Abstract
Melanin-concentrating hormone (MCH), a neuropeptide expressed in central and peripheral nervous systems, plays an important role in the control of feeding behaviors and energy metabolism. An orphan G protein-coupled receptor (SLC-1/GPR24) has recently been identified as a receptor for MCH (MCHR1). We report here the identification and characterization of a G protein-coupled receptor as the MCH receptor subtype 2 (MCHR2). MCHR2 has higher protein sequence homology to MCHR1 than any other G protein-coupled receptor. The expression of MCHR2 has been detected in many regions of the brain. In contrast to MCHR1, which is intronless in the coding region and is located at the chromosomal locus 22q13.3, the MCHR2 gene has multiple exons and is mapped to locus 6q21. MCHR2 is specifically activated by nanomolar concentrations of MCH, binds to MCH with high affinity, and signals through Gq protein. This discovery is important for a full understanding of MCH biology and the development of potential therapeutics for diseases involving MCH, including obesity.
Melanin-concentrating hormone (MCH) is a cyclic neuropeptide originally isolated from the salmon pituitary gland (1). It regulates skin color in teleost fish by the induction of melanin aggregation in the melanocytes. Mammalian MCH is present in neurons of both the central and peripheral nervous systems, with predominant expression in the lateral hypothalamus and zona incerta (2, 3). Although MCH has been implicated in a variety of physiological functions, its central role in the control of feeding and energy balance has been the focus of previous studies (reviewed in ref. 4). When administered centrally, MCH promotes food intake. Its expression in the lateral hypothalamus is induced by starvation (5). MCH knockout mice are lean and hypophagic and have increased metabolic rates and reduced body weight (6). Overexpression of MCH in transgenic mice leads to obesity (7). MCH and components of MCH signaling pathways have therefore become very attractive as potential antiobesity drug targets.
Recently, an orphan G protein-coupled receptor (GPCR) named SLC-1/GPR24 (8) was identified as a receptor for MCH (MCHR1) (9–13). Its expression has been detected in many regions of the brain (8, 9, 11, 13–15), including the cortex, hippocampus, thalamus, midbrain, and pons, with relatively high expression levels in the ventromedial, dorsomedial, and arcuate nuclei of the hypothalamus. The distribution of MCHR1 in these areas of the central nervous system is consistent with the involvement of the system in the control of feeding behaviors. Furthermore, MCH has been indicated in a variety of physiological processes in addition to feeding, and circumstantial evidence exists for the possible presence of an additional MCH receptor(s) (16, 17). In this study, we isolated and identified a MCH receptor (MCHR2). Its expression pattern is similar to that of MCHR1, with high levels in many areas of the brain. It binds to MCH with high affinity and is specifically activated by nanomolar concentrations of the hormone.
Materials and Methods
Sigma-RBI Peptide Library.
The library was purchased from Sigma. It contains 60 different bioactive peptides.
GPCR Sequence Searches.
A sequence alignment of the protein sequences of 204 known human GPCRs was used to construct a hidden Markov model (search date was October 19, 2000; refs. 18 and 19) with the use of hmmbuild of the hmmer 2.1.1 software package (Sean Eddy, Washington University, St. Louis). High-throughput human genomic sequences deposited in the GenBank database were searched with this hidden Markov model, with the use of hframe 2.1.5 software (Paracel) on a hardware accelerator (GeneMatcher, Paracel, Pasadena, CA).
Cloning of the Full-Length cDNA for MCHR2.
The MCHR2 cDNA was cloned by 5′/3′ rapid amplification of cDNA ends (RACE) followed by full-length PCR amplification. For RACE, the Marathon cDNA Racing Kit and human fetal brain Marathon Ready cDNA (CLONTECH) were used according to the kit instructions. For the 5′ RACE, three primers were used: 5′-GATGTTGCCAACCAGCCCTGTTGAA-3′, 5′-CCCAATCATGGAAGGGAGGATGACTG-3′, and 5′-TTCGGCAGAGGTGTTCCAACAAGATG-3′. For the 3′ RACE, the following 3 primers were used: 5′-TCATGCATCTTGTTGGAACACCTCTGC-3′, 5′-CCAGTGTGGTAGATACAGTCATCCTCCCTTC-3′, and 5′-GGCTGGTTGGCAACATCCTCATTGTA-3′. The RACE products were sequenced to find the start and stop codons for MCHR2. The start codon was confirmed by the presence of an in-frame stop codon preceding the ATG. The full-length cDNA encoding MCHR2 was then PCR amplified from the same cDNA library with the use of an Advantage 2 High Fidelity PCR Kit (CLONTECH) and primers 5′-CACCATGAATCCATTTCATGCATCTTG-3′ and 5′-CAATCATGTCTAGACTCATGGTG-3′. The full-length cDNA was cloned into the expression vector pEF6/V5-His-TOPO or pcDNA3.1/V5-His-TOPO with the use of a TOPO cloning kit from Invitrogen.
Reverse Transcription–PCR Analysis of MCHR2 Expression.
