Abstract
Mas-related G-protein coupled receptor member D (MRGPRD) is a G protein-coupled receptor (GPCR) which belongs to the Mas-related GPCRs expressed in the dorsal root ganglia (DRG). In this study, we investigated two novel ligands in addition to beta-alanine: (1) beta-aminoisobutyric acid, a physiologically active substance, with which possible relation to tumors has been seen together with beta-alanine; (2) diethylstilbestrol, a synthetic estrogen hormone. In addition to the novel ligands, we found that transfection of MRGPRD leads fibroblast cells to form spheroids, which would be related to oncogenicity. To understand the MRGPRD novel character, oncogenicity, a large chemical library was screened in order to obtain MRGPRD antagonists to utilize in exploring the character. The antagonist in turn inhibited the spheroid proliferation that is dependent on MRGPRD signaling as well as MRGPRD signals activated by beta-alanine. The antagonist, a small-molecule compound we found in this study, is a potential anticancer agent.
1. Introduction
G protein-coupled receptors (GPCRs) compose the largest family of membrane-bound receptors and many members still have unrevealed functions. Efforts have been made to find ligands for GPCRs, and a broad range of ligands, including small organic compounds [1, 2], eicosanoids [3], peptides [4], and proteins [5], have been identified. Members of the Mas-related GPCR (Mrgpr) family are known to be expressed mainly in the subpopulations of sensory neurons [6]. This has resulted in their having another name sensory neuron specific receptors (SNSRs) [7]. Mas-related G-protein coupled receptor member D (MRGPRD), formerly called Mas-related gene D (MRGD) and also referred to as hGPCR45 [8] or TGR7 [9], belongs to the Mrgpr family and is known to be mainly expressed in the dorsal root ganglia (DRG) [6, 9–12]. When phylogenic trees of the hMRGPR family and the mouse MRGPR family are compared, variation of the members found in those phylogenic trees are not parallel [6]. Members of the human MRGPR family may cover other functions from the ones revealed by studies with the mouse MRGPR family. We hypothesized MRGPRD could possess functions besides the ones in the DRG and found a spheroid forming activity. At this time, for MRGPRD, beta-alanine (βAla) is the only known ligand that would be physiological [9]. When we screened a physiologically active substance library and a known small compound library, we found two novel ligands for MRGPRD, that is, beta-aminoisobutyric acid (βAIBA) and diethylstilbestrol (DES). As βAla promoted spheroid growth of MRGPRD stably expressing cells, it would be useful to obtain antagonists in order to analyze the tumorigenic character of MRGPRD. We obtained MU-6840 as an antagonist of MRGPRD by high-throughput-screening at a large small chemical library. Since it specifically inhibits the growth of spheroids formed with cells stably expressing MRGPRD, MU-6840 is a potential anticancer agent. In this paper, we will discuss the novel ligand, the antagonist, and the novel character of MRGPRD.
2. Materials and Methods
2.1. Materials
Dulbecco's Modified Eagle Medium (DMEM), Minimum Essential Medium Alpha without nucleic acids (α-MEM), RPMI1640, Opti-MEM I reduced serum media (Opti-MEM), Grace's Insect Medium, Penicillin-Streptomycin, Hanks' Balanced Salt Solution (HBSS), and LipofectAMINE 2000 were purchased from Invitrogen Corporation, USA. DMEM inositol-free was purchased from MP Biomedicals, USA. pSV2-dhfr was purchased from ATCC, USA. Fetal bovine serum (FBS) was purchased from Sanko Junyaku, Japan. PathDetect NFAT cis-Reporting System and PathDetect Elk1 trans-Reporting System were purchased from Agilent Technologies, USA. pRL-TK was purchased from Promega, USA. FBS dialyzed was purchased from JHR Biosciences, USA. Guanosine diphosphate (GDP), guanosine 5′-O-(3-thiotriphosphate) (GTPγS), saponin, LiCl, formic acid, and bovine serum albumin (BSA) were purchased from Sigma-Aldrich Inc., USA. Complete EDTA-free was purchased from Roche Diagnostics. LEADseeker beads, RNA binding YSi, myo-[2-3H] inositol and GTPγS [35S] were purchased from GE Healthcare, UK.
2.2. Cell Culture
HEK293 cells (DS Pharma Biomedical Corporation, Japan) were maintained in DMEM with 10% FBS and Penicillin-Streptomycin at 37°C under 5% CO2. CHO/dhfr− cells (DS Pharma Biomedical Corporation, Japan) expressing hMRGPRD (described in this paper) were maintained in α-MEM with 10% FBS dialyzed and Penicillin-Streptomycin at 37°C under 5% CO2. NIH/3T3 cells (ATCC, USA) were maintained in RPMI1640 with 10% FBS and Penicillin-Streptomycin at 37°C under 5% CO2.
2.3. Endogenous Signal Assay
Human adrenergic alpha 1A receptor (ADRA1A, AY389505.1) gene and human MRGPRD (hMRGPRD, NM_198923.2) gene were cloned into pcDNA3.1 (Invitrogen, USA) (hereafter referred to as pcDNA-ADRA1A and pcDNA-hMRGPRD, resp.). Using LipofectAMINE 2000, 20000 HEK293 cells in each well of a 96-well multiplate were transfected with a set of 3 plasmids, consisting of one from each category as follows: (1) pNFAT-Luc plasmid (PathDetect NFAT cis-Reporting System), or pFR-Luc plasmid with pFA2-Elk1 plasmid (PathDetect Elk1 trans-Reporting System); (2) pRL-TK; (3) pcDNA3.1, pcDNA-ADRA1A, or pcDNA-hMRGPRD. Twenty-four hours after the transfection, the cells were lysed and transferred to OptiPlate-96. Samples were prepared in accordance with the manufacturer's instructions for the Dual-Luciferase Reporter Assay System (Promega) and measured with the ARVO SX 1420 Multilabel Counter (Wallac). The signal intensity of each sample was calculated by dividing the value of firefly luciferase luminescent by the value of Renilla luciferase luminescent within the same sample. The cell culture media used in this assay was DMEM with 10% FBS, which may contain βAla less than 0.5 μM, at least 40 times lower than the EC50 of βAla to MRGPRD.
