Abstract
Invertebrate species possess one or two Na+ channel genes, yet there are 10 in mammals. When did this explosive growth come about during vertebrate evolution? All mammalian Na+ channel genes reside on four chromosomes. It has been suggested that this came about by multiple duplications of an ancestral chromosome with a single Na+ channel gene followed by tandem duplications of Na+ channel genes on some of these chromosomes. Because a large-scale expansion of the vertebrate genome likely occurred before the divergence of teleosts and tetrapods, we tested this hypothesis by cloning Na+ channel genes in a teleost fish. Using an approach designed to clone all of the Na+ channel genes in a genome, we found six Na+ channel genes. Phylogenetic comparisons show that each teleost gene is orthologous to a Na+ channel gene or gene cluster on a different mammalian chromosome, supporting the hypothesis that four Na+ channel genes were present in the ancestors of teleosts and tetrapods. Further duplications occurred independently in the teleost and tetrapod lineages, with a greater number of duplications in tetrapods. This pattern has implications for the evolution of function and specialization of Na+ channel genes in vertebrates. Sodium channel genes also are linked to homeobox (Hox) gene clusters in mammals. Using our phylogeny of Na+ channel genes to independently test between two models of Hox gene evolution, we support the hypothesis that Hox gene clusters evolved as (AB) (CD) rather than {D[A(BC)]}.
The evolution of voltage-dependent Na+ channels in metazoans permitted long-distance propagation of action potentials (1). For many years this was still believed to be their main function. Recently, it has been shown that Na+ currents participate in shaping and filtering synaptic inputs, back-propagation of dendritic action potentials (which can have enormous consequences for associative synaptic plasticity), and initiation and maintenance of cellular oscillations and burst generation (2–5). Sodium currents also have been implicated in various forms of developmental and regulatory plasticity (6, 7). Last, they are the locus of a number of mutations resulting in a variety of muscle, cardiac, and neural diseases (8).
Sodium channel genes have been cloned and sequenced in a number of phyla, and an interesting pattern has emerged: whereas only a single or a few genes are reported in invertebrate species (9, 10), 10 distinct Na+ channel genes have been identified in mammals (8, 11, 12). In situ hybridization and channel-specific antibodies (13–17) have revealed cell-specific expression and subcellular localization of many of these mammalian Na+ channels leading to a more complex picture of Na+ channel function in the mammalian brain. When this proliferation of Na+ channel genes and the differential localization of their products occurred during vertebrate evolution is unknown.
In humans, all 10 Na+ channel genes reside on four chromosomes with clusters of three and five Na+ channel genes respectively on human chromosomes 3 and 2. Plummer and Meisler (11) suggested that this pattern arose when an ancestral chromosome with a single Na+ channel gene underwent a series of duplications during a hypothetical large-scale expansion of the genome (18–20). Such a series of duplications would have resulted in four orthologous genes, each on a different chromosome. The clusters of Na+ channel genes on chromosomes 2 and 3 likely resulted from tandem duplications on those chromosomes that occurred later (11).
These hypotheses can only be tested by analyzing Na+ channel genes in pivotal nonmammalian species. However, little information is available on Na+ channel genes in nonmammalian vertebrates. Toward this end, we systematically cloned as many Na+ channels as possible in a single species of teleost fish, the weakly electric fish Sternopygus macrurus. We selected a teleost because the wide-scale chromosomal duplication events likely occurred before the divergence of teleosts and tetrapods (18, 19). We selected this species because there is good background literature on sodium currents in electric fish (3, 21–23).
Methods
This study was carried out by using the National Institutes of Health and University of Texas guidelines for animal care. All procedures were reviewed by the University of Texas Institutional Animal Care and Use Committee.
Tissue was excised under a dissecting microscope, immediately placed in liquid nitrogen, and then stored at −80°C until nucleic acid extraction. Genomic DNA was isolated from ≈100 mg of brain (24). Total RNA was extracted as described in Ausubel et al. (24) “The single-step RNA isolation method.” Extracts were analyzed by electrophoresis and spectrophotometry (OD260/OD280) to determine concentration and evaluate quality. The following primers were used for PCR analysis: primer 1, TGCTSGTGTGYYTGATYTTCTGG; primer 2, TACATRATWTCCATCCARCCTTT; primer 3, AAAGGYTGGATGGASATYATGTA; primer 4, GTGAAGAAKGAKCCRAAGATGATG; modified primer 1, TRYTSGTGTGYYTSATHTTCTGG; and modified primer 4, GTRAARAAKGAKCCRAAKRTGATG. Both strands were sequenced for all clones on an Applied Biosystems 377A DNA sequencer or an 3700 DNA analyzer by using Big Dye cycle sequencing chemistry according to the manufacturer's specifications (Applied Biosystems).