A panel of cDNAs synthesized from RNA from different human tissues (CLONTECH) was used. For the PCR amplification reactions the Taq polymerase kit (Qiagen, Chatsworth, CA) was used under the following PCR conditions: 94°C, 40 s; 60°C, 30 s; 72°C, 2 min; 35 cycles. The primers were 5′-CACCATCATCACATCCCTGGATAC-3′ and 5′-GTATCACATGATAAGGGGCAGCAC-3′.
Northern Blot and RNA Array Blot Hybridization.
The human 12-lane mRNA blots were purchased from CLONTECH. Each lane of the blots contains 2 μg of mRNA isolated from different human tissues. The multiple tissue RNA array blots used in this study were also from CLONTECH. It contains normalized loadings of mRNA from 76 different human tissues and control RNAs. The human brain RNA blots were purchased from the Biochain Institute. Each lane of the Biochain blots contains 20 μg of total RNA isolated from various regions of the human brain. The MCHR2 probe used for Northern and array blot hybridization was a PCR product that includes the entire MCHR2 coding region. The primers used for the PCR amplification of MCHR2 probe were 5′-ATGAATCCATTTCATGCATCTTG-3′ and 5′-CAATCATGTCTAGACTCATGGTG-3′. The probe for MCHR1 was a restriction fragment from the MCHR1 expression plasmid pEF6/V5-MCHR1 that contains the full-length human MCHR1 cDNA. The random primer labeling kit Prime-it II (Stratagene) was used to label the probes for MCHR1 and MCHR2. The hybridization was carried out with the use of the Rapid-hyb buffer system (Amersham Pharmacia).
Cell Transfection.
Chinese hamster ovary (CHO) and HEK293 cells were transfected with MCHR2- or MCHR1-expressing plasmids (4 μg per 5–8 × 106 cells) with the use of Lipofectamine 2000 (GIBCO/BRL).
Measurement of Intracellular Ca2+ Mobilization.
An aequorin luminescent assay was used to measure the intracellular Ca2+ concentration (20). A plasmid expressing mitochondria-targeted apoaequorin pcDNA3-AEQ was cotransfected with the receptor-expressing plasmids at a ratio of 1:10. After 24 h of incubation, the transfected cells were incubated for 1 h with Opti-MEM containing 2 μM coelenterazine f (Molecular Probes). The cells were then dispersed by repeated pipetting or by trypsin-EDTA, and washed once with and resuspended (106 cells per milliliter) in Hanks' balanced saline solution supplemented with 10 mM Hepes and 0.1% BSA. One hundred microliters of the cell suspension was stimulated with an equal volume of each peptide ligand, and aequorin luminescence elicited by intracellular Ca2+ in the cells was recorded with a microplate luminometer (EG & G Bertholt, Gaithersburg, MD).
Measurements of Inositol Phosphates.
MCH-stimulated formation of inositol phosphates was measured as described (21). Briefly, HEK293 or CHO cells transiently transfected with MCHR1 or MCHR2 were labeled with [3H]myoinositol (Amersham Pharmacia) for 16 h. After pretreatment with 20 mM LiCl for 30 min, cells were stimulated with 100 nM MCH. [3H]Inositol phosphates were extracted with 20 mM formic acid, isolated by Dowex AG1 × 8 (Bio-Rad) columns, and counted on a scintillation counter. To assess the involvement of Gi/o proteins, 100 ng/ml pertussis toxin (PTX) was added to the labeling medium for 16 h before inositol phosphates were measured.
MCH Binding.
Binding of 125I-labeled MCH to MCHR1 and MCHR2 was measured by filtration binding assay. Membranes (10 μg protein) from transiently transfected HEK293-MCHR1 and HEK293-MCHR2 cells were mixed with 0–9.8 nM 125I-labeled MCH (specific activity 2000 Ci/mmol; Amersham Pharmacia) in the binding buffer (50 mM Hepes/10 mM MgCl2/2 mM EGTA; Roche Molecular Biochemicals protease inhibitors, 0.1% BSA, pH 7.6). After incubation for 1 h at room temperature, membrane-bound 125I-labeled MCH was separated from the free 125I-labeled MCH by filtration through a 96-well GF/B plate on a Packard Filtermate Cell Harvester and washed with ice-cold binding buffer supplemented with 80 mM NaCl. Eighty microliters of scintillation liquid (Packard MicroScint) was added, and the radioactivity was counted on a Packard Microplate Topcount.
Results
Isolation of Full-Length cDNA for MCHR2.
