Abstract
Selenium is an essential trace element for which both beneficial and toxic effects in human health have been described. It is now clear that the importance of having adequate amounts of this micronutrient in the diet is primarily due to the fact that selenium is required for biosynthesis of selenocysteine, the twenty first naturally occurring amino acid in protein. In this review, we provide an overview of eukaryotic selenoproteins and selenoproteomes, which are sets of selenoproteins in these organisms. In eukaryotes, selenoproteins show a mosaic occurrence, with some organisms, such as vertebrates and algae, having dozens of these proteins, while other organisms, such as higher plants and fungi, having lost all selenoproteins during evolution. We also discuss selenoprotein functions and evolutionary trends in the use of these proteins in eukaryotes. Functional analysis of selenoproteins is critical for better understanding of the role of selenium in human health and disease.
Keywords: Selenocysteine, Selenoprotein, Selenoproteome, SECIS element
1. Introduction
Selenium (Se) is best known for its roles in various redox processes, attributed to its presence in proteins as the 21st naturally occurring amino acid, selenocysteine (Sec), encoded by the UGA codon [1,2]. Proteins containing Sec are called selenoproteins (proteins containing selenomethionine are not regarded as selenoproteins due to the non-specific nature of Se utilization in these proteins). Most selenoproteins act as oxidoreductases that prevent damage to cellular components, repair this damage, regulate redox state of proteins or have other redox functions. This class of proteins plays a major role in human health and disease [3,4]. For example, low dietary Se may result in selenoprotein deficiency leading to such human diseases as Keshan disease, Kashin-Beck disease, Myxedematous Endemic Cretinism and male infertility [3].
Sec insertion into protein is dictated by the codon, UGA, and requires the presence of a conserved stem–loop structure, known as the Sec insertion sequence (SECIS) element [5]. In eukaryotes, this structure is located in the 3′-UTR [6,7]. Two forms of SECIS elements are found in mammalian selenoprotein mRNAs: Type I and Type II elements, the latter possessing an additional mini-stem and being more widespread [8]. Analysis of coding and recoding potentials of UGA codons in various organisms led to a recent discovery of a new feature in the genetic code — in a ciliate Euplotes crassus, UGA codes for both cysteine and Sec, depending on the presence and availability of the SECIS element and on the location of the UGA within selenoprotein mRNAs [9]. Both amino acids encoded by UGA may even be present within the same protein. This finding raises questions regarding evolution of additional protein amino acids and demonstrates that one codon can code for multiple amino acids at internal positions of proteins. In addition, this finding suggested that Sec may not be inserted at any position in a selenoprotein, and that insertion may occur only at specific positions, depending on the availability of the SECIS element within the overall mRNA structure for translation machinery.
Eukaryotes have highly variable sets of selenoproteins (selenoproteomes), which vary from zero selenoproteins in higher plants and fungi to more than 30 in some fishes and algae. Prokaryotes feature similarly diverse use of selenoproteins. Eukaryotic and prokaryotic selenoproteomes only partially overlap. Selenoproteomes of closely related species are generally similar to each other, but at larger evolutionary distances selenoprotein use becomes sporadic wherein some selenoproteins were lost numerous times in various phyla, and some evolved only in small sets of organisms. The mosaic occurrence of selenoproteins in eukaryotes, as opposed to limited or uniform use of these proteins, provides opportunities for linking patterns of selenoproteome occurrence and composition to environmental and other factors that shape the evolution of eukaryotic selenoproteomes.
2. Selenoproteins
Selenoproteins are present in all three domains of life: bacteria, archaea and eukaryota. However, some organisms do not use Sec. For example, yeast and higher plants lost the Sec insertion machinery during evolution and, therefore, do not possess selenoproteins; in these organisms, cysteine-containing homologs of some selenoproteins are utilized instead. It has been shown that selenoproteins can be a thousand times more effective in catalysis than their cysteine homologs [10], even though exceptions are also known. The greater effectiveness of Sec in catalysis is likely one of the major reasons that nature has invested in evolving Se-dependent pathways and the specialized machinery used for Se insertion into protein.
