Abstract
X-linked muscular dystrophy is a highly progressive disease of childhood and characterized by primary genetic abnormalities in the dystrophin gene. Senescent mdx specimens were used for a large-scale survey of potential age-related alterations in the dystrophic phenotype, because the established mdx animal model of dystrophinopathy exhibits progressive deterioration of muscle tissue with age. Since the mdx tibialis anterior muscle is a frequently used model system in muscular dystrophy research, we employed this particular muscle to determine global changes in the dystrophic skeletal muscle proteome. The comparison of mdx mice aged 8 weeks versus 22 months by mass-spectrometry-based proteomics revealed altered expression levels in 8 distinct protein species. Increased levels were shown for carbonic anhydrase, aldolase, and electron transferring flavoprotein, while the expressions of pyruvate kinase, myosin, tropomyosin, and the small heat shock protein Hsp27 were found to be reduced in aged muscle. Immunoblotting confirmed age-dependent changes in the density of key muscle proteins in mdx muscle. Thus, segmental necrosis in mdx tibialis anterior muscle appears to trigger age-related protein perturbations due to dystrophin deficiency. The identification of novel indicators of progressive muscular dystrophy might be useful for the establishment of a muscle subtype-specific biomarker signature of dystrophinopathy.
1. Introduction
Voluntary muscle fibres are one of the most abundant cellular units in the body. Skeletal muscle tissues are responsible for the provision of postural control, the coordination of excitation-contraction-relaxation cycles for voluntary movements, the integration of key metabolic and biochemical pathways, and the regulation of heat homeostasis. Under normal physiological conditions, these highly complex cellular tasks require a large and diverse number of protein interactions. Hence, supramolecular protein complexes with specialized functions, structures, and connections represent a major biochemical feature of muscle fibres. An excellent example of a large protein assembly present in skeletal muscle is the dystrophin-glycoprotein complex of the sarcolemma [1–5]. The crucial importance of the dystrophin-associated protein complex is exemplified by the pathophysiological fact that primary genetic abnormalities in the dystrophin gene result in progressive muscle wasting diseases, such as Duchenne or Becker muscular dystrophy [6–8]. In normal muscle, the dystrophin-glycoprotein complex provides a trans-sarcolemmal linkage between the actin membrane cytoskeleton and the extracellular matrix component laminin [2]. The subsarcolemmal dystrophin matrix and the molecular connection between the basal lamina structure and the muscle interior is believed to prevent damage to the muscle surface from potential membrane-distorting forces during contraction-relaxation cycles [5].
In X-linked muscular dystrophy, dystrophin deficiency results in a drastic reduction of sarcolemmal glycoproteins that triggers a loss of plasmalemmal integrity [9]. Dystrophic muscle fibres are more susceptible to contraction-induced injury [10] and their lateral transmission of force is impaired [11]. Cycles of sarcolemmal microrupturing and natural membrane repair mechanisms appear to cause the introduction of Ca2+-leak channels [12] that in turn elevate cytosolic Ca2+-levels [13] and disturb Ca2+-fluxes through the sarcoplasmic reticulum in dystrophic fibres [14–16]. Interestingly, a recent study on the therapeutic effect of upregulating the intramuscular heat shock protein Hsp72 to ameliorate the dystrophic phenotype revealed that the SERCA-type Ca2+-ATPase is dysfunctional in severely dystrophic muscle [17]. These findings strongly indicate that impaired Ca2+-homeostasis plays a key role in X-linked muscular dystrophy. However, it is not well understood how many molecular and cellular factors are involved in the overall process leading to the highly complex pathology of dystrophinopathy. Thus, in order to determine the hierarchy of secondary pathobiochemical effects that render a dystrophic muscle more susceptible to necrosis, it is crucial to elucidate global alterations due to the disintegration of the dystrophin-glycoprotein complex [18]. Mass-spectrometry-based proteomics suggests itself as a suitable analytical tool for such large-scale and high-throughput approaches to study the effects of dystrophin deficiency.
In contrast to hypothesis-based and targeted bioresearch, proteomics can be considered an unbiased and technology-driven approach for the comprehensive cataloging of entire protein complements [19–21]. Skeletal muscle proteomics in particular is concerned with the global identification and detailed cataloguing of the protein constituents of voluntary contractile fibres in health and disease [22–24]. In the long term, comparative proteomics promises to be instrumental for the establishment of comprehensive biomarker signatures of myogenesis, muscle repair mechanisms, physiological adaptations and pathological changes [25], as well as the natural aging process [26]. Most gel electrophoresis-based proteomic studies use high-resolution two-dimensional gel electrophoresis in combination with advanced mass spectrometric analysis for the unequivocal identification of muscle proteins of interest [27–29]. In the case of X-linked muscular dystrophy, a variety of mass spectrometric investigations have attempted to determine proteome-wide changes in dystrophin-deficient muscle tissue in order to establish a dystrophy-specific biomarker signature [30, 31].
Large-scale proteomic profiling studies have included investigations of serum [32, 33], cardiac muscle [34, 35], and various skeletal muscle tissues [16, 36–43] from the mdx mouse model of Duchenne muscular dystrophy, as well as a proteomic analysis of dystrophic GRMD dog skeletal muscle [44]. Although the findings from individual studies do not agree on the exact number and extent of protein alterations within the dystrophic muscle proteome, all investigations concur that dystrophin-deficient fibres exhibit a generally perturbed protein expression pattern [30]. Previous gel electrophoresis-based proteomic profiling studies of dystrophic samples have focused on the cytosolic fraction from 1, 3, and 6 months old hind limb muscle covering a pI range of 4–7 and using Coomassie and silver-staining methods [36, 37], crude extracts from gastrocnemius muscle from 9 weeks old hind limb tissue covering a pI range of 3–10 and using Stains-All labeling [16], preparations from 6 weeks old gastrocnemius muscle covering a pI range of 3–10 and using fluorescence 2D-DIGE labeling [38], crude extracts from 9 weeks old diaphragm tissue covering a pI range of 3–10 and using hot Coomassie staining [39], crude extracts from 9 weeks old diaphragm muscle covering a pI range of 3–10 and using fluorescence 2D-DIGE labeling [40], crude extracts from 10 weeks old antisense oligomer-treated diaphragm tissue covering a pI range of 3–10 and using 2D-DIGE labeling [41], crude extracts from 9 weeks old extraocular muscle covering a pI range of 3–10 and using fluorescence 2D-DIGE labeling [42], and crude extracts from aged diaphragm muscle covering a pI range of 3–10 and using fluorescence RuBPs labeling [43].
In analogy to the above-outlined proteomic studies, this report has focused on aged tibialis anterior muscle from dystrophic mdx mice. The tibialis anterior is one of the most active lower leg muscles [45], which exhibits a relatively high degree of resistance to fatigue during periods of intense running [46], making it an interesting contractile system to study with respect to secondary effects of dystrophinopathy. In addition, previous experimental gene therapy studies have focused on mdx tibialis anterior muscle [47]. Labeling of proteins with fluorescent dyes has been extensively applied in proteomic investigations [48] and we have used here fluorescent RuBPs staining [49] for a comparative proteomic survey of dystrophic leg muscle from 8 weeks, 12 months, and 22 months old mdx mice. The densitometric analysis of two-dimensional gels, covering a pI range of 3–10, in combination with mass spectrometry identified significant age-related changes in carbonic anhydrase, aldolase, electron transferring flavoprotein, pyruvate kinase, myosin, tropomyosin, and the small heat shock protein Hsp27 in dystrophic mdx tibialis anterior muscle.