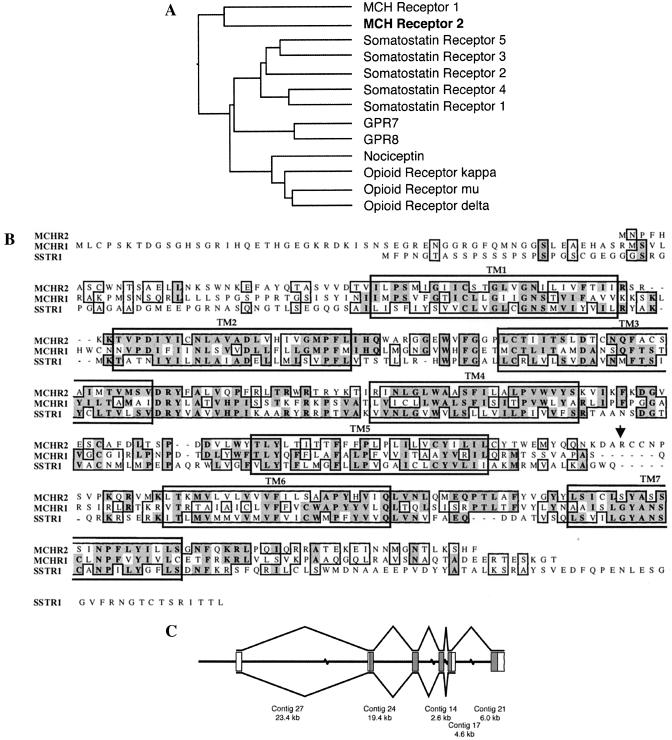
By searching human genomic sequences with a GPCR hidden Markov model (Materials and Methods), we identified a sequence encoding part of a putative GPCR. This sequence was found in a BAC clone with the GenBank accession number AC027643. It was later shown to be a MCH receptor, so we designate it as MCHR2. blast searches with this partial MCHR2 sequence revealed that it was a gene with no expressed sequence tag or protein matches. The closest known homolog was the initially described MCH receptor, followed by the somatostatin and opioid receptors (Fig. 1A).
Figure 1.
Analysis of the sequences, genomic structure, and splicing of the MCHR2 gene. (A) Sequence similarity between MCHR2 and related GPCRs. The protein sequence of MCHR2 was aligned with the GPCRs most similar to it, including all of the members of the somatostatin and opioid receptor families and two orphan GPCRs, GPR7 and GPR8. The GCG programs distances and growtree were used to generate a similarity tree based on the alignment. The lengths of the horizontal line segments connecting two GPCRs are inversely proportional to the similarities between their protein sequences. (B) Alignment of the protein sequences of MCHR1, MCHR2, and SSTR1. Identical and similar amino acid residues are boxed, with identical ones in shadowed boxes. The large boxes indicate predicted transmembrane domains. The arrow indicates the end of the putative truncated form of the MCHR2 protein as a result of alternative splicing. (C) MCHR2 exon structure. The MCHR2 mRNA sequence was used to order and orient five of the unordered contigs in the GenBank BAC sequence AC027643. The central line represents the BAC sequence, with discontinuities representing gaps of unknown length between contigs. Boxes represent regions of the genomic sequence containing MCRH2 exons; filled boxes are translated, and open boxes contain 5′ and 3′ untranslated region sequences. Lines above and below the exons indicate the two splicing patterns observed. The position of the 3′ end of the 3′ untranslated region of the longer transcript is unknown. The exon lengths are drawn out of scale with the contig lengths.
The full-length MCHR2 cDNA was cloned by 5′ and 3′ RACE. After identifying the start and stop codons of MCHR2 by sequencing the RACE products, we cloned the full-length cDNA by PCR, with the use of a human brain cDNA library as the template. Analysis of the protein sequence encoded by the full-length MCHR2 cDNA confirmed that it was most similar to the MCH receptor (Fig. 1A). MCHR2 has an overall amino acid identity of 36% with MCH receptor protein and a 44% amino acid identity in the transmembrane regions. It shares 39% amino acid identity with somatostatin receptor 1 (SSTR1) in the transmembrane regions and a 32% identity to SSTR1 over 312 aa (Fig. 2B). It is worth noting that the second and third intracellular loops of MCHR2 are rather divergent from those of MCHR1 and SSTR1. There is an insertion of six amino acids in the third intracellular loop of MCHR2 compared with MCHR1 and SSTR1. The gene structure and the chromosome location were determined by comparison of the cloned cDNA sequence to the genomic sequence in the BAC clone (AC027643). In contrast to MCHR1, which is intronless in the coding region located at chromosomal locus 22q13.3, MCHR2 has five coding exons and one noncoding exon that are at locus 6q21 (Fig. 1C). Furthermore, two representatives of an alternatively spliced MCHR2 mRNA were found by sequencing RACE products. This splice form has an extended fifth exon and lacks the sixth exon, giving rise to a predicted protein sequence that lacks the last two transmembrane domains (Fig. 1 B and C; the arrow in B indicates the end of the putative truncated protein).
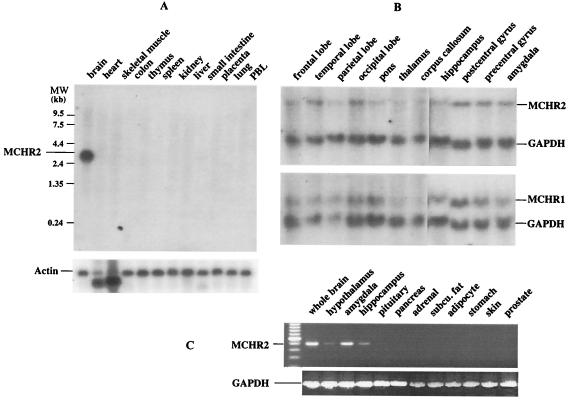
Figure 2.