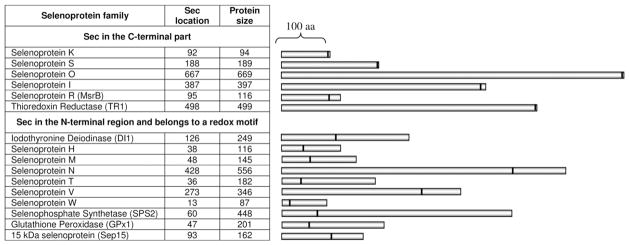
Most selenoproteins belong to one of two main groups according to the location of the Sec residue (Fig. 1). Selenoproteins of the first group contain Sec very close to the C-terminus of the protein. Mammalian selenoproteins K, S, O, I, R (methionine-R-sulfoxide reductase, MsrB) and thioredoxin reductases (TRs) are examples of such proteins. Other proteins, such as mammalian selenoproteins H, M, T, V, W, Sep15, selenophosphate synthetase 2 (SPS2), thyroid hormone deiodinases (DIs) and glutathione peroxidases (GPxs), have Sec in the N-terminal segments of proteins (almost all are thioredoxin fold proteins), often as part of the CxxU motif that corresponds to the CxxC motif (two cysteines separated by two residues) in the active site of thioredoxin.
Fig. 1.
Mammalian selenoproteins. Mammalian selenoproteins can roughly be divided into two groups. In the first group, Sec is located very close to the C-terminus, often in the C-terminal penultimate position. Selenoproteins of the second group possess Sec in their N-terminal or middle regions, often in a redox motif within a thioredoxin fold and have an α-helix downstream of Sec. Selenoprotein P, which has both an N-terminal redox Sec and multiple Sec residues in the C-terminal region, is not shown in the figure. Sec positions and selenoprotein lengths refer to human proteins. Close homologs of GPx1 (four selenoproteins), DI1 and TR1 (two each) are not shown.
Whereas functions of several selenoprotein families, including GPxs, TRs, and DIs, have been known for some time, recent studies have led to a significant progress in the functional analysis of several other selenoproteins, such as MsrB, Selenoprotein P (SelP), Selenoprotein N and SPS2. In addition, the function of Selenoprotein H in regulating expression levels of genes for glutathione synthesis was described [11]. However, the functions of most other selenoprotein families remain unknown. These include Selenoprotein K, a short protein with a single transmembrane domain; Selenoprotein I, a recently evolved membrane selenoprotein; Selenoprotein O, the largest mammalian selenoprotein that has cysteine homologs in bacteria, yeast and plants; Selenoprotein T, a protein with a predicted redox motif; two related selenoproteins, Selenoproteins W and V (the C-terminal part of SelV is highly homologous to SelW); and Selenoprotein M, which is a distant homolog of the 15 kDa selenoprotein, Sep15.
3. Selenoproteomes
The size of selenoproteome varies significantly among eukaryotes. The highest content of selenoproteins is observed in aquatic organisms, whether they are animals (e.g., fish) or plants (e.g., algae). Some algae are especially rich in selenoproteins. Blue-green algae are thought to be among the first photosynthetic organisms, and it has been hypothesized that an antioxidant function offered by selenoproteins is required to protect cells from oxygen [12]. Availability of steady supplies of Se in the sea water could have made it possible to increase the use of this element for various oxidoreductases.
The change from aquatic to terrestrial habitats posed a challenge to plants and animals, as the availability of some trace elements greatly diminished, and these organisms were now exposed to higher oxygen levels. As a result, many terrestrial organisms might have lost selenoproteins or replaced them with cysteine-containing homologs. However, it also became apparent that some selenoproteins evolved in animals, while other selenoproteins, such as Fep15 [13], SelJ [14] and SelL [15], manifested a quite narrow distribution among aquatic eukaryotes. For example, Fep15 [13], a protein distantly related to the Sep15 selenoprotein family, was only detected in fish. Another rare selenoprotein, SelJ [14], shows significant similarity to jellyfish J1-crystallins and appears to be restricted to actinopterygian fishes and sea urchin. SelJ is expressed in the lens in the early stages of development, and its potential role as a structural crystallin is in contrast to the functions of the majority of known selenoenzymes.