2. Materials and Methods
2.1. Materials
Materials and electrophoresis-grade chemicals for the proteomic analysis of muscle proteins were purchased from Amersham Biosciences/GE Healthcare, Little Chalfont, Buckinghamshire, UK. For protein digestion, sequencing grade-modified trypsin was obtained from Promega (Madison, WI, USA). Chemiluminescence substrate and protease inhibitors were from Roche Diagnostics (Mannheim, Germany). Primary antibodies were obtained from Vision Biosystems Novocastra, Newcastle upon Tyne, UK (mAb NCL-b-DG to the dystrophin-associated glycoprotein β-dystroglycan) and Abcam, Cambridge, UK (ab54913 to the CA3 isoform of carbonic anhydrase; and ab12351 to the small heat shock protein Hsp27). Secondary antibodies were purchased from Chemicon International (Temecula, CA). All other chemicals used were of analytical grade and purchased from Sigma Chemical Company, Dorset, UK.
2.2. Dystrophic mdx Animal Model
The mdx mouse is an established model system of X-linked muscular dystrophy and widely used in basic research and for the evaluation of novel therapeutic options to treat diseases of progressive skeletal muscle wasting [50]. The mdx mouse is a naturally occurring mutant [51] that is missing the Dp427 protein isoform of the membrane cytoskeletal protein dystrophin due to a point mutation in the dmd gene [52]. In analogy to the etiology of patients suffering from Duchenne muscular dystrophy [9], deficiency in full-length dystrophin results in a drastic reduction of all dystrophin-associated glycoproteins in mdx skeletal muscle [53], making it a suitable animal model for studying secondary pathobiochemical changes due to dystrophin deficiency. Dystrophic tibialis anterior muscle from 8 weeks, 12 months, and 22 months old mdx mice and normal tissues from age-matched C57 mice were obtained from the bioresource unit of the University of Bonn [54]. Mice were kept under standard conditions and all procedures were performed in accordance with German guidelines on the use of animals for scientific experiments. Animals were sacrificed by cervical dislocation and muscle tissues quickly removed and quick-frozen in liquid nitrogen.
2.3. Preparation of Skeletal Muscle Extracts from Aged mdx Mice
For the mass spectrometry-based proteomic survey of aged mdx skeletal muscle tissue, tibialis anterior specimens were shipped to Ireland on dry ice and stored at −80°C prior to usage. In order to obtain muscle protein extracts, 4 dystrophic muscle specimens from each age group were pulverized by grinding tissue pieces in liquid nitrogen using a mortar and pestle. Ground muscle powder was solubilized in lysis buffer with the ratio of 100 mg wet weight to 1 mL lysis buffer (7 M urea, 2 M thiourea, 4% CHAPS, 2% IPG buffer pH 3–10, 2% (w/v) DTT). To prevent excess protein degradation, the lysis buffer was supplemented with a freshly prepared protease inhibitor cocktail [35]. Following gentle rocking for 30 minutes, suspensions were centrifuged at 4°C for 20 min at 20,000 ×g and the protein concentration determined [55].
2.4. Two-Dimensional Fluorescence Gel Electrophoretic Analysis
For the separation of muscle proteins, standard two-dimensional gel electrophoresis was carried out by previously optimized methodology using first dimension isoelectric focusing with pH 3–10 strips, and second dimension slab gel electrophoresis with 500 μg protein per gel [55]. Twelve slab gels were run in parallel at 0.5 W/gel for 60 min and then 15 W/gel until the blue dye front had disappeared from the bottom of the gel. Postelectrophoretic staining for the total protein profile was performed with the fluorescent dye ruthenium II tris bathophenanthroline disulfonate (RuBPs). As described previously by Rabilloud and colleagues [56], a stock solution of RuBPs dye was prepared. Following washing twice for 5 min, gels were stained for 6 hours in 20% (v/v) ethanol containing 200 nM of ruthenium chelate. Gels were re-equilibrated twice for 10 min in distilled water prior to imaging [57]. Fluorescently labelled proteins from aged mdx tibialis anterior muscle were visualised using a Typhoon Trio variable mode imager. Gel analysis was performed with Progenesis 2D analysis software and protein spots with significantly altered expression levels were identified by mass spectrometry.
2.5. Mass Spectrometric Identification of Proteins
Protein identification was performed with 2D protein spots from Coomassie-stained pick gels, following counter-staining of RuBPs-labelled analytical gels. Excised protein spots were treated by standardized in-gel tryptic digestion for the generation of representative peptide mixtures. Excision, washing, destaining, and treatment with sequencing-grade trypsin were performed by a previously optimized method [55]. Peptide populations were harvested by removing supernatants from digested gel plugs after centrifugation. Further recovery was achieved by adding 30% acetonitrile/0.2% trifluoroacetic acid to the gel plugs for 10 min at 37°C with gentle agitation. Resulting supernatants were pooled with the initially recovered peptides following trypsin digestion. Samples were dried through vacuum centrifugation and concentrated peptide fractions were then suspended in mass spectrometry-grade distilled water and 0.1% formic acid, spun down through spin filters and added to LC-MS vials for identification by ion trap LC-MS analysis. The mass spectrometric analysis of peptides was carried out with a Model 6340 Ion Trap LC/MS apparatus from Agilent Technologies (Santa Clara, CA, USA). Separation of peptides was performed with a nanoflow Aglient 1200 series system equipped with a Zorbax 300SB C18 analytical reversed phase column using HPLC-Chip technology. Mobile phases used were A: 0.1% formic acid, B: 50% acetonitrile and 0.1% formic acid. Samples were loaded into the enrichment part of the chip at a capillary flow rate set to 4 μL/min with a mix of solvent A and solvent B at a ratio of 19 : 1. Tryptic digests were eluted with a linear gradient of 5% to 70% solvent B over 6 min, 70% to 100% solvent B over 1 min, 100% to 5% over 1 min. A 5 min post-time of solvent A was used to remove any potential carry over. The capillary voltage was set to 2000 V. The flow and temperature of the drying gas were 4L/min and 300°C, respectively. Database searches were carried out with Mascot MS/MS ion search (Matrix Science, London, UK; NCBI database, release 20100212). All searches used “Mus musculus” as taxonomic category and the following parameters: (1) two missed cleavages by trypsin, (2) mass tolerance of precursor ions ±2.5 Da and product ions ±0.7 Da, (3) carboxymethylated cysteins fixed modification, (4) oxidation of methionine as variable modifiaction, (5) percentage coverage was set at over 10%, and (6) at least 2 matched distinct peptides. Mascot scores over 50 are listed in Tables 1 and 2. All pI-values and molecular masses of identified proteins were compared to the relative position of their corresponding 2D spots on analytical slab gels.
Table 1.
List of unchanged landmark 2D protein spots from normal mouse tibialis anterior muscle.
| Spot No. | Protein name | Accession No. | Isoelectric point (pI) | Molecular mass (Da) | Number of peptides | Coverage | Mascot score |
|---|---|---|---|---|---|---|---|
| (%) | |||||||
| 1 | Mitochondrial ATP synthase | AAH37127 | 5.24 | 56,632 | 23 | 64 | 456 |
| 2 | Unnamed protein [Mus musculus] | BAC34145 | 5.75 | 70,730 | 12 | 21 | 230 |
| 3 | Pyruvate kinase, isozymes M1/M2 | NP_001240812 | 6.69 | 58,461 | 25 | 51 | 1149 |
| 4 | Enolase, beta, isoform 1 | NP_031959 | 6.73 | 47,337 | 21 | 56 | 450 |
| 5 | Creatine Kinase, M-type | NP_031736 | 6.58 | 43,250 | 23 | 47 | 509 |
| 6 | Actin, beta | CAA27396 | 5.78 | 39,446 | 12 | 41 | 103 |
| 7 | Tropomyosin, beta chain | NP_033442 | 4.66 | 32,931 | 22 | 59 | 395 |
| 8 | Tropomyosin, beta chain | NP_033442 | 4.66 | 32,933 | 32 | 15 | 241 |
| 9 | Malate dehydrogenase, cytosolic | AAA37423 | 6.16 | 36,625 | 9 | 36 | 190 |
| 10 | Aldolase A, isoform 2 | NP_031464 | 8.31 | 39,795 | 24 | 75 | 498 |
| 11 | Glyceraldehyde-3-phosphate dehydrogenase | NP_032110 | 8.44 | 36,072 | 14 | 57 | 238 |
| 12 | Carbonic anhydrase CA3 | NP_031632 | 6.89 | 29,638 | 12 | 43 | 431 |
| 13 | Triosephosphate isomerase | AAB48543 | 5.62 | 22,720 | 11 | 62 | 248 |
| 14 | Triosephosphate isomerase | AAB48543 | 5.62 | 22,720 | 11 | 68 | 638 |
| 15 | Troponin TnI, fast skeletal muscle | NP_033431 | 8.65 | 21,515 | 5 | 20 | 150 |
| 16 | Adenylate kinase, isoenzyme 1 | NP_067490 | 5.7 | 23,330 | 14 | 70 | 184 |
| 17 | Myosin light chain MLC1/3 | NP_067260 | 4.98 | 20,697 | 17 | 79 | 426 |
| 18 | Myosin light chain MLC2 | NP_058034 | 4.82 | 19,057 | 15 | 61 | 216 |
| 19 | Myosin light chain MLC2 | NP_058034 | 4.82 | 19,059 | 19 | 81 | 391 |
| 20 | Myosin light chain MLC3 | AAH59087 | 4.63 | 18,968 | 11 | 60 | 222 |
| 21 | Parvalbumin, alpha | NP_038673 | 5.02 | 11,923 | 12 | 89 | 622 |
| 22 | Myoglobin | NP_038621 | 7.07 | 17,116 | 5 | 37 | 98 |
Table 2.