MCHR2 expression in the brain. (A) Northern hybridization of the human multiple tissue mRNA blot. MCHR2 expression was detected only in the brain. (B) Northern hybridization of human brain blots. MCHR2, like MCHR1, was present in many brain regions.
Tissue Distribution of MCHR2.
The expression of MCHR2 was analyzed by Northern hybridization, RNA dot array blot hybridization, and reverse transcription–PCR. By Northern hybridization with MCHR2 cDNA as a probe, a single species of mRNA with an estimated size of 3.0 kb was detected specifically in the brain (Fig. 2A). MCHR2, like MCHR1, is widely distributed in the brain. Its expression was detected in most of the brain regions checked (Fig. 2 B and C and RNA dot array hybridization, result not shown), including frontal lobe, temporal lobe, occipital lobe, precentral gyrus, pons, hippocampus, amygdala, putamen, and the caudate nucleus, with relatively high levels in the cerebral cortex. Lower levels of expression were also detected in thalamus, corpus callosum, and medulla oblongata. No clear signal was detected in cerebellum or spinal cord. Expression of MCHR2 was also detected in hypothalamus (Fig. 2C), adipocytes (very weak band), pituitary, and pancreas (data not shown). We detected the alternatively spliced short form of MCHR2 mRNA only by RACE-PCR and not by Northern hybridization, indicating a very low level of expression. In summary, MCHR2 appears to be expressed predominantly in the brain, with no dramatic differences observed from the expression pattern of MCHR1 in the central nervous system.
Identification of MCH as a Natural Ligand for MCHR2.
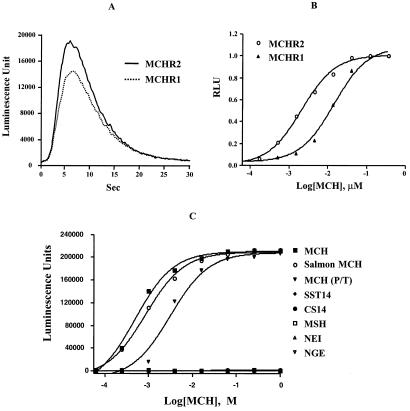
To identify its natural ligand, MCHR2 was cloned into the expression plasmid pEF6/V5-His. The resulting plasmid, pEF6/V5-MCHR2, was cotransfected with pcDNA3-AEQ into HEK293 and CHO cells. The Sigma-RBI biologically active peptide library was screened with the transfected cells, and the intracellular Ca2+ levels were measured with the use of an aequorin assay (Materials and Methods). Among the 60 peptides included in the library, only MCH stimulated a sharp transient increase of Ca2+ in either HEK293 or CHO cells transfected with pEF6/V5-MCHR2 (Fig. 3A). Cells transfected with the control vector or plasmids expressing other GPCRs did not show any response to MCH (data not shown).
Figure 3.
MCHR2 is a receptor for MCH. (A) MCH stimulates a transient increase in intracellular Ca2+ similar to that resulting from MCH activation of MCHR1. CHO cells were transfected with MCHR2 or MCHR1 expression plasmids. The intracellular Ca2+ levels were measured with an aequorin luminescent assay (Materials and Methods). (B) MCH activated MCHR2 with an EC50 comparable to that observed with MCHR1 in transiently transfected HEK293 cells. (C) Specific activation of MCHR2 by MCH. MCH or MCH-related peptides were tested for activation of MCHR2 receptor by the aequorin assay. Only MCH, salmon MCH, and the MCH analog [Phe13, Tyr19]-MCH [MCH (P/T)] induced a Ca2+ increase in MCHR2-transfected HEK293 cells. SST14, somatostatin-14; CS14, cortistatin-14; αMSH, α-melanotropin; NEI, neuropeptide-EI (MCH-precursor-derived peptide).
The stimulation of a transient increase of Ca2+ in the MCHR2 transfected cells is MCH concentration dependent (Fig. 3 A and B), with an EC50 of 2.10 nM in HEK293 cells. The EC50 for MCHR1 under similar conditions was 14.4 nM. To clearly determine the specificity of MCHR2, we tested several MCH-related peptides for agonist activity. Salmon MCH has a high degree of homology to the human MCH (4) and activates MCHR1 (9). It also activates MCHR2 (Fig. 3C). [Phe13, Tyr19]-MCH, a synthetic analog of MCH known to bind to and activate MCHR1 (8, 12, 22), also activates MCHR2, with a slightly lower potency compared with MCH. Other peptides from the same MCH precursor neuropeptide-EI and neuropeptide GE (23, 24) were inactive for MCHR2. Because MCHR1 and MCHR2 share relatively high homology with the somatostatin receptors, we tested somatostatin-14, somatostatin-28, somatostatin analog, RC-160, and two peptides structurally very similar to somatostatin, cortistatin-14 and cortistatin-29 (25). None of these peptides showed any activity on MCHR2 at concentrations up to 1 μM. Finally, α-melanotropin, a functional antagonist of MCH (26–29), was also inactive on MCHR2 as an agonist.
