Abstract
Background: TCF7L2 gene variants have been associated with increased risk of type 2 diabetes and higher adiposity. Observational studies and short-term trials have suggested that macronutrients may modify these effects. However, to our knowledge, this has yet to be verified in long-term interventions.
Objective: In a long-term intervention setting, we investigated the effects of TCF7L2 polymorphisms rs7903146 and rs12255372 and dietary total fat on changes in body composition and subsequent glycemic control.
Design: Data were analyzed for 591 participants in the Preventing Overweight Using Novel Dietary Strategies (Pounds Lost) trial, which is a 2-y weight-loss randomized clinical trial of diets that differed in macronutrient proportions. Adjusted means for changes in body composition at 6 and 24 mo were obtained for gene main effects and interactions with a low-fat diet (20% from energy) compared with a high-fat diet (40% from energy). Interactions with protein and carbohydrate intakes were also tested. Predicted changes in glycemic control from changes in adiposity were determined by genotype and diet type.
Results: Significant interactions were observed for rs12255372 TT (risk genotype) and fat intake for changes in BMI, total fat mass, and trunk fat mass (all P/q < 0.05) at 6 mo, with nonsignificant larger decreases for TT carriers on a low-fat diet. No significant associations were observed at 24 mo or for other macronutrients. Changes in body composition for TT carriers predicted reductions in plasma glucose and insulin only on the low-fat diet.
Conclusions: Individuals with the TCF7L2 rs12255372 risk genotype may reduce body adiposity by consuming a diet lower in total fat. These reductions may induce better glycemic control for such individuals predisposed to type 2 diabetes. The Pounds Lost trial was registered at clinicaltrials.gov as NCT00072995.
INTRODUCTION
Population genetic studies have consistently shown that common variants in the TCF7L2 gene significantly predict type 2 diabetes risk (1–4). The deleterious effect is hypothesized to be related to β cell function (2–5). In addition, compelling evidence has suggested important roles of TCF7L2 in the regulation of body weight and adiposity including 1) taking part in the Wnt signaling cascade, which inhibits adipogenesis (6), 2) gene expression in adipose tissue differing by genotype after caloric restriction (7, 8), 3) the association with hunger-satiety hormones that influence weight loss (9), and 4) the promotion of the transcription of proglucagon, which induces the synthesis of glucagon-like peptide, which is a regulator of insulin and glucagon secretion, appetite, and food intake (6).
Despite this evidence, reports on the association of TCF7L2 variants with weight and body composition have been contradictory (2, 8, 10). One possible explanation for the discrepancies might be interactions between TCF7L2 genotypes and environmental factors such as dietary intake. Such interactions have been reported in observational studies for dietary fat (11–13), carbohydrate intake (14, 15), and animal protein (16), but results from intervention settings are limited. A recent randomized trial reported that obese individuals with the TCF7L2 single-nucleotide polymorphism (SNP)4 rs7903146 risk allele have better responses in weight loss and adiposity outcomes after 10 wk of consumption of a low-fat, but not high-fat, dietary intervention (17). The study suggested that individuals who carry TCF7L2 risk alleles are more sensitive to low-fat than high-fat weight-loss diets; however, the intervention period was short. Thus, we aimed to investigate whether TCF7L2 genotypes of the well-recognized SNPs rs7903146 and rs12255372, which are in moderate linkage disequilibrium (LD) in European populations (r2 = 0.73) (18), modulate the response to diets of different fat content in relation to long-term changes in anthropometric measures and body composition in a 2-y randomized, weight-loss intervention trial and whether changes in adiposity by gene variant and diet type predict subsequent changes in glycemic control. A secondary aim was to test interactions with dietary carbohydrate and protein.
SUBJECTS AND METHODS
Study participants
The Preventing Overweight Using Novel Dietary Strategies (Pounds Lost) trial (www.clinicaltrials.gov; NCT00072995) was a 2-y randomized clinical trial for weight loss conducted at Boston, Massachusetts, and Baton Rouge, Louisiana, in 2004–2007 (19, 20). The study randomly assigned 811 participants into 4 energy-reduced diets of varying macronutrient proportions. Participants had to be 30–70 y of age and have BMI (in kg/m2) of 25–40. Exclusion criteria included the presence of diabetes or unstable cardiovascular disease. The 2-y retention was 80%; the analysis was conducted as an intention-to-treat analysis. All participants provided informed consent, and the study was approved and monitored by the human subjects committee at Harvard School of Public Health, Brigham and Women's Hospital, Pennington Biomedical Research Center, and the National Heart, Lung and Blood Institute.
