercular MtMetAP1c in complex with the bengamides 5-8 (pdb codes 3PKA, 3PKB, 3PKC, 3PKD, 3PKE).20, 23 Together, the structural information of bengamides at the active site of different MetAP enzymes provides guidance of using bengamides as a template for designing potent and selective MetAP inhibitors for antibacterial and anticancer therapeutics.
Results and Discussion
X-ray structures of HsMetAP1 in complex with 5, 7, 8 or 9
Four X-ray structures of HsMetAP1 in complex with four different bengamide derivatives 5, 7, 8 and 9 in the Mn(II) form were solved at high resolution (1.47-1.75 Å) (Table 1, Fig. 2B and Figs. 3A-3D), from crystals obtained individually by co-crystallization. As seen before on MtMetAP1c,20, 23 and on HsMetAP2,5 these bengamides bound to HsMetAP1 similarly for part of the molecules including the triol moiety and the t-butylalkene chain. The triol moiety coordinated with the two active site metal ions, and the t-butylalkene chain occupied the S1 site. However, each of the bengamide derivatives bound significantly differently from 4 at the amide moiety, exploring different interactions at the opening of the active site pocket due to different amide structures.
Table 1.
X-ray data collection and refinement statistics
| Inhibitor | 5 | 7 | 8 | 9 |
|---|---|---|---|---|
| Inhibitor code | Y16 | Y08 | Y10 | YZ6 |
| PDB code | 4FLI | 4FLJ | 4FLK | 4FLL |
| Cell Parameters | ||||
| space group | P21 | P21 | P21 | P21 |
| a (Å) | 47.8 | 48.0 | 47.8 | 47.8 |
| b (Å) | 77.6 | 77.4 | 77.5 | 77.1 |
| c (Å) | 48.8 | 48.5 | 48.6 | 48.6 |
| α (deg) | 90 | 90 | 90 | 90 |
| β (deg) | 90.2 | 90.0 | 90.4 | 90.2 |
| γ (deg) | 90 | 90 | 90 | 90 |
| X-ray Data Collection | ||||
| Resolution range (Å) a | 50-1.56 (1.59-1.56) | 50-1.75 (1.78-1.75) | 50-1.47 (1.50-1.47) | 50-1.50 (1.53-1.50) |
| Collected reflections | 185,568 | 130,604 | 215,352 | 209,851 |
| Unique reflections | 50,632 | 35,919 | 60,021 | 55,956 |
| Completeness (%) a | 99.8 (99.3) | 99.9 (99.5) | 99.5 (96.1) | 99.9 (100) |
| I/σ (I) a | 36.1 (2.5) | 30.1 (2.2) | 41.9 (4.0) | 34.2 (3.0) |
| Rmerge (%) a | 9.6 (53.4) | 8.9 (56.2) | 9.5 (38.5) | 9.5 (53.9) |
| Refinement Statistics | ||||
| R (%) | 18.3 | 19.8 | 19.9 | 18.0 |
| Rfree (%) | 21.1 | 23.2 | 22.1 | 21.3 |
| R.m.s.d. bonds (Å) | 0.030 | 0.025 | 0.031 | 0.030 |
| R.m.s.d. angles (°) | 2.38 | 2.02 | 2.43 | 2.50 |
| No. of solvent molecules |
184 | 137 | 154 | 213 |
| <B> protein (Å2) | 22.4 | 27.9 | 22.3 | 21.1 |
| <B> inhibitor (Å2) | 17.5 | 30.6 | 22.7 | 27.6 |
| <B> water (Å2) | 28.6 | 30.8 | 27.3 | 29.3 |
Values given in parentheses correspond to the outer shell of data.
Figure 3.
Binding of the bengamide derivatives 5, 7, 8, and 9 at the active site pocket of HsMetAP1 (A-D, respectively). The inhibitors and protein residues (sticks) and the Mn ions (spheres) are colored in the same scheme as in Fig. 2. The semi transparent surface formed by protein residues is colored grey for carbon, red for oxygen, and blue for nitrogen. Fobs-Fcalc omit maps (inhibitors were omitted in the models) are shown superimposed on the refined structures as light green meshes contoured at 3 σ.