List of identified proteins that exhibit a drastic change in abundance during aging of the dystrophic mdx tibialis anterior muscle.
| Spot No. | Protein name | Accession No. | Isoelectric point (pI) | Molecular mass (Da) | Number of peptides | Coverage | Mascot | Fold change 8w-22 m |
|---|---|---|---|---|---|---|---|---|
| (%) | Score | |||||||
| 1 | Carbonic anhydrase CA3 | NP031632 | 6.89 | 29,638 | 4 | 26 | 181 | 2.9 |
| 2 | Aldolase A isoform 2 | NP031464 | 8.31 | 39,795 | 2 | 14 | 85 | 2.3 |
| 3 | Electron transferring flavoprotein, beta | EDL22660 | 8.50 | 29,177 | 13 | 42 | 190 | 1.9 |
| 4 | Carbonic anhydrase CA3 | NP031632 | 6.89 | 29,638 | 12 | 53 | 182 | 1.7 |
| 5 | Pyruvate kinase | NP035229 | 7.18 | 58,388 | 7 | 18 | 440 | 0.7 |
| 6 | Myosin 3 | NP001078847 | 5.62 | 224,755 | 9 | 5 | 141 | 0.7 |
| 7 | Tropomyosin, beta chain | NP033442 | 4.66 | 32,933 | 3 | 14 | 126 | 0.6 |
| 8 | Heat shock protein Hsp27 | AAA18335 | 6.45 | 22,945 | 2 | 15 | 55 | 0.5 |
2.6. Immunoblot Analysis
One-dimensional immunoblotting was employed to verify key findings from the proteomic profiling of aged mdx tibialis anterior muscle. Gel electrophoretic separation and transfer was carried out with a Mini-Protean II electrophoresis and transfer system from BioRad Laboratories (Hemel-Hempstead, Herts, UK). Muscle proteins were transferred to nitrocellulose for 70 minutes at 100 V and at 4°C. Blocking of membranes was achieved with a milk protein solution (5% (w/v) fat-free milk powder in 0.9% (w/v) NaCl, 50 mM sodium phosphate, pH 7.4) for 1 hour. Incubation with sufficiently diluted primary antibody was carried out overnight with gentle agitation. Nitrocellulose sheets were washed and then incubated for 1 hour with secondary peroxidase-conjugated antibodies, diluted in blocking solution [55]. Immunodecorated bands were visualized using chemiluminescence substrate (Roche Diagnostics, Mannheim, Germany). Densitometric scanning of immunoblots was performed using ImageJ (NIH, USA) software.
3. Results
3.1. Proteomic Profiling of Tibialis Anterior Muscle
Prior to the proteomic analysis of differently aged dystrophic tibialis anterior muscle preparations, the protein complement from extracts of normal muscle samples was gel electrophoretically separated and key proteins identified by mass spectrometry. This procedure established a select number of reliable landmark protein spots of a typical proteomic muscle map for control purposes. Figure 1 shows a representative fluorescent RuBPs-labelled gel with the electrophoretically separated protein spot pattern of normal mouse tibialis anterior muscle. Major 2D protein spots were treated by in-gel digestion and the most abundant constituent of this area of the gel identified by mass spectrometry. Table 1 lists the names of identified muscle marker proteins, their international accession number, pI-values, their relative molecular masses, number of matched peptide sequences, percentage sequence coverage, and Mascot scores. Identified proteins ranged in molecular mass from 17.1 kDa (spot 22, myoglobin) to 70.7 kDa (spot 2, unknown protein), and covered a pI-range from pI 4.6 (spot 20, myosin light chain MLC3) to pI 8.7 (spot 15, fast troponin subunit TnI). Spots 1 to 22 represent major muscle-associated protein species with apparent molecular mass to isoelectric point ratios of 57 kDa/pI 5.2, 71 kDa/pI 5.8, 59 kDa/pI 6.7, 47 kDa/pI 6.7, 43 kDa/pI 6.6, 40 kDa/pI 5.8, 33 kDa/pI 4.7, 33 kDa/pI 4.7, 37 kDa/pI 6.2, 40 kDa/pI 8.3, 36 kDa/pI 8.4, 30 kDa/pI 6.9, 23 kDa/pI 5.6, 23 kDa/pI 5.6, 22 kDa/pI 8.7, 23 kDa/pI 5.7, 21 kDa/pI 5.0, 19 kDa/pI 4.8, 19 kDa/pI 4.8, 19 kDa/pI 4.6, 12 kDa/pI 5.0, and 17 kDa/pI 7.1, respectively (Figure 1). Electrospray ionization mass spectrometry identified these marker proteins as isoforms of mitochondrial ATP synthase, pyruvate kinase, enolase, creatine kinase, actin, tropomyosin, malate dehydrogenase, aldolase, glyceraldehyde-3-phosphate dehydrogenase, carbonic anhydrase, triosephosphate isomerase, troponin, adenylate kinase, parvalbumin, myoglobin, and various myosin light chains (Table 1).
Figure 1.
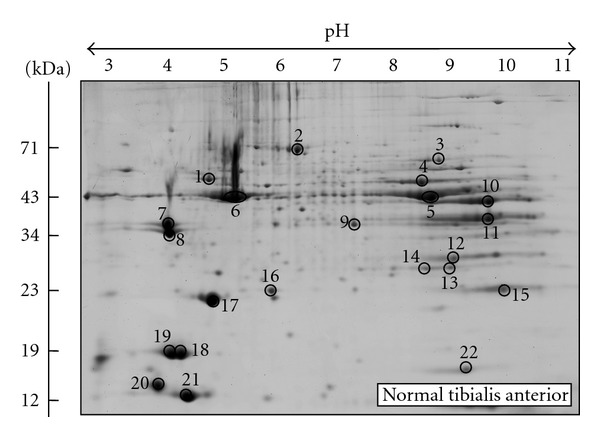
Two-dimensional gel electrophoretic analysis of normal mouse tibialis anterior muscle. Shown is a fluorescent RuBPs-stained gel of total extracts from 8 weeks old tibialis anterior muscle. Major protein spots are marked by circles and are numbered 1 to 22. See Table 1 for the mass spectrometric identification of 2D landmark proteins that do not change during aging of the mdx animal model of Duchenne muscular dystrophy. The pH values of the first dimension gel system and molecular mass standards of the second dimension are indicated on the top and on the left of the panels, respectively.
