Summary
Chloroplast genetic engineering offers several advantages, including high levels of transgene expression, transgene containment via maternal inheritance and multigene engineering in a single transformation event. Entamoeba histolytica infects 50 million people, causing about 100 000 deaths annually, but there is no approved vaccine against this pathogen. LecA, a potential target for blocking amoebiasis, was expressed for the first time in transgenic plants. Stable transgene integration into chloroplast genomes and homoplasmy were confirmed by polymerase chain reaction and Southern blot analyses. LecA expression was evaluated by Western blots and quantified by enzyme-linked immunosorbent assay (up to 6.3% of total soluble protein or 2.3 mg LecA/g leaf tissue). Subcutaneous immunization of mice with crude extract of transgenic leaves resulted in higher immunoglobulin G titres (up to 1 : 10 000) than in previous reports. An average yield of 24 mg of LecA per plant should produce 29 million doses of vaccine antigen per acre of transgenic plants. Such high levels of expression and immunogenicity should facilitate the development of a less expensive amoebiasis vaccine.
Keywords: amoebiasis vaccine, genetically modified crops, plant-made vaccines, tobacco
Introduction
Amoebiasis caused by Entamoeba histolytica, an enteric protozoan parasite, ranks only second to malaria as a protozoan cause of death. The World Health Organization estimates that there are about 50 million cases of colitis and liver abscess annually and about 100 000 deaths each year from E. histolytica infection (Dodson et al., 1998; Huston and Petri, 1998; Petri et al., 2002). This infection occurs throughout the world, but predominantly in the developing countries of Central and South America, Africa and Asia.
Amoebic adherence and contact-dependent cytolysis of target cells are mediated by amoebic galactose/N-acetyl-d-galactosamine (Gal/GalNAc)-inhibitable adhesin (Mann and Lockhart, 1998). The Gal/GalNAc lectin is a heterodimer with disulphide-linked heavy (170-kDa) and light (35/31-kDa) subunits, which are non-covalently associated with an intermediate subunit of 150 kDa (Huston and Petri, 1998; Mann and Lockhart, 1998; Mann, 2002; Petri et al., 2002). The light subunit is encoded by multiple genes encoding isoforms with different post-translational modifications. The 35-kDa isoform is highly glycosylated and lacks the acylglycosylphosphatidylinositol (GPI) anchor present in the 31-kDa isoform (McCoy et al., 1993; Ramakrishnan et al., 2000). The carbohydrate recognition domain (CRD) was identified in the heavy subunit of the Gal/GalNAc lectin, and it has been demonstrated that an adherence-inhibitory antibody response against this domain protects against amoebic liver abscess in an animal model (Dodson et al., 1998). Preliminary studies have shown that the recombinant fragments of the cysteine-rich region of lectin (termed ‘LecA’) containing the CRD of Gal/GalNAc lectin confer protection against amoebiasis (Mann et al., 1993; Houpt et al., 2004).
Chloroplast genetic engineering offers several unique advantages, including high levels of transgene expression, multigene engineering in a single transformation event, appropriate post-translational modifications with the exception of glycosylation, low cost of production and maternal inheritance of the transgenes or containment via cytoplasmic male sterility (Daniell, 2002; Grevich and Daniell, 2005; Quesada-Vargas et al., 2005; Ruiz and Daniell, 2005). In addition, some of the challenges encountered in nuclear transgene expression, including the position effect (Daniell et al., 2001; Grevich and Daniell, 2005) and gene silencing at both the level of transcription (Lee et al., 2003; Dhingra et al., 2004) and translation (DeCosa et al., 2001), can be overcome in transgenic chloroplasts.
Tobacco is an ideal choice because of its ease of genetic manipulation, large biomass, prolific seed production and self-pollination (this provides the advantages of maternal inheritance and containment of the transgene via the chloroplast genome) (Watson et al., 2004; Koya et al., 2005). Vaccine antigens that have already been expressed via the chloroplast genome include the cholera toxin B-subunit (Daniell et al., 2001), the F1-V fusion antigen for plague (Singleton, 2003), the 2L21 peptide from the canine parvovirus (Molina et al., 2004, 2005), anthrax protective antigen (Watson et al., 2004; Koya et al., 2005), the C-terminus of Clostridium tetani (Tregoning et al., 2003), rotavirus VP6 protein (Birch-Machin et al., 2004) and NS3 protein as vaccine antigen for hepatitis C (Bhati, 2005).
Using the chloroplast transformation approach, several proteins have been expressed, including human elastin-derived polymers for various biomedical applications (Guda et al., 2000; Daniell et al., 2005a), magainin, a broad-spectrum topical agent, systemic antibiotic, wound-healing stimulant and potential anticancer agent (DeGray et al., 2001), human blood proteins, including human serum albumin (Fernandez-San Millan et al., 2003), interferon and insulin-like growth factor (Daniell et al., 2004a), human somatotropin (Staub et al., 2000) and interferon–β-glucuronidase (GUS) fusion proteins (Leelavathi and Reddy, 2003). In addition, the transformation of plastids in non-green tissue in cotton, soybean and carrot has recently been achieved (Dufourmantel et al., 2004; Kumar et al., 2004a; Daniell et al., 2005b). In particular, plastid transformation in carrot and lettuce (Kumar et al., 2004a,b; Lelivelt et al., 2005; Kanamoto et al., 2006) opens the door for the oral delivery of vaccine antigens.
The aforementioned observations suggest that chloroplast transformation may be an ideal system for the expression of vaccine antigens and other therapeutic proteins in large quantities. In this study, we report the expression of LecA, a surface antigen of E. histolytica, in transgenic chloroplasts, and the evaluation of the immunogenicity of the vaccine antigen in mice. This is the first report of LecA expression in any cellular compartment of transgenic plants.
Results
Construction of vectors
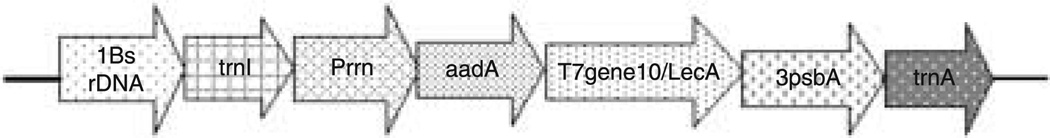
The pLD-SC vector (Figure 1) was derived from the universal transformation vector pLD-CtV (Daniell et al., 1998). The pLD-SC chloroplast transformation vector, containing the aadA gene, LecA coding region and 3′ psbA, integrates the transgene cassette into the trnI–trnA region of the chloroplast genome via homologous recombination. Integration of the transgene into one inverted repeat region facilitates integration into another inverted repeat via the copy correction mechanism. The T7 bacteriophage gene10 5′ untranslated region (UTR) containing the ribosome binding site (rbs) enhances the translation of LecA, and the psbA 3′ UTR present in the transgene cassette confers transcript stability. The chimeric, aminoglycoside-3′-adenlyl transferase (aadA) gene, conferring resistance to spectinomycin, was used as a selectable marker; its expression is driven by the 16S (Prrn) promoter (Svab and Maliga, 1993; Daniell, 1997; Kumar and Daniell, 2004).