To avoid population stratification (3, 15), we restricted analysis to individuals who self-identified as white (n = 643). Stratified analysis for other racial-ethnic groups could not be conducted because of low sample sizes (n = 127 African-Americans, n = 29 Hispanics, and n = 12 other races). Of the 811 enrolled participants, 8 subjects did not provide consent for DNA sampling. There were 745 samples available for genotyping at the time of this analysis, of which 597 samples indicated white race. The final sample size was 588 for rs7903146 and 591 for rs12255372.
Dietary intervention
Participants were randomly assigned to 4 diet groups with the following goals for total fat, protein, and carbohydrate as percentages of total energy: 20%, 15%, 65%; 20%, 25%, 55%; 40%, 15%, 45%; and 40%, 25%, 35%, respectively. The following 2 diet categories were defined to test interactions with dietary fat: low fat (2 diets with an aim of 20% from total energy) and high fat (2 diets with an aim of 40% from total energy). Dietary fat was the primary macronutrient considered for gene-diet interactions on the basis of previous reports and plausible mechanisms. A secondary analysis was conducted to test interactions with other macronutrients as follows: 2 low-protein (15% of total energy) compared with 2 high-protein (25%) groups and the 2 assigned diets with extreme carbohydrate composition (35% compared with 65% of total energy). All participants, independent of diet group, were instructed to consume >20 g dietary fiber, ≤8% saturated fat, and ≤150 mg cholesterol/100 kcal and to choose foods low in glycemic index. The energy deficit was 750 kcal from baseline.
To enhance compliance, participants were initially screened for any potential eating disorder, medical condition, or psychological behavior that could interfere with acceptability or adherence to any of the diets and were further interviewed to ensure sufficient suitability and enthusiasm for the study before enrolling. Assigned diets were introduced during an initial individual orientation with a trained dietitian. Adherence was encouraged through periodic group and individual behavioral counseling sessions with reinforced instructions on following meal plans and through daily self-monitoring by using diet records and Web-based tools. Dietary compliance was formally measured with biomarkers of nutrient intake (HDL cholesterol for carbohydrate, urinary nitrogen excretion for protein, and respiratory quotient for fat) and with 24-h recalls at 6 and 24 mo to determine adherence to macronutrient targets.
Measurements and genotyping
Anthropometric measures were collected at baseline and 6, 12, and 18 mo and 2 y according to standard protocols (19, 20). BMI was calculated by dividing weight by the squared height of a participant. Dual-energy X-ray absorptiometry (DXA) scans were performed on a random subset of 50% of participants (n = 326 for this analysis) to assess body fat at baseline, 6 mo, and 2 y. The DXA protocol has been described in detail previously (20). Briefly, measurements were taken after an overnight fast with the participant wearing a hospital gown in the supine position on a Hologic QDR-4500A bone densitometer (Hologic). Digital files were analyzed by using a single reader at the Pennington site. The accuracy of instruments at the 2 sites was assessed by taking 3-point body fat phantoms; the instruments were well matched and stable. A comparison of fat mass with lean mass was determined as the percentage from body fat and body weight.
Fasting blood samples were collected at baseline and 6 and 24 mo. Analyses for glucose and insulin were done from aliquotted serum at the Pennington Laboratory with an immunoassay with chemiluminescent detection on an Immukite analyzer (Diagnostics Products Corp).
Genotyping has been described in detail previously (21). Briefly, DNA was extracted by using a QIAmp Blood Kit (Qiagen). Polymorphisms rs7903146 and rs12255372 were genotyped by using the OpenArray SNP Genotyping System (BioTrove). The genotype success rate and replication concordance were ≥99%.
Statistical analysis
Outcomes included changes at 6, 12, 18, and 24 mo (determined as the difference from baseline) for anthropometric measures (weight, waist circumference, and BMI) and at 6 mo and 2 y for DXA body fat measures (total body fat, lean mass, and trunk fat). The body-composition outcomes were selected to depict various adipose depots (ie, abdominal or central adiposity by using waist circumference and overall body fat by using BMI). Weight loss was the primary outcome of the original study. Although waist circumference and BMI have shown strong correlations with DXA-measured body fat, especially in whites, DXA measurements were included in our study as a validation of body adiposity outcomes by using more precise estimates of fat mass because DXA distinguishes fat mass from lean mass (22). Thus, DXA-measured total fat mass indicates overall body fat (as BMI), whereas trunk fat mass reflects central adiposity (as waist circumference). Although the outcome measures overlap to some degree, they may differentiate fat-mass depots and may help establish consistency and accuracy of results.