3.2. Proteomic Analysis of Dystrophic Tibialis Anterior Muscle during Aging
Following the optimization and initial mass spectrometric identification of muscle marker proteins in normal mouse tibialis anterior muscle, fluorescence high-resolution two-dimensional gel electrophoresis was employed to detect potential differences in aging-related protein expression patterns in mdx tibialis anterior muscle. Figure 2 summarizes analytical gels with 4 biological repeats of 8 weeks, 12 months, and 22 months old total mdx muscle extracts. Panels TA MDX 1 to 4, TA MDX 5 to 8 and TA MDX 9 to 12 represent 8 weeks, 12 months, and 22 months old muscle preparations, respectively. Since the overall 2D spot patterns of normal versus dystrophic tibialis anterior muscle were relatively comparable, a detailed denitometric analysis was carried out in order to evaluate potential differences in individual protein species. Densitometric scanning was performed with a Typhoon Trio variable imager and Progenesis 2D analysis software was used to establish differential expression patterns during muscle aging. The detailed proteomic survey of dystrophic tibialis anterior revealed distinct age-related changes in 8 muscle protein species between 8 weeks and 22 months old total mdx muscle preparations.
Figure 2.
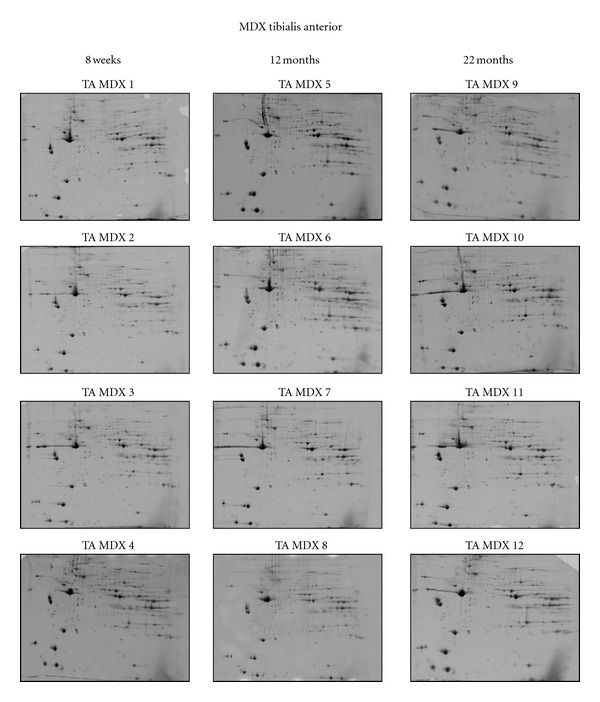
Two-dimensional gel electrophoretic analysis of aging mdx tibialis anterior muscle. Shown are RuBPs-stained gels of total extracts from 8 weeks (TA MDX 1 to 4), 12 months (TA MDX 5 to 8), and 22 months (TA MDX 9 to 12) old tibialis anterior muscle. Fluorescent images are shown for the pH 3–10 range.
3.3. Mass Spectrometric Identification of Protein Alterations in Aged mdx Tibialis Anterior Muscle
A representative fluorescent 2D master gel of mdx tibialis anterior muscle is shown in Figure 3. As compared to a recent study on senescent mdx diaphragm muscle, which showed drastic age-dependent changes in 11 proteins in this severely necrotic tissue [43], the more mildly affected mdx tibialis anterior muscle showed less pronounced proteome-wide changes during the aging process. This finding agrees with the differing pathology of mdx leg muscle versus mdx diaphragm muscle. Skeletal muscle proteins that exhibited significant alterations in expression levels are marked by circles and are numbered 1 to 8 in the 2D gel representing the urea-soluble proteome from mdx tibialis anterior muscle. The mass spectrometric identification of these altered protein species is listed in Table 2. Besides listing the names of identified proteins, their accession number, pI-values, their relative molecular masses, the number of matched peptide sequences, percentage sequence coverage, and MS/MS scores, this table also shows the fold change of individual proteins affected in dystrophin-deficient mdx tibialis anterior muscle during aging. Proteins species with a changed concentration in mdx tibialis anterior muscle ranged in molecular mass from 23 kDa (heat shock protein Hsp27) to 224 kDa (myosin 3) and covered a pI-range from pI 4.7 (tropomyosin) to pI 8.5 (electron transferring flavoprotein). An increased abundance was shown for the CA3 isoform of carbonic anhydrase (spots 1 and 4), the glycolytic enzyme aldolase (spot 2), and electron transferring flavoprotein (spot 3). The key cytosolic enzyme pyruvate kinase (spot 5), myosin 3 (spot 6), tropomyosin (spot 7), and the molecular chaperone Hsp27 (spot 8) were found to be decreased in mdx tissue.
Figure 3.
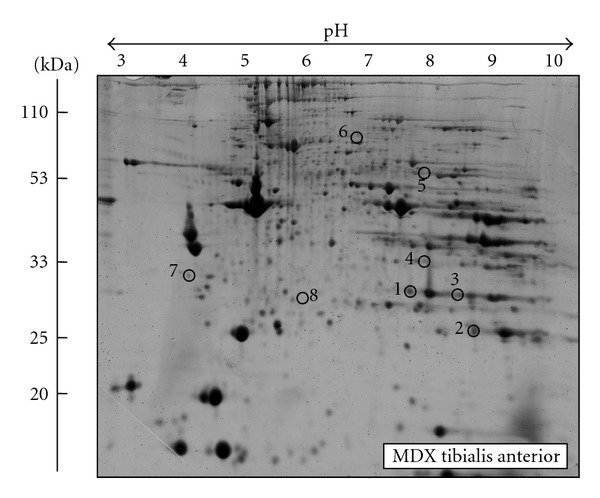
Fluorescence gel electrophoretic analysis of aged mdx tibialis anterior muscle. Shown is a representative RuBPs-labelled master gel of crude tissue extracts from mdx tibialis anterior muscle. Protein spots with an age-related change in expression levels are marked by circles and are numbered 1 to 8. See Table 2 for the mass spectrometric identification of individual muscle-associated proteins. The pH values of the first dimension gel system and molecular mass standards of the second dimension are indicated on the top and on the left of the panels, respectively.
3.4. Immunoblot Analysis of Novel Proteomic Markers of Muscular Dystrophy
Following the mass spectrometric establishment of age-related changes in the urea-soluble mdx tibialis anterior muscle proteome, immunoblotting was used to investigate the concentration of the two most extensively changed new markers CA3 and Hsp27 in normal versus dystrophic preparations. Antibodies to the dystrophin-associated glycoprotein β-dystroglycan (β-DG), which forms the main trans-sarcolemmal linker between the extracellular matrix and the cortical actin cytoskeleton in the fibre periphery, were employed to verify the dystrophic status of mdx tissue samples during aging. Figure 4(a) illustrates the drastic reduction of β-DG in both 8 weeks and 22 months old mdx tibialis anterior muscle, which is characteristic of dystrophinopathy. Equal loading of lanes was ensured by silver staining of gel electrophoretically separated protein preparations (not shown). Immunoblotting of young versus old muscle samples with antibodies to the CA3 isoform of carbonic anhydrase (Figure 4(b)) and the molecular chaperone Hsp27 (Figure 4(c)) showed an increased abundance of the metabolic enzyme and a decreased concentration of the small heat shock protein in dystrophin-deficient muscle. Thus, both the fibre type-specific protein CA3 and the stress protein Hsp27 represent suitable candidate biomarkers of the dystrophic phenotype.
Figure 4.