Figure 1.
Schematic representation of pLD-SC. The pLD-SC tobacco transformation vector contains the trnI and trnA genes as flanking sequences for homologous recombination. The constitutive 16S rRNA promoter regulates the expression of the aadA gene (aminoglycoside-3′-adenylyltransferase) that confers resistance to spectinomycin– streptomycin and the gene10-LecA gene encoding the Entamoeba histolytica lectin antigen. Upstream of trnA, the vector contains the 3′ untranslated region (UTR), which is a transcript stabilizer, derived from the chloroplast psbA gene.
Polymerase chain reaction (PCR) confirmation of transgene integration in chloroplasts
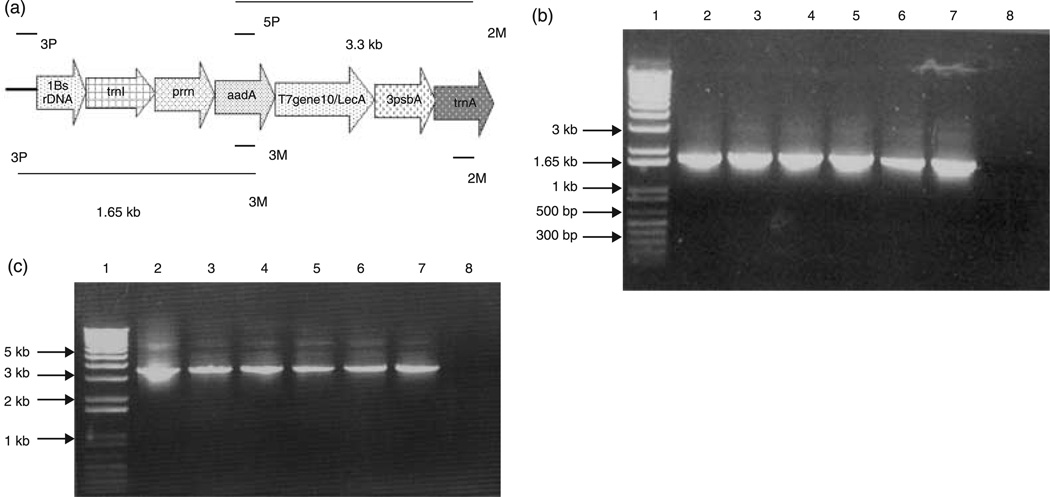
After the bombardment of tobacco leaves with pLD-SC plasmid-coated gold particles, about five shoots/plate appeared after a period of 5–6 weeks. True chloroplast transformants were distinguished from nuclear transformants and mutants by PCR. Two primers, 3P and 3M, were used to test for the chloroplast integration of transgenes (Daniell, 1997; Daniell et al., 2004b; Kumar and Daniell, 2004). The primer 3P landed on the native chloroplast DNA within the 16S rDNA gene and the primer 3M landed on the aadA gene, as shown in Figure 2a, thereby eliminating nuclear transformants and mutants. The primers 3P and 3M, on chloroplast integration of the transgene, yielded a 1.65-kb product as shown in Figure 2b.
Figure 2.
Polymerase chain reaction (PCR) analysis of wild-type and putative transformants of pLD-gene10-LecA. (a) Primers land within the native chloroplast genome (3P) or the aadA gene (3M) to yield a 1.65-kb product, and 5P/2M primers yield a 3.3-kb product. (b) Lane 1, 1-kb plus DNA ladder; lane 2, positive control (interferon clone); lanes 3–7, transgenic lines pLD-gene10-LecA (2, 6, 8*, 14, 17); lane 8, negative control (wild-type). (c) Lane 1, 1-kb plus DNA ladder; lane 2, positive control (pLD-gene10-LecA plasmid); lanes 3–7, transgenic lines pLD-gene10-LecA (2, 6, 8*, 14, 17); lane 8, negative control (wild-type).
The integration of the aadA gene, gene10-LecA gene and 3′ psbA cassette was confirmed using the 5P and 2M primer pair for PCR analysis. The 5P and 2M primers annealed to the internal regions of the aadA gene and the trnA gene, respectively, as shown in Figure 2a. The product size of the positive clone was 3.3 kb for LecA; the mutants and control should not show any product. Figure 2c shows the result of the 5P/2M PCR analysis. As spectinomycin was used to select for the true chloroplast transformants, any mutants obtained were spontaneous. After PCR analysis using both primer pairs, the transgenic plants were subsequently transferred through different rounds of selection to obtain a mature plant and to reach homoplasmy (containing only transformed chloroplasts).
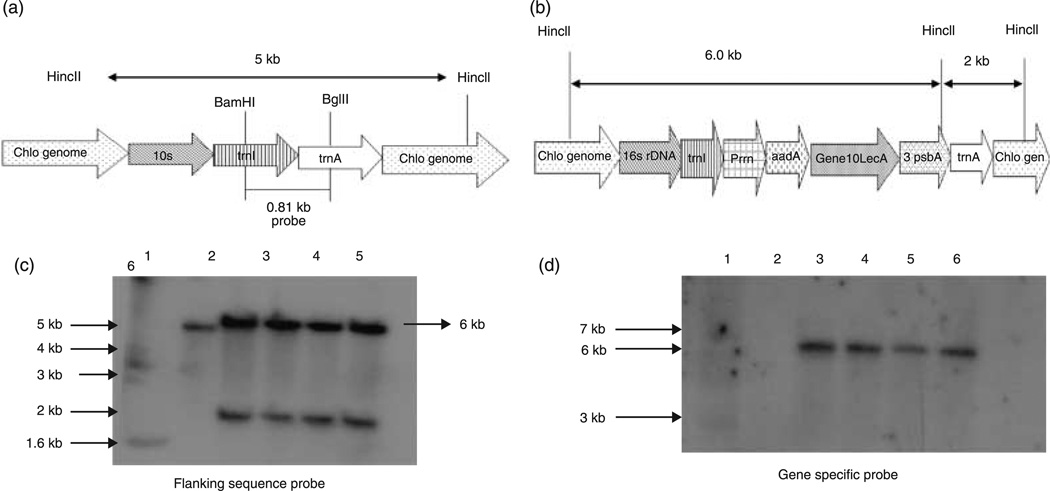
Evaluation of homoplasmy
The plants that tested positive for PCR analysis were moved through three rounds of selection, and were then tested by Southern blot analysis for site-specific integration of the transgene and homoplasmy. DNA of the fully regenerated clones growing in jars (third selection) was extracted and used for Southern blot analysis. A flanking sequence probe of 0.81 kb in size allowed the detection of the site-specific integration of the gene cassette into the chloroplast genome (Figure 3a). Figure 3b shows the HincII sites present outside the recombination region used for the restriction digestion of the plant DNA. The transformed chloroplast genome digested with HincII produced fragments of 6.0 and 2.0 kb for pLD-SC (Figure 3c), whereas the untransformed chloroplast genome that had been digested with HincII generated a 5.0-kb fragment. The flanking sequence probe also indicated whether homoplasmy of the chloroplast genome had been achieved through three rounds of selection. If homoplasmy (containing only transformed chloroplasts) had not been achieved, heteroplasmy (containing both transformed and untransformed chloroplasts) could result in changes in the relative ratios of the two genomes on cell division. The plants expressing LecA showed homoplasmy, as no hybridizing wild-type fragment was seen in transgenic lines. The gene-specific probe showed transgene integration, resulting in a fragment of 6 kb, as illustrated in Figure 3d.