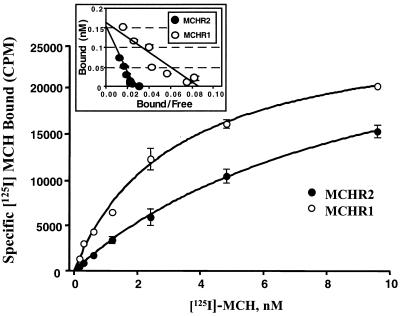
We analyzed the binding of MCHR2 to radiolabeled MCH in a receptor-binding assay (Materials and Methods). Membranes were prepared from HEK293 cells that were transiently transfected with pEF6/V5-MCHR2. Specific binding of 125I-labeled MCH was calculated as the total binding minus the nonspecific binding (inhabitable with 1 μM cold MCH). As a control, 125I-labeled MCH binding to membrane prepared from MCHR1-transfected cells was also measured in parallel. As shown in Fig. 4, MCHR2 bound to MCH with an affinity (Kd = 9.6 ± 0.5 nM) comparable to that of MCHR1 (Kd = 3.1 ± 0.4 nM). The maximum binding (Bmax ≈ 1.6 nmol/g membrane protein) for both MCHR2 and MCHR1 indicates a similar moderate level of expression of the receptors in the cells. Based on the specific and high-affinity MCH binding we observed for MCHR2 and the functional assay results, we conclude that MCHR2 is, in fact, a bona fide high-affinity MCH receptor.
Figure 4.
Binding of 125I-labeled MCH to MCHR2. Radioligand binding was performed as described in Materials and Methods. Binding of MCHR1 to 125I-labeled MCH was also performed for comparison. Smooth lines are a nonlinear regression fit. MCHR2 bound to MCH with an affinity (Kd = 9.6 ± 0.5 nM) comparable to that of MCHR1 (Kd = 3.1 ± 0.4 nM). (Inset) Scatchard plots. The Bmax was about 0.16 nM for both MCHR2 and MCHR1, corresponding to 1.6 nmol/g membrane protein. Data are expressed as mean ± SD (n = 3).
Signal Transduction of MCHR2 Through Gq.
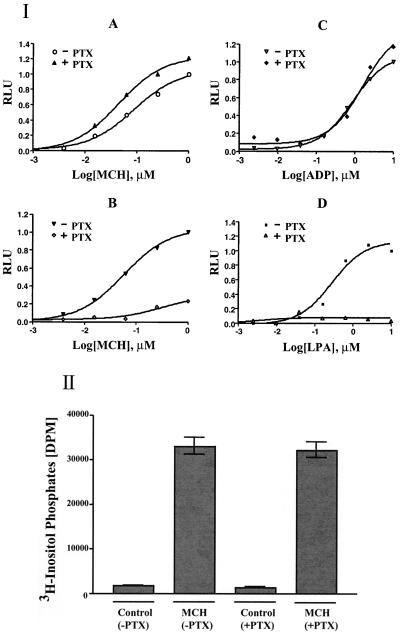
In HEK293 cells transfected with pEF6/V5-MCHR2, MCH stimulated increased levels of both inositol phosphates and Ca2+, and this effect was insensitive to PTX treatment (Fig. 5 A and B). In contrast, the Ca2+ response in cells transfected with MCHR1 was mostly inhibited, which is consistent with previous reports (30). Lysophosphatidic acid (LPA) (31) and ADP (32), which stimulate Ca2+ increase through endogenous Gi and Gq receptors, respectively, were tested as controls. As expected, the LPA-induced Ca2+ response was PTX sensitive, whereas the ADP-induced response was resistant. Similar results were also obtained with CHO cells. These results suggested that MCHR2 is primarily coupled to Gq, which is in contrast with the findings for MCHR1, which has been shown to couple to multiple G proteins, including Gi, Go, and Gq (30).
Figure 5.
MCHR2 is primarily coupled to Gq. (I) HEK293 cells transfected with MCHR2 or MCHR1 were stimulated with MCH, ADP, or lysophosphatidic acid (LPA) at the indicated concentrations with or without pretreatment with PTX (100 ng/ml) for 16 h. The Ca2+ responses were measured with the aequorin assay. Relative luminescence (RLU) was the ratio of luminescence units from cells with different treatments to that from cells treated with the highest concentration of ligand in the absence of PTX. The cells were transfected with MCHR2 (A) or MCHR1 (B) and stimulated with MCH. (C and D) HEK293 cells transiently transfected with MCHR2 were also stimulated with ADP (C) or LPA (D) to activate their endogenous receptors in the cells. (II) MCH increased the production of inositol phosphates in the MCHR2-transfected cells, and this effect is insensitive to PTX. HEK293 cells transfected with MCHR2 were stimulated with 100 nM MCH with or without PTX. Controls did not receive MCH.