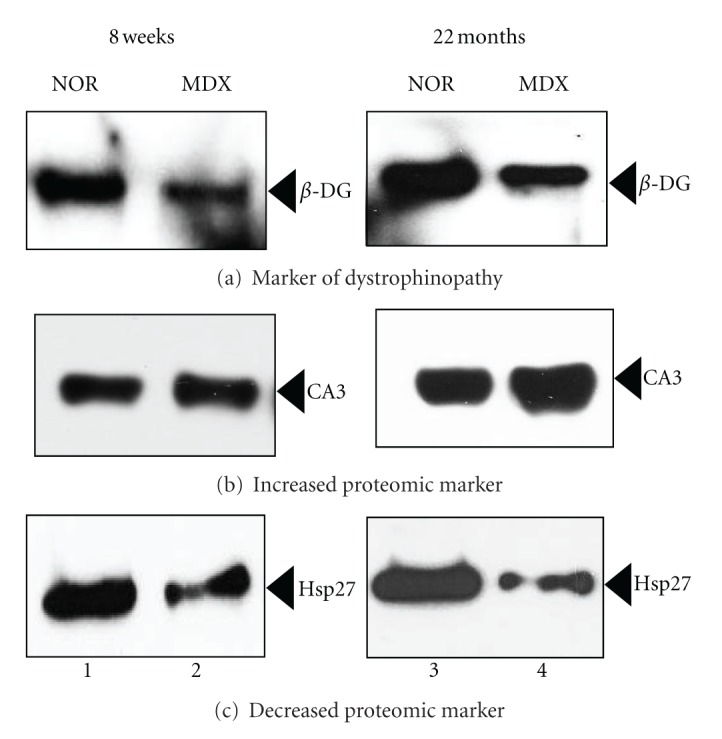
Immunoblot analysis of novel marker proteins in normal versus mdx tibialis anterior muscle during aging. Shown are representative immunoblots with expanded views of antibody-decorated protein bands. Panels (a, b, c) were labeled with antibodies to the dystrophin-associated glycoprotein β-dystroglycan (β-DG), the CA3 isoform of the fibre type-specific enzyme carbonic anhydrase, and the molecular chaperone Hsp27, respectively. Lanes 1 and 2, and lanes 3 and 4, represent 8 weeks versus 22 months old normal wild type versus dystrophic mdx tibialis anterior muscle, respectively. Immuno-decorated protein bands are indicated by arrowheads.
4. Discussion
Duchenne muscular dystrophy is one of the most crippling neuromuscular disorders of childhood [58], therefore warranting detailed large-scale studies into the establishment of comprehensive biomarker signatures of dystrophinopathy [30, 31]. In dystrophinopathy, the almost complete absence of the Dp427 isoform of the membrane cytoskeletal protein dystrophin causes a drastic reduction of a large number of surface glycoproteins that in turn triggers a loss of sarcolemmal integrity. The mdx mouse is a widely used model system for the evaluation of novel treatment options to counter-act the symptoms of X-linked muscular dystrophy [59] and basic biomedical research promises to provide the basis of evidence for the development of novel treatment regimes, such as stem cell therapy, myoblast transfer, or exon skipping therapy [60]. Previous proteomic studies have established a considerable number of novel biomarkers of secondary changes in dystrophin-deficient organisms [30]. Besides the cataloging of generally perturbed protein expression patterns, individual proteomic surveys of mdx muscles of differing subtype and age have demonstrated a drastically altered abundance of adenylate kinase isoform AK1 [36], the luminal Ca2+-binding protein calsequestrin of the terminal cisternae [16, 41], the cytosolic Ca2+-buffering element regucalcin [39], mitochondrial isocitrate dehydrogenase [38], and the muscle-specific molecular chaperone cvHsp [40, 41]. A recent aging study of the severely dystrophic mdx diaphragm has demonstrated a drastic increase in the extracellular matrix proteins collagen and dermatopontin, the molecular chaperone αB-crystallin, and the intermediate filament protein vimentin, suggesting increased accumulation of connective tissue, an enhanced cellular stress response and compensatory stabilization of the weakened membrane cytoskeleton in severely dystrophic muscle tissue [43].
In the present report, proteomic profiling showed that during the natural aging of the moderately dystrophic tibialis anterior muscle a number of key skeletal muscle proteins change in abundance. We have studied aged mdx muscle, because dystrophic mouse muscle tissue progressively deteriorates with age and thus more closely resembles the neuromuscular pathology seen in Duchenne patients [61]. The age-related pathogenesis of mdx muscle is characterized by a drastic loss of myofibres and concomitant replacement by connective tissue [62–64], progressive motor weakness [65], the presence of branched fibres that trigger mechanical weakening of the muscle periphery [66], a reduced life span and increased susceptibility to spontaneous rhabdomyosarcoma [67], a decline in regenerative potential and alterations in the crucial mTOR signaling pathway [68], and impaired functional and structural recovery after injury [54]. Hence, senescent mdx muscle represents a suitable dystrophic phenotype for determining potential global changes in the protein complement during aging. This report has summarized the findings of a comparative proteomic analysis of mildly affected mdx tibialis anterior muscle from 8 weeks versus 22 months old mice. The identification of aldolase, pyruvate kinase, carbonic anhydrase, tropomyosin, myosin, electron transferring flavoprotein and small heat shock protein Hsp27 as new indicators of progressive muscular dystrophy might be useful for the establishment of a more comprehensive biomarker signature of dystrophinopathy.
The protein with the highest age-related increase was identified as carbonic anhydrase isoform CA3 [69]. In general, carbonic anhydrases catalyze the reversible hydration of CO2 and are widely distributed throughout the body [70]. Skeletal muscles express several isoforms of this crucial metabolic enzyme in a fibre-type-specific manner. The predominant CA3 isoform is mostly present in the cytosolic fraction of type I and IIa fibers [69]. Interestingly, metabolic adaptations, altered neuromuscular activity patterns, stretch-induced hypertrophy, and disuse atrophy greatly influence the expression of muscle carbonic anhydrases [71–73]. The higher concentration of the CA3 isoform of carbonic anhydrase in aged mdx muscle, as shown here by mass spectrometry-based proteomics, could be an indication of an increased demand for efficient CO2 removal during mdx fibre aging. On the other hand, since the CA3 isoform is predominantly located in slower-twitching fibre populations, its altered density could also be due to age-related fibre-type shifting in the mdx tibialis anterior muscle. This would agree with the findings of a recent proteomic survey of middle aged versus aged vastus lateralis muscle, which revealed increased levels of CA3 in senescent human skeletal muscle [74].
The greatest reduction in a muscle-associated protein during aging of the mdx tibialis anterior was shown to be the small heat shock protein Hsp27. This indicates a potentially blunted cellular stress response in dystrophic tibialis anterior fibres and demonstrates that marked differences exist with respect to expression levels of small heat shock proteins in moderately affected hind limb muscles versus severely dystrophic diaphragm muscle in the mdx model of dystrophinopathy [40, 41]. While changes in elements of the contractile apparatus suggest downstream effects of dystrophin deficiency on myosin and tropomyosin organization, altered expression levels in electron transferring flavoprotein and glycolytic enzymes indicate perturbed mdx muscle metabolism. The beta-polypeptide chain of the electron transferring flavoprotein is involved in mitochondrial fatty acid and amino acid catabolism and mediates the shuttling of electrons between flavoprotein dehydrogenases [75]. The enzymes aldolase and pyruvate kinase catalyze the reversible break-down of fructose-1,6-biphosphate into dihydroxyacetone phosphate and glyceraldehyde-3-phosphate and the critical oxidoreduction-phosphorylation step that converts ADP and phosphoenolpyruvate to ATP and pyruvate, respectively [76]. Alterations in glycolytic enzymes and mitochondrial proteins in mdx tibialis anterior muscle indicate altered flux rates through key metabolic pathway. With respect to the glycolytic pathway, the activity of four muscle proteins is central to its regulation on the enzymatic level, i.e. the metabolic flux through hexokinase, phosphofructokinase, glycogen phosphorylase, and pyruvate kinase, whereby metabolic silencing of muscle glycolysis is probably mediated by the inactivation of pyruvate kinase [76]. This central role of pyruvate kinase in muscle metabolism makes its changed abundance in the mdx tibialis anterior muscle a crucial finding. Pyruvate kinase was previously shown to be a suitable biomarker of the general aging process in skeletal muscle tissues [77, 78]. Interestingly, the change of metabolic enzymes in aged mdx tibialis anterior muscle, such as pyruvate kinase and aldolase, agrees with the proteomic analysis of golden retriever muscular dystrophy. In the grmd dog model of dystrophinopathy, targets of the transcriptional control factor of energy metabolism PGC-1α, that is, various glycolytic and oxidative enzymes, were found to be reduced [44].