Clear electron density was seen surrounding the whole molecule of inhibitor 5 (Fig. 3A). However, for inhibitors 7, 8 and 9, although the density is clear on the t-butylalkene moiety and the triol moiety, the density is not as clear on the amide moiety at this contour level (Figs. 3B-3D). This is consistent with the smaller average B factor value (17.5) for inhibitor 5, in comparison with those for 7-9 (30.6, 22.7, and 27.6, respectively). However, this observation of lack of electron density to cover the part of amide moieties is unique to HsMetAP1, because, in the recently solved structures of MtMetAP1c in complex with 7 or 8, the density was clear all over the inhibitor molecules.23 Therefore, this lack of density indicates mobility and poor binding of the amide moiety at the opening of the pocket. Indeed, all of these bengamide derivatives have similarly potencies in inhibiting the Mn(II)-form of HsMetAP1 (IC50: 5, 3.59 IM; 7, 7.45 IM, 8, 5.58 IM, 9, 5.14 IM), suggesting that the major contributors of the bengamide binding to HsMetAP1 are from the t-butylalkene moiety and the triol moiety. It is interesting to note that in the structure of HsMetAP2 (pdb code 1QZY), the average B factor (23.4 ± 1.7) for non-hydrogen atoms in the amide moiety of the inhibitor molecule 4 is clearly larger than that (19.8 ± 1.4) for atoms in other parts. Compound 4 is the one used in structural determination, and comparing with the clinically tested 3, it lacks the cyclohexanecarboxyl group, which may be needed for enhanced binding and activity. Clearly, the substitution on the seven-membered caprolactam ring had significant influence on in vitro and in vivo activities of these bengamide derivatives.2 Similarly for our bengamide derivatives 5-9, the X-ray structures revealed their binding modes on MetAPs and provided the directions for further modifications. Substitutions to extend the amide moiety will likely explore additional interactions and enhance their binding to their targeting MetAPs.
Structurally mobile loop in HsMetAP1 at the vicinity of the binding pocket
When we built the structures, it was difficult to model the loop of K132, G133 and T134 in all of these HsMetAP1 structures. Considering the closeness of this loop to the active site, it is questionable whether this difficulty is related to the lack of density around the amide moiety of inhibitors mentioned above. The final structures have much larger B-factors for these three residues (Figs. 4A and 4C, indicated by an arrow). This led us to reexamine the seven previously obtained HsMetAP1 structures (2B3H, 2B3K, 2B3L, 2G6P, 2GZ5, 2NQ6 and 2NQ7). Indeed, in all HsMetAP1 structures, with or without a ligand, these residues have larger B-factors, indicating that their mobility is a common feature in HsMetAP1 structures.
Figure 4.
A flexible K132-G133-T134 loop in HsMetAP1 at the junction of the catalytic domain and the N-terminal extension. The structure of HsMetAP1 with 5 is shown as ribbon drawing (A) and as putty drawing (C) with the tube diameters proportional to B factors. For comparison, the structures of MtMetAP1c with 5 (D) and HsMetAP2 with 4 (E) are also shown as putty drawings. The active site is shown as semitransparent surfaces with the inhibitor and metal ions. The flexible loop in human MetAP1 is indicated by an arrow in A and C. The junction residues in HsMetAP2 are invisible in 1QZY and indicated by a dotted line in E. All of the structures were adjusted to similar orientations with the same color scheme as in Fig. 2. HsMetAP1 belongs to type 1 MetAP, as EcMetAP1 and MtMetAP1c, while HsMetAP2 belongs to type 2 MetAP with a typical insert for MetAP2, and their domain structures are shown in B.