Sequences were aligned by using the predicted amino acid sequences and then reverse-translated to the nucleotide sequences. Gaps were treated as missing characters. Maximum likelihood phylogeny estimation was performed by using paup* (version 4.0b3) with model settings corresponding to HKY + gamma + estimated proportion of invariant sites (PINVAR) with four discrete Γ categories (25–28). Initial model parameters were estimated on a maximum parsimony tree and refined by using successive approximations on subsequent maximum likelihood estimates. Heuristic searches were performed with five random stepwise taxa additions. Branch support was estimated from 100 bootstrap replicate samples by using the final model parameters.
Alternative trees compatible with the two scenarios of homeobox (Hox) gene evolution were found by conducting the above analyses with the heuristic searches constrained to only consider trees compatible with the respective hypotheses. We tested for significant differences between the alternative phylogenetic hypotheses by using the Kashino-Hasegawa test.
Results
Cloning of Teleost Na+ Channel Genes.
We used two methods to clone the initial fragments of Na+ channel genes. First, reverse-transcribed RNA isolated from four tissues—brain, muscle, electric organ (which is developmentally derived from muscle), and heart—was amplified with degenerate primers flanking the highly conserved intracellular loop between domains III and IV (Fig. 1A). This generated three different fragments whose sequences were subsequently expanded with PCR to differing extents. These were termed SterNa1 (amplified from the electric organ and muscle), SterNa2 (amplified from heart), and SterNa3 (amplified from brain).
Figure 1.
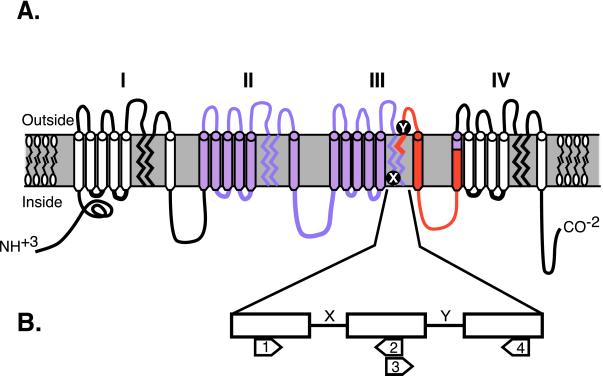
Schematic representation of primer positions for RT-PCR of RNA and PCR of genomic DNA. (A) Schematic Na+ channel in the membrane with four repeating domains (I–IV). Primers for RT-PCR were directed to the highly conserved regions of domains III and IV spanning the inactivation loop (loop III-IV) that links them (shown in red). The region of the gene used in phylogenetic analysis is shown in purple. The positions of the highly conserved intron/exon borders are denoted as X and Y. (B) Depiction of primer sites and intron locations for Na+ channels in the conserved pore region of domain III. Three semi-independent primers sets were used to amplify ≈380 bp of coding sequence and introns X and Y (primers 1 and 4); 255 bp of coding sequence and intron X (primers 1 and 2); and 125 bp of coding sequence and intron Y (primers 3 and 4).
To ensure that all of the Na+ channel genes in the genome were represented, we amplified Na+ channel genes from genomic DNA. We designed a set of degenerate primers based on published sequences and sequences from the three Na+ channels that we had cloned. The initial primer set targeted a highly conserved region of the pore in domain III (Fig. 1B). These primers targeted a genome fragment known to contain two introns (introns 19 and 20 in Nav1.4) in mammalian Na+ channel genes (29, 30). Introns at these positions are conserved from jellyfish to mammals (31). Because we do not know the intronic structure of the teleost Na+ channel genes, we refer to them as introns “X” and “Y.” We reasoned that because the lengths of introns vary, the resulting PCR products would be separable on a gel, which would facilitate identification of all PCR fragments.