2.4. Guanosine 5′-O-(3-Thiotriphosphate) Binding Assay
pFast-Bac-hMRGPRD-Giα, in which bovine Giα (NM_174324) was fused to hMRGPRD in frame and cloned into pFast-Bac (Invitrogen Corporation), was served to obtain baculovirus possessing hMRGPRD-Giα, using the Bac-to-Bac Baculovirus Expression System with pFast-Bac1 in accordance with the manufacturer's instructions. Sf9 cells in suspension culture were infected with the baculovirus possessing hMRGPRD-Giα at MOI = 2 to 5 and maintained for 72 hours at 27°C. Cells were harvested and fractured using the general method for the N2 cavitation system. Briefly, cells were suspended in a membrane preparation buffer (20 mM Hepes-KOH, 1 mM EDTA, 2 mM MgCl2, Complete EDTA-free, pH7.4) before being put in the N2 cavitation apparatus (Fike Metal Products), and then pressurized at 600 psi for 30 minutes on ice. Burst cells were collected and separated from the nuclear fraction by centrifugal separation (1,800 xvg for 10 minutes). The supernatant was centrifuged at 100,000 x g for 30 minutes to obtain a membrane fraction. The pellet was then resuspended in a membrane stock buffer (20 mM Tris-HCl, 10% glycerol, Complete EDTA-free, pH7.4) and kept as a hMRGPRD-Giα expressing membrane at −80°C. One microgram of the hMRGPRD-Giα expressing membrane, 0.44 nM GTPγS [35S], 5 μM GDP, the compound of interest, and 3.65 mg/mL LEADseeker beads were prepared with an assay buffer (20 mM Hepes, pH7.3, 100 mM NaCl, 10 mM MgCl2, 1 mM EDTA, 100 μg/mL saponin) in standard white 384 well assay microplates (Corning Inc.) at final volume of 60 μL. After incubating the microplates at room temperature for 1 hour, 10 μL of 100 μM GTPγS was added to each well. The microplates were measured using LEADseeker (GE Healthcare). The assay buffer used in this assay did not contain any βAla.
2.5. Calcium Flux Assay
CHO/dhfr− cells were transfected with pSV2-dhfr and pSR-hMRGPRD, where hMRGPRD was cloned into an original vector containing SRα promoter, using LipofectAMINE2000 to establish hMRGPRD/CHO/dhfr− cells, stable CHO/dhfr− cell lines which overexpress hMRGPRD. Three thousand six hundred hMRGPRD/CHO/dhfr− cells were plated onto each well of black 384 well cell culture microplates (Corning Inc.) and cultured for 2 days. In accordance with the manufacturer's instructions for the FLIPR Calcium 3 Assay Kit (Molecular Devices), the loading buffer, in which Calcium 3 Assay Solution was diluted with a HH buffer (HBSS, 25 mM Hepes, pH7.4, 2.5 mM probenecid), was added to each well after media disposal and kept at 37°C for 1 hour under 5% CO2. For the agonist screening, the HH buffer containing a test compound was added to each well and then the fluorescence transition caused by 488 nm excitation light was detected using FLIPR (Molecular Devices). For the antagonist screening, the HH buffer containing the test compound was added and incubated for 10 minutes and then the fluorescence transition was detected as the HH buffer containing βAla was added to each well. The assay buffer used in this assay did not contain any βAla.
2.6. IP Assay
Ten thousand hMRGPRD/CHO/dhfr− cells were plated onto each well of 96-well cell culture microplates and cultured for 6 hours, which were then labeled using labeling media (DMEM inositol-free, 10% FBS dialyzed, 1 nCi/mL myo-[2-3H]inositol) and cultured overnight. Labeling media was replaced with an IP assay buffer (200 mM HBSS, 25 mM Hepes, pH7.0, 0.1% BSA, 20 mM LiCl) that contained test compounds and was incubated for 1 hour for the agonists screening and incubated for 15 minutes for the antagonists screening. For the antagonist screening, in addition to the test compounds, βAla was added and incubated for 1 hour. After removing the assay buffer, 20 mM formic acid was added and incubated for 3 hours at 4°C. Formic acid lysate was added to RNA binding YSi and shaken vigorously in OptiPlate-96 (Perkin Elmer). The microplates were incubated at room temperature overnight and examined for radioactivity using TopCount (Perkin Elmer). The assay buffer used in this assay did not contain any βAla.
2.7. Spheroid Assay
The Retro-X System (Clontech Laboratories, Mountain View, CA) was used to generate NIH/3T3-hMRGPRD cells, which are NIH/3T3 cells stably expressing hMRGPRD and NIH/3T3-RasV12 cells, which are NIH/3T3 cells stably expressing RasV12. Briefly, cDNA encoding hMRGPRD or RasV12 was cloned into a pLNCX vector. The viral solution obtained from 293-10A1 cells (established in our laboratory), which were transformed with either pLNCX-hMRGPRD or pLNCX-RasV12, was used to establish those overexpressing cells. Five thousand NIH/3T3-hMRGPRD cells or NIH/3T3-RasV12 cells were plated onto each well of a CellTightSpheroid (Sumitomo Bakelite Co., Ltd.) and cultured overnight in RPMI1640 medium supplemented with 10% FBS, which may contain βAla less than 0.5 μM, at least 40 times lower than the EC50 of βAla to MRGPRD. Test compounds were added to each well and incubated for 72 hours. No agonists were used in this assay. In accordance with the manufacturer's instructions for the CellTiter-Glo Luminescent Cell Viability Assay (Promega), an assay solution was added to each well, and the cell lysate solution was then transferred to OptiPlate-96 to be measured with the ARVO SX 1420 Multilabel Counter (Wallac).