All MetAPs are homologous in sequence in the catalytic domain, and E. coli MetAP1 (EcMetAP1) is a typical bacterial MetAP and the smallest with only the catalytic domain (Fig. 4B). On the other hand, mammalian MetAPs, including HsMetAP1 and HsMetAP2, have an extension at the N-terminus.10, 24 S. cerevisiae MetAP1 has two zinc fingers in the extension, and the zinc fingers are essential for normal MAP function in vivo, even though the in vitro enzyme assays indicate that they are not involved in catalysis.25 It is interesting to note that when bacterial MetAPs and mammalian MetAPs are aligned by sequence, this mobile loop is located right at the junction of the catalytic domain and the N-terminal extension. When HsMetAP1 was expressed in E. coli, the full length enzyme was obtained but not stable during purification and storage, and N-terminal truncations were observed.26 All HsMetAP1 structures so far, including ours, start at the N-terminus around Y90, and the structure of a full length HsMetAP1 remain elusive. The mobility of the K132-G133-T134 loop may partially contribute to the instability of protein. Similarly, in all HsMetAP2 structures, N-terminus is missing before K110, and the junction (around T151) of the N-terminal extension and the catalytic domain was either invisible (Fig. 4E, indicated by a dotted line) or showed much larger B factors.
The function of the N-terminal extension in MetAP1 and MetAP2 is not clear, and neither is clear whether the truncated forms exist in vivo. Previously, we prepared and characterized truncated HsMetAP1(Δ1-66) and HsMetAP1(Δ1-135) in E. coli.26 Although the N-terminal extension in HsMetAP1 is not required for enzyme activity, it has a significant impact on interaction of the enzyme with its substrates and inhibitors.26 Mycobacterium tuberculosis is a mycobacterium, and MtMetAP1c has a shorter N-terminal extension. Comparing with HsMetAP1, similarly located residues in MtMetAP1c showed no such mobility as indicated by much tighter B factors in the region (Fig. 4D). Addlagatta et al. noticed that despite the structural and sequence changes in this extension, there are two spatially conserved residues (Y117 and E128 in HsMetAP1, and Y27 and E35 in MtMetAP1c) that form hydrogen bonds and hydrophobic interactions with residues that help form the substrate binding pocket (S191, Y195, F309, and H310 in HsMetAP1, and S93, Y97, and F211 in MtMetAP1c).27 The mobility of the K132-G133-T134 loop extends at least to E128, and their mobility may affect the binding of substrates and inhibitors directly or indirectly. We observed significant differences in catalysis and inhibition between full-length and truncated HsMetAP1s.26
Similarities and differences in binding of bengamide derivatives on HsMetAP1, HsMetAP2 and MtMetAP1c
Five residues that coordinate to the two metal ions at the active site (D97, D108, H171, E204 and E235 in EcMetAP1, Table 2) and two catalytically involved histidine residues are conserved in all MetAPs. When the structures of HsMetAP1, HsMetAP2 and MtMetAP1c are superimposed, the bengamides 4 and 5 showed the same binding mode on the three different MetAP enzymes (Fig. 5). Apparently, the two metal ions dictate the conformation of the metal coordinating residues and the triol moiety of the bengamide inhibitors. With the triol moiety in the middle of the linear inhibitors fixed at the metal site, other parts of the inhibitors had little variation in the bound conformation.
Table 2.
Residues within 4A of the bengamide inhibitors in the active site pocket.
| Type 1 | Type 2 | ||
|---|---|---|---|
| HsMetAP1 | MtMetAP1c | EcMetAP1 a | HsMetAP2 b |
| Residues of metal ligation | |||
| H303 | H205 | H171 | H331 |
| D240 | D142 | D108 | D262 |
| E367 | E269 | E235 | E459 |
| E336 | E238 | E204 | E364 |
| D229 | D131 | D97 | D251 |
| Two conserved histidines | |||
| H310 | H212 | H178 | H339 |
| H212 | H114 | H79 | H231 |
| Other active site residues | |||
| W353 | W255 | W221 | |
| Y195 | Y97 | Y62 | |
| F309 | F211 | F177 | |
| T231 | T133 | T99 | |
| C203 | C105 | C70 | |
| C211 | C113 | C78 | A230 |
| C301 | T203 | C169 | N329 |
| Y300 | F202 | Y168 | L328 |
| P192 | T94 | C59 | |
| N314 | V216 | Q182 | T343 |
Because no structure of EcMetAP1 with a bengamide has been obtained, listed are the similarly located residues.