A family of selenoproteins designated SelL [15] contains two Sec residues organized in a form of a UxxU motif. SelL proteins were found in organisms from diverse aquatic organisms, such as fish, invertebrates and marine bacteria. A single SECIS element is used for insertion of both Sec residues in this protein. Distantly related SelL-like proteins are present in a variety of organisms, with one or both Sec residues replaced by Cys. The formation of a diselenide bond between the two Sec moieties in SelL was confirmed experimentally [15]. This makes SelL the first example of a protein with a natural diselenide bond.
Another selenoprotein with multiple Sec residues is SelP. In animals with larger selenoproteomes, SelP usually contains more Sec residues than in organisms with few selenoproteins, which is consistent with its role in selenium transport and utilization. Currently, SelP is the only known selenoprotein that contains more than 2 Sec [16,17]. Unlike SelL, where a single SECIS element serves both UGA codons [15], Sec incorporation into SelP is carried out by two SECIS elements, with proximal Type 2 SECIS responsible for insertion of multiple Sec residues in the C-terminal part of the protein, and a more distal Type 1 SECIS serving the first UGA codon. A significant variation in Sec content was observed for SelPs [18], with only 7 Sec residues present in guinea pig and 28 Sec in sea urchin SelPs. A distant Cys-containing SelP homolog containing 23 Cys was identified in nematodes, and the lack of Sec in this protein suggested that its function is unrelated to Se transport. Selenoproteomes of fishes, birds and early mammals (such as platypus) contain two forms of SelP — a Sec-rich SelPa and a SelPb containing a single Sec [18]. We further discuss the selenoproteomes of humans and model organisms.
3.1. Humans
Humans contain 25 known selenoprotein genes [19]. The human selenoproteome consists of 5 Sec-containing GPxs, including cytosolic GPx1, gastrointestinal GPx2, plasma GPx3, its close homolog GPx6, and phospholipid hydroperoxide GPx, GPx4; three TRs, including cytosolic TR1, mitochondrial TR3 and thioredoxin/glutathione reductase TGR; and three DIs. Other selenoproteins are Selenoprotein W (originally isolated from muscles, the smallest mammalian selenoprotein), Selenoprotein V, SPS2 (participates in Sec biosynthesis by providing a selenium donor compound, selenophosphate), Sep15, SelM, MsrB1, and Selenoproteins I, N, O, H, T, K and S. The remaining human selenoprotein is SelP, which has 10 Sec residues.
3.2. Rodents
Mouse and rat were among the first mammals for which completely sequenced genomes were determined. Evolutionary distance between rodents and primates allowed the use of these genomes as references for the analysis of the human selenoproteome. This analysis utilized the fact that functionally important sequences (such as SECIS elements) remained conserved between primates and rodents, whereas the flanking sequences differed significantly. The rodent selenoproteomes were found to be quite similar to the human selenoproteome. The only difference is GPx6: in humans it is a selenoprotein, but the rodent GPx6 replaced Sec with Cys, decreasing the rodent selenoproteome to 24 selenoproteins [19].
3.3. Other mammals
Generally, mammalian selenoproteomes are very similar to those of human, rat and mouse. To date, no unique selenoprotein families were found in individual or closely related mammals. Of interest is the observation that early mammals, such as armadillo, opossum and platypus still possess a short form of SelP (i.e., SelPb) containing a single Sec residue. In addition, platypus retained a Sec-containing form of SelU, which was replaced in other mammals with a Cys homolog.
3.4. Fishes
Fish selenoproteomes are larger than those of mammals; however, with few exceptions, the same core selenoprotein families are found in mammals and fish. In addition, fish have several selenoproteins (Fep15, SelJ, SelL) that are missing in mammals, as well as several Sec-containing copies of Selenoproteins T, U and W, and two forms of SelP. Interestingly, the number of Sec residues in SelPa is also higher: 16–17 in fish, as opposed to 7–15 in mammals. At 30–37 selenoproteins, fish selenoproteomes are among the largest known.