Discussion
MCH plays roles far beyond the one originally discovered in the regulation of skin color in teleost fish (1). It has been shown to regulate the hypothalamo—pituitary–adrenal axis (33), modulate water and electroyte fluxes in the gastrointestinal tract (34), stimulate oxytocin secretion (35), regulate sensory processing (36), and modulate the activity of monoaminergic systems (37). Its critical role in feeding behaviors and energy balance, however, has been the most and the best characterized so far (4–7, 26). One MCH receptor, MCHR1, has recently been identified, and it is believed to be the key mediator of MCH activities (9–13). MCHR1 mRNA is present in many different regions of the brain. Relatively high levels of expression of MCHR1 have been observed in the ventromedial, dorsalmedial, and arcuate nuclei of the hypothalamus, regions that are known to play an important role in feeding behaviors (15, 38, 39). Recently, it has been shown that MCHR1 is a target of leptin, and overexpression of MCH in transgenic mice causes obesity (7, 15). Because MCH exerts a wide variety of effects, and GPCRs often have multiple subtypes that are involved in different processes, there was a possibility of additional MCH receptor(s). Circumstantial evidence did support such a possibility. For example, MCH seems to specifically bind to several cell lines, B16F1 melanoma cells and the SVK14 keratinocytes, which do not express MCHR1 (4, 16, 17). We searched the genomic databases (search date was October 19, 2000) for sequences that have high similarity to that of the MCHR1 and identified a MCH receptor. It would be interesting to determine whether MCHR2 is expressed in these cells.
The tissue distribution of MCHR2 mRNA is very similar to that of MCHR1 (Fig. 2 A and B). Both are expressed in many regions of the brain, including the hypothalamus. MCHR2 expression has also been detected in a few other tissues that have been reported to express MCHR1, including adipose tissue (40) and pituitary (14). With such similar expression patterns for these two receptors, they are likely to both be important in mediating MCH effects, including modulation of feeding and energy balance. Despite the similarities, there may still be unobserved differences in the expression patterns of the two receptors. For example, it is quite possible that MCHR1 and MCHR2 might be expressed in different types of cells within the same tissue and/or have polarized localization in the same cell (41). A detailed comparative analysis of the localization of MCHR1 and MCHR2 by in situ hybridization and immunohistochemistry is needed. Furthermore, expression of the truncated MCHR2 splice form could also modulate the activity of the MCHR1 or MCHR2 receptors through heterodimerization. The expression pattern of this alternative splice form of MCHR2 is unclear, however. It is also noteworthy that the overall homology between MCHR2 and MCHR1 (36%) is relatively low for a family of receptors that bind to the same ligand and that the genomic structures of the two genes are very different. These indicate that they are more divergent in evolution than some other GPCR family members. It should not be surprising that additional ligand(s) might be found for MCHR2 and that MCHR2 might play a biological role rather different from that played by MCHR1.
MCHR1 has been shown to couple to Gi, Go, and Gq, whereas MCHR2 primarily couples to Gq. In our assays, the Ca2+ response and production of inositol phosphates upon stimulation with MCH in cells transfected with MCHR2 is completely resistant to PTX. Activation of different G proteins triggers different downstream responses in cells. Thus the difference in G protein coupling of MCHR1 and MCHR2 may mediate different effects on cells. The relative expression levels or activities of the two receptors in a cell may determine how it responds to MCH. The second and third intracellular loops of MCHR2 are rather divergent from that of MCHR1 and SSTRs, both of which are similarly coupled to multiple G proteins. This sequence divergence may be responsible for the difference in G protein coupling of MCHR2, because the intracellular loops, the third loop in particular, have been implicated in G protein coupling in other GPCRs (42).
The discovery of a MCH receptor provides a tool essential to a full understanding of the complex biology of MCH. It will also play an important role in the development of subtype-specific agonists and antagonists that is useful for the study of physiological functions and therapeutic applications involving MCH.
Acknowledgments
We thank W. W. Zhong, J. Lu, and J. Gupte for their excellent technical assistance. We thank Tim Hoey for his advice on the preparation of the manuscript. We also thank U. Schindler, R. Schwandner, H, Beckmann, D. Lin, S.-Y. Namgoong, G. Peterson, M. Learned, Y. Li, K. Amaral, and S. Lynch for helpful discussions.