In conclusion, the comparative proteomic survey of dystrophic tibialis anterior muscle from 8 weeks versus 22 months old mdx mice has revealed altered expression levels in a number of critical proteins during skeletal muscle aging. However, the degree of concentration changes was less pronounced in the moderately dystrophic tibialis anterior muscle as compared to the recently analyzed aged mdx diaphragm [43]. These differing proteomic findings agree with the pathophysiological concept that the aged mdx diaphragm muscle is more severely affected as compared to moderately necrotic mdx hind limb muscle. In the long-term, the proteomic identification of new biomarkers of dystrophinopathy might be useful for the establishment of a comprehensive and muscle subtype-specific signature of Duchenne muscular dystrophy. This would be useful for improving diagnostic procedures, monitor disease progression, identify novel therapeutic targets and aid in the evaluation of novel treatments, such as exon-skipping therapy or stem cell therapy.
Acknowledgments
Research was supported by project grants from Muscular Dystrophy Ireland and Duchenne Ireland and a Hume scholarship from NUI Maynooth, as well as equipment grants from the Irish Health Research Board and the Higher Education Authority.
References
- 1.Campbell KP. Three muscular dystrophies: loss of cytoskeleton-extracellular matrix linkage. Cell. 1995;80(5):675–679. doi: 10.1016/0092-8674(95)90344-5. [DOI] [PubMed] [Google Scholar]
- 2.Ohlendieck K. Towards an understanding of the dystrophin-glycoprotein complex: linkage between the extracellular matrix and the membrane cytoskeleton in muscle fibers. European Journal of Cell Biology. 1996;69(1):1–10. [PubMed] [Google Scholar]
- 3.Michele DE, Campbell KP. Dystrophin-glycoprotein complex: post-translational processing and dystroglycan function. Journal of Biological Chemistry. 2003;278(18):15457–15460. doi: 10.1074/jbc.R200031200. [DOI] [PubMed] [Google Scholar]
- 4.Ervasti JM, Sonnemann KJ. Biology of the striated muscle dystrophin-glycoprotein complex. International Review of Cytology. 2008;265:191–225. doi: 10.1016/S0074-7696(07)65005-0. [DOI] [PubMed] [Google Scholar]
- 5.Gumerson JD, Michele DE. The dystrophin-glycoprotein complex in the prevention of muscle damage. Journal of Biomedicine and Biotechnology. 2011;2011 doi: 10.1155/2011/210797.210797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffman EP, Brown RH, Kunkel LM. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51(6):919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- 7.Culligan KG, Mackey AJ, Finn DM, Maguire PB, Ohlendieck K. Role of dystrophin isoforms and associated proteins in muscular dystrophy. International Journal of Molecular Medicine. 1998;2(6):639–648. doi: 10.3892/ijmm.2.6.639. [DOI] [PubMed] [Google Scholar]
- 8.Dalkilic I, Kunkel LM. Muscular dystrophies: genes to pathogenesis. Current Opinion in Genetics and Development. 2003;13(3):231–238. doi: 10.1016/s0959-437x(03)00048-0. [DOI] [PubMed] [Google Scholar]
- 9.Ohlendieck K, Matsumura K, Ionasescu VV, et al. Duchenne muscular dystrophy: deficiency of dystrophin-associated proteins in the sarcolemma. Neurology. 1993;43(4):795–800. doi: 10.1212/wnl.43.4.795. [DOI] [PubMed] [Google Scholar]
- 10.Lynch GS, Rafael JA, Chamberlain JS, Faulkner JA. Contraction-induced injury to single permeabilized muscle fibers from mdx, transgenic mdx, and control mice. American Journal of Physiology. 2000;279(4):C1290–C1294. doi: 10.1152/ajpcell.2000.279.4.C1290. [DOI] [PubMed] [Google Scholar]
- 11.Ramaswamy KS, Palmer ML, Van Der Meulen JH, et al. Lateral transmission of force is impaired in skeletal muscles of dystrophic mice and very old rats. Journal of Physiology. 2011;589(5):1195–1208. doi: 10.1113/jphysiol.2010.201921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alderton JM, Steinhardt RA. Calcium influx through calcium leak channels is responsible for the elevated levels of calcium-dependent proteolysis in dystrophic myotubes. Journal of Biological Chemistry. 2000;275(13):9452–9460. doi: 10.1074/jbc.275.13.9452. [DOI] [PubMed] [Google Scholar]
- 13.Mallouk N, Jacquemond V, Allard B. Elevated subsarcolemmal Ca2+ in mdx mouse skeletal muscle fibers detected with Ca2+-activated K+ channels. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(9):4950–4955. doi: 10.1073/pnas.97.9.4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Culligan K, Banville N, Dowling P, Ohlendieck K. Drastic reduction of calsequestrin-like proteins and impaired calcium binding in dystrophic mdx muscle. Journal of Applied Physiology. 2002;92(2):435–445. doi: 10.1152/japplphysiol.00903.2001. [DOI] [PubMed] [Google Scholar]
- 15.Dowling P, Doran P, Ohlendieck K. Drastic reduction of sarcalumenin in Dp427 (dystrophin of 427 kDa)-deficient fibres indicates that abnormal calcium handling plays a key role in muscular dystrophy. Biochemical Journal. 2004;379(2):479–488. doi: 10.1042/BJ20031311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doran P, Dowling P, Lohan J, McDonnell K, Poetsch S, Ohlendieck K. Subproteomics analysis of Ca2+-binding proteins demonstrates decreased calsequestrin expression in dystrophic mouse skeletal muscle. European Journal of Biochemistry. 2004;271(19):3943–3952. doi: 10.1111/j.1432-1033.2004.04332.x. [DOI] [PubMed] [Google Scholar]
- 17.Gehrig SM, van der Poel C, Sayer TA, et al. Hsp72 preserves muscle function and slows progression of severe muscular dystrophy. Nature. 2012;484(7394):394–398. doi: 10.1038/nature10980. [DOI] [PubMed] [Google Scholar]
- 18.Lewis C, Doran P, Ohlendieck K. Proteomic analysis of dystrophic muscle. Methods in Molecular Biology. 2012;798(1):357–369. doi: 10.1007/978-1-61779-343-1_20. [DOI] [PubMed] [Google Scholar]
- 19.de Hoog CL, Mann M. Proteomics. Annual Review of Genomics and Human Genetics. 2004;5:267–293. doi: 10.1146/annurev.genom.4.070802.110305. [DOI] [PubMed] [Google Scholar]
- 20.Cravatt BF, Simon GM, Yates JR. The biological impact of mass-spectrometry-based proteomics. Nature. 2007;450(7172):991–1000. doi: 10.1038/nature06525. [DOI] [PubMed] [Google Scholar]
- 21.Walther TC, Mann M. Mass spectrometry-based proteomics in cell biology. Journal of Cell Biology. 2010;190(4):491–500. doi: 10.1083/jcb.201004052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Isfort RJ. Proteomic analysis of striated muscle. Journal of Chromatography B. 2002;771(1-2):155–165. doi: 10.1016/s1570-0232(02)00056-9. [DOI] [PubMed] [Google Scholar]
- 23.Ohlendieck K. Skeletal muscle proteomics: current approaches, technical challenges and emerging techniques. Skeletal Muscle. 2011;1(1):p. 6. doi: 10.1186/2044-5040-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gelfi C, Vasso M, Cerretelli P. Diversity of human skeletal muscle in health and disease: contribution of proteomics. Journal of Proteomics. 2011;74(6):774–795. doi: 10.1016/j.jprot.2011.02.028. [DOI] [PubMed] [Google Scholar]