3.5. Chlamydomonas and other green algae
Chlamydomonas reinhardtii was the first representative of the plant kingdom, in which selenoproteins were found [20]. Most selenoproteins in C. reinhardtii (TR, SelK, SelM, SelW1–2, SelH, GPx1–2, SelT) are also present in mammals. Since these selenoproteins are absent in higher plants, as well as in fungi and most insects and nematodes, three possibilities were considered: (i) mammalian/algae selenoproteins were independently lost in higher plants, nematodes and insects, (ii) they independently evolved in mammals and algae; and (iii) they were acquired through horizontal gene transfer. In addition, three selenoproteins set the Chlamydomonas selenoproteome apart —a novel membrane selenoprotein MSP, selenoprotein U (it mostly occurs in aquatic organisms) and a Sec-containing MsrA [21]. While horizontal gene transfer plays an important role in the gene transfer between bacteria and archaea [22], and examples of gene transfer from eukaryotes (including humans) were also found, no evidence was found for selenoprotein gene transfer between Chlamydomonas and other organisms [20]. Further analyses of completed genomes and EST databases also revealed selenoproteins that are common to algae and mammals in numerous unicellular eukaryotes. Altogether, these data suggest that the majority of Chlamydomonas selenoproteins evolved in the common ancestor of plants and animals.
Various species of Ostreococcus (i.e., O. tauri and O. lucimarinus) represent selenoprotein-rich algae. The high numbers of selenoproteins identified (26 and 29, respectively) [23] fit well with the hypothesis that organisms with aquatic habitat tend to possess larger selenoproteomes. As discussed above, the availability of trace elements in sea water may facilitate Se utilization. In addition, the use of Se may be to be linked to the levels of oxygen: i.e., highly reactive selenoproteins may be less susceptible to oxidation when oxygen levels are lower.
3.6. Amoebae
A slime mold, Dictyostelium discoideum, was also examined for the occurrence of selenoproteins. Five proteins, SPS2, SelK, Sep15, MSP and a DI homolog were found to contain Sec [23]. The presence of the DI homolog was unexpected (thyroid hormones are not known to occur in the amoebae), but other DI sequences were later found in additional lower eukaryotes as well as in some bacteria. MSP homologs were found only in Dictyostelium, Chlamydomonas, Volvox and Ostreococcus, and this protein currently has the narrowest distribution among eukaryotes. An interesting feature of D. discoideum is a higher than usual conservation of its SECIS elements at the sequence level: all detected selenoproteins had SECIS elements with a highly conserved UGUA quartet preceding the conserved SECIS core, as well as a U–U mismatch immediately after it. The unrelated evolutionary histories of detected proteins suggest that the stronger primary sequence conservation of these SECIS elements is the result of convergent evolution.
3.7. Nematodes
Nematodes, or roundworms, are among the most diverse animals, with more than 80,000 species described [24]. In contrast to other animals, only one selenoprotein, TR, was described in Caenorhabditis elegans [25,26]. A Cys-containing homolog of this protein is also present in C. elegans. Since some animals (e.g., D. melanogaster) only possess Cys-containing thioredoxin reductases, a possibility was suggested that the use of Sec could be completely lost in some animals, and this prediction was recently confirmed in insects (see below). On the other hand, the finding of only one selenoprotein in C. elegans raised a possibility that additional selenoproteins exist in this organism. However, extensive searches did not result in the identification of additional selenoproteins [27]. Three independent algorithms were used, including a SECISearch-based approach, a search for Cys- and Sec-containing homologs of potential selenoproteins, and a search for Sec-containing homologs of annotated proteins. Thus, C. elegans and C. briggsae represent an interesting case of selenoproteome reduction, wherein in a single UGA codon in the entire genome codes for Sec.
3.8. Apicomplexan parasites
Apicomplexans possess reduced selenoproteomes. Four unique selenoproteins were found in Plasmodium [28,29], including two predicted to be located in the apicoplast (a cell organelle that contains several unique metabolic pathways and is absent in the host). In contrast, the selenoproteome of Toxoplasma consists of five selenoproteins [30], four of which have homologs in mammals. Two Toxoplasma selenoproteins are expressed with the help of an unusual, but highly effective SECIS element that utilizes a GGGA quartet instead of the canonical AUGA. The complete lack of selenoproteins in Cryptosporidium parvum represents a further compelling example of massive selenoprotein loss [23].