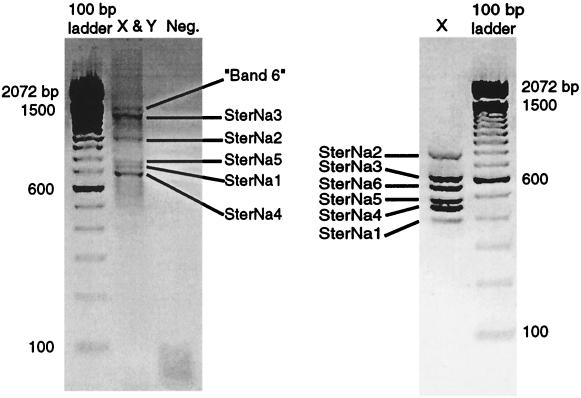
Six bands were resolved during the initial genomic PCR reaction that spanned introns X and Y, although only five of them could be cloned (Fig. 2A). They were all found to be sodium channel genes upon sequencing. Three of the fragments were identical to the genes that we had previously cloned with RT-PCR (SterNa1–3). The two new sodium channel genes were named SterNa4 and SterNa5.
Figure 2.
Results of separate genomic DNA surveys across both introns X and Y (X and Y), and intron X alone (X). Each band corresponds to a distinct Na+ channel gene. Negative controls without template (Neg.) were included in the assay (the clean negative control lane is not shown in Left).
To ensure that we had not missed any genes because of the limitations of PCR amplification length or primer bias, we then used additional primer sets to amplify smaller fragments spanning only intron X (Fig. 2B) or intron Y (data not shown). The banding pattern of the PCR products generated by amplifying across introns X or Y separately met the pattern predicted by the sequence of the five clones, except that an additional band was present in the PCR across intron X (Fig. 2B). This sixth band, which likely corresponded to the uncloneable “band 6” from the PCR across X and Y, was also cloned and determined to be yet another sodium channel gene (SterNa6).
By using primers to highly conserved sequences and cloning across introns, we believe that we have identified all, or certainly most, of the Na+ channel genes in this species. The independent recovery from genomic DNA of the three Na+ channel genes that we had initially cloned from RNA confirms the general approach because we targeted different regions of the gene with different primer sets.
Two technical problems that might have led us to miss genes were excessive intron length and primer mismatches. The introns of mammalian Na+ channel genes may be a few kilobases in length (29, 32), making this method problematic for the recovery of mammalian genes. However, the average length of the introns we encountered in the fish Na+ channel genes was 270 bp and the longest was 580 bp. Because we easily amplified fragments as large as 1,325 bp that spanned both introns, it is unlikely that the length of a single intron was limiting.
A post hoc analysis revealed a few mismatches in the sequence of the initial degenerate primer sets used in the PCR-based genomic census and that we successfully cloned channels with up to three mismatches (in a 23-mer). To reduce the possibility that primer mismatches were introducing a bias prohibiting the amplification of further channel types, we redesigned the primer sets based on the sequence of all six cloned channels so that there were no mismatches. When the PCR reactions were repeated with the modified primers (see Methods), only bands corresponding to the six cloned channels fragments were produced (data not shown). Finally, we determined the expected number of mismatches among our primer sets and all known mammalian Na+ channel genes to estimate whether any genes would have been unlikely to emerge in our census. We noted three or fewer total mismatches with at least one primer set to all of the mammalian Na+ channel genes except for the highly unusual Nax (33). Thus, our primers would have likely detected orthologs of all but one type of Na+ channel.
The sequences of the six genomic fragments were used to design specific primers for further tissue-specific RT-PCR reactions to extend the sequences of each gene, mainly in the 5′ direction, providing complete sequence from segment 1 (S1) in domain II, to S1 in domain IV (Fig. 1A, in purple). Due to the evolutionary history of Na+ channels, domains I and III and domains II and IV are most closely related to each other (1, 34). By focusing on domains II and III, we included sequence from both domain types. Domains II and III alone were sufficient to reconstruct the known topology for the mammalian Na+ channels based on domains I–IV (11, 12).
Phylogenetic Analysis.
The Sternopygus Na+ channel sequences were aligned with those of mammalian Na+ channels and the few other nonmammalian vertebrate Na+ channels that have been sequenced. A maximum likelihood phylogenetic analysis indicates that each of the six Sternopygus Na+ channels is orthologous to a mammalian Na+ channel gene or gene cluster on each of the four mammalian chromosomes (Fig. 3). These relationships are also supported by amino acid analysis (data not shown).
Figure 3.