3. Results
3.1. Signaling Pathways and Constitutive Activity of MRGPRD
MRGPRD is a GPCR that induces intracellular calcium ion influx and inhibits adenylyl cyclase activity by G protein signaling and cAMP production in response to βAla [6, 9, 13, 14]. To confirm these signaling pathways, we performed reporter systems using NFAT as a downstream signal of the Gqα subunit and Elk1 as a downstream signal of the Giα subunit. Together with either of those reporters, hMRGPRD or ADRA1A, which is known to couple to both the Gqα and Giα subunits and used as a positive control, were transfected to the cells to measure their signals. The plasmid which constitutively expresses Renilla luciferase by thymidine kinase promoter was also transfected simultaneously as an internal control of the assay to normalize transfection efficacy and cell growth of each sample. The endogenous signaling activities of the GPCRs were observed as relative units which were the quotients of either NFAT or Elk1 signal divided by constitutive signal. As shown in Figures 1(a) and 1(b), both ADRA1A and hMRGPRD showed signals depending on the Gqα subunit as well as the Giα subunit. No signal was seen for either pathways when no GPCR was added (Figures 1(a) and 1(b)). We found very high signals of hMRGPRD for both NFAT and Elk1 compared to those of ADRA1A: 19 times for NFAT and 28 times for Elk1, respectively (Figures 1(a) and 1(b)). The difference can be explained by assuming the “two-state model,” where GPCRs exist in equilibrium between an inactive (R) state and an active (R*) state [15]. As we used the same molar concentration of plasmid for hMRGPRD and ADRA1A and no ligand was added to this assay, these results indicate that the equilibrium of hMRGPRD is much more shifted to the R* state than that of ADRA1A, in other words, hMRGPRD exhibits strong constitutive activity. These results indicate that expressing hMRGPRD in the cells can reinforce downstream signals without any ligand.
Figure 1.
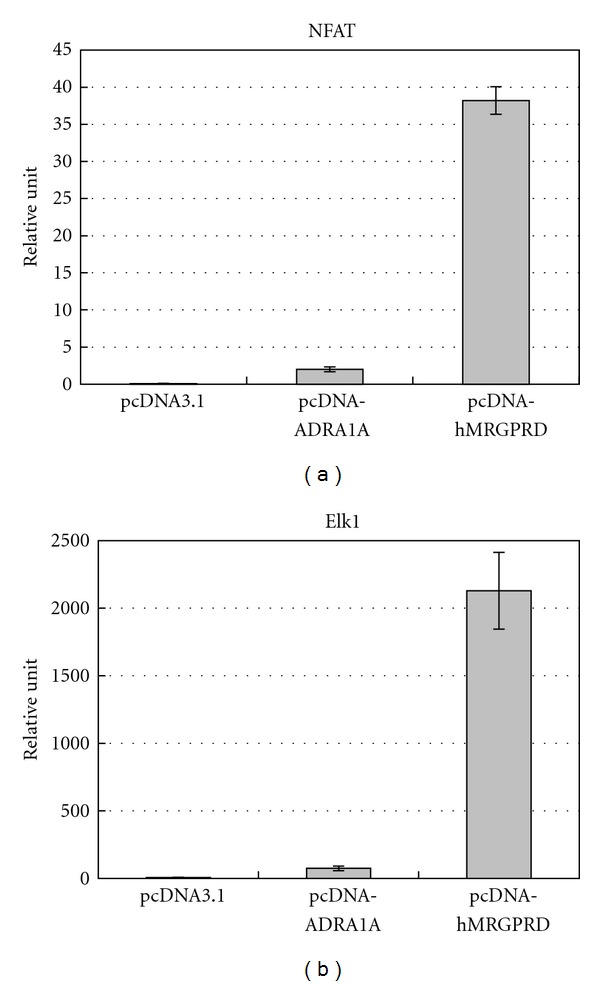
Signaling pathways of MRGPRD. Representative graphs of relative luciferase intensity of (a) NFAT and (b) Elk1. The results are the average of three independent samples.
3.2. Screening a Physiologically Active Substance Library
Currently, βAla and gamma-aminobutyric acid (GABA) are commonly regarded as physiological ligands of MRGD and both molecules are deeply related to neurotransmission [9]. When phylogenic trees of the hMRGPR family and the mouse MRGPR family are compared, variation of the members found in those phylogenic trees are not parallel [6]. This difference may cause the hMRGPR family members to have functions besides the ones revealed by the studies with mice. Therefore, we hypothesized hMRGPRD may have other functions besides neurotransmission. Since finding physiological ligands may help us deduce possible functions, we executed GTPγS binding assay using Giα fused hMRGPRD to screen a physiologically active substance library. For GTPγS binding assay, as the membrane fraction of hMRGPRD-Giα expressing Sf9 cells was used and no βAla was contained in the assay buffer (20 mM Hepes, pH7.3, 100 mM NaCl, 10 mM MgCl2, 1 mM EDTA, 100 μg/mL saponin) (see Section 2.4), the signals we detected were completely dependent on the compounds added to be tested. Compounds considered as hits were tested with a calcium flux assay. For the calcium flux assay, only transient signals were detected and compounds were added with a buffer (HBSS, 25 mM Hepes, pH7.4, 2.5 mM probenecid) that did not contain βAla (see Section 2.5); therefore, the signals detected were again completely dependent on the compounds added for testing. We found 3 compounds that were positive, each of which in turn underwent an IP assay. For the IP accumulation assay, DMEM inositol-free media with 10% FBS dialyzed was used to label myo-[2, 3]inositol, then the labeling media was replaced with an IP assay buffer (200 mM HBSS, 25 mM Hepes, pH7.0, 0.1% BSA, 20 mM LiCl), that did not contain any βAla (see Section 2.6); therefore, accumulated IPs were attributed to the compound added for testing. The 3 compounds which we considered as hits were βAla (Figure 2(a)), βAIBA (Figure 2(b)), and DES (Figure 2(d)) whose EC50s are shown in Table 1. Representative dose-response curves of these compounds in each assay are shown in Figure 3. In all assays, the activity value of hMRGPRD with the saturated concentration of βAla was set as 100% for each assay system, and activity values are shown in percentages. For the IP assay, since the cells were kept in the assay buffer containing compound for an hour, a much higher DES concentration than the cells could tolerate was needed to calculate EC50; therefore, we could not obtain EC50 of DES in the IP assay. βAIBA was a newly found agonist, which was as potent as βAla, and had a structure similar to βAla (Figure 2). We also tested gamma-aminobutyric acid (GABA), which shares the same moiety with βAla and βAla (Figure 2) and had showed some activity for hMRGPRD in previous study [9]. Although with much weaker activity, GABA also worked as an MRGPRD agonist (Table 1, Figure 3).
Figure 2.