Figure 3.
Southern blot analysis of pLD-gene10-LecA. Schematic diagram of the products expected from digestion of wild-type untransformed plants (a) and plants transformed with pLD-SC (b). (c) Southern blot with the flanking sequence probe of pLD-gene10-LecA transgenic plants showing homoplasmy. Lane 1, 1-kb plus DNA ladder; lane 2, wild-type; lanes 3–6, pLD-SC transgenic lines (8*, 17). (d) LecA gene-specific probe showing the presence of LecA in the transgenic plants. Lane 1, 1-kb plus DNA ladder; lane 2, wild-type; lanes 3–6, pLD-SC transgenic lines (8*, 17).
Expression of LecA in transgenic chloroplasts
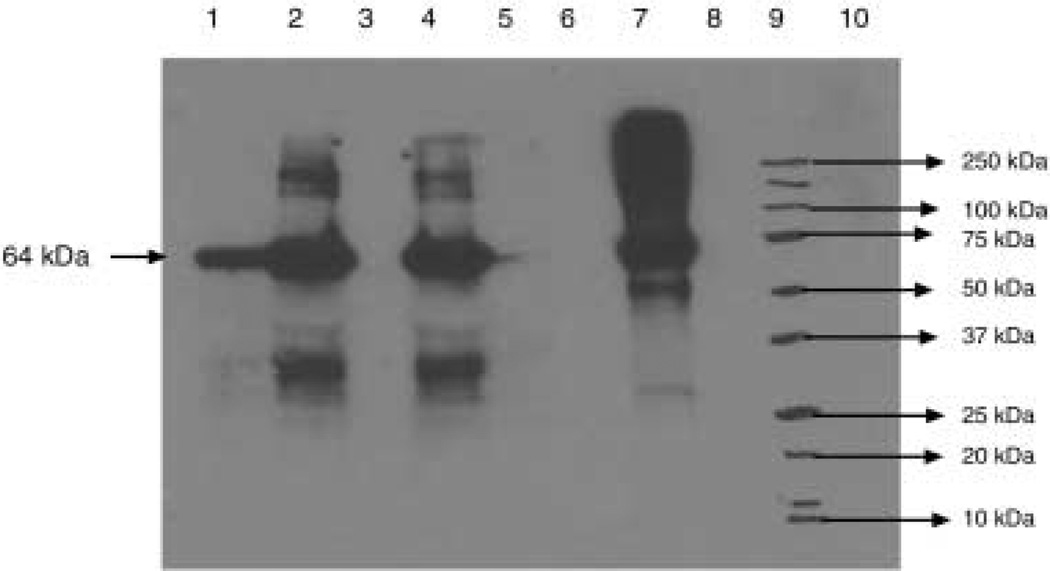
Goat anti-LecA polyclonal antibodies were used to detect the 64-kDa protein. The wild-type plant (‘Petit havana’) did not show any bands, indicating that the anti-LecA antibodies did not cross-react with any other proteins in the crude extract. The T1 generation plants showed good levels of expression (Figure 4). Each of the lanes contained around 1.5 µg of the LecA protein detected by the LecA antibodies. The lower bands seen are probably cleaved LecA protein and the higher bands are probably LecA protein aggregates.
Figure 4.
Immunoblot analysis of crude plant extracts expressing LecA. Lane 1, T1 generation transgenic plant (28 µg crude plant extract loaded); lanes 2 and 4, T0 generation transgenic plant (28 µg crude plant extract loaded); lane 6, wild-type; lane 7, standard protein (1 µg); lane 9, marker; lanes 3, 5, 8, 10, empty.
Quantification of transgenic plant-derived LecA protein
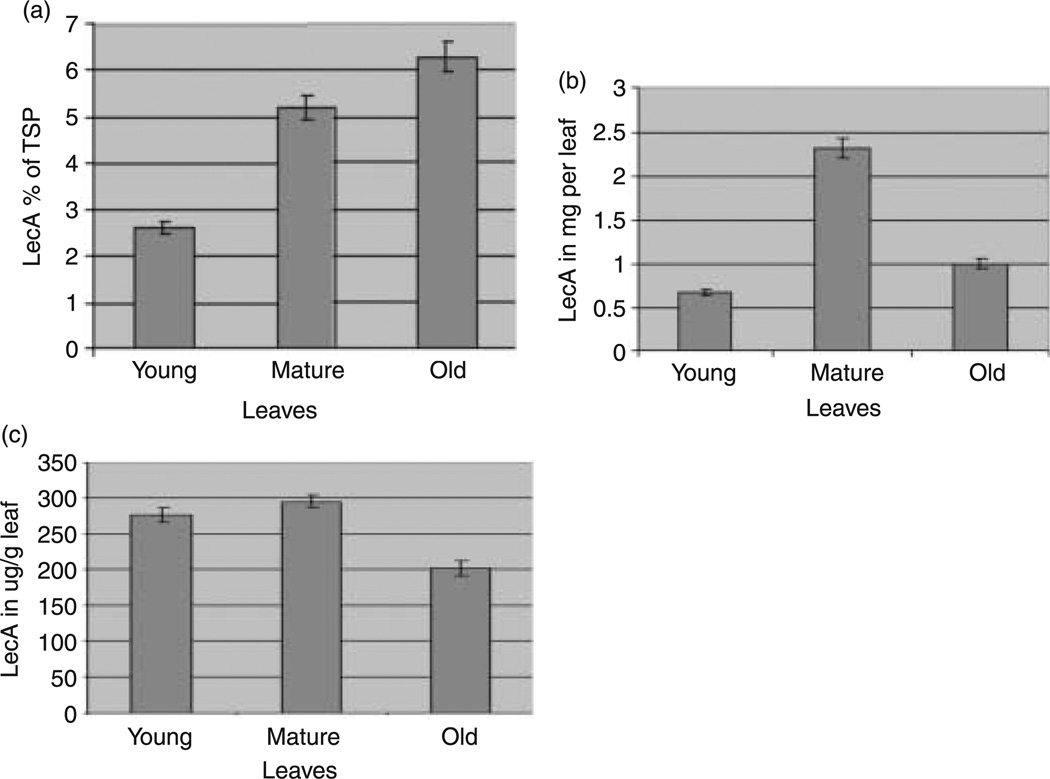
Different dilutions of purified LecA were used to obtain a standard curve. Goat polyclonal antibodies against LecA were used as the primary antibody, and the secondary antibody was peroxidase-conjugated rabbit anti-goat immunoglobulin G (IgG). The percentage of LecA, expressed as a percentage of total soluble protein (TSP), was calculated using the Bradford assay, i.e. the LecA percentage was inversely proportional to the TSP values. LecA expression reached maximum levels of 6.3% of TSP in old leaves, 2.6% of TSP in young leaves and 5.2% of TSP in mature leaves. Maximum LecA expression was observed in old leaves (Figure 5a). On the basis of the fresh weight calculations, the amounts of LecA obtained from young, mature and old leaves were 0.67, 2.32 and 1 mg per leaf, respectively (Figure 5b). Figure 5c shows the amount of LecA (µg) per milligram of leaf.