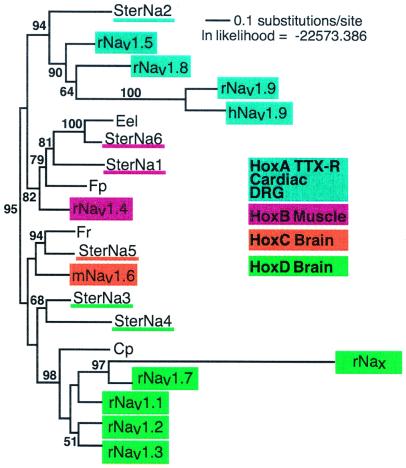
Phylogenetic analysis of vertebrate Na+ channel nucleotide sequences based on an unrooted maximum likelihood tree. The analysis was done without rooting because rooting with sequences of distantly related animals, as invertebrates in this case, are not strongly supported and potentially misleading due to long-branch attraction (25). The longest available nucleotide sequences common to the pufferfishes, Fugu pardalis (Fugu p.) (AB030482) and Fugu rubripes (Fugu r.) (D37977), the electric eel, Electrophorus electricus (Eel) (M22252), the knife fish, S. macrurus, the Japanese newt, Cynops pyrrhogaster (Newt) (AF123593), and rat were aligned by using megalign software DNAstar, Madison, WI. The mammalian sequences are the same as used by Plummer and Meisler (11), except for hNav1.9 (AF109737) (53), which was thought to be a novel channel and which is now believed to be an ortholog of rNav1.9 (53). The common segment corresponds to the available SterNa5 sequence, which runs from just past IISI to IVS1. This region contains 2,190 nucleotides or ≈40% of the nucleotides in the coding sequence of the E. electricus channel. From the preliminary alignment, nine blocks, totaling 1,545 nucleotides, were unambiguously aligned across the taxa being considered. The final alignment represents an estimated 55% of what might have been included, had complete cDNAs been available for all 22 channels. The aligned blocks correspond to the nucleotide positions from the E. electricus channel 2146–2319, 2350–2529, 2572–2661, 2680–2775, 2788–2832, 3166–3192, 3259–3618, 3673–3858, and 3949–4335. Mammalian Na+ channel genes are in colored blocks; genes on the same chromosome are the same color. Sternopygus Na+ channel genes (GenBank accession nos. AF378139–AF378144) are underlined with the color used for their mammalian orthologs. To the right of the tree are boxes indicating the Hox gene cluster with which each mammalian Na+ channel gene is linked as well as the main tissue in which these genes are expressed.
SterNa6 and SterNa1 group with Nav1.4, which is located on human chromosome 17. This Na+ channel gene is exclusively expressed in muscle in mammals (35). Similarly, SterNa1 and 6 are expressed in muscle and electric organ (data not shown) in the electric fish. These genes also group with other teleost Na+ channel genes found in muscle or electric organ (36, 37). SterNa2 groups with a cluster of mammalian Na+ channel genes, which map to chromosome 3 and encode channels that are resistant to the neurotoxin tetrodotoxin (TTX) (15, 32, 38). Based on its sequence and its expression in the fish heart (data not shown), SterNa2 is closest to Nav1.5, the mammalian cardiac Na+ channel. SterNa5 groups with Nav1.6, which is on human chromosome 12 (39, 40), and a pufferfish brain Na+ channel gene (41). The electric fish Na+ channel genes SterNa3 and SterNa4 group with five Na+ channel genes that map to human chromosome 2, most of which are mainly expressed in the brain. A newt brain Na+ channel gene also clusters with this group (42). Thus, each mammalian gene or gene cluster has a teleostean ortholog.
Our analysis of Na+ channel genes supports the hypothesis of Plummer and Meisler (11) that four chromosomes, each with a single Na+ channel gene, likely evolved from a series of duplications of an ancestral chromosome with a single Na+ channel gene. Our data show that these duplications occurred before the divergence of teleosts and tetrapods. It further reinforces the concept of the “tetraploidization” of the vertebrate genome (18, 19), although this formulation is not universally accepted (43).
Our analysis also supports the premise that further duplications occurred after these initial events. One interesting observation is that these duplications seem to have occurred independently in fish and mammals. For example, SterNa3 and 4 are orthologs of the cluster of five Na+ channel genes on human chromosome 2. Because they branch outside of the cluster of mammalian genes, the most parsimonious interpretation is that a single gene was present in a common ancestor of fish and mammals, and that this gene independently duplicated once in fish and four times in the tetrapod lineage. Similarly, two orthologs of a single mammalian muscle Na+ channel gene (Nav1.4) were found in Sternopygus. In this case, gene duplication occurred in fish but not in mammals.
Sodium Channels and Hox Genes.