Hardy-Weinberg equilibrium was tested by using the chi-square test. The LD between the 2 polymorphisms was determined with PowerMarker 3.25 software (23). Linear mixed models were used to compare means for continuous outcomes across genotypes at baseline with adjustment for age, sex, and center as well as the change at 6 mo and 2 y and with additional adjustments for the baseline measure and diet intervention. Main additive effects for each variant were assessed separately for each diet group. Interactions between diet group and genotypes were tested by including an interaction term in the models as independent predictors of each outcome. Tukey's test for multiple-comparison adjustment was applied to the reported adjusted means. To minimize false-positive findings for gene-diet interaction tests, a positive false discovery rate correction was applied, with the corresponding adjusted q value reported (24). Linear mixed models adjusted for age, sex, site, and baseline measure were conducted by diet type and genotype for continuous changes in blood glucose and insulin to determine whether any observed significant changes in body composition by genotype-diet effect predicted additional changes in glycemic control. Statistical significance was deemed at P < 0.05 (and q < 0.05 for interaction terms). Statistical analyses were performed with SAS 9.3 software (SAS Institute).
RESULTS
The frequency for the TCF7L2 rs7903146 risk allele was 42.2% for heterozygous (CT) and 8.3% for homozygous (TT), whereas the frequency for the rs12255372 risk allele was 40.6% for heterozygous (GT) and 7.8% for homozygous (TT) (Table 1). Genotype frequencies did not differ by age, sex, center, or randomized diet. Both variants were in Hardy-Weinberg equilibrium (P = 0.736 for rs7903146 and P = 0.886 for rs12255372). The 2 SNPs were in moderate LD (r2 = 0.71). There were no differences in baseline or the change at 6 mo or 2 y by TCF7L2 genotype for any outcome (Table 1).
TABLE 1.
Characteristics by TFC7L2 gene variant at baseline and changes in 6 mo and 2 y in participants in the Preventing Overweight Using Novel Dietary Strategies study1
| rs7903146 |
rs12255372 |
|||||||
| CC | CT | TT | P | GG | GT | TT | P | |
| Full cohort [n (%)] | 291 (49.5) | 248 (42.2) | 49 (8.3) | — | 305 (51.6) | 240 (40.6) | 46 (7.8) | — |
| Age (y) | 52.5 ± 8.52 | 51.6 ± 9.5 | 51.6 ± 8.8 | — | 52.4 ± 8.9 | 51.4 ± 9.2 | 52.6 ± 8.4 | — |
| Sex (F) (%) | 55.7 | 58.9 | 53.1 | — | 58.0 | 55.0 | 58.7 | — |
| Low-fat diet group (%) | 47.4 | 52.8 | 40.8 | — | 46.9 | 52.5 | 45.7 | — |
| Low-protein group (%) | 52.2 | 50.0 | 49.0 | — | 53.1 | 47.9 | 47.8 | — |
| Lowest-carbohydrate group (%) | 25.4 | 23.4 | 30.6 | — | 24.6 | 24.6 | 30.4 | — |
| Center (Boston) (%) | 47.8 | 47.6 | 42.9 | — | 45.3 | 49.2 | 41.3 | — |
| Weight (kg) | ||||||||
| Baseline | 95.0 ± 0.7 | 94.8 ± 0.8 | 93.3 ± 1.8 | 0.450 | 94.8 ± 0.7 | 95.2 ± 0.8 | 90.8 ± 1.8 | 0.272 |