Figure 5.
Quantification of LecA expression levels in transgenic plants (T0 generation). (a) Expression levels, as percentage of total soluble protein (TSP), of LecA in young, mature and old leaves under regular illumination conditions (16 h light and 8 h dark period). (b) Amount of LecA (mg) obtained from young, mature and old leaves based on the fresh weight. (c) Amount of LecA (µg) obtained per milligram of leaf. The error bars shown in all figures indicate the deviation of the amount of LecA from the mean in leaves of different stages, where n = 5 is the number of samples used.
Evaluation of immunogenicity
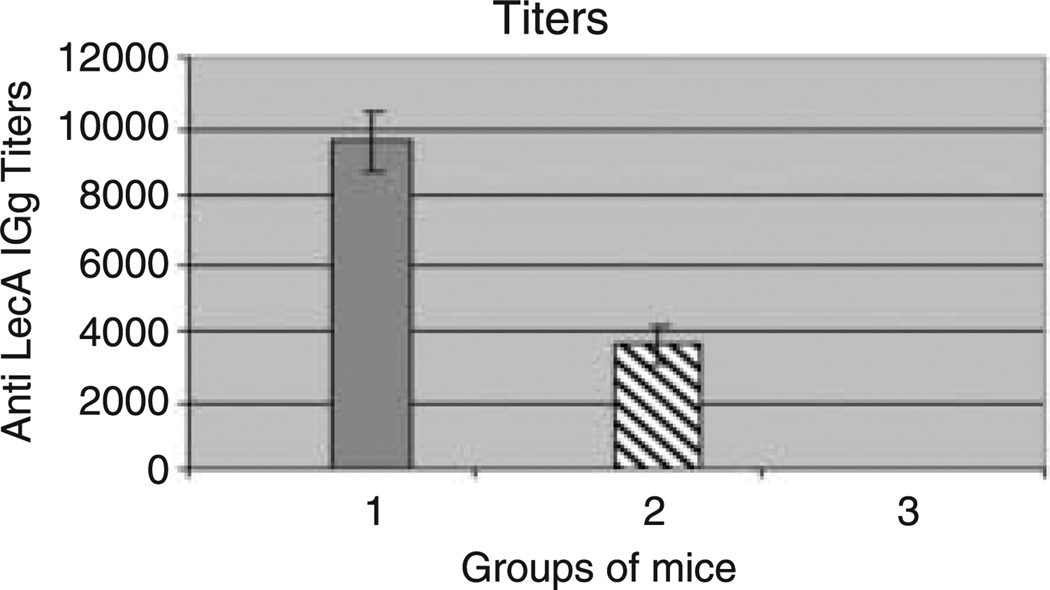
Having confirmed the expression of lectin in transgenic plants, we tested the ability of plant-derived lectin to be functional in vivo. For this, mice were immunized with crude extracts of the lectin-expressing plant. Mice groups immunized with crude extracts of the lectin-expressing plant together with adjuvant showed immunization titres up to 1 : 10 000, and mice groups immunized with crude extracts of the lectin-expressing plant with no adjuvant showed immunization titres up to 1 : 4000 (Figure 6).
Figure 6.
Comparison of immune responses in serum samples from mice after subcutaneous administration of: (1) leaf crude extract of lectin-expressing plant together with adjuvant (mean titres of 1 : 9600); (2) leaf crude extract of lectin-expressing plant with no adjuvant (mean titres of 1 : 3600); (3) leaf crude extract of wild-type plant (no immune titres). The error bars and statistical analysis represent the deviation of the titres from the mean values of the different groups of mice (n = 5).
Discussion
The pLD-SC vector was derived from the universal transformation vector pLD-CtV. The pLD-SC transgene cassette was integrated into the trnI–trnA region of the chloroplast genome via homologous recombination. Expression of the LecA recombinant protein in the chloroplast depends on several factors. First, the pLD-SC vector was designed to integrate into the inverted repeat region of the chloroplast genome via homologous recombination. The copy number of the transgene is thus doubled when integrated at this site. The increased copy number leads to increased transcript levels, resulting in higher protein accumulation (Grevich and Daniell, 2005; Quesada-Vargas et al., 2005). Second, the T7 bacteriophage gene10 5′ UTR containing the rbs and psbA-3′ UTR has been successfully used to enhance the translation of foreign proteins (Guda et al., 2000; Staub et al., 2000; Dhingra et al., 2004). Third, homoplasmy of the transgene is a condition in which all of the chloroplast genomes contain the transgene cassette. There are 100–1000 chloroplasts per cell and 100–1000 chloroplast genomes per chloroplast (Daniell et al., 2005b); for optimal production of the recombinant protein and transgene stability, it is essential that homoplasmy be achieved through several rounds of selection on medium containing spectinomycin. Fourth, expression can depend on the source of the gene and its relative AT/GC content. The prokaryotic-like chloroplast favours AT-rich sequences, which reflects the respective tRNA abundance. Therefore, the LecA gene having 67% AT was expected to express well in the chloroplast, similar to human serum albumin (66% AT). High-level expression of several other human proteins, including human somatotropin, interferon-α2b, interferon-α and insulin-like growth factor, shows that eukaryotic genes can be expressed well in transgenic chloroplasts (Staub et al., 2000; Fernandez-San Millan et al., 2003; Leelavathi and Reddy, 2003; Daniell et al., 2005a). Thus, genetic engineering of the chloroplast genome to express LecA achieves high expression levels and offers transgene containment.