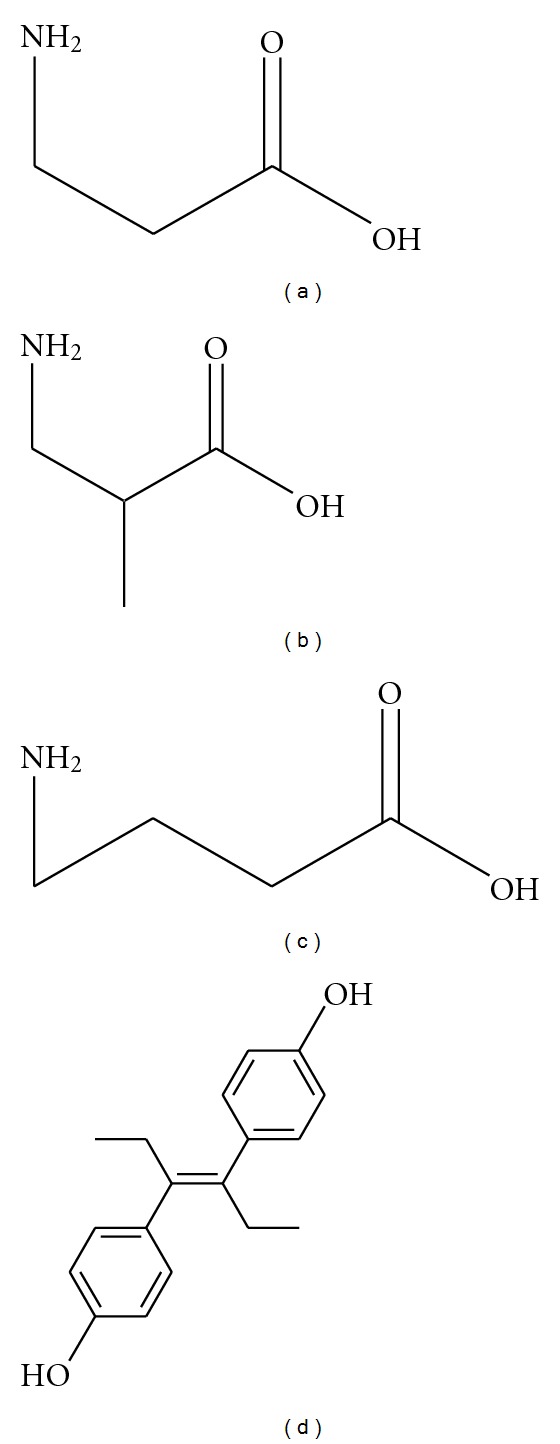
Structures of physiologically active substances which can activate MRGPRD: (a) beta-alanine (βAla), (b) beta-aminoisobutyric acid (βAIBA), (c) gamma-aminobutyric acid (GABA), and (d) diethylstilbestrol (DES).
Table 1.
Agonistic potency of physiologically active substances to induce signals of hMRGPRD.
| EC50 (μM) | GTPγS | Calcium flux | IP |
|---|---|---|---|
| βAla | 219.8 ± 115.7 | 22.6 ± 11.4 | 353.3 ± 217.0 |
| βAIBA | 821.7 ± 120.9 | 53.4 ± 27.3 | 378.3 ± 290.5 |
| GABA | Not determined | 4385 ± 871.1 | >1000 |
| DES | 50.7 ± 18.1 | 10.4 ± 3.0 | not available |
EC50 (sample concentration producing 50% of the maximal response) was calculated from dose-response curves and shown in μM. Multiple assays were executed to calculate EC50s: calcium flux assay (Calcium flux), GTPγS binding assay (GTPγS), and IP assay (IP).
Figure 3.
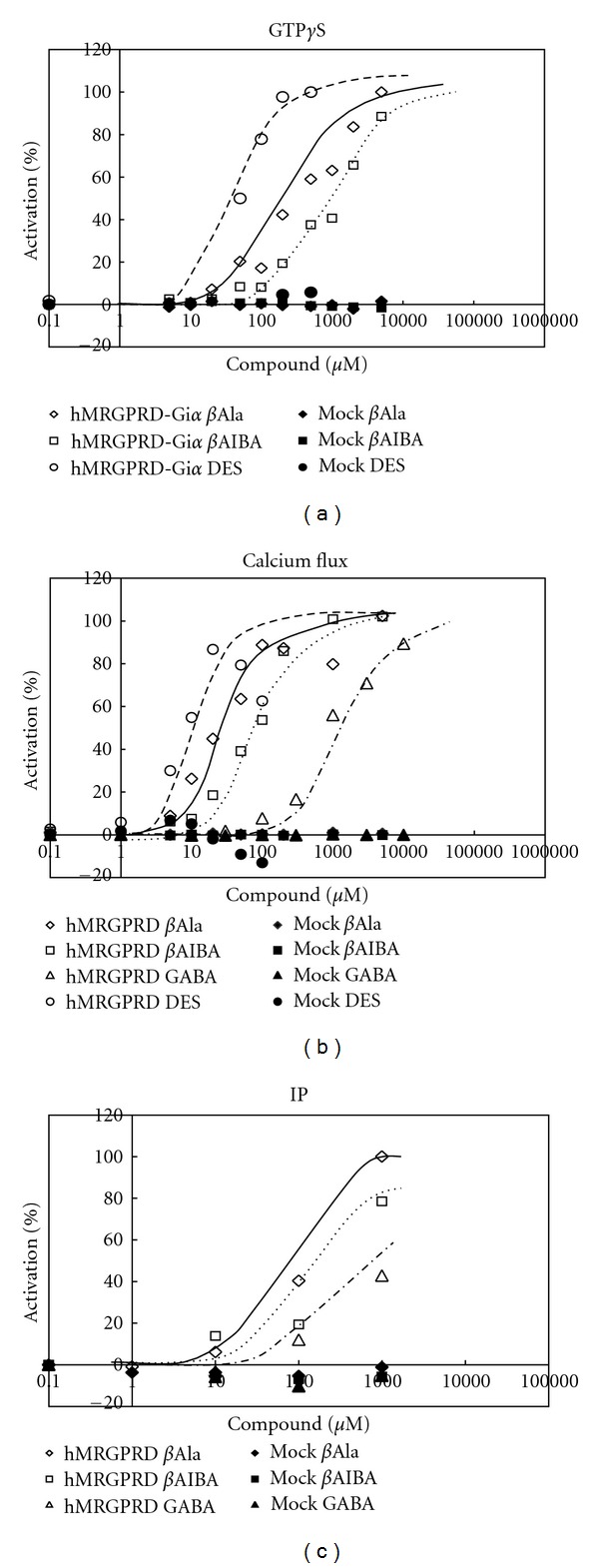
Agonistic effects of physiologically active substances on MRGPRD. Representative dose-dependent curves of physiologically active substances in (a) GTPγS binding assay, (b) calcium flux assay, and (c) IP assay are shown. Each value expresses the percentage of control with maximum efficacy in each assay. βAla on MRGPRD (open diamond), βAIBA on MRGPRD (open square), GABA on MRGPRD (open triangle). DES on MRGPRD (open circle), βAla on mock (closed diamond), βAIBA on mock (closed square), GABA on mock (closed triangle), DES on mock (closed circle).