For HsMetAP2, only residues that occur at corresponding positions in space in HsMetAP1 are listed in the same horizontal line.
Figure 5.
Stereo view of the superimposed bengamide derivatives 5 (with HsMetAP1, carbon green; with MtMetAP1c, carbon grey) and 4 (with HsMetAP2, carbon yellow) at the active site with coordination to catalytic metal ions (shown as green or orange spheres). The left panel and the right panel are a stereo pair, and one was rotated six degree to the other along z axis for viewing in stereo. The inhibitors are thicker sticks, and the protein residues are thinner sticks. Only five metal coordinating residues and two catalytically important histidine residues (See Table 2) are shown.
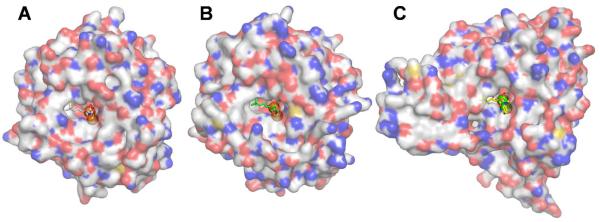
Type 1 MetAPs are more homologous in the active site pocket, and most residues that form the pocket are either identical or conserved (Table 2). This presents a challenge in developing selective inhibitors for antibacterial purposes that can differentiate bacterial MetAPs and HsMetAP1, because they all belong to type 1 MetAP. However, at the opening and outside of the pocket, more structural variations exist for type 1 MetAPs (Fig. 6), and extending the bengamide derivatives to explore additional interactions will potentially increase not only potency on bacterial MetAPs but also selectivity. The carbonyl group used to form the amide moiety in bengamides is properly located and provides a chemical handle for chemical modifications by attaching different amino containing substitutions.
Figure 6.
Overall structures of MtMetAP1c with 5 bound (A), HsMetAP1 with 5 bound (B) and HsMetAP2 with 4 bound (C), shown as surface drawing (carbon, grey; nitrogen blue; and oxygen, red). Inhibitors are colored the same as in Fig. 5.
Both HsMetAP1 and HsMetAP2 are potential targets for developing anticancer drugs. However, because the important function played by MetAP in normal cells, it is possible but unknown whether selective inhibition of one of the two human MetAP enzymes will provide any therapeutic or toxicological benefits. Besides the two well-studied human MetAPs, there is another less-known human MetAP named MetAP1D, which is of mitochondrial origin.28 It was only biochemically characterized,29 and its structural information is not available. Therefore, selective inhibitors of human MetAPs are desired to provide the research tools to elucidate the mechanism of individual human MetAP in cancer pathogenesis and to generate novel leads as anticancer agents. Due to the insertion in the catalytic domain in HsMetAP2 (Fig. 4B), the residues forming the active site pocket are not paired well between HsMetAP1 and HsMetAP2 (Table 2), except the metal coordinating residues and two conserved histidine residues. This variation in the active site pocket provides opportunities to explore the differences for selectivity. In addition, significant differences exist outside the pocket (Figs. 6B-6C) for exploration for increased potency and selectivity.
In conclusion, natural bengamides and synthetic bengamide derivatives are inhibitors of MetAPs, and their unique binding modes make them an ideal template for developing potent and selective MetAP inhibitors as therapeutics. The X-ray structures of MetAP enzymes in complex with different bengamide derivatives provide the valuable structural information for such inhibitor development.