PCR analysis was used to distinguish between the chloroplast transformants, nuclear transformants and mutants. Southern blot analysis was utilized to confirm the site-specific integration of the gene cassette and also to determine homo-or heteroplasmy. High protein expression levels were obtained in mature and old leaves of up to 6.3% of TSP, which was quantified using enzyme-linked immunosorbent assay (ELISA). The difference in LecA expression levels when calculated on the basis of the percentage of TSP vs. fresh weight is a result of the low TSP in old leaves relative to mature leaves. This may possibly be caused by the greater degradation of native soluble proteins relative to LecA in old leaves. On the basis of fresh weight, mature leaves showed higher expression levels, as TSP was not taken into account. A greater number of chloroplasts in mature leaves, their large size and the greater number of mature leaves per plant contribute to the higher expression. The correlation between leaf age and expression levels is very important. Plant cells in young leaves have fewer chloroplast genomes than those in mature leaves, and this is reflected in their expression levels of LecA. Plant cells in older leaves have higher levels of protease activity. Because of the lower levels of leaf protein in older leaves, it appears that the LecA expression level is high when calculated on the basis of TSP, but it is actually lower than that of mature leaves when calculated on the basis of fresh weight. When evaluating the total amount of LecA produced in a plant, the age of the leaf is very important.
An average yield of 24 mg of LecA (Table 1) per plant should produce 29 million doses of vaccine antigen per acre of transgenic plants. The dosages per acre were calculated on the basis of subcutaneous delivery only. In the case of the anthrax vaccine, the same amount of vaccine antigen (5 µg) is used for subcutaneous delivery for mice and humans (Koya et al., 2005). It is possible that, in the amoebiasis vaccine, a higher dosage may be used for humans. The use of plants for the production of vaccine antigens could therefore result in low-cost vaccine when compared with other expression systems.
Table 1.
Yield of LecA expressed in pLD-SC tobacco T0 transgenic lines relative to the biomass
| Leaves per plant |
Average weight (mg) of leaf |
LecA (mg/g) in fresh leaf |
Amount of LecA (mg)/leaf |
Amount of LecA (mg) per age group |
|
| Young | 3.2 | 2.5 | 0.27 | 0.67 | 2.144 |
| Mature | 7.8 | 8 | 0.29 | 2.32 | 18.096 |
| Old | 4.5 | 5 | 0.20 | 1 | 4.05 |
| Total recombinant LecA/plant | 24.29 |
Calculations: 8000 plants are grown, giving 8000 × 24.29 = 194 320 mg. Based on three cuttings per year, the total yield would be 582 960 mg. With an average loss of 50% during purification, the net protein yield would be 291 480 mg. The amount of lectin for a single dose of vaccine is 10 µg. At this dose, 291 480 mg should give 29 148 000 doses or about 29 million doses. Therefore, 29 million doses may be obtained from one acre of tobacco.
Differences in the titre values of the groups of mice receiving the extract with and without adjuvant were caused by a depot effect (Audibert, 2003) and by alhydrogel’s non-specific priming of the immune system. Control mice immunized with wild-type plant leaf crude extract did not show any immune response, demonstrating the specificity of the recombinant lectin-elicited immune response in the case of transgenic plant crude extracts.
Having confirmed the expression of lectin in transgenic plants, we tested the ability of plant-derived lectin to be functional in vivo. Therefore, mice were immunized with crude extracts of the lectin-expressing plant. Mice groups immunized with crude extracts of the lectin-expressing plant together with adjuvant showed immunization titres up to 1 : 10 000, and mice groups immunized with crude extracts of the lectin-expressing plant with no adjuvant showed immunization titres up to 1 : 4000 (Figure 6). Previous reports of vaccination with full-length native lectin antigen in gerbils through the subcutaneous route yielded titres up to 1 : 1024 (Lotter et al., 2000). Similarly, vaccination in gerbils with a 25-mer peptide derived from the cysteine-rich region of lectin yielded IgG titres up to 1 : 200 (Lotter et al., 2000). In this study, vaccination with crude extracts of LecA transgenic lines resulted in titres up to 1 : 4000. This represents a 4–20-fold higher immunogenicity than that obtained with purified full-length native lectin antigen or peptide derived from the cysteine-rich region of lectin, although care should be taken when comparing results between different animal models. However, the authors would like to stress that the antibody titres are meaningful. The antibody titres raised against lectin should confer protective immunity. Lectin helps E. histolytica to bind to the host cell receptor. The antibody raised against lectin should prevent E. histolytica from entering the host tissue by blocking the lectin ligand. Therefore, it can be anticipated that the antibody titres should be protective when challenged with the pathogen. We could not perform pathogen challenge tests because BALB/c mice are not susceptible to infection with E. histolytica. Our aim of immunization in this study was to test the ability of plant-derived LecA to elicit the IgG immune response.
In addition, for pathogen challenge studies, the production of IgA antibodies would be more effective than the production of IgG antibodies, because infection occurs predominantly in the intestinal mucosa, where secretory IgA plays a predominant role in effectively neutralizing the infection. Therefore, future investigations should be performed to determine the immunogenicity of orally administered (in order to elicit the IgA response) LecA-expressing plant, followed by pathogen challenge. In this context, the development of plastid transformation systems in lettuce and carrot (Kumar et al., 2004a, b; Lelivelt et al., 2005; Kanamoto et al., 2006) and the demonstration of receptor-mediated oral delivery of foreign proteins expressed in transgenic chloroplasts into the circulatory system (Limaye et al., 2006) are significant developments in this field. The development of transgenic carrot or lettuce expressing LecA should open the door for the oral delivery of the vaccine and the development of a mucosal immune response. An ideal vaccine for amoebiasis should induce both mucosal and systemic protection. If both subcutaneous and oral delivery proves to be immunoprotective, priming both the mucosal and systemic systems may prove to be the cheapest and most effective method of vaccination against any pathogen that attacks both the mucosal and systemic systems.
Experimental procedures
Construction of vectors for the transformation of tobacco chloroplasts
The plasmid pcDNA 3.1 containing the LecA gene, provided by Dr Barbara Mann (University of Virginia Health System, Charlottesville, VA, USA), was used as the template to introduce start and stop codons at the N-terminal and C-terminal of the LecA gene. The primers used were as follows: forward, 5′-GGAATTGAATTCCATATGTGTGAGAACAGA-3′ reverse, 5′-AGAATTGCCTCTAGACTATTCTGAAAC-3′. The PCR product was purified using a PCR purification kit (Qiagen, Valencia, CA) and was subcloned into the TOPO vector, pCR2.1-LecA. The PCR product was digested from the pCR2.1-LecA vector with NdeI and NotI enzymes and subcloned into pBluescript (Stratagene, La Jolla, CA, USA) containing gene10 T7 bacteriophage UTR, designated pBS-g10-LecA. The final product containing gene10 and the LecA gene (~1.8 kb) was digested with HincII and NotI enzymes and subcloned into the tobacco universal vector pLD-Ctv (Daniell et al., 1998) between EcoRV and NotI sites.