One of the pieces of evidence supporting the concept of a large-scale chromosomal duplication of the vertebrate genome comes from analyses of Hox genes (19, 20, 44). Whereas one cluster of up to 13 paralogous genes on a single chromosome is observed in invertebrates and nonvertebrate chordates, four clusters (labeled A–D) are found in mammals, each on a different chromosome. Various scenarios have been proposed for the expansion of Hox genes in the chordate and basal vertebrate lineages (19, 44–46).
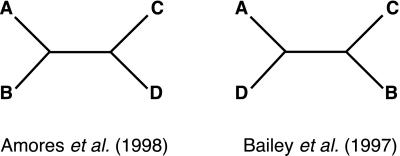
There are three possible unrooted trees by which four Hox gene clusters may have evolved. One tree, supported by analysis of Hox clusters and other linked genes in zebrafish, is that Hox genes evolved in two sets of duplication events such that a proto-Hox gene gave rise to two (proto-AB and proto-CD) genes, each of which duplicated again (46). A different scenario has been proposed based on an analysis of collagen genes, which are also linked to Hox genes. This scenario proposes three duplication events in which the proto-Hox gene gave rise to the Hox D cluster and a proto-ABC gene. The proto-ABC gene gave rise to HoxA and a proto-BC gene, which eventually duplicated again resulting in the B and C clusters (45).
Because the mammalian Na+ channel genes are also linked to the Hox gene clusters, we can independently test these two hypotheses. Inspection of the phylogenetic tree of vertebrate Na+ channel genes shows that the Na+ channel genes associated with Hox A and B are most closely related, and those genes associated with the C and D Hox genes are more closely related (Fig. 4). When formally tested, the (AB) (CD) topology is strongly supported whereas the other topology can be rejected (Kishino-Hasegawa log-likelihood ratio test, P < 0.03) (Fig. 4).
Figure 4.
Two unrooted trees for the four Hox gene clusters. Topology no. 1 is proposed by Amores et al. (46) and is supported by our analysis of Na+ channel genes, topology no. 2 is proposed by Bailey et al. (45) and not supported.
Discussion
Na+ Channel Gene Duplications Occurred Early in Vertebrate History.
Our data suggest that the common ancestor of teleost fish and mammals had four Na+ channel genes and that these underwent further duplications independently in the teleost and tetrapod lineages. From chromosome mapping studies, it is clear that the second wave of Na+ channel gene duplications in mammals are tandem duplications. Whether the duplications in teleosts are also tandem duplications on the same chromosome or whether they are on different chromosomes and reflect an additional large-scale genome-wide expansion of the teleost genome (46) is not known. This difference could be resolved by mapping these genes in zebrafish for which linkage maps exist.
It is likely that the duplications of the Na+ channel genes occurred close to the emergence of the first vertebrates. Only two presumptive Na+ channel genes have been cloned in an ascidian chordate (47), and one of these genes is highly unusual and not in the same lineage as vertebrate Na+ channel genes. Furthermore, ascidians and amphioxus have only one Hox cluster (19) indirectly suggesting that they would only possess only one or a few Na+ channel genes.
Significance of Na+ Channel Gene Evolution for Function.
Our finding of the independent duplications of Na+ channel genes in fish and mammals raises the question of whether genes that have independently diverged in these two lineages evolve similar or different functions. For example, in mammals Nav1.1, 1.2, and 1.3 arose as duplications of a founder gene. Nav1.1 and Nav1.3 channels are localized in the somata of many kinds of neurons whereas Nav1.2 is mainly found in axons of unmyelinated neurons (13, 14, 16). Presumably the ancestral gene was expressed generally in neurons and the current distribution occurred as a refinement of this pattern. Teleosts have two orthologs of these genes (SterNa3 and 4), which presumably arose from the same ancestral gene that gave rise to these three genes in mammals. It will be intriguing to know whether the teleostean orthologs of these genes have evolved comparable subcellular distributions as in mammals.
On the other hand, the mammalian gene Nav1.6 occurs in a number of subcellular loci, including nodes of Ranvier of myelinated axons (17). Fish have an ortholog of that gene, SterNa5. We predict that this Na+ channel is localized to nodes of Ranvier in fish as well, which would suggest this is an ancient function of the Nav1.6 precursor gene.