Abbreviations
- GPCR
G protein-coupled receptor
- MCH
melanin-concentrating hormone
- MCHR
melanin-concentrating hormone receptor
- PTX
pertussis toxin
- RACE
rapid amplification of cDNA ends
- CHO
Chinese hamster ovary
- SSTR
somatostatin receptor
References
- 1.Kawauchi H, Kawazoe I, Tsubokawa M, Kishida M, Baker B I. Nature (London) 1983;305:321–323. doi: 10.1038/305321a0. [DOI] [PubMed] [Google Scholar]
- 2.Skofitsch G, Jacobowitz D M, Zamir N. Brain Res Bull. 1985;15:635–649. doi: 10.1016/0361-9230(85)90213-8. [DOI] [PubMed] [Google Scholar]
- 3.Bittencourt J C, Presse F, Arias C, Peto C, Vaughan J, Nahon J L, Vale W, Sawchenko P E. J Comp Neurol. 1992;319:218–245. doi: 10.1002/cne.903190204. [DOI] [PubMed] [Google Scholar]
- 4.Saito Y, Nothacker H-P, Civelli O. Trends Endocrinol Metabol. 2000;11:299–303. doi: 10.1016/s1043-2760(00)00290-3. [DOI] [PubMed] [Google Scholar]
- 5.Qu D, Ludwig D S, Gammeltoft S, Piper M, Pelleymounter M A, Cullen M J, Mathes W G, Przypek J, Kanarek R, Maratos-Flier E. Nature (London) 1996;380:243–247. doi: 10.1038/380243a0. [DOI] [PubMed] [Google Scholar]
- 6.Shimada M, Tritos N A, Lowell B B, Flier J S, Maratos-Flier E. Nature (London) 1998;396:243–247. doi: 10.1038/25341. [DOI] [PubMed] [Google Scholar]
- 7.Ludwig D S, Tritos N A, Mastaitis J W, Kulkarni R, Kokkotou E, Elmquist J, Lowell B, Flier J S, Maratos-Flie E. J Clin Invest. 2001;107:379–386. doi: 10.1172/JCI10660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kolakowski L F, Jr, Jung B P, Nguyen T, Johnson M P, Lynch K R, Cheng R, Heng H H Q, George S R, O'Dowd B. FEBS Lett. 1996;398:253–258. doi: 10.1016/s0014-5793(96)01160-x. [DOI] [PubMed] [Google Scholar]
- 9.Lembo P M C, Grazzini E, Cao J, Hubatsch D A, Pelletier M, Hoffert C, St-Onge S, Pou C, Labrecque J, Groblewski T, et al. Nat Cell Biol. 1999;1:267–271. doi: 10.1038/12978. [DOI] [PubMed] [Google Scholar]
- 10.Chambers J, Ames R S, Bergsma D, Muir A, Fitzgerald L R, Hervieu G, Dytko G M, Foley J J, Martin J, Liu W S, et al. Nature (London) 1999;400:261–265. doi: 10.1038/22313. [DOI] [PubMed] [Google Scholar]
- 11.Saito Y, Nothacker H P, Wang Z, Lin S H S, Leslie F, Civelli O. Nature (London) 1999;400:265–269. doi: 10.1038/22321. [DOI] [PubMed] [Google Scholar]
- 12.Shimomura Y, Mori M, Sugo T, Ishibashi Y, Abe M, Kurokawa T, Onda H, Nishimura O, Sumino Y, Fujino M. Biochem Biophys Res Commun. 1999;261:622–626. doi: 10.1006/bbrc.1999.1104. [DOI] [PubMed] [Google Scholar]
- 13.Bachner D, Kreienkamp H J, Weise C, Buck F, Richter D. FEBS Lett. 1999;457:522–524. doi: 10.1016/s0014-5793(99)01092-3. [DOI] [PubMed] [Google Scholar]
- 14.Hervieu G J, Cluderay J E, Harrison D, Meakin J, Maycox P, Nasir S, Leslie R A. Eur J Neurosci. 2000;12:1194–1216. doi: 10.1046/j.1460-9568.2000.00008.x. [DOI] [PubMed] [Google Scholar]
- 15.Kokkotou E G, Tritos N A, Mastaitis J W, Slieker L, Maratos-Flier E. Endocrinology. 2001;142:680–686. doi: 10.1210/endo.142.2.7981. [DOI] [PubMed] [Google Scholar]
- 16.Drozdz R, Siegrist W, Baker B I, Chluba-de Tapia J, Eberle A N. FEBS Lett. 1995;359:199–202. doi: 10.1016/0014-5793(95)00043-9. [DOI] [PubMed] [Google Scholar]
- 17.Burgaud J L, Poosti R, Fehrentz J A, Martinez J, Nahon J L. Biochem Biophys Res Commun. 1997;241:622–629. doi: 10.1006/bbrc.1997.7849. [DOI] [PubMed] [Google Scholar]
- 18.Baldi P, Chauvin Y. J Comput Biol. 1994;1:311–336. doi: 10.1089/cmb.1994.1.311. [DOI] [PubMed] [Google Scholar]