Bombardment and selection of transgenic shoots
Bombardment and selection of transgenic shoots were performed as described previously (Svab and Maliga, 1993; Daniell, 1997; Kumar and Daniell, 2004). A Bio-Rad PDS-1000/He biolistic device (Bio-Rad, Hercules, CA), which is a particle delivery system, was used to bombard leaves of Nicotiana tabacum var. Petit havana. Bombarded leaves, after 2 days of incubation, were transferred to RMOP (Regeneration Medium of Plants) medium containing 500 µg/mL of spectinomycin. After 4–6 weeks, the shoots were transferred to fresh RMOP plus spectinomycin for the second round of selection. Finally, after 4 weeks on the second round of selection, the shoots were transferred to jars containing MSO (Murashige and Skoog medium with no hormones) medium with 500 µg/mL spectinomycin.
Confirmation of transgene integration into the chloroplast genome
To confirm the transgene cassette integration into the chloroplast genome, PCR was performed using the primer pairs 3P (5′-AAAACCCGTCCTCGTTCGGATTGC-3′) and 3M (5′-CCGCGTTGTTTCATCAAGCCTTACG-3′). To confirm the integration of the gene of interest, PCR was performed using the primer pairs 5P (5′-CTGTAGAAGTCACCATTGTTGTGC-3′) and 2M (5′-GACTGCCCACCTGAGAGCGGACA-3′). Positive (known transgenic plant DNA sample) and negative (wild-type Petit havana DNA sample) controls were used to monitor PCR. The PCR conditions used have been described previously (Daniell, 1997; Daniell et al., 2004b; Kumar and Daniell, 2004). The PCR product was analysed on 0.8% agarose gel.
Southern blot analysis
Total plant DNA was extracted from transgenic T0 plants and from untransformed tobacco plants using a Qiagen DNeasy Plant Mini Kit. Total plant DNA was digested with HincII and probed by the flanking sequence probe, which was obtained from the pUC-Ct vector by digesting with BamHI and BglII to obtain a 0.81-kb fragment. The gene-specific probe (length, 400 bp) was obtained by digesting pLD-SC with BglII and PvuII. The probes were prepared by random primed 32P-labelling (Ready-to-go DNA labelling beads, Amersham Pharmacia, Arlington Heights, IL). The probes were hybridized to the membrane using Stratagene Quick-hyb solution and protocol, as described previously (Daniell, 1997; Daniell et al., 2004b; Kumar and Daniell, 2004). The radiolabelled blots were exposed to X-ray films and developed in an X-ray film processor.
Western blot analysis
Protein was extracted from 100 mg of plant leaf tissue from both untransformed and transformed plants, as described previously (Daniell, 1997; Daniell et al., 2004b; Kumar and Daniell, 2004). Protein extracts mixed with sample loading buffer containing BME (beta mercaptoethanol) were boiled for 5 min and loaded into 10% sodium dodecylsulphate-polyacrylamide gel electrophoresis (SDS-PAGE) gel. The Chloroplast-derived vaccine antigen for amoebiasis separated proteins were transferred to a membrane and blocked with P-T-M [phosphate-buffered saline (PBS): 12 mm Na2HPO4, 3.0 mm NaH2PO4·H2O, 145 mm NaCl, pH 7.2; 0.5% Tween 20; 3% dry milk], as described previously (Daniell, 1997; Daniell et al., 2004b; Kumar and Daniell, 2004). LecA was detected by goat anti-LecA antibody, and the secondary antibody was anti-goat IgG antibody conjugated with horseradish peroxidase (Sigma, St. Louis, MO, USA).
Estimation of TSP
The Bradford assay was used to determine the total protein from the plant extracts. Ground leaf tissue (100 mg) from transformed and untransformed plants was used to extract proteins. Bovine serum albumin (BSA) standards ranged from 0.05 to 0.5 µg/µL. Plant extracts were diluted 1 : 10 and 1 : 20, and 10 µL of each standard and 10 µL of each plant dilution were added to the wells of a 96-well microtitre plate (Cell star, Greiner Bio-one, Stonehouse, UK) in duplicate. Bradford reagent (Bio-Rad protein assay) was diluted 1 : 4 with distilled water as specified, and 200 µL was added to each well and assayed as described previously (Daniell, 1997; Daniell et al., 2004b; Kumar and Daniell, 2004). The absorbance was read at 630 nm. Comparison of the absorbance of known amounts of BSA with that of the samples was used to estimate the amount of total protein.
ELISA
The quantification of LecA in the plant crude extract was performed using ELISA (Daniell, 1997; Daniell et al., 2004b; Kumar and Daniell, 2004). Transgenic (100 mg, young, mature, old) and wild-type (young, mature, old) leaf samples exposed to a regular lighting pattern (16 h light and 8 h dark) were used. Standards ranging from 100 to 1000 ng/mL were prepared from purified LecA. Standards and protein samples (100 µL) were coated on to a 96-well polyvinylchloride micro-titre plate (Cell star) and assayed as described previously (Daniell, 1997; Daniell et al., 2004b; Kumar and Daniell, 2004). Goat anti-LecA primary antibody (provided by Dr Mann, University of Virginia) and horseradish peroxidase-conjugated rabbit anti-goat IgG secondary antibody (1 : 5000; American Qualex, San Clemente, CA) were used.
Immunization of mice with plant-derived lectin antigen
Three groups of five female 6–7-week-old BALB/c mice were injected subcutaneously with plant crude extracts on days 0, 15, 30 and 45. Group 1 mice were injected with lectin (10 µg)-expressing plant crude extracts together with 50 µL of alhydrogel adjuvant. Group 2 mice were injected with lectin (10 µg)-expressing plant crude extracts with no adjuvant. Group 3 mice were injected with plant crude extracts of wild-type tobacco. Blood was drawn from the retro-orbital plexus 15 days after the final dose (i.e. on day 60). The blood samples were stored undisturbed for 2 h at room temperature, and were then centrifuged at 3000 g for 10 min to extract the serum.
ELISA to detect anti-LecA IgG antibodies in the serum samples
Ninety-six-well microtitre ELISA plates were coated with 100 µL/well of purified Escherichia coli-derived lectin standard obtained at a concentration of 2.0 µg/mL in PBS, pH 7.4. The plates were stored overnight at 4 °C. Mouse serum samples were serially diluted (1 : 100–1 : 20 000). Plates were incubated with 100 µL of diluted serum samples for 1 h at 37 °C, followed by washing with PBS-Tween. The plates were then incubated for 1 h at 37 °C with 100 µL of horseradish peroxidase-conjugated goat anti-mouse IgG (1 : 5000 dilution of 1 mg/mL stock). TMB (3,3′,5,5′-tetramethyl benzidine; American Qualex) was used as the substrate and the reaction was stopped by the addition of 50 µL of 2 m sulphuric acid. The plates were read on a plate reader (Dynex Technologies, Chantilly, VA) at 450 nm. Titre values were calculated using a cut-off value equal to an absorbance difference of 0.5 between immunized and non-immunized mice.