The cluster of three TTX-resistant genes on human chromosome 3 includes Nav1.5, mainly expressed in heart, Nav1.8, and Nav1.9, mainly expressed in small diameter dorsal root ganglion nociceptors. The genes in this cluster likely evolved from an ancestral gene (32). Although it is always possible that we have missed a gene in our census, we have only uncovered a single ortholog of this cluster in teleosts: it is most similar to Nav1.5 and it is expressed in the heart. This expression pattern lends credence to the idea that the cardiac gene Nav1.5 is possibly the ancestral gene of this cluster in tetrapods. One implication of our data is that the specialized small-diameter dorsal root nociceptive pathway with its specialized Na+ channels may be a tetrapod innovation. Indeed, cartilaginous fish have uniform diameter dorsal root ganglion neurons and little physiological indications of nociceptive fibers (48). However, TTX-resistant Na+ currents have been observed in small diameter dorsal root ganglion neurons in frogs (49–51), suggesting that one or both of these TTX-resistant genes, and a specialized nociceptive pathway, may have appeared early in the tetrapod lineage. Cloning of Na+ channel genes in frogs will address this question.
Hox Gene Evolution.
Because Na+ channel genes are linked to Hox gene clusters, we used a Na+ channel gene phylogeny to test theories of Hox gene evolution and were able to support one theory (46). The relationship of Hox gene clusters at the base of the vertebrate tree is still cloudy. Hox gene evolution early in the vertebrate lineage is best inferred from lampreys. However, establishing relationships among short conserved Hox genes in lampreys is proving difficult (P. Holland, personal communication). Our results suggest that a phylogeny of lamprey Na+ channels, which have longer stretches of conserved regions, could shed light on Hox gene evolution in basal vertebrates.
Conclusions
Many ion channel genes have been cloned in mammals, and hypotheses on their evolution have been advanced (11, 34, 52). These hypotheses can only be tested when all of the ion channels in a family are cloned in pivotal nonmammalian vertebrates, as we did with Na+ channel genes in teleosts. We believe that this type of analysis is critical to an understanding of the evolution of the vertebrate brain.
We support the hypothesis that Na+ channel genes evolved in vertebrates initially by chromosomal duplications followed by a later set of tandem duplications, and we show here that these later duplications occurred independently in teleosts and mammals. Similar analyses of Na+ channel genes in key vertebrates will allow us to better visualize how this gene family evolved early in vertebrate evolution (i.e., lamprey, elasmobranchs) and at the base of the mammalian lineage (i.e., lungfish, frogs).
Acknowledgments
We thank Gwen Gage and Katie Kendall for artwork, Jason Rall for help with the PCR, Rob Reinauer for fish care, and Klaus Linse at the Institute for Cellular and Molecular Biology Core Facility for sequencing assistance. This work was supported by National Institutes of Health grants (to H.H.Z. and N.S.A.) and by University of Texas in-house grants (to G.F.L.).
Abbreviations
- TTX
tetrodotoxin
- HOX
homeobox
Footnotes
References
- 1.Hille B. Q J Exp Physiol. 1989;74:785–804. doi: 10.1113/expphysiol.1989.sp003349. [DOI] [PubMed] [Google Scholar]
- 2.Bell C, Han V, Sugawara Y, Grant K. Nature (London) 1997;387:278–281. doi: 10.1038/387278a0. [DOI] [PubMed] [Google Scholar]
- 3.Lemon N, Turner R W. J Neurophysiol. 2000;84:1519–1530. doi: 10.1152/jn.2000.84.3.1519. [DOI] [PubMed] [Google Scholar]