3.9. Kinetoplastidan parasites
Similar to other eukaryotic parasites, the selenoproteomes of Leishmania and Trypanosoma are small. Three selenoproteins were reported in Kinetoplastida, two being homologs of SelK and SelT, and a third a novel selenoprotein with two rhodanese and one rubredoxin: oxygen oxidoreductase domains that appears to be a Kinetoplastida-specific selenoprotein. The functions of any of these selenoproteins are not known.
3.10. Insects
The Drosophila melanogaster selenoproteome was the first fully characterized eukaryotic selenoproteome. It consists of three selenoproteins, SelH (also known as BthD), SelK (also known as G-rich selenoprotein) and SPS2. Other analyzed insects, such as Anopheles gambiae, Apis mellifera, D. pseudoobscura, also possess small selenoproteomes, having 1–3 selenoproteins. Recently, the first animals (D. willistoni, B. mori and T. castaneum) that completely lack selenoproteins were found [31,32]. These findings make these organisms an extreme example of selenoprotein loss and a potentially useful model to study the role of selenium in a selenoproteinless background.
3.11. Crustacea
Unlike their terrestrial counterparts (insects), aquatic arthropods (such as shrimp and daphnia) possess many selenoprotein genes. For example, at least 7 selenoproteins were detected in Daphnia magna EST sequences, and this number will likely increase when the full genome sequence becomes available. The depletion of selenoproteomes in insects and their abundance in Crustacea could be explained by several factors, such as the presence of bioavailable Se in the water or higher concentration of oxygen in the air; however, further research is needed to address these possibilities.
4. Lack of selenoproteins in fungi and higher plants
No selenoproteins or Sec insertion machinery were detected in completely sequenced fungal genomes, which is consistent with the idea that selenoprotein genes were lost at the base of the kingdom of fungi. Similarly, while large selenoproteomes occur in Chlamydomonas, Volvox and Ostreococcus, higher plants such as A. thaliana and O. sativa completely lost both selenoproteins and Sec insertion machinery [23]. Analysis of known selenoprotein families indicates that they were either lost or replaced with Cys-containing homologs. It is interesting to note that both selenoproteins and Sec insertion machinery are present in an early Streptophyte, green alga Mesostigma viride. Therefore, selenoproteins may have been lost in Viridiplantae after changing their habitat from aquatic to terrestrial.
5. Selenoproteome evolution
The identification of selenoproteins with restricted occurrence showed that the mammalian selenoproteome which, until recently, was thought to accurately represent all eukaryotic selenoproteins, is, in fact, a reduced version of the vertebrate selenoproteome. Several rare selenoproteins were also found in lower eukaryotes, and several independent selenoprotein loss events were described. Such mosaic selenoprotein distribution enabled us to examine directions and reversibility of Sec to Cys (and vice versa) replacements. It appears that the environment in which organisms live and the availability of Se may shape selenoprotein evolution.
The reason for replacement of selenoproteins with cysteine homologs may be higher oxygen levels, which could have made selenoproteins more susceptible to oxidative damage. On the other hand, tissue specialization, organism size and protective cover of the skin in multicellular organisms allows these organisms to control (and restrict) oxygen delivery to internal sites, thus reducing negative influence of this gas on selenoprotein use. While it is highly unlikely that the entire multicellular organization of organisms was adopted merely for the protection of selenoproteins from excess oxygen, it definitely helped, and larger organisms (like mammals) now utilize selenoproteins to a much greater extent than, for instance, nematodes or insects. The recent findings of the complete loss of selenoproteins in some insects [31,32] demonstrate that some small animals can survive even in the absence of Sec-containing proteins.
Detailed information on selenoprotein distribution can help uncover selenoproteome evolution and provide insights into advantages and disadvantages of Sec utilization. One approach has been suggested [33] for high-throughput identification of catalytic redox-active Cys in proteins by searching for sporadic Sec–Cys pairs. Thus, information about selenoproteomes may also be of great importance for researchers in various other fields.
6. Conclusions
Considerable progress has been made in characterizing the mechanisms and regulation of selenoprotein synthesis in eukaryotes. Significant discoveries were made recently that helped us better understand Se metabolic pathways, mechanism of Sec biosynthesis in eukaryotes, identities of selenoproteins, and functions of some of these proteins. However, the biological functions of most Sec-containing proteins remain unknown. Analysis of sequenced genomes for selenoprotein occurrence and functions as well as for selenoproteome composition and evolution offers a powerful tool to help uncover the evolutionary trends in selenium utilization and function, which may ultimately lead to a better understanding of the role of Se in human health and disease.