Acknowledgements
This investigation was supported in part by United States Department of Agriculture (USDA) 3611-21000-017-00D and National Institutes of Health (NIH) R01GM63879 awards to Henry Daniell. The authors are grateful to Dr Barbara J. Mann (Departments of Internal Medicine and Microbiology, University of Virginia Health System, Charlottesville, VA 22908-1340, USA) for her support throughout this investigation, and for the plasmid pcDNA 3.1 containing the LecA gene, purified LecA protein and goat anti-LecA primary antibody. The technical assistance provided by Mr Vijay Koya for mice immunization studies is also gratefully acknowledged.
References
- Audibert F. Adjuvants for vaccines, a quest. Int. Immunopharmacol. 2003;3:1187–1193. doi: 10.1016/S1567-5769(03)00011-0. [DOI] [PubMed] [Google Scholar]
- Bhati A. Master’s Thesis. Orlando, FL: University of Central Florida; 2005. Expression of hepatitis C viral non-structural 3 protein in transgenic chloroplasts. [Google Scholar]
- Birch-Machin I, Newell CA, Hibberd JM, Gray JC. Accumulation of rotavirus VP6 protein in chloroplasts of transplastomic tobacco is limited by protein stability. Plant Biotechnol. J. 2004;2:261–270. doi: 10.1111/j.1467-7652.2004.00072.x. [DOI] [PubMed] [Google Scholar]
- Daniell H. Transformation and foreign gene expression in plants mediated by micro projectile bombardment. Methods Mol. Biol. 1997;62:453–488. doi: 10.1385/0-89603-480-1:463. [DOI] [PubMed] [Google Scholar]
- Daniell H. Molecular strategies for gene containment in transgenic crops. Nat. Biotechnol. 2002;20:581–587. doi: 10.1038/nbt0602-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Datta R, Varma S, Gray S, Lee SB. Containment of herbicide resistance through genetic engineering of the chloroplast genome. Nat. Biotechnol. 1998;16:345–348. doi: 10.1038/nbt0498-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Lee SB, Panchal T, Wiebe PO. Expression of cholera toxin B subunit gene and assembly as functional oligomers in transgenic tobacco chloroplasts. J. Mol Biol. 2001;311:1001–1009. doi: 10.1006/jmbi.2001.4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Carmona-Sanchez O, Burns B. Chloroplast derived antibodies, biopharmaceuticals and edible vaccines. In: Fischer R, Schillberg S, editors. Molecular Farming. Weinheim: Wiley-VCH-Verlag; 2004a. pp. 113–133. [Google Scholar]
- Daniell H, Dhingra A, Ruiz ON. Chloroplast genetic engineering to confer desired plant traits. Methods Mol. Biol. 2004b;286:111–137. doi: 10.1385/1-59259-827-7:111. [DOI] [PubMed] [Google Scholar]
- Daniell H, Chebolu S, Kumar S, Singleton M, Falconer R. Chloroplast-derived vaccine antigens and other therapeutic proteins. Vaccine. 2005a;23:1779–1783. doi: 10.1016/j.vaccine.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Daniell H, Kumar S, Dufourmantel N. Breakthrough in chloroplast genetic engineering of agronomically important crops. Trends Biotechnol. 2005b;23:238–245. doi: 10.1016/j.tibtech.2005.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCosa B, Moar W, Lee SB, Miller M, Daniell H. Over expression of the Btcry2Aa2 operon in chloroplasts leads to formation of insecticidal crystals. Nat. Biotechnol. 2001;19:71–74. doi: 10.1038/83559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGray G, Rajasekaran K, Smith F, Sanford J, Daniell H. Expression of an antimicrobial peptide via the chloroplast genome to control phytopathogenic bacteria and fungi. Plant Physiol. 2001;127:852–862. [PMC free article] [PubMed] [Google Scholar]
- Dhingra A, Portis AR, Daniell H. Enhanced translation of a chloroplast-expressed RbcS gene restores small subunit levels and photosynthesis in nuclear antisense RbcS plants. Proc. Natl. Acad. Sci. USA. 2004;101:6315–6320. doi: 10.1073/pnas.0400981101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson JM, Lenkowski PW, Jr, Eubanks AC, Jackson TFGH, Napodano J, Lyerly DM, Lockhart LA, Mann BJ, Petri WA., Jr Infection and immunity mediated by the carbohydrate recognition domain of the Entamoeba histolytica Gal/GalNAc lectin. J. Infect. Dis. 1998;179:460–466. doi: 10.1086/314610. [DOI] [PubMed] [Google Scholar]
- Dufourmantel N, Pelissier B, Garcon F, Peltier G, Ferullo JM, Tissot G. Generation of fertile transplastomic soybean. Plant Mol. Biol. 2004;55:479–489. doi: 10.1007/s11103-004-0192-4. [DOI] [PubMed] [Google Scholar]
- Fernandez-San Millan A, Mingeo-Castel AM, Miller M, Daniell H. A chloroplast transgenic approach to hyper-express and purify human serum albumin, a protein highly susceptible to proteolytic degradation. Plant Biotechnol. J. 2003;1:71–79. doi: 10.1046/j.1467-7652.2003.00008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grevich JJ, Daniell H. Chloroplast genetic engineering: recent advances and future perspectives. Crit. Rev. Plant Sci. 2005;24:83–108. [Google Scholar]
- Guda C, Lee SB, Daniell H. Stable expression of biodegradable protein based polymer in tobacco chloroplasts. Plant Cell Rep. 2000;19:257–262. doi: 10.1007/s002990050008. [DOI] [PubMed] [Google Scholar]
- Houpt E, Barroso L, Lockhart L, Wright R, Cramer C, Lyerly D, Petri WA., Jr Prevention of intestinal amebiasis by vaccination with Entamoeba histolytica Gal/GalNAc lectin. Vaccine. 2004;22:611–617. doi: 10.1016/j.vaccine.2003.09.003. [DOI] [PubMed] [Google Scholar]
- Huston CD, Petri WA., Jr Host–pathogen interaction in amebiasis and progress of vaccine development. Eur. J. Clin. Microbiol. Infect. Dis. 1998;17:601–614. doi: 10.1007/BF01708342. [DOI] [PubMed] [Google Scholar]
- Kanamoto H, Yamashita A, Asao H, Takase OS, Hattori H, Yokata A, Tomizawa K. Efficient and stable transformation of Lactuca sativa L. cv. Cisco (lettuce) plastids. Transgenic Res. 2006;15:205–217. doi: 10.1007/s11248-005-3997-2. [DOI] [PubMed] [Google Scholar]