3.3. Antagonists Screening
One of the nearest neighbors of hMRGPRD is MAS1 oncogene, which shows focus formation when transfected into NIH-3T3 cells [6, 16]. Also, there is information that βAla, βAIBA, and GABA are related to some kinds of tumors [17–22]. We took notice of oncogenicity and found that hMRGPRD-transformed fibroblast cells could form spheroids. The details of this feature will be reported elsewhere. We next screened a large chemical library to obtain MRGPRD antagonists which would help us examine the novel character of MRGPRD, the spheroid forming activity. Using βAla as a ligand for MRGPRD, a high-throughput screening (HTS) for antagonists was executed by the calcium flux method utilizing FLIPR, and MU-6840 (Figure 4) was selected as a hit. MU-6840 did not inhibit signals of the following GPCRs: CCR4, CCR5, CCR7, CCR8, CXCR3, S1PR1, P2Y12R, OGR1, OXER1, GPR84, GPR14, or GHS-R (data not shown). As shown in Table 2, MU-6840 inhibited βAla signals through hMRGPRD dose-dependently in all methods tested here, namely, calcium flux (Figure 5(a)), GTPγS binding (Figure 5(b)), IP accumulation (Figure 5(c)), and spheroid proliferation (Figure 5(d)). We did not use the endogenous signaling assay mentioned above (Section 2.3) because it is not suitable for detecting the effects of compounds due to the following reasons: (a) the transient transfection assay requires 24 hours to garner enough luciferase expression to be measured quantitatively; (b) cells cannot tolerate compound addition at the same time as transfection, as this would cause cell damage; (c) if the compound is added after suitable luciferase expression has been reached, the translated luciferase prior to compound addition remains and only the luciferase being transcribed/translated during compound addition would be inhibited, which would make the basal signal high enough to mask the inhibitory effect of the compound.
Figure 4.
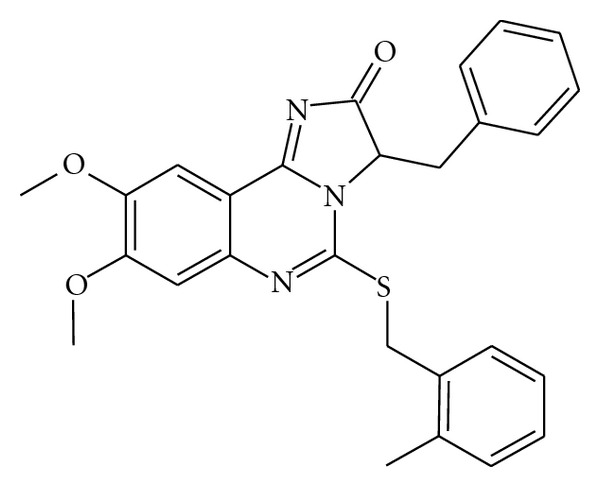
Structure of hMRGPRD antagonist, MU-6840.
Table 2.
Antagonistic activity of MU-6840 for hMRGPRD.
| Calcium flux | GTPγS | IP | Spheroid | |
|---|---|---|---|---|
| IC50 (nM) | 691.7 ± 120.9 | 1703.3 ± 353.9 | 473.8 ± 160.3 | 655.9 ± 204.3 |
IC50 (sample concentration producing 50% of the maximal inhibition) was calculated from dose-response curves and shown in nM. Multiple assays were used to calculate IC50s: calcium flux assay (Calcium flux), GTPγS binding assay (GTPγS), IP assay (IP), and spheroids growth assay (Spheroid).
Figure 5.
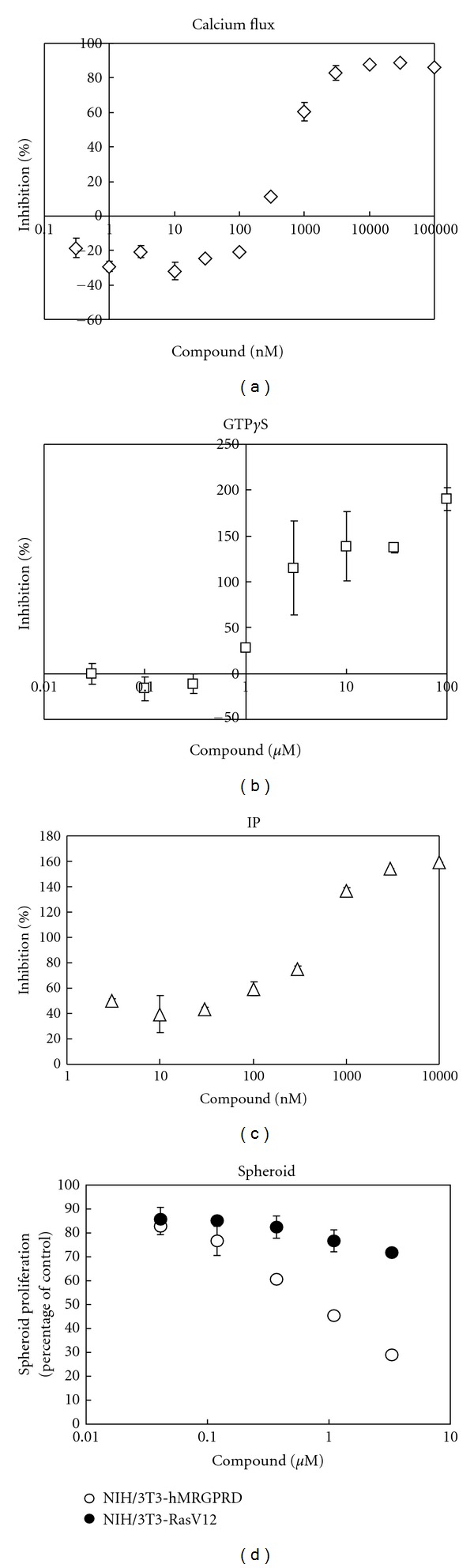
Inhibitory effects of MU-6840 on MRGPRD. Representative dose-dependent inhibition curves in (a) calcium flux assay, (b) GTPγS binding assay, (c) IP assay, and (d) spheroid assay by MU-6840 are shown. Each value expresses the percentage of control with no compound present. The results are the average of three independent tests.