| 6-mo change | −6.3 ± 0.3 | −6.9 ± 0.4 | −7.0 ± 0.8 | 0.210 | −6.3 ± 0.3 | −6.9 ± 0.4 | −7.3 ± 0.8 | 0.125 |
| 2-y change | −3.9 ± 0.4 | −3.6 ± 0.5 | −4.9 ± 1.0 | 0.760 | −3.8 ± 0.4 | −4.0 ± 0.5 | −5.2 ± 1.1 | 0.360 |
| Waist circumference (cm) | ||||||||
| Baseline | 105.2 ± 0.6 | 105.3 ± 0.7 | 104.5 ± 1.5 | 0.779 | 105.2 ± 0.6 | 105.5 ± 0.7 | 102.8 ± 1.6 | 0.420 |
| 6-mo change | −6.6 ± 0.4 | −7.1 ± 0.4 | −6.5 ± 0.9 | 0.563 | −6.6 ± 0.4 | −6.9 ± 0.4 | −7.4 ± 0.9 | 0.350 |
| 2-y change | −4.8 ± 0.4 | −4.8 ± 0.5 | −5.4 ± 1.1 | 0.724 | −4.8 ± 0.4 | −5.0 ± 0.5 | −6.1 ± 1.1 | 0.377 |
| BMI (kg/m2) | ||||||||
| Baseline | 32.6 ± 0.2 | 32.7 ± 0.2 | 32.5 ± 0.5 | 0.914 | 32.5 ± 0.2 | 32.8 ± 0.2 | 32.1 ± 0.6 | 0.999 |
| 6-mo change | −2.2 ± 0.1 | −2.4 ± 0.1 | −2.4 ± 0.3 | 0.187 | −2.2 ± 0.1 | −2.4 ± 0.1 | −2.5 ± 0.3 | 0.120 |
| 2-y change | −1.3 ± 0.2 | −1.3 ± 0.2 | −1.7 ± 0.4 | 0.670 | −1.3 ± 0.1 | −1.4 ± 0.2 | −1.8 ± 0.4 | 0.350 |
| DXA3 subset [n (%)] | 159 (49.3) | 136 (42.2) | 27 (8.4) | — | 162 (49.7) | 135 (41.4) | 29 (8.9) | — |
| Total fat mass (%) | ||||||||
| Baseline | 36.2 ± 0.3 | 36.6 ± 0.4 | 37.1 ± 0.8 | 0.258 | 36.1 ± 0.3 | 36.9 ± 0.4 | 37.1 ± 0.8 | 0.073 |
| 6-mo change | −2.9 ± 0.3 | −3.3 ± 0.3 | −3.4 ± 0.6 | 0.364 | −3.0 ± 0.3 | −3.2 ± 0.3 | −3.5 ± 0.6 | 0.449 |
| 2-y change | −2.4 ± 0.4 | −2.3 ± 0.4 | −1.8 ± 0.8 | 0.583 | −2.5 ± 0.4 | −2.3 ± 0.4 | −2.5 ± 0.8 | 0.904 |
| Lean mass (kg) | ||||||||
| Baseline | 61.1 ± 0.5 | 61.4 ± 0.6 | 58.6 ± 1.2 | 0.256 | 61.1 ± 0.5 | 61.7 ± 0.5 | 57.1 ± 1.2 | 0.100 |
| 6-mo change | −2.3 ± 0.2 | −2.3 ± 0.2 | −2.0 ± 0.4 | 0.615 | −2.3 ± 0.2 | −2.3 ± 0.2 | −2.3 ± 0.4 | 0.976 |
| 2-y change | −2.3 ± 0.3 | −1.9 ± 0.3 | −2.4 ± 0.7 | 0.601 | −2.2 ± 0.3 | −2.0 ± 0.3 | −2.5 ± 0.7 | 0.987 |
| Trunk fat (%) | ||||||||
| Baseline | 37.2 ± 0.4 | 37.9 ± 0.4 | 38.3 ± 0.9 | 0.116 | 37.1 ± 0.4 | 38.3 ± 0.4 | 38.2 ± 0.9 | 0.050 |
| 6-mo change | −3.7 ± 0.3 | −4.3 ± 0.4 | −4.0 ± 0.8 | 0.496 | −3.9 ± 0.3 | −4.2 ± 0.4 | −4.0 ± 0.8 | 0.662 |
| 2-y change | −3.1 ± 0.5 | −3.2 ± 0.5 | −2.0 ± 1.1 | 0.512 | −3.3 ± 0.5 | −3.1 ± 0.5 | −2.9 ± 1.0 | 0.707 |
Linear mixed models were used to determine mean baseline outcome measures adjusted for age, sex, and center and 6-mo and 2-y changes adjusted for age, sex, center, baseline measure, and diet intervention. Macronutrients from 4 randomized intervention diets were dichotomized as follows: low fat (2 diets with a target for total fat of 20% from total energy) compared with high fat (2 diets with a target for total fat of 40% from total energy); low protein (2 diets with a target of 15% from energy) compared with high protein (2 diets with a target of 25% from energy); and low carbohydrate (one diet with the lowest carbohydrate composition of 35% from energy) compared with high carbohydrate (one diet with a carbohydrate composition of 65%).