- 25.Ohlendieck K. Proteomics of skeletal muscle differentiation, neuromuscular disorders and fiber aging. Expert Review of Proteomics. 2010;7(2):283–296. doi: 10.1586/epr.10.2. [DOI] [PubMed] [Google Scholar]
- 26.Ohlendieck K. Proteomic profiling of fast-to-slow muscle transitions during aging. Frontiers in Physiology. 2011;2(1):p. 105. doi: 10.3389/fphys.2011.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422(6928):198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 28.Cañas Montalvo B, López-Ferrer D, Ramos-Fernández A, Camafeita E, Calvo E. Mass spectrometry technologies for proteomics. Briefings in Functional Genomics and Proteomics. 2006;4(4):295–320. doi: 10.1093/bfgp/eli002. [DOI] [PubMed] [Google Scholar]
- 29.Yates JR, Ruse CI, Nakorchevsky A. Proteomics by mass spectrometry: approaches, advances, and applications. Annual Review of Biomedical Engineering. 2009;11:49–79. doi: 10.1146/annurev-bioeng-061008-124934. [DOI] [PubMed] [Google Scholar]
- 30.Lewis C, Carberry S, Ohlendieck K. Proteomic profiling of x-linked muscular dystrophy. Journal of Muscle Research and Cell Motility. 2009;30(7-8):267–279. doi: 10.1007/s10974-009-9197-6. [DOI] [PubMed] [Google Scholar]
- 31.Griffin JL, Rosiers CD. Applications of metabolomics and proteomics to the mdx mouse model of Duchenne muscular dystrophy: lessons from downstream of the transcriptome. Genome Medicine. 2009;1(3, article 32) doi: 10.1186/gm32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alagaratnam S, Mertens BJA, Dalebout JC, et al. Serum protein profiling in mice: identification of factor XIIIa as a potential biomarker for muscular dystrophy. Proteomics. 2008;8(8):1552–1563. doi: 10.1002/pmic.200700857. [DOI] [PubMed] [Google Scholar]
- 33.Colussi C, Banfi C, Brioschi M, et al. Proteomic profile of differentially expressed plasma proteins from dystrophic mice and following suberoylanilide hydroxamic acid treatment. Proteomics. 2010;4(1):71–83. doi: 10.1002/prca.200900116. [DOI] [PubMed] [Google Scholar]
- 34.Gulston MK, Rubtsov DV, Atherton HJ, et al. A combined metabolomic and proteomic investigation of the effects of a failure to express dystrophin in the mouse heart. Journal of Proteome Research. 2008;7(5):2069–2077. doi: 10.1021/pr800070p. [DOI] [PubMed] [Google Scholar]
- 35.Lewis C, Jockusch H, Ohlendieck K. Proteomic profiling of the dystrophin-deficient MDX heart reveals drastically altered levels of key metabolic and contractile proteins. Journal of Biomedicine and Biotechnology. 2010;2010 doi: 10.1155/2010/648501.648501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ge Y, Molloy MP, Chamberlain JS, Andrews PC. Proteomic analysis of mdx skeletal muscle: great reduction of adenylate kinase 1 expression and enzymatic activity. Proteomics. 2003;3(10):1895–1903. doi: 10.1002/pmic.200300561. [DOI] [PubMed] [Google Scholar]
- 37.Ge Y, Molloy MP, Chamberlain JS, Andrews PC. Differential expression of the skeletal muscle proteome in mdx mice at different ages. Electrophoresis. 2004;25(15):2576–2585. doi: 10.1002/elps.200406013. [DOI] [PubMed] [Google Scholar]
- 38.Gardan-Salmon D, Dixon JM, Lonergan SM, Selsby JT. Proteomic assessment of the acute phase of dystrophin deficiency in mdx mice. European Journal of Applied Physiology. 2011;111(11):2763–2773. doi: 10.1007/s00421-011-1906-3. [DOI] [PubMed] [Google Scholar]
- 39.Doran P, Dowling P, Donoghue P, Buffini M, Ohlendieck K. Reduced expression of regucalcin in young and aged mdx diaphragm indicates abnormal cytosolic calcium handling in dystrophin-deficient muscle. Biochimica et Biophysica Acta. 2006;1764(4):773–785. doi: 10.1016/j.bbapap.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 40.Doran P, Martin G, Dowling P, Jockusch H, Ohlendieck K. Proteome analysis of the dystrophin-deficient MDX diaphragm reveals a drastic increase in the heat shock protein cvHSP. Proteomics. 2006;6(16):4610–4621. doi: 10.1002/pmic.200600082. [DOI] [PubMed] [Google Scholar]
- 41.Doran P, Wilton SD, Fletcher S, Ohlendieck K. Proteomic profiling of antisense-induced exon skipping reveals reversal of pathobiochemical abnormalities in dystrophic mdx diaphragm. Proteomics. 2009;9(3):671–685. doi: 10.1002/pmic.200800441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lewis C, Ohlendieck K. Proteomic profiling of naturally protected extraocular muscles from the dystrophin-deficient mdx mouse. Biochemical and Biophysical Research Communications. 2010;396(4):1024–1029. doi: 10.1016/j.bbrc.2010.05.052. [DOI] [PubMed] [Google Scholar]
- 43.Carberry S, Zweyer M, Swandulla D, Ohlendieck K. Proteomic profiling of diaphragm muscle during aging of the mdx model of Duchenne muscular dystrophy. International Journal of Molecular Medicine. 2012;30(2):229–234. doi: 10.3892/ijmm.2012.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guevel L, Lavoie JR, Perez-Iratxeta C, et al. Quantitative proteomic analysis of dystrophic dog muscle. Journal of Proteome Research. 2011;10(5):2465–2478. doi: 10.1021/pr2001385. [DOI] [PubMed] [Google Scholar]
- 45.Mesin L, Merlo E, Merletti R, Orizio C. Investigation of motor unit recruitment during stimulated contractions of Tibialis anterior muscle. Journal of Electromyography and Kinesiology. 2010;20(4):580–589. doi: 10.1016/j.jelekin.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 46.Jones DA, Turner DL, McIntyre DB, Newham DJ. Energy turnover in relation to slowing of contractile properties during fatiguing contractions of the human anterior tibialis muscle. Journal of Physiology. 2009;587(17):4329–4338. doi: 10.1113/jphysiol.2009.175265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gilbert R, Dudley RWR, Liu AB, Petrof BJ, Nalbantoglu J, Karpati G. Prolonged dystrophin expression and functional correction of mdx mouse muscle following gene transfer with a helper-dependent (gutted) adenovirus-encoding murine dystrophin. Human Molecular Genetics. 2003;12(11):1287–1299. doi: 10.1093/hmg/ddg141. [DOI] [PubMed] [Google Scholar]
- 48.Westermeier R, Marouga R. Protein detection methods in proteomics research. Bioscience Reports. 2005;25(1-2):19–32. doi: 10.1007/s10540-005-2845-1. [DOI] [PubMed] [Google Scholar]
- 49.Aude-Garcia C, Collin-Faure V, Luche S, Rabilloud T. Improvements and simplifications in in-gel fluorescent detection of proteins using ruthenium II tris-(bathophenanthroline disulfonate): the poor man’s fluorescent detection method. Proteomics. 2011;11(2):324–328. doi: 10.1002/pmic.201000370. [DOI] [PubMed] [Google Scholar]
- 50.Durbeej M, Campbell KP. Muscular dystrophies involving the dystrophin-glycoprotein complex: an overview of current mouse models. Current Opinion in Genetics and Development. 2002;12(3):349–361. doi: 10.1016/s0959-437x(02)00309-x. [DOI] [PubMed] [Google Scholar]