In Figures 5(a), 5(b), and 5(c), values are shown in percentages: the value at adding only βAla with no antagonist is set as 0%; the value at adding neither ligand nor antagonist is set as 100%. For the calcium flux assay (Figure 5(a)), as no endogenous activity could be measured, the maximum inhibition never exceeded 100%. On the other hand, for the GTPγS binding assay (Figure 5(b)) and IP accumulation assay (Figure 5(c)), the maximum inhibitions exceeded 100% as these assays are able to detect hMRGPRD's strong constitutive activity. These excesses indicate that MU-6840 can act as an inverse agonist.
Owing to hMRGPRD's endogenous activity (Figures 1(a) and 1(b)), when hMRGPRD is stably expressed, NIH/3T3, which would not form spheroids by itself, can form spheroids with no ligand, such as βAla. In this spheroid assay (see Section 2.7), the culture media, RPMI1640, does not contain βAla. In addition, βAla which would come from FBS, taking up 10% of the media, could be negligible for the following reasons: the βAla concentration of normal serum is around 4 μM [23, 24] which makes βAla concentration in this media less than 0.5 μM; EC50s of βAla to hMRGPRD is higher than 20 μM (Table 1). When βAla was added to this assay system, spheroid growth of NIH/3T3-hMRGPRD was promoted, although no effect was seen with the spheroid growth of NIH/3T3-RasV12 cells, overexpressing oncogenic gene RasV12 (data not shown). It shows that the spheroid proliferation of NIH/3T3-hMRGPRD is dependent on MRGPRD signaling pathways. In order to determine the effect of the compounds, a spheroid proliferation assay is more suitable than a focus formation assay. For a spheroid assay, it is common to use RasV12 as a positive control [25, 26]. In the spheroid assay, MU-6840 inhibited NIH/3T3-hMRGPRD spheroid proliferation, which was dependent on basal activity of MRGPRD, that is, independent of any agonist. This result indicates that MU-6840 exhibits inverse agonism. Furthermore, MU-6840 did not inhibit spheroid proliferation of NIH/3T3-RasV12 cells (Figure 5(d)). Taking all things into account, MU-6840 shows inhibitory activity specific to MRGPRD.
4. Discussion
MRGPRD belongs to the Mas-related GPCR family, which has been shown to be expressed in the DRG [6, 9–12]. When phylogenic trees of the hMRGPR family and the mouse MRGPR family are compared, variation of the members found in those phylogenic trees are not parallel [6]. This difference may cause the hMRGPR family members to have functions besides the ones revealed by the studies with mice. Being one of the members of hMRGPR, we hypothesized that MRGPRD could have other functions besides the ones in DRG and found a spheroid forming activity.
In this study, we found 2 physiologically active substances as novel ligands for MRGPRD: βAIBA and DES (Figures 2(b) and 2(d)). βAla has long been widely known to act as a ligand for glycine receptors and GABA receptors in the central nervous system [27–32]. Since MRGPRD is known to be expressed in the DRG [6, 9–12], it is reasonable that βAla acts as a ligand of MRGPRD to have some roles in the nervous system. Besides the nervous system, MRGPRD may also have roles in renin-angiotensin system by releasing arachidonic acid in response to angiotensin (Ang) II metabolites, although no direct signals, that is, calcium flux, IP accumulation, and so forth, of MRGPRD in response to Ang II metabolites are shown [33]. On the other hand, besides being one of the nearest members of MAS1 oncogene [6, 16], there is some information that βAla, as well as βAIBA, is related to some types of tumors. Rat or mouse mammary tumors contain high levels of taurine, βAla, and GABA, which are enriched in neural tissue and which are not seen in normal rat or mouse mammary tissue [17]. βAla concentrations in nerve-related tissue are shown to be about 50 μM in rat sciatic nerve and 60 μM in cat brain [24, 34], which are higher than EC50 of βAla to MRGPRD in a calcium flux assay (Table 1). Cancer patients were more prone to have increased urinary βAIBA excretion than patients with different diseases [18]. The percentage of high urinary βAIBA excreter was higher in urinary bladder carcinoma patients compared to patients with urinary bladder papilloma or cystitis, or healthy volunteers [19]. The massive amounts of βAIBA in the urine of Burkitt's lymphoma patients were measured, and the degree of elevation of βAIBA excretion seemed to be related to the amount of tumor mass present [20, 21]. βAIBA excretion in patients with chronic myeloid leukemia was directly correlated to the leukocyte count, the indicator of tumor cell mass [22]. The average βAIBA level in the urine of patients with Burkitt's lymphoma is about 170 times higher than that of normal subjects [21]. Since the serum concentrations of βAIBA in normal subjects are about 2.5 μM [35], even if the difference of the serum βAIBA level is not as conspicuous as that of in urine, only 30 times difference is enough to exceed the EC50 of βAIBA to MRGPRD in a calcium flux assay (Table 1). All together, these suggest that βAla and βAIBA may be involved in tumorigenicity by enhancing MRGPRD oncogenic signals. However, it does not exclude the possibility that increasing concentrations of βAla and βAIBA is just a sign of the existence of cells with high metabolic rates, such as cancer cells, as βAla and βAIBA are degradation products of nucleic acids.