- Koya V, Moayeri M, Leppla S, Daniell H. Plant based vaccine: mice immunized with chloroplast-derived anthrax protective antigen survive anthrax lethal toxin challenge. Infect. Immun. 2005;73:8266–8274. doi: 10.1128/IAI.73.12.8266-8274.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Daniell H. Engineering the chloroplast genome for hyper-expression of human therapeutic proteins and vaccine antigens in recombinant protein protocols. Methods Mol. Biol. 2004;267:365–383. doi: 10.1385/1-59259-774-2:365. [DOI] [PubMed] [Google Scholar]
- Kumar S, Dhingra A, Daniell H. Plastid-expressed betaine aldehyde dehydrogenase gene in carrot cultured cells, roots, and leaves confers enhanced salt tolerance. Plant Physiol. 2004a;136:2843–2854. doi: 10.1104/pp.104.045187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Dhingra A, Daniell H. Stable transformation of the cotton plastid genome and maternal inheritance of transgenes. Plant Mol. Biol. 2004b;56:203–216. doi: 10.1007/s11103-004-2907-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SB, Kwon HB, Kwon SJ, Park SC, Jeong MJ, Han SE, Byun MO, Daniell H. Accumulation of trehalose within transgenic chloroplasts confers drought tolerance. Mol. Breed. 2003;11:1–13. [Google Scholar]
- Leelavathi S, Reddy VS. Chloroplast expression of His-tagged GUS fusions: a general strategy to overproduce and purify foreign proteins using transplastomic plants as bioreactors. Mol. Breed. 2003;11:49–58. [Google Scholar]
- Lelivelt CL, McCabe MS, Newell CA, Desnoo CB, van Dun KM, Birch-Machin I, Gray JC, Mills KH, Nugent JM. Stable plastid transformation in lettuce (Lactuca sativa L.) Plant Mol. Biol. 2005;58:763–774. doi: 10.1007/s11103-005-7704-8. [DOI] [PubMed] [Google Scholar]
- Limaye A, Koya V, Samsam M, Daniell H. Receptor mediated oral delivery of green fluorescent protein expressed in transgenic chloroplasts into the mouse circulatory system. FASEB J. 2006;20:1–10. doi: 10.1096/fj.05-5134fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotter H, Khajawa F, Stanley SL, Jr, Tannich E. Protection of gerbils from amebic liver abscess by vaccination with a 25-mer peptide derived from the cysteine-rich region of Entamoeba histolytica galactose-specific adherence lectin. Infect. Immun. 2000;68:4416–4421. doi: 10.1128/iai.68.8.4416-4421.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann BJ. Structure and function of the Entamoeba histolytica Gal/GalNAc lectin. Int. Rev. Cyt. 2002;216:59–80. doi: 10.1016/s0074-7696(02)16003-7. [DOI] [PubMed] [Google Scholar]
- Mann BJ, Lockhart LA. Molecular analysis of the Gal/GalNAc adhesin of Entamoeba histolytica. J. Euk. Microbiol. 1998;45:13S–16S. doi: 10.1111/j.1550-7408.1998.tb04518.x. [DOI] [PubMed] [Google Scholar]
- Mann BJ, Chung CY, Dodson JM, Ashley LS, Braga LL, Snodgrass TL. Neutralizing monoclonal antibody epitopes of the Entamoeba histolytica galactose adhesin map to the cysteine-rich extra cellular domain of the 170-kilodalton subunit. Infect. Immun. 1993;61:1772–1778. doi: 10.1128/iai.61.5.1772-1778.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy JJ, Mann BJ, Vedvick TS, Pak Y, Heimark DB, Petri WA., Jr Structural analysis of the light subunit of the Entamoeba histolytica galactose-specific adherence lectin. J. Biol. Chem. 1993;268:24 223–24 231. [PubMed] [Google Scholar]
- Molina A, Herva-Stubbs S, Daniell H, Mingo-Castel AM, Veramendi J. High yield expression of a viral peptide animal vaccine in transgenic tobacco chloroplasts. Plant Biotechnol. J. 2004;2:141–153. doi: 10.1046/j.1467-7652.2004.00057.x. [DOI] [PubMed] [Google Scholar]
- Molina A, Veramendi J, Hervas-Stubbs S. Induction of neutralizing antibodies by a tobacco chloroplast-derived vaccine based on a B cell epitope from canine parvovirus. Virology. 2005;342:266–275. doi: 10.1016/j.virol.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Petri WA, Jr, Haque R, Mann BJ. The bittersweet interface of parasite and host: lectin–carbohydrate interactions during human invasion by the parasite Entamoeba histolytica. Annu. Rev. Microbiol. 2002;56:39–64. doi: 10.1146/annurev.micro.56.012302.160959. [DOI] [PubMed] [Google Scholar]
- Quesada-Vargas T, Ruiz ON, Daniell H. Characterization of heterologous multigene operons in transgenic chloroplasts: transcription, processing, and translation. Plant Physiol. 2005;138:1746–1762. doi: 10.1104/pp.105.063040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan G, Lee S, Mann BJ, Petri WA., Jr Entamoeba histolytica: deletion of the GPI anchor signal sequence on the Gal/GalNAc lectin light subunit prevents its assembly into the lectin heterodimer. Exp. Parasitol. 2000;96:57–60. doi: 10.1006/expr.2000.4543. [DOI] [PubMed] [Google Scholar]
- Ruiz ON, Daniell H. Engineering cytoplasmic male sterility via the chloroplast genome by the expression of β-ketothiolase. Plant Physiol. 2005;138:1232–1246. doi: 10.1104/pp.104.057729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton ML. Master’s Thesis. Orlando, FL: University of Central Florida; 2003. Expression of CaF1 and LcrV as a fusion protein for development of a vaccine against Yersisnia pestis via chloroplast genetic engineering. [Google Scholar]
- Staub JM, Garcia B, Graves J, Hajduklewicz PTJ, Hunter P, Nehra N, Paradkar V, Schlitter M, Carroll JA, Spatola L, Ward D, Ye G, Russell DA. High-yield production of a human therapeutic protein in tobacco chloroplasts. Nat. Biotechnol. 2000;8:333–338. doi: 10.1038/73796. [DOI] [PubMed] [Google Scholar]
- Svab Z, Maliga P. High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc. Natl. Acad. Sci. USA. 1993;90:913–917. doi: 10.1073/pnas.90.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tregoning JS, Nixon P, Kuroda H, Svab Z, Clare S, Bowe F, Fairweather N, Ytterberg J, Wijk KJV, Dougan G, Maliga P. Expression of tetanus toxin Fragment C in tobacco chloroplasts. Nucleic Acids Res. 2003;31:1174–1179. doi: 10.1093/nar/gkg221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson J, Koya V, Leppla S, Daniell H. Expression of Bacillus anthracis protective antigen in transgenic chloroplasts of tobacco, a non-food/feed crop. Vaccine. 2004;22:4374–4384. doi: 10.1016/j.vaccine.2004.01.069. [DOI] [PMC free article] [PubMed] [Google Scholar]