Experimental Section
Compounds used in the experiments were synthesized in this laboratory and were at least 95% pure on the basis of HPLC–MS.20 Proteins were expressed in E. coli and purified to homogeneity as previously described with minor modifications.26 Crystals of the enzyme inhibitor complexes were obtained independently by microbath method under paraffin oil at room temperature. Each of the inhibitors (5, 7, 8 and 9; 100 mM in DMSO) was added to concentrated metalated enzyme (10 mg/mL, 0.27 mM protein; 0.675 mM metal) in 20 mM Tris, pH 8.0, and 100 mM NaCl, 2 mM DTT, and the molar ratio of inhibitor to HsMetAP1 was 5:1. The enzyme/inhibitor mixture was mixed with a reservoir buffer in a 1:1 ratio. The reservoir buffer was 200 mM sodium acetate, pH 8.0, and 20% PEG 3350 for 5, 7 and 9, and 200 mM sodium formate, pH 7.2, and 20% PEG 3350 for 8. Diffraction data were collected at the Advanced Photon Source, Argonne National Laboratory (beamlines 19BM and 19ID) and processed with HKL3000.30 The crystals belong to space group P21. In each case, one molecule is in the asymmetric unit. The structures were solved by molecular replacement with MolRep 31 in CCP4 32 with CCP4i interface,33 using the previously published HsMetAP1 structure (PDB code 2NQ6) 34 as the search model. The structure was refined with REFMAC5 35 with iterative model building using WinCoot.36 The refinement was monitored with 5% of the reflections set aside for Rfree factor analysis throughout the whole refinement process. Electron density was clear for all residues except a few residues at the N-terminus, and residues from Y90 in the native protein to the S393, third residue from the C-terminus, were modeled. Comparison of structures and generation of structural drawings were carried out by using PyMOL.37 Statistic parameters in data collection and structural refinement are shown in Table 1. Atomic coordinates and structure factors for the three structures were deposited in the Protein Data Bank.
Acknowledgments
This work was supported by National Institutes of Health Grants R01 AI065898 and R56 AI065898, and by Indiana University School of Medicine (BRG), Indiana University and Purdue University at Indianapolis (RSFG), and the Experimental and Developmental Therapeutics Program at the IU Simon Cancer Center (ITRAC) (to Q.-Z.Y.). We thank the staffs at Structural Biology Center of the Advanced Photon Source, Argonne National Laboratory (beamlines 19BM and 19ID) for assistance with data collection.
Abbreviations
- MetAP
methionine aminopeptidase
- HsMetAP1
human MetAP type 1
- HsMetAP2
human MetAP type 2
- MtMetAP1a
M. tuberculosis MetAP type 1a
- MtMetAP1c
M. tuberculosis MetAP type 1c
- EcMetAP1
E. coli MetAP type 1
- IC50
concentration of 50% inhibition.
Footnotes
PDB ID: 4FLI, 4FLJ, 4FLK and 4FLL
References
- 1.Quinoa E, Adamczeski M, Crews P, Bakus GJ. Bengamides, heterocyclic anthelmintics from a Jaspidae marine sponge. J Org Chem. 1986;51:4494–4497. [Google Scholar]
- 2.Kinder FR, Jr., Versace RW, Bair KW, Bontempo JM, Cesarz D, Chen S, Crews P, Czuchta AM, Jagoe CT, Mou Y, Nemzek R, Phillips PE, Tran LD, Wang RM, Weltchek S, Zabludoff S. Synthesis and antitumor activity of ester-modified analogues of bengamide B. J Med Chem. 2001;44:3692–3699. doi: 10.1021/jm010188c. [DOI] [PubMed] [Google Scholar]
- 3.Thale Z, Kinder FR, Bair KW, Bontempo J, Czuchta AM, Versace RW, Phillips PE, Sanders ML, Wattanasin S, Crews P. Bengamides revisited: new structures and antitumor studies. J Org Chem. 2001;66:1733–1741. doi: 10.1021/jo001380+. [DOI] [PubMed] [Google Scholar]
- 4.Phillips PE, Bair KW, Bontempo J, Crews P, Czuchta M,A, Kinder FR, Vattay A, Versace RW, Wang B, Wang J, Wood A, Zabludoff S. Bengamide E arrests cells at the G1/S restriction point and within the G2/M phase of the cell cycle. Proc. Am. Assoc. Cancer Res. 2000;41:59. [Google Scholar]