- 51.Bulfield G, Siller WG, Wight PAL, Moore KJ. X chromosome-linked muscular dystrophy (mdx) in the mouse. Proceedings of the National Academy of Sciences of the United States of America. 1984;81(4):1189–1192. doi: 10.1073/pnas.81.4.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sicinski P, Geng Y, Ryder-Cook AS, Barnard EA, Darlison MG, Barnard PJ. The molecular basis of muscular dystrophy in the mdx mouse: a point mutation. Science. 1989;244(4912):1578–1580. doi: 10.1126/science.2662404. [DOI] [PubMed] [Google Scholar]
- 53.Ohlendieck K, Campbell KP. Dystrophin-associated proteins are greatly reduced in skeletal muscle from mdx mice. Journal of Cell Biology. 1991;115(6):1685–1694. doi: 10.1083/jcb.115.6.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Irintchev A, Zweyer M, Wernig A. Impaired functional and structural recovery after muscle injury in dystrophic mdx mice. Neuromuscular Disorders. 1997;7(2):117–125. doi: 10.1016/s0960-8966(96)00422-1. [DOI] [PubMed] [Google Scholar]
- 55.Staunton L, Jockusch H, Wiegand C, Albrecht T, Ohlendieck K. Identification of secondary effects of hyperexcitability by proteomic profiling of myotonic mouse muscle. Molecular BioSystems. 2011;7(8):2480–2489. doi: 10.1039/c1mb05043e. [DOI] [PubMed] [Google Scholar]
- 56.Rabilloud T, Strub JM, Luche S, Van Dorsselaer A, Lunardi J. A comparison between Sypro Ruby and ruthenium ii tris (bathophenanthroline disulfonate) as fluorescent stains for protein detection in gels. Proteomics. 2001;1(5):699–704. doi: 10.1002/1615-9861(200104)1:5<699::AID-PROT699>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 57.Gannon J, Staunton L, O’Connell K, Doran P, Ohlendieck K. Phosphoproteomic analysis of aged skeletal muscle. International Journal of Molecular Medicine. 2008;22(1):33–42. [PubMed] [Google Scholar]
- 58.Emery AEH. The muscular dystrophies. The Lancet. 2002;359(9307):687–695. doi: 10.1016/S0140-6736(02)07815-7. [DOI] [PubMed] [Google Scholar]
- 59.Spurney CF, Gordish-Dressman H, Guerron AD, et al. Preclinical drug trials in the mdx mouse: assessment of reliable and sensitive outcome measures. Muscle and Nerve. 2009;39(5):591–602. doi: 10.1002/mus.21211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Partridge TA. Impending therapies for Duchenne muscular dystrophy. Current Opinion in Neurology. 2011;24(5):415–422. doi: 10.1097/WCO.0b013e32834aa3f1. [DOI] [PubMed] [Google Scholar]
- 61.Lefaucheur JP, Pastoret C, Sebille A. Phenotype of dystrophinopathy in old mdx mice. Anatomical Record. 1995;242(1):70–76. doi: 10.1002/ar.1092420109. [DOI] [PubMed] [Google Scholar]
- 62.Wieneinger MA, Ted Abresch R, Walsh SA, Carter GT. Effects of aging and voluntary exercise on the function of dystrophic muscle from mdx mice. American Journal of Physical Medicine and Rehabilitation. 1998;77(1):20–27. doi: 10.1097/00002060-199801000-00004. [DOI] [PubMed] [Google Scholar]
- 63.Pastoret C, Sebille A. Age-related differences in regeneration of dystrophic (mdx) and normal muscle in the mouse. Muscle and Nerve. 1995;18(10):1147–1154. doi: 10.1002/mus.880181011. [DOI] [PubMed] [Google Scholar]
- 64.Pastoret C, Sebille A. mdx mice show progressive weakness and muscle deterioration with age. Journal of the Neurological Sciences. 1995;129(2):97–105. doi: 10.1016/0022-510x(94)00276-t. [DOI] [PubMed] [Google Scholar]
- 65.Lynch GS, Hinkle RT, Chamberlain JS, Brooks SV, Faulkner JA. Force and power output of fast and slow skeletal muscles from mdx mice 6–28 months old. Journal of Physiology. 2001;535(2):591–600. doi: 10.1111/j.1469-7793.2001.00591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Head SI. Branched fibres in old dystrophic mdx muscle are associated with mechanical weakening of the sarcolemma, abnormal Ca2+ transients and a breakdown of Ca2+ homeostasis during fatigue. Experimental Physiology. 2010;95(5):641–656. doi: 10.1113/expphysiol.2009.052019. [DOI] [PubMed] [Google Scholar]
- 67.Chamberlain JS, Metzger J, Reyes M, Townsend D, Faulkner JA. Dystrophin-deficient mdx mice display a reduced life span and are susceptible to spontaneous rhabdomyosarcoma. Federation of American Societies for Experimental Biology Journal. 2007;21(9):2195–2204. doi: 10.1096/fj.06-7353com. [DOI] [PubMed] [Google Scholar]
- 68.Mouisel E, Vignaud A, Hourdé C, Butler-Browne G, Ferry A. Muscle weakness and atrophy are associated with decreased regenerative capacity and changes in mtor signaling in skeletal muscles of venerable (18–24-month-old) dystrophic mdx mice. Muscle and Nerve. 2010;41(6):809–818. doi: 10.1002/mus.21624. [DOI] [PubMed] [Google Scholar]
- 69.Fremont P, Charest PM, Cote C, Rogers PA. Carbonic anhydrase III in skeletal muscle fibers: an immunocytochemical and biochemical study. Journal of Histochemistry and Cytochemistry. 1988;36(7):775–782. doi: 10.1177/36.7.3133407. [DOI] [PubMed] [Google Scholar]
- 70.Geers C, Gros G. Carbon dioxide transport and carbonic anhydrase in blood and muscle. Physiological Reviews. 2000;80(2):681–715. doi: 10.1152/physrev.2000.80.2.681. [DOI] [PubMed] [Google Scholar]
- 71.Brownson C, Isenberg H, Brown W, Salmons S, Edwards Y. Changes in skeletal muscle gene transcription induced by chronic stimulation. Muscle and Nerve. 1988;11(11):1183–1189. doi: 10.1002/mus.880111113. [DOI] [PubMed] [Google Scholar]
- 72.Brownson C, Loughna PT. Alterations in the mRNA levels of two metabolic enzymes in rat skeletal muscle during stretch-induced hypertrophy and disuse atrophy. Pflugers Archiv European Journal of Physiology. 1996;431(6):990–992. doi: 10.1007/s004240050097. [DOI] [PubMed] [Google Scholar]
- 73.Côté CH, Ambrosio F, Perreault G. Metabolic and contractile influence of carbonic anhydrase III in skeletal muscle is age dependent. American Journal of Physiology. 1999;276(2):R559–R565. doi: 10.1152/ajpregu.1999.276.2.R559. [DOI] [PubMed] [Google Scholar]
- 74.Staunton L, Zweyer M, Swandulla D, Ohlendieck K. Mass spectrometry-based proteomic analysis of middle-aged versus aged vastus lateralis reveals increased levels of carbonic anhydrase isoform 3 in senescent human skeletal muscle. doi: 10.3892/ijmm.2012.1056. International Journal of Molecular Medicine. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Toogood HS, Leys D, Scrutton NS. Dynamics driving function—new insights from electron transferring flavoproteins and partner complexes. Federation of European Biochemical Societies Journal. 2007;274(21):5481–5504. doi: 10.1111/j.1742-4658.2007.06107.x. [DOI] [PubMed] [Google Scholar]
- 76.Ohlendieck K. Proteomics of skeletal muscle glycolysis. Biochimica et Biophysica Acta. 2010;1804(11):2089–2101. doi: 10.1016/j.bbapap.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 77.Doran P, O’Connell K, Gannon J, Kavanagh M, Ohlendieck K. Opposite pathobiochemical fate of pyruvate kinase and adenylate kinase in aged rat skeletal muscle as revealed by proteomic DIGE analysis. Proteomics. 2008;8(2):364–377. doi: 10.1002/pmic.200700475. [DOI] [PubMed] [Google Scholar]
- 78.Doran P, Donoghue P, O’Connell K, Gannon J, Ohlendieck K. Proteomics of skeletal muscle aging. Proteomics. 2009;9(4):989–1003. doi: 10.1002/pmic.200800365. [DOI] [PubMed] [Google Scholar]