Mean ± SD (all such values).
DXA, dual-energy X-ray absorptiometry.
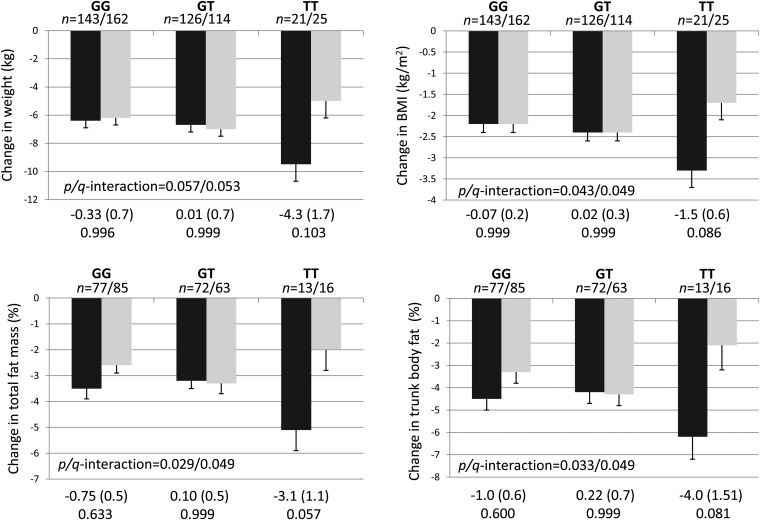
After adjustment for age, sex, center, and baseline measure, significant interactions with the randomly assigned dietary fat groups were observed for rs12255372 on changes in BMI (P/q-interaction = 0.043/0.049), total fat mass (P/q = 0.029/0.049), and trunk fat mass (P/q = 0.033/0.049) at 6 mo (Figure 1). In general, individuals with the TT genotype showed a greater reduction in each of those outcomes when the consumed a low-fat diet than did individuals with the same genotype but who consumed a high-fat diet; however, none of these differences were significant after a multiple-comparison adjustment. The interaction for change in weight at 6 mo was a not significant (P/q = 0.057/0.053) but suggested a similar direction as the other reported interactions. When subjects consumed a low-fat diet, individuals with the risk allele of rs12255372 had a greater decrease in BMI (P = 0.046) than did carriers of the C allele (see Table 1 under “Supplemental data” in the online issue). There were no differences by genotype with the high-fat diet.
FIGURE 1.
Adjusted mean (±SE) gene-nutrient interactions between TCF7L2 rs12255372 and total dietary fat intake as assigned at random to participants in the Preventing Overweight Using Novel Dietary Strategies trial for changes in body-composition measures at 6 mo. Values were from linear mixed models adjusted for age, sex, center, and baseline value. The mean difference (±SE) and adjusted P value by using Tukey's test for multiple comparisons are indicated at the bottom of each panel. The low-fat group (black bars) included individuals who were randomly assigned to consume diets with a target for total fat of 20% from energy; the high-fat group (gray bars) included individuals who were randomly assigned to consume diets with a target for total fat of 40% from energy. Significance for interactions is reported as the P value/adjusted q value from a positive false discovery rate correction.
A significant interaction with the randomized diet was observed for rs7903146 and the change in lean mass at 6 mo (P/q = 0.035/0.049; see Table 2 under “Supplemental data” in the online issue), with no difference by dietary fat group for TT individuals but a greater loss of lean mass for CC carriers who consumed the low-fat diet. No significant interactions were observed beyond the 6-mo time point for changes in weight, waist circumference, or BMI (see Table 3 under “Supplemental data” in the online issue) nor for changes in DXA-measured body fat when assessed at 24 mo (see Table 1 under “Supplemental data” in the online issue).
An interaction between protein intake and rs12255372 for changes in lean mass was observed at 6 and 24 mo (P = 0.004 and P = 0.049, respectively); however, these interactions were no longer significant after a positive false discovery rate correction (q = 0.096 and q = 0.588; see Table 4 under “Supplemental data” in the online issue). Carriers of the rs7903146 risk variant showed less loss of lean mass in the highest-carbohydrate diet only (P = 0.003); the interactive term was not significant (see Table 5 under “Supplemental data” in the online issue). No other main effect or interactive terms were detected as significant for the TCF7L2 SNPs and dietary protein or carbohydrate intake.