- 5.Towbin H, Bair KW, DeCaprio JA, Eck MJ, Kim S, Kinder FR, Morollo A, Mueller DR, Schindler P, Song HK, van Oostrum J, Versace RW, Voshol H, Wood J, Zabludoff S, Phillips PE. Proteomics-based target identification: bengamides as a new class of methionine aminopeptidase inhibitors. J Biol Chem. 2003;278:52964–52971. doi: 10.1074/jbc.M309039200. [DOI] [PubMed] [Google Scholar]
- 6.Giglione C, Boularot A, Meinnel T. Protein N-terminal methionine excision. Cell Mol Life Sci. 2004;61:1455–1474. doi: 10.1007/s00018-004-3466-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang SY, McGary EC, Chang S. Methionine aminopeptidase gene of Escherichia coli is essential for cell growth. J Bacteriol. 1989;171:4071–4072. doi: 10.1128/jb.171.7.4071-4072.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller CG, Kukral AM, Miller JL, Movva NR. pepM is an essential gene in Salmonella typhimurium. J Bacteriol. 1989;171:5215–5217. doi: 10.1128/jb.171.9.5215-5217.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vaughan MD, Sampson PB, Honek JF. Methionine in and out of proteins: targets for drug design. Curr Med Chem. 2002;9:385–409. doi: 10.2174/0929867023371102. [DOI] [PubMed] [Google Scholar]
- 10.Li X, Chang YH. Amino-terminal protein processing in Saccharomyces cerevisiae is an essential function that requires two distinct methionine aminopeptidases. Proc Natl Acad Sci U S A. 1995;92:12357–12361. doi: 10.1073/pnas.92.26.12357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griffith EC, Su Z, Turk BE, Chen S, Chang YH, Wu Z, Biemann K, Liu JO. Methionine aminopeptidase (type 2) is the common target for angiogenesis inhibitors AGM-1470 and ovalicin. Chem Biol. 1997;4:461–471. doi: 10.1016/s1074-5521(97)90198-8. [DOI] [PubMed] [Google Scholar]
- 12.Hu X, Addlagatta A, Lu J, Matthews BW, Liu JO. Elucidation of the function of type 1 human methionine aminopeptidase during cell cycle progression. Proc Natl Acad Sci U S A. 2006;103:18148–18153. doi: 10.1073/pnas.0608389103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yeh JR, Ju R, Brdlik CM, Zhang W, Zhang Y, Matyskiela ME, Shotwell JD, Crews CM. Targeted gene disruption of methionine aminopeptidase 2 results in an embryonic gastrulation defect and endothelial cell growth arrest. Proc Natl Acad Sci U S A. 2006;103:10379–10384. doi: 10.1073/pnas.0511313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ingber D, Fujita T, Kishimoto S, Sudo K, Kanamaru T, Brem H, Folkman J. Synthetic analogues of fumagillin that inhibit angiogenesis and suppress tumour growth. Nature. 1990;348:555–557. doi: 10.1038/348555a0. [DOI] [PubMed] [Google Scholar]
- 15.Sin N, Meng L, Wang MQ, Wen JJ, Bornmann WG, Crews CM. The anti-angiogenic agent fumagillin covalently binds and inhibits the methionine aminopeptidase, MetAP 2. Proc Natl Acad Sci U S A. 1997;94:6099–6103. doi: 10.1073/pnas.94.12.6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dumez H, Gall H, Capdeville R, Dutreix C, van Oosterom AT, Giaccone G. A phase I and pharmacokinetic study of LAF389 administered to patients with advanced cancer. Anticancer Drugs. 2007;18:219–225. doi: 10.1097/CAD.0b013e328010ef5b. [DOI] [PubMed] [Google Scholar]
- 17.Bhargava P, Marshall JL, Rizvi N, Dahut W, Yoe J, Figuera M, Phipps K, Ong VS, Kato A, Hawkins MJ. A Phase I and pharmacokinetic study of TNP-470 administered weekly to patients with advanced cancer. Clin Cancer Res. 1999;5:1989–1995. [PubMed] [Google Scholar]
- 18.Logothetis CJ, Wu KK, Finn LD, Daliani D, Figg W, Ghaddar H, Gutterman JU. Phase I trial of the angiogenesis inhibitor TNP-470 for progressive androgen-independent prostate cancer. Clin Cancer Res. 2001;7:1198–1203. [PubMed] [Google Scholar]