References
- 1.Cone JE, Del Rio RM, Davis JN, Stadtman TC. Chemical characterization of the selenoprotein component of clostridial glycine reductase: identification of selenocysteine as the organoselenium moiety. Proc Natl Acad Sci U S A. 1976;73:2659–2663. doi: 10.1073/pnas.73.8.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stadtman TC. Selenocysteine. Annu Rev Biochem. 1996;65:83–100. doi: 10.1146/annurev.bi.65.070196.000503. [DOI] [PubMed] [Google Scholar]
- 3.Surai PF. Selenium in Nutrition and Health. Nottingham University Press; Nottingham: 2006. [Google Scholar]
- 4.Hatfield D, Berry M, Gladyshev V. Selenium: Its Molecular Biology and Role in Human Health. Springer; 2006. [Google Scholar]
- 5.Berry MJ, Banu L, Chen YY, Mandel SJ, Kieffer JD, Harney JW, Larsen PR. Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3′untranslated region. Nature. 1991;353:273–276. doi: 10.1038/353273a0. [DOI] [PubMed] [Google Scholar]
- 6.Walczak R, Westhof E, Carbon P, Krol A. A novel RNA structural motif in the selenocysteine insertion element of eukaryotic selenoprotein mRNAs. RNA. 1996;2:367–379. [PMC free article] [PubMed] [Google Scholar]
- 7.Berry MJ, Banu L, Harney JW, Larsen PR. Functional characterization of the eukaryotic SECIS elements which direct selenocysteine insertion at UGA codons. EMBO J. 1993;12:3315–3322. doi: 10.1002/j.1460-2075.1993.tb06001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chapple CE, Guigo R, Krol A. SECISaln, a web-based tool for the creation of structure-based alignments of eukaryotic SECIS elements. Bioinformatics. 2009;25:674–675. doi: 10.1093/bioinformatics/btp020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turanov AA, Lobanov AV, Fomenko DE, Morrison HG, Sogin ML, Klobutcher LA, Hatfield DL, Gladyshev VN. Genetic code supports targeted insertion of two amino acids by one codon. Science. 2009;323:259–261. doi: 10.1126/science.1164748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim HY, Fomenko DE, Yoon YE, Gladyshev VN. Catalytic advantages provided by selenocysteine in methionine-S-sulfoxide reductases. Biochemistry. 2006;45:13697–13704. doi: 10.1021/bi0611614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Panee J, Stoytcheva ZR, Liu W, Berry MJ. Selenoprotein H is a redox-sensing high mobility group family DNA-binding protein that up-regulates genes involved in glutathione synthesis and phase II detoxification. J Biol Chem. 2007;282:23759–23765. doi: 10.1074/jbc.M702267200. [DOI] [PubMed] [Google Scholar]
- 12.Venturi S, Venturi M. Evolution of dietary antioxidant defences. European EPI-Marker. 2007;11:1–12. [Google Scholar]
- 13.Novoselov SV, Hua D, Lobanov AV, Gladyshev VN. Identification and characterization of Fep15, a new selenocysteine-containing member of the Sep15 protein family. Biochem J. 2006;394:575–579. doi: 10.1042/BJ20051569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castellano S, et al. Diversity and functional plasticity of eukaryotic selenoproteins: identification and characterization of the SelJ family. Proc Natl Acad Sci U S A. 2005;102:16188–16193. doi: 10.1073/pnas.0505146102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shchedrina VA, Novoselov SV, Malinouski M, Gladyshev V. Identification and characterization of a novel selenoprotein family containing a diselenide bond in a redox motif. Proc Natl Acad Sci U S A. 2007 doi: 10.1073/pnas.0703448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burk RF, Hill KE. Selenoprotein P: an extracellular protein with unique physical characteristics and a role in selenium homeostasis. Annu Rev Nutr. 2005;25:215–235. doi: 10.1146/annurev.nutr.24.012003.132120. [DOI] [PubMed] [Google Scholar]
- 17.Herrman JL. The properties of a rat serum protein labelled by the injection of sodium selenite. Biochim Biophys Acta. 1977;500:61–70. doi: 10.1016/0304-4165(77)90046-0. [DOI] [PubMed] [Google Scholar]