Because we detected a significant interaction between TCF7L2 rs12255372 and dietary fat on body fat outcomes, we determined whether those gene- and diet-induced changes predicted glycemic control. Reductions in weight and BMI at 6 mo were associated with significant reductions in plasma glucose and insulin for subjects with the rs12255372 G allele in both the low- and high-fat diet groups but only in the low-fat diet group for subjects with the TT risk genotype (Table 2; see Table 6 under “Supplemental data” in the online issue for baseline values). Subjects with the TT genotype showed stronger changes in glycemic markers per unit change of BMI or weight than did G carriers. Changes in trunk mass and total fat mass did not significantly predict changes in glucose except for subjects with the dominant homozygous genotype in the high-fat diet group. For TT carriers, plasma insulin decreased with reductions in total mass and trunk fat mass only in the low-fat diet group [β coefficient (95% CI): 1.01 (0.30, 1.71) and 0.88 (0.36, 1.40), respectively].
TABLE 2.
β coefficients (95% CIs) of changes in body composition as predictors of changes in plasma glucose and insulin at 6 mo by diet type and TCF7L2 rs12255372 genotype1
| GG |
GT |
TT |
||||
| Predictor | Low fat | High fat | Low Fat | High fat | Low fat | High fat |
| Change in plasma glucose (mg/dL) | ||||||
| Δ Weight | 0.32 (0.11, 0.53) | 0. 29 (0.08, 0.50) | 0.26 (0.01, 0.52) | 0.34 (0.08, 0.60) | 0.58 (0.20, 0.97) | 0.39 (−0.50, 1.28) |
| P | 0.003 | 0.006 | 0.045 | 0.010 | 0.006 | 0.362 |
| Δ BMI | 0.90 (0.29, 1.52) | 0.90 (0.28, 1.52) | 0.82 (0.07, 1.60) | 0.92 (0.18, 1.66) | 1.78 (0.63, 2.92) | 1.30 (−1.42, 4.01) |
| P | 0.004 | 0.005 | 0.031 | 0.016 | 0.005 | 0.326 |
| Δ Total fat | 0.52 (−0.16, 1.19) | 0.68 (0.19, 1.16) | 0.48 (−0.28, 1.23) | 0.44 (−0.27, 1.15) | 1.06 (−0.27, 2.39) | 2.45 (−0.81, 5.71) |
| P | 0.131 | 0.007 | 0.211 | 0.221 | 0.100 | 0.119 |
| Δ Trunk fat | 0.30 (−0.21, 0.81) | 0.49 (0.13, 0.85) | 0.41 (−0.16, 0.67) | 0.31 (−0.20, 0.83) | 0.92 (−0.11, 1.95) | 1.58 (−0.59, 3.75) |
| P | 0.248 | 0.009 | 0.157 | 0.226 | 0.071 | 0.129 |
| Change in plasma insulin (mg/dL) | ||||||
| Δ Weight | 0.44 (0.22, 0.65) | 0.28 (0.13, 0.42) | 0.33 (0.17, 0.49) | 0.29 (0.16, 0.43) | 0.47 (0.14, 0.81) | 0.27 (−0.22, 0.76) |
| P | <0.0001 | 0.0003 | <0.0001 | <0.0001 | 0.009 | 0.265 |
| Δ BMI | 1.28 (0.65, 1.91) | 0.86 (0.43, 1.28) | 0.96 (0.49, 1.44) | 0.84 (0.45, 1.23) | 1.46 (0.48, 2.44) | 1.02 (−0.46, 2.49) |
| P | 0.0001 | 0.0001 | 0.0001 | <0.0001 | 0.008 | 0.163 |
| Δ Total fat | 0.32 (−0.46, 1.09) | 0.33 (−0.84, 1.50) | 0.74 (0.29, 1.20) | 0.68 (0.28, 1.07) | 1.01 (0.30, 1.71) | 0.31 (−1.41, 2.03) |
| P | 0.419 | 0.172 | 0.002 | 0.001 | 0.017 | 0.683 |
| Δ Trunk fat | 0.20 (−0.38, 0.78) | 0.27 (0.01, 0.53) | 0.52 (0.17, 0.86) | 0.47 (0.18, 0.76) | 0.88 (0.36, 1.40) | 0.17 (−0.97, 1.31) |
| P | 0.497 | 0.042 | 0.004 | 0.002 | 0.009 | 0.735 |
Values were obtained from linear mixed models by diet type and genotype for continuous changes in blood glucose and insulin adjusted for age, sex, center, and baseline measure. Low-fat randomized diets had a target for total fat of 20% from total energy; high-fat randomized diets had a target for total fat of 40% from total energy. n = 591 for models with weight and BMI as predictors, and n = 326 for models with dual-energy X-ray absorptiometry total fat and trunk fat measures as predictors.