Besides βAla, βAIBA, GABA, and DES, 5 synthetic agonists for MRGPRD are reported [14, 36]. At least two of them are specific to MRGPRD, in other words they do not activate other Mas-related GPCRs [36]. Considering EC50s and the concentrations in vivo, we assume that βAla and βAIBA are the ones which can contribute to the physiological functions of MRGPRD.
Another new aspect we would like to focus on is the high endogenous signaling activity of MRGPRD. There are publications that some GPCRs, such as human β2 adrenoceptor, exhibit significant basal, agonist-independent G protein activation, which is associated with the number or ratio of the active (R*) state receptors [15, 37, 38]. In the report of Ajit et al., they noted that “our studies indicate that MrgD RNA expression inversely correlated with larger beta-alanine induced calcium transient functional responses as assessed by FLIPR responses” in the explanation of their selection of the cloned MRGPRD expressing cell for a calcium flux assay [14]. We also had a similar experience with this episode, and we assume this phenomenon clearly implies MRGPRD's high basal activity: if there are many MRGPRDs in the cells, the signal has already reached the maximum without any ligand, and no transient signal with the ligand could be detected, since FLIPR can only detect transient signal. Regarding MRGPRD's high endogenous signaling activity, just expressing a suitable amount of MRGPRD may add cells of a tumorigenic phenotype. High MRGPRD expression might attribute to some types of cancer, as we have detected a very high MRGPRD expression in some clinical cancer tissues (data not shown). MU-6840, a specific MRGPRD antagonist and possibly acting as an inverse agonist, has the potential ability to become an anticancer agent.
References
- 1.Dixon RAF, Kobilka BK, Strader DJ. Cloning of the gene and cDNA for mammalian β-adrenergic receptor and homology with rhodopsin. Nature. 1986;321(6065):75–79. doi: 10.1038/321075a0. [DOI] [PubMed] [Google Scholar]
- 2.Kobilka BK, MacGregor C, Daniel K, Kobilka TS, Caron MG, Lefkowitz RJ. Functional activity and regulation of human beta 2-adrenergic receptors expressed in Xenopus oocytes. Journal of Biological Chemistry. 1987;262(32):15796–15802. [PubMed] [Google Scholar]
- 3.Felder CC, Briley EM, Axelrod J, Simpson JT, Mackie K, Devane WA. Anandamide, an endogenous cannabimimetic eicosanoid, binds to the cloned human cannabinoid receptor and stimulates receptor-mediated signal transduction. Proceedings of the National Academy of Sciences of the United States of America. 1993;90(16):7656–7660. doi: 10.1073/pnas.90.16.7656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Masu Y, Nakayama K, Tamaki H, Harada Y, Kuno M, Nakanishi S. cDNA cloning of bovine substance-K receptor through oocyte expression system. Nature. 1987;329(6142):836–838. doi: 10.1038/329836a0. [DOI] [PubMed] [Google Scholar]
- 5.Parmentier M, Libert F, Maenhaut C, et al. Molecular cloning of the thyrotropin receptor. Science. 1989;246(4937):1620–1622. doi: 10.1126/science.2556796. [DOI] [PubMed] [Google Scholar]
- 6.Dong X, Han SK, Zylka MJ, Simon MI, Anderson DJ. A diverse family of GPCRs expressed in specific subsets of nociceptive sensory neurons. Cell. 2001;106(5):619–632. doi: 10.1016/s0092-8674(01)00483-4. [DOI] [PubMed] [Google Scholar]
- 7.Lembo PMC, Grazzini E, Groblewski T, et al. Proenkephalin A gene products activate a new family of sensory neuron-specific GPCRs. Nature Neuroscience. 2002;5(3):201–209. doi: 10.1038/nn815. [DOI] [PubMed] [Google Scholar]
- 8.Takeda S, Kadowaki S, Haga T, Takaesu H, Mitaku S. Identification of G protein-coupled receptor genes from the human genome sequence. FEBS Letters. 2002;520(1-3):97–101. doi: 10.1016/s0014-5793(02)02775-8. [DOI] [PubMed] [Google Scholar]
- 9.Shinohara T, Harada M, Ogi K, et al. Identification of a G protein-coupled receptor specifically responsive to β-alanine. Journal of Biological Chemistry. 2004;279(22):23559–23564. doi: 10.1074/jbc.M314240200. [DOI] [PubMed] [Google Scholar]
- 10.Luo W, Wickramasinghe SR, Savitt JM, Griffin JW, Dawson TM, Ginty DD. A hierarchical NGF signaling cascade controls ret-dependent and ret-independent events during development of nonpeptidergic DRG neurons. Neuron. 2007;54(5):739–754. doi: 10.1016/j.neuron.2007.04.027. [DOI] [PubMed] [Google Scholar]
- 11.Zhang L, Taylor N, Xie Y, et al. Cloning and expression of MRG receptors in macaque, mouse, and human. Molecular Brain Research. 2005;133(2):187–197. doi: 10.1016/j.molbrainres.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 12.Zylka MJ, Rice FL, Anderson DJ. Topographically distinct epidermal nociceptive circuits revealed by axonal tracers targeted to Mrgprd. Neuron. 2005;45(1):17–25. doi: 10.1016/j.neuron.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 13.Fredriksson R, Lagerström MC, Lundin LG, Schiöth HB. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Molecular Pharmacology. 2003;63(6):1256–1272. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- 14.Ajit SK, Pausch MH, Kennedy JD, Kaftan EJ. Development of a FLIPR assay for the simultaneous identification of MrgD agonists and antagonists from a single screen. Journal of Biomedicine and Biotechnology. 2010;2010:8 pages. doi: 10.1155/2010/326020.326020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seifert R, Wenzel-Seifert K. Constitutive activity of G-proteins-coupled receptors: cause of disease and common property of wild-type receptors. Naunyn-Schmiedeberg’s Archives of Pharmacology. 2002;366(5):381–416. doi: 10.1007/s00210-002-0588-0. [DOI] [PubMed] [Google Scholar]