- 4.Markram H, Lubke J, Frotscher M, Sakmann B. Science. 1997;275:213–215. doi: 10.1126/science.275.5297.213. [DOI] [PubMed] [Google Scholar]
- 5.Raman I, Sprunger L, Meisler M, Bean B. Neuron. 1997;19:881–891. doi: 10.1016/s0896-6273(00)80969-1. [DOI] [PubMed] [Google Scholar]
- 6.Nick T, Ribera A. Nat Neurosci. 2000;3:142–149. doi: 10.1038/72082. [DOI] [PubMed] [Google Scholar]
- 7.Cantrell A, Scheuer T, Catterall W. J Neurosci. 1999;19:5301–5310. doi: 10.1523/JNEUROSCI.19-13-05301.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Catterall W. Neuron. 2000;26:13–25. doi: 10.1016/s0896-6273(00)81133-2. [DOI] [PubMed] [Google Scholar]
- 9.Hong C, Ganetzky B. J Neurosci. 1994;14:5160–5169. doi: 10.1523/JNEUROSCI.14-09-05160.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Littleton J, Ganetzky B. Neuron. 2000;26:35–43. doi: 10.1016/s0896-6273(00)81135-6. [DOI] [PubMed] [Google Scholar]
- 11.Plummer N, Meisler M. Genomics. 1999;57:323–331. doi: 10.1006/geno.1998.5735. [DOI] [PubMed] [Google Scholar]
- 12.Goldin A, Barchi R, Caldwell J, Hofmann F, Howe J, Hunter J, Kallen R, Mandel G, Meisler M, Berwald Netter Y, et al. Neuron. 2000;28:365–368. doi: 10.1016/s0896-6273(00)00116-1. [DOI] [PubMed] [Google Scholar]
- 13.Westenbroek R, Merrick D, Catterall W. Neuron. 1989;3:695–704. doi: 10.1016/0896-6273(89)90238-9. [DOI] [PubMed] [Google Scholar]
- 14.Westenbroek R, Noebels J, Catterall W. J Neurosci. 1992;12:2259–2267. doi: 10.1523/JNEUROSCI.12-06-02259.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dib-Hajj S, Tyrell L, Cummins T, Black J, Wood P, Waxman S. FEBS Lett. 1999;462:117–120. doi: 10.1016/s0014-5793(99)01519-7. [DOI] [PubMed] [Google Scholar]
- 16.Gong B, Rhodes K, Bekele-Arcuri Z, Trimmer J. J Comp Neurol. 1999;412:342–352. [PubMed] [Google Scholar]
- 17.Caldwell J, Schaller K, Lasher R, Peles E, Levinson S. Proc Natl Acad Sci USA. 2000;97:5616–5620. doi: 10.1073/pnas.090034797. . (First Published April 25, 2000, 10.1073/pnas.090034797) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohno S. Evolution by Gene Duplication. Berlin: Springer; 1970. [Google Scholar]
- 19.Holland P, Garcia-Fernandez J, Williams N, Sidow A. Development (Cambridge, U.K.) 1994. , Suppl., 125–133. [PubMed] [Google Scholar]
- 20.Shimeld S, Holland P. Proc Natl Acad Sci USA. 2000;97:4449–4452. doi: 10.1073/pnas.97.9.4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith G, Zakon H. J Neurbiol. 2000;42:270–286. doi: 10.1002/(sici)1097-4695(20000205)42:2<270::aid-neu10>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 22.McAnelly M L, Zakon H H. J Neurosci. 1996;16:4383–4388. doi: 10.1523/JNEUROSCI.16-14-04383.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferrari M B, McAnelly M L, Zakon H H. J Neurosci. 1995;15:4023–4032. doi: 10.1523/JNEUROSCI.15-05-04023.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Smith J, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1994. [Google Scholar]
- 25.Swofford D, Olsen G, Waddell P, Hillis D. In: Molecular Systematics. Hillis D M, Moritz C, Mable B K, editors. Sunderland, MA: Sinauer; 1996. pp. 407–514. [Google Scholar]
- 26.Swofford D. paup* Phylogenetic Analysis Using Parsimony (*and other methods) Sunderland, MA: Sinauer; 1998. , Ver. 4. [Google Scholar]
- 27.Yang Z. Trends Ecol Evol. 1996;11:367–372. doi: 10.1016/0169-5347(96)10041-0. [DOI] [PubMed] [Google Scholar]
- 28.Rogers J, Swofford D. Mol Biol Evol. 1999;16:1079–1085. doi: 10.1093/oxfordjournals.molbev.a026197. [DOI] [PubMed] [Google Scholar]
- 29.McClatchey A, Lin C, Wang J, Hoffman E, Rojas C, Gusella J. Hum Mol Genet. 1992;1:521–527. doi: 10.1093/hmg/1.7.521. [DOI] [PubMed] [Google Scholar]