DISCUSSION
We report that overweight individuals with the risk genotype of TCF7L2 rs12255372 may reduce body adiposity by consuming a diet low in total fat, and those changes may translate into additional reductions in blood glucose and insulin. The changes were observed after 6 mo with an energy-restricted intervention when maximum weight-loss was achieved. The genotype-by-intervention interaction disappeared after 6 mo of intervention, which was likely because of diminished adherence to randomized dietary targets from 6 mo to 2 y (19). The Pounds Lost trial measured adherence by assessing the reported intake of macronutrient targets and biomarkers of nutrient intake (19, 20). However, the self-reported dietary-assessment tools used to determine target proximity might have been be prone to misreporting, and biomarker data suggested that individuals in the diet groups might have been consuming comparable macronutrient content as the study progressed, which thus concealed any detectable differences (20, 25). The achievement of full compliance in dietary trials is not viable beyond 6–12 mo (25); thus, any possible benefits from low-fat diets for individuals with the risk TCF7L2 genotype on long-term changes in weight and body adiposity may be limited and would need to be evaluated in trials with stronger adherence.
Grau et al (17) reported that obese, white individuals with the rs7903146 risk genotype had significantly greater decreases in fat-free mass and fat mass after 10 wk of consumption of a reduced-calorie low-fat (20–25%) diet than did individuals who consumed a high-fat (40–45%) diet. We also report a significant interaction for rs7903146 and randomized diet on the change in lean mass, but the greater decrease was observed for subjects with the common genotype. We report interactions with TCF7L2 rs12255372, whereby BMI and total and trunk body fat percentages decreased more in participants with the TT risk genotype when assigned to a diet with ∼20% than when assigned to a diet with ∼40% of calories from fat. Although these results were reported for rs7903146, Grau et al (17) observed similar results because individuals with the TCF7L2 risk variant responded better for changes in body weight and composition when consuming a low-fat diet of comparable composition as in the Pounds Lost trial. However, Haupt et al (10) reported that carriers of the risk allele for both SNPs had significantly less reduction in total body fat and visceral and nonvisceral fat after 9 mo of a restricted-calorie intervention in which all participants consumed <30% fat. The authors did not analyze interactions with macronutrients. The Diabetes Prevention Program did not find an interaction between genotype and a lifestyle intervention with recommended 25% fat on weight changes (26) and suggested that TCF7L2 may not determine the ability to lose weight. Differences in dietary composition, time frame, other lifestyle changes, and populations may have accounted for discrepancies between studies.
Reductions in markers of glycemic control by changes in body fat were observed for rs12255372 C carriers in either the low- or high-fat diet group but for TT carriers only in the low-fat diet group; these results were consistent with the observation that TT carriers have a greater benefit on body composition with this diet. Inconsistencies in the results for DXA measurements might have been due to limited statistical power. Other general lifestyle–intervention studies have shown improved plasma glucose and insulin homeostasis and insulin sensitivity mediated by decreases in adiposity in overweight men (27) and obese postmenopausal women (28). Our results suggested that such glycemic benefits may not occur for individuals with the TT risk variant when they consume diets with a higher fat content, which is an observation that emphasizes the importance of following diabetes-prevention dietary strategies and should be further examined in intervention studies.
A recent article by Fisher et al (16) showed that TCF7L2 HapA (which is a haplotype derived from rs7903146 and rs10885406) attenuated a positive association between animal-protein intake and long-term weight changes in middle-aged Europeans. In our study, the interaction between protein intake and rs12255372 on changes in lean mass was not significant after adjustment for multiple tests. Pounds Lost participants were not instructed to consume a specific type of protein source, and our observation was with the rs12255372 SNP; thus, we may not be able to draw direct comparisons between studies. However, we observed a significant interaction between rs7903146 and dietary fat, whereby carriers of risk variant lost similar lean mass in both diet groups, and these subjects showed less loss of lean mass when consuming the highest-carbohydrate diet. Because the highest carbohydrate content corresponded to the diet assigned for a lower fat content in our study, and animal -protein sources may have a higher fat content, it may be difficult to determine which nutrient influences lean mass in conjunction with this SNP.