- 18.Lobanov AV, Hatfield DL, Gladyshev VN. Reduced reliance on the trace element selenium during evolution of mammals. Genome Biol. 2008;9:R62. doi: 10.1186/gb-2008-9-3-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, Guigo R, Gladyshev VN. Characterization of mammalian selenoproteomes. Science. 2003;300:1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 20.Novoselov SV, Rao M, Onoshko NV, Zhi H, Kryukov GV, Xiang Y, Weeks DP, Hatfield DL, Gladyshev VN. Selenoproteins and selenocysteine insertion system in the model plant cell system, Chlamydomonas reinhardtii. EMBO J. 2002;21:3681–3693. doi: 10.1093/emboj/cdf372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merchant SS, et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koonin EV, Makarova KS, Aravind L. Horizontal gene transfer in prokaryotes: quantification and classification. Annu Rev Microbiol. 2001;55:709–742. doi: 10.1146/annurev.micro.55.1.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lobanov AV, Fomenko DE, Zhang Y, Sengupta A, Hatfield DL, Gladyshev VN. Evolutionary dynamics of eukaryotic selenoproteomes: large selenoproteomes may associate with aquatic life and small with terrestrial life. Genome Biol. 2007;8:R198. doi: 10.1186/gb-2007-8-9-r198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cobb NA. Nematodes and Their Relationships. Government Printing Office; Washington, DC: 1914. [Google Scholar]
- 25.Gladyshev VN, Krause M, Xu XM, Korotkov KV, Kryukov GV, Sun QA, Lee BJ, Wootton JC, Hatfield DL. Selenocysteine-containing thioredoxin reductase in C. elegans. Biochem Biophys Res Commun. 1999;259:244–249. doi: 10.1006/bbrc.1999.0765. [DOI] [PubMed] [Google Scholar]
- 26.Buettner C, Harney JW, Berry MJ. The Caenorhabditis elegans homologue of thioredoxin reductase contains a selenocysteine insertion sequence (SECIS) element that differs from mammalian SECIS elements but directs selenocysteine incorporation. J Biol Chem. 1999;274:21598–21602. doi: 10.1074/jbc.274.31.21598. [DOI] [PubMed] [Google Scholar]
- 27.Taskov K, Chapple C, Kryukov GV, Castellano S, Lobanov AV, Korotkov KV, Guigo R, Gladyshev VN. Nematode selenoproteome: the use of the selenocysteine insertion system to decode one codon in an animal genome? Nucleic Acids Res. 2005;33:2227–2238. doi: 10.1093/nar/gki507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mourier T, Pain A, Barrell B, Griffiths-Jones S. A selenocysteine tRNA and SECIS element in Plasmodium falciparum. RNA. 2005;11:119–122. doi: 10.1261/rna.7185605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lobanov AV, Delgado C, Rahlfs S, Novoselov SV, Kryukov GV, Gromer S, Hatfield DL, Becker K, Gladyshev VN. The Plasmodium selenoproteome. Nucleic Acids Res. 2006;34:496–505. doi: 10.1093/nar/gkj450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Novoselov SV, Lobanov AV, Hua D, Kasaikina MV, Hatfield DL, Gladyshev VN. A highly efficient form of the selenocysteine insertion sequence element in protozoan parasites and its use in mammalian cells. Proc Natl Acad Sci U S A. 2007;104:7857–7862. doi: 10.1073/pnas.0610683104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clark AG, et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- 32.Lobanov AV, Hatfield DL, Gladyshev VN. Selenoproteinless animals: selenophosphate synthetase SPS1 functions in a pathway unrelated to selenocysteine biosynthesis. Protein Sci. 2008;17:176–182. doi: 10.1110/ps.073261508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fomenko DE, Xing W, Adair BM, Thomas DJ, Gladyshev VN. High-throughput identification of catalytic redox-active cysteine residues. Science. 2007;315:387–389. doi: 10.1126/science.1133114. [DOI] [PubMed] [Google Scholar]