- 19.Lowther WT, Orville AM, Madden DT, Lim S, Rich DH, Matthews BW. Escherichia coli methionine aminopeptidase: implications of crystallographic analyses of the native, mutant, and inhibited enzymes for the mechanism of catalysis. Biochemistry. 1999;38:7678–7688. doi: 10.1021/bi990684r. [DOI] [PubMed] [Google Scholar]
- 20.Lu JP, Yuan XH, Yuan H, Wang WL, Wan B, Franzblau SG, Ye QZ. Inhibition of Mycobacterium tuberculosis methionine aminopeptidases by bengamide derivatives. ChemMedChem. 2011;6:1041–1048. doi: 10.1002/cmdc.201100003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu X, Addlagatta A, Matthews BW, Liu JO. Identification of pyridinylpyrimidines as inhibitors of human methionine aminopeptidases. Angew Chem Int Ed Engl. 2006;45:3772–3775. doi: 10.1002/anie.200600757. [DOI] [PubMed] [Google Scholar]
- 22.Addlagatta A, Matthews BW. Structure of the angiogenesis inhibitor ovalicin bound to its noncognate target, human Type 1 methionine aminopeptidase. Protein Sci. 2006;15:1842–1848. doi: 10.1110/ps.062278006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu JP, Yuan XH, Ye QZ. Structural analysis of inhibition of Mycobacterium tuberculosis methionine aminopeptidase by bengamide derivatives. Eur J Med Chem. 2012;47:479–484. doi: 10.1016/j.ejmech.2011.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arfin SM, Kendall RL, Hall L, Weaver LH, Stewart AE, Matthews BW, Bradshaw RA. Eukaryotic methionyl aminopeptidases: two classes of cobalt dependent enzymes. Proc Natl Acad Sci U S A. 1995;92:7714–7718. doi: 10.1073/pnas.92.17.7714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zuo S, Guo Q, Ling C, Chang YH. Evidence that two zinc fingers in the methionine aminopeptidase from Saccharomyces cerevisiae are important for normal growth. Mol Gen Genet. 1995;246:247–253. doi: 10.1007/BF00294688. [DOI] [PubMed] [Google Scholar]
- 26.Li JY, Chen LL, Cui YM, Luo QL, Gu M, Nan FJ, Ye QZ. Characterization of full length and truncated type I human methionine aminopeptidases expressed from Escherichia coli. Biochemistry. 2004;43:7892–7898. doi: 10.1021/bi0360859. [DOI] [PubMed] [Google Scholar]
- 27.Addlagatta A, Hu X, Liu JO, Matthews BW. Structural basis for the functional differences between type I and type II human methionine aminopeptidases. Biochemistry. 2005;44:14741–14749. doi: 10.1021/bi051691k. [DOI] [PubMed] [Google Scholar]
- 28.Leszczyniecka M, Bhatia U, Cueto M, Nirmala NR, Towbin H, Vattay A, Wang B, Zabludoff S, Phillips PE. MAP1D, a novel methionine aminopeptidase family member is overexpressed in colon cancer. Oncogene. 2006;25:3471–3478. doi: 10.1038/sj.onc.1209383. [DOI] [PubMed] [Google Scholar]
- 29.Hu XV, Chen X, Han KC, Mildvan AS, Liu JO. Kinetic and mutational studies of the number of interacting divalent cations required by bacterial and human methionine aminopeptidases. Biochemistry. 2007;46:12833–12843. doi: 10.1021/bi701127x. [DOI] [PubMed] [Google Scholar]
- 30.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 31.Vagin A, Teplyakov A. MOLREP: an Automated Program for Molecular Replacement. J. Appl. Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 32.Collaborative Computational Project Number 4 The CCP4 Suite: Programs for Protein Crystallography. Acta Cryst. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 33.Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr D Biol Crystallogr. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- 34.Lu JP, Chai SC, Ye QZ. Catalysis and inhibition of Mycobacterium tuberculosis methionine aminopeptidase. J Med Chem. 2010;53:1329–1337. doi: 10.1021/jm901624n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 36.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 37.DeLano WL. The PyMOL Molecular Graphics System. 2002 on World Wide Web http://www.pymol.org. [Google Scholar]