- 19.Baldi P, Chauvin Y, Hunkapiller T, McClure M A. Proc Natl Acad Sci USA. 1994;91:1059–1063. doi: 10.1073/pnas.91.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stables J, Green A, Marshall F, Fraser N, Knight E, Sautel M, Milligan G, Lee M, Rees S. Anal Biochem. 1997;252:115–126. doi: 10.1006/abio.1997.2308. [DOI] [PubMed] [Google Scholar]
- 21.Hung D T, Vu T H, Nelken N A, Congkli S R. J Cell Biol. 1992;116:827–831. doi: 10.1083/jcb.116.3.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drozdz R, Eberle A N. J Pept Sci. 1995;1:58–65. doi: 10.1002/psc.310010108. [DOI] [PubMed] [Google Scholar]
- 23.Nahon J-L, Presse F, Bittencourt J C, Swachenko P, Wale W. Endocrinology. 1989;125:2056–2065. doi: 10.1210/endo-125-4-2056. [DOI] [PubMed] [Google Scholar]
- 24.Toumaniantz G, Bittencourt J C, Nahon J-L. Endocrinology. 1996;137:4518–4521. doi: 10.1210/endo.137.10.8828517. [DOI] [PubMed] [Google Scholar]
- 25.De Lecea L, Criado J R, Prospero-Garcia O, Gautvik K M, Schweitzer P, Danielson P E, Dunlop C L, Siggins G R, Henriksen S J, Sutcliffe J G. Nature (London) 1996;381:242–245. doi: 10.1038/381242a0. [DOI] [PubMed] [Google Scholar]
- 26.Miller C L, Hruby V, Matsunaga T, Bickford P. Peptides (Tarrytown, NY) 1993;14:1–10. doi: 10.1016/0196-9781(93)90128-4. [DOI] [PubMed] [Google Scholar]
- 27.Gonzalez M, I, Vaziri S, Vilson C A. Peptides (Tarrytown, NY) 1996;17:171–177. doi: 10.1016/0196-9781(95)02092-6. [DOI] [PubMed] [Google Scholar]
- 28.Sanchez M, Baker B I, Celis M. Peptides (Tarrytown, NY) 1997;18:393–396. doi: 10.1016/s0196-9781(96)00327-0. [DOI] [PubMed] [Google Scholar]
- 29.Ludwig D S, Mountjoy K G, Tatro J B, Gillette J A, Frederich R C, Flier J S, Maratos-Flier E. Am J Physiol. 1998;274:E627–E633. doi: 10.1152/ajpendo.1998.274.4.E627. [DOI] [PubMed] [Google Scholar]
- 30.Hawes B E, Kil E, Green B, O'Neill K, Fried S, Graziano M P. Endocrinology. 2000;141:4524–4532. doi: 10.1210/endo.141.12.7833. [DOI] [PubMed] [Google Scholar]
- 31.Moolenaar W H. Ann NY Acad Sci. 2000;905:1–10. doi: 10.1111/j.1749-6632.2000.tb06532.x. [DOI] [PubMed] [Google Scholar]
- 32.Fields T A, Casey P J. Biochem J. 1997;321:561–571. doi: 10.1042/bj3210561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jezova D, Bartanusz V, Westergren I, Johansson B B, Riview J, Vale W, Rivier C. Endocrinology. 1992;130:1024–1029. doi: 10.1210/endo.130.2.1310274. [DOI] [PubMed] [Google Scholar]
- 34.Hervien G, Volant K, Grishina O, Descroix-Vagne M, Nahon J L. Endocrinology. 1996;137:561–571. doi: 10.1210/endo.137.2.8593803. [DOI] [PubMed] [Google Scholar]
- 35.Parkes D G, Vale W. In: Melanotropic Peptides. Eberle A, Vaudry H, editors. New York: N.Y. Acad. Sci.; 1993. p. 558. [Google Scholar]
- 36.Miller C L, Hruby V S, Matsungaga T O, Bickford P C. Peptides (Tarrytown, NY) 1993;14:431–440. doi: 10.1016/0196-9781(93)90128-4. [DOI] [PubMed] [Google Scholar]
- 37.Gonzalez M I, Kalia V, Hole D R, Wilson C A. Peptides (Tarrytown, NY) 1997;18:387–392. doi: 10.1016/s0196-9781(96)00337-3. [DOI] [PubMed] [Google Scholar]
- 38.Fulwiller C E, Saper C B. Brain Res. 1984;319:229–259. doi: 10.1016/0165-0173(84)90012-2. [DOI] [PubMed] [Google Scholar]
- 39.Fulwiller C E, Saper C B. Neurosci Lett. 1985;53:289–296. doi: 10.1016/0304-3940(85)90553-1. [DOI] [PubMed] [Google Scholar]
- 40.Bradley R L, Kokkotou E G, Maratos-Flier E, Cheatham B. Diabetes. 2000;49:1073–1077. doi: 10.2337/diabetes.49.7.1073. [DOI] [PubMed] [Google Scholar]
- 41.Wozniak M, Keefer J R, Saunders C, Limbird L E, Recept J. Signal Transduction Res. 1997;17:373–383. doi: 10.3109/10799899709036615. [DOI] [PubMed] [Google Scholar]
- 42.Wess J, Liu J, Blin N, Yun J, Lerche C, Kostenis E. Life Sci. 1997;60:1007–1014. doi: 10.1016/s0024-3205(97)00041-6. [DOI] [PubMed] [Google Scholar]