- 16.Young D, Waitches G, Birchmeier C. Isolation and characterization of a new cellular oncogene encoding a protein with multiple potential transmembrane domains. Cell. 1986;45(5):711–719. doi: 10.1016/0092-8674(86)90785-3. [DOI] [PubMed] [Google Scholar]
- 17.Pine MJ, Kim U, Ip C. Free amino acid pools of rodent mammary tumors. Journal of the National Cancer Institute. 1982;69(3):729–735. [PubMed] [Google Scholar]
- 18.Abe M, Takahashi M, Nishidai T. The significance of urinary beta aminoisobutyric acid in cancer patients. International Journal of Radiation Biology. 1973;24(1):73–79. doi: 10.1080/09553007314550831. [DOI] [PubMed] [Google Scholar]
- 19.Nielsen HR, Nyholm K, Sjolin KE. Beta-aminoisobutyric acid in urine from patients with bladder tumours. Acta Pathologica et Microbiologica Scandinavica. Section A. 1970;78(3):p. 368. doi: 10.1111/j.1699-0463.1970.tb03315.x. [DOI] [PubMed] [Google Scholar]
- 20.Waalkes TP, Gehrke CW, Bleyer WA. Potential biologic markers in Burkitt’s lymphoma. Cancer Chemotherapy Reports. 1975;59(4):721–727. [PubMed] [Google Scholar]
- 21.Waalkes TP, Gehrke CW, Lakings DB. Beta aminoaciduria in patients with Burkitt’s lymphoma. Journal of the National Cancer Institute. 1976;57(2):435–438. doi: 10.1093/jnci/57.2.435. [DOI] [PubMed] [Google Scholar]
- 22.Nielsen HR, Killmann SA. Urinary excretion of β-aminoisobutyrate and pseudouridine in acute and chronic myeloid leukemia. Journal of the National Cancer Institute. 1983;71(5):887–891. [PubMed] [Google Scholar]
- 23.Van Kuilenburg ABP, Stroomer AEM, Van Lenthe H, Abeling NGGM, Van Gennip AH. New insights in dihydropyrimidine dehydrogenase deficiency: A pivotal role for β-aminoisobutyric acid? Biochemical Journal. 2004;379(1):119–124. doi: 10.1042/BJ20031463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tallan HH, Moore S, Stein WH. Studies on the free amino acids and related compounds in the tissues of the cat. The Journal of Biological Chemistry. 1954;211(2):927–939. [PubMed] [Google Scholar]
- 25.Fukazawa H, Mizuno S, Uehara Y. A microplate assay for quantitation of anchorage-independent growth of transformed cells. Analytical Biochemistry. 1995;228(1):83–90. doi: 10.1006/abio.1995.1318. [DOI] [PubMed] [Google Scholar]
- 26.Kunz-Schughart LA, Groebe K, Mueller-Klieser W. Three-dimensional cell culture induces novel proliferative and metabolic alterations associated with oncogenic transformation. International Journal of Cancer. 1996;66(4):578–586. doi: 10.1002/(SICI)1097-0215(19960516)66:4<578::AID-IJC25>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 27.Calvo DJ, Miledi R. Activation of GABA(ρ1) receptors by glycine and β-alanine. NeuroReport. 1995;6(8):1118–1120. doi: 10.1097/00001756-199505300-00011. [DOI] [PubMed] [Google Scholar]
- 28.Choquet D, Korn H. Does β-alanine activate more than one chloride channel associated receptor? Neuroscience Letters. 1988;84(3):329–334. doi: 10.1016/0304-3940(88)90530-7. [DOI] [PubMed] [Google Scholar]
- 29.Horikoshi T, Asanuma A, Yanagisawa K, Anzai K, Goto S. Taurine and beta-alanine act on both GABA and glycine receptors in Xenopus oocyte injected with mouse brain messenger RNA. Brain research. 1988;464(2):97–105. doi: 10.1016/0169-328x(88)90002-2. [DOI] [PubMed] [Google Scholar]
- 30.Sandberg M, Jacobson I. β-Alanine, a possible neurotransmitter in the visual system? Journal of Neurochemistry. 1981;37(5):1353–1356. doi: 10.1111/j.1471-4159.1981.tb04691.x. [DOI] [PubMed] [Google Scholar]
- 31.Schmieden V, Kuhse J, Betz H. Mutation of glycine receptor subunit creates β-alanine receptor responsive to GABA. Science. 1993;262(5131):256–258. doi: 10.1126/science.8211147. [DOI] [PubMed] [Google Scholar]
- 32.Wu FS, Gibbs TT, Farb DH. Dual activation of GABA(A) and glycine receptors by β-alanine: Inverse modulation by progesterone and 5α-pregnan-3α-ol-20-one. European Journal of Pharmacology. 1993;246(3):239–246. doi: 10.1016/0922-4106(93)90037-a. [DOI] [PubMed] [Google Scholar]
- 33.Gembardt F, Grajewski S, Vahl M, Schultheiss HP, Walther T. Angiotensin metabolites can stimulate receptors of the Mas-related genes family. Molecular and Cellular Biochemistry. 2008;319(1-2):115–123. doi: 10.1007/s11010-008-9884-4. [DOI] [PubMed] [Google Scholar]
- 34.Marks N, Datta RK, Lajtha A. Distribution of amino acids and of exo- and endopeptidases along vertebrate and invertebrate nerves. Journal of Neurochemistry. 1970;17(1):53–63. doi: 10.1111/j.1471-4159.1970.tb00501.x. [DOI] [PubMed] [Google Scholar]
- 35.Gejyo F, Kinoshita Y, Ikenaka T. Elevation of serum levels of β-aminoisobutyric acid in uremic patients and the toxicity of the amino acid. Clinical Nephrology. 1977;8(6):520–525. [PubMed] [Google Scholar]
- 36.Zhang R, Yan PK, Zhou CH, Liao JY, Wang MW. Development of a homogeneous calcium mobilization assay for high throughput screening of mas-related gene receptor agonists. Acta Pharmacologica Sinica. 2007;28(1):125–131. doi: 10.1111/j.1745-7254.2007.00451.x. [DOI] [PubMed] [Google Scholar]
- 37.Cotecchia S. Constitutive activity and inverse agonism at the α1a and α1b adrenergic receptor subtypes. Methods in Enzymology. 2010;485(C):123–138. doi: 10.1016/B978-0-12-381296-4.00007-5. [DOI] [PubMed] [Google Scholar]
- 38.Rasmussen SGF, Choi HJ, Rosenbaum DM, et al. Crystal structure of the human β2 adrenergic G-protein-coupled receptor. Nature. 2007;450(7168):383–388. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]