- 30.Wang Q, Li Z-Z, Shen J-X, Keating M T. Genomics. 1996;34:9–16. doi: 10.1006/geno.1996.0236. [DOI] [PubMed] [Google Scholar]
- 31.Spafford J, Spencer A, Galin W. Recept Channels. 1999;6:493–506. [PubMed] [Google Scholar]
- 32.Dib-Hajj S, Tyrell L, Escayg A, Wood P, Meisler M, Waxman S. Genomics. 1999;59:309–318. doi: 10.1006/geno.1999.5890. [DOI] [PubMed] [Google Scholar]
- 33.Gautron S, Dos Santos G, Pinto-Henrique D, Koulakoff A, Gros F, Berwald-Netter Y. Proc Natl Acad Sci USA. 1992;89:7272–7276. doi: 10.1073/pnas.89.15.7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strong M, Chandy K, Gutman G. Mol Biol Evol. 1993;10:221–242. doi: 10.1093/oxfordjournals.molbev.a039986. [DOI] [PubMed] [Google Scholar]
- 35.Barchi R. Ann NY Acad Sci. 1986;479:179–185. doi: 10.1111/j.1749-6632.1986.tb15569.x. [DOI] [PubMed] [Google Scholar]
- 36.Noda M, Shimizu S, Tanabe T, Takai T, Kayano T, Ikeda T, Takahashi H, Nakayama H, Kanaoka Y, Minamino N, et al. Nature (London) 1984;312:121–127. doi: 10.1038/312121a0. [DOI] [PubMed] [Google Scholar]
- 37.Yotsu-Yamashita M, Nishimori K, Nitanai Y, Isemura M, Sugimoto A, Yasumoto T. Biochem Biophys Res Commun. 2000;267:403–412. doi: 10.1006/bbrc.1999.1974. [DOI] [PubMed] [Google Scholar]
- 38.Akopian A, Sivilotti L, Wood J. Nature (London) 1996;379:257–262. doi: 10.1038/379257a0. [DOI] [PubMed] [Google Scholar]
- 39.Schaller K, Krzemien D, Yarowsky P, Krueger B, Caldwell J. J Neurosci. 1995;15:3231–3242. doi: 10.1523/JNEUROSCI.15-05-03231.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kohrman D, Plummer N, Schuster T, Jones J, Jang W, Burgess D, Galt J, Spear B, Meisler M. Genomics. 1995;26:171–177. doi: 10.1016/0888-7543(95)80198-u. [DOI] [PubMed] [Google Scholar]
- 41.Shahjahan M, Yamada M, Nagaya M, Kawai M, Kakazawa A. Ann NY Acad Sci. 1993;707:346–348. doi: 10.1111/j.1749-6632.1993.tb38066.x. [DOI] [PubMed] [Google Scholar]
- 42.Kaneko Y, Matsumoto G, Hanyu Y. Biochem Biophys Res Commun. 1997;240:651–656. doi: 10.1006/bbrc.1997.7696. [DOI] [PubMed] [Google Scholar]
- 43.Hughes A. J Mol Evol. 1999;48:565–576. doi: 10.1007/pl00006499. [DOI] [PubMed] [Google Scholar]
- 44.Pendleton J, Nagai B, Murtha M, Ruddle F. Proc Natl Acad Sci USA. 1993;90:6300–6304. doi: 10.1073/pnas.90.13.6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bailey W, Kim J, Wagner G, Ruddle F. Mol Biol Evol. 1997;14:843–853. doi: 10.1093/oxfordjournals.molbev.a025825. [DOI] [PubMed] [Google Scholar]
- 46.Amores A, Force A, Yan Y-L, Joly L, Amemiya C, Wang Y-L, Westerfield M, Ekker M, Postlehwait J. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- 47.Nagahora H, Kokada T, Yahagi N, Chong J, Mandel G, Okamura Y. Biochem Biophys Res Commun. 2000;275:558–564. doi: 10.1006/bbrc.2000.3290. [DOI] [PubMed] [Google Scholar]
- 48.Snow P, Plenderleith M, Wright L. J Comp Neurol. 1993;334:97–103. doi: 10.1002/cne.903340108. [DOI] [PubMed] [Google Scholar]
- 49.Campbell D. Proc Natl Acad Sci USA. 1992;89:9569–9573. doi: 10.1073/pnas.89.20.9569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kobayashi J, Ohta M, Terada Y. Neurosci Lett. 1993;162:93–96. doi: 10.1016/0304-3940(93)90568-6. [DOI] [PubMed] [Google Scholar]
- 51.Buchanan S, Harper A, Elliott J. Neurosci Lett. 1996;219:131–134. doi: 10.1016/s0304-3940(96)13189-x. [DOI] [PubMed] [Google Scholar]
- 52.Street V, Tempel B. Genomics. 1997;44:110–117. doi: 10.1006/geno.1997.4799. [DOI] [PubMed] [Google Scholar]
- 53.Jeong S, Goto J, Hashida H, Suzuki T, Ogata K, Masuda N, Hirai M, Isahara K, Uchiyama Y, Kanazawa I. Biochem Biophys Res Commun. 2000;267:262–270. doi: 10.1006/bbrc.1999.1916. [DOI] [PubMed] [Google Scholar]