Fisher et al (16) suggested that, because the rs7903146 HapA variant arose during a transition from hunter-gatherer to agricultural practices (and, therefore, a reduction in protein sources), carriers of the variant were selectively adapted to maintain weight stability under low-protein conditions. It may be possible that concurrent environmental pressures of less availability of fat but more plant-based carbohydrates, selected for lean mass preservation in addition to weight stability for individuals with this variant. This speculation and the potential mechanisms by which TCF7L2 rs7903146 interacts with macronutrients to affect lean mass warrant additional scrutiny.
How dietary fats and TCF7L2 modulate responses in weight and adiposity remains unclear. In light of the available literature, it may be possible that all 3 macronutrients modulate the effect of TCF7L2 through different pathways. In our study, TCF7L2 variants influenced fat depots only in response to dietary fat intake, whereas the interaction with protein intake affected lean mass. Collectively, previous reports as well as our study may encourage additional molecular studies to help elucidate the mechanisms of TCF7L2 under various dietary conditions.
In our cohort, TCF7L2 rs12255372 and rs7903146 were in moderate LD, which was similar to what has been reported in HapMap European populations (18). Most studies referenced in the current study that analyzed both polymorphisms for gene-diet interactions reported parallel results between SNPs. The reason for the lack of replication with rs7903146 in our study is unclear. Although the use of regression methods to analyze multiple correlated loci as the predictor variable is appropriate for markers that are not in strong LD, it is difficult to discriminate the individual variation likely to be causally associated to the outcome (29).
Alternative splicing of TCF7L2 expression in adipose tissue has been reported, although there was no difference by variants (30). However, a recent study suggested that the TCF7L2 risk alleles analyzed in our study might act through gene-expression levels rather than alternative splicing, with the expression levels of TCF7L2 mRNA being between 1.5- and 3-fold higher for individuals homozygous for both rs7903146 and rs12255372 risk alleles compared with individuals homozygous for the common alleles, depending on the tested cell line (31). In addition, these SNPs were in strong LD with 5 additional variants in the region, and some of them showed differential enhancer activity. It is possible that multiple loci in TCF7L2 with varied functionality may act together or additively to affect the associations with adiposity and diabetes outcomes. Additional tissue-specific functional studies may help elucidate these discrepancies.
An important strength of our study was that Pounds Lost is one of the largest and longest weight-loss trials to date. The design allowed for control of behaviors that may confound associations, such as the use of medications, smoking, and alcohol consumption because these did not differ by randomization group (19). One limitation of our study was that compliance to randomized diet goals was not fully achieved, which may have created some misclassification, as previously discussed. In addition, we could not explore the effect of specific types of fat or fatty acids. Studies have reported interactions between TCF7L2 variants and saturated fat specifically (11, 12) in the same direction as our results for total fat. Although all Pounds Lost participants were instructed to consume ≤8% of saturated fat, it is likely that subjects in the low-fat diet group consumed less saturated fat or had a different ratio of types of fat. However, confirmation of this would imply the use of reported intake, which would invalidate the random assignment. Because gene interactions with other macronutrients have been reported, it might be difficult to distinguish the effects of these nutrients compared with those of fat. We also restricted analysis to white individuals as other researchers have done (3, 15) to avoid bias from population stratification. Replication in other racial-ethnic groups is warranted, because inconsistent evidence for the effect of TCF7L2 across ethnic groups has been reported (2, 4, 32, 33).
In conclusion, our results suggest that overweight and obese individuals with TCF7L2 risk variants may obtain greater benefit of reducing body fat and mass by consuming diets low in dietary fat (∼20% from total energy), and the changes may improve glycemic control. Our results may encourage individuals with increased genetic susceptibility from TCF7L2 to further reduce risk of type 2 diabetes by adhering to current prevention guidelines of limiting dietary fat intake.
Acknowledgments
We thank participants of the Pounds Lost trial for their contribution to the study.
The authors’ responsibilities were as follows—JM: designed the research question, researched data, and wrote the manuscript; QQ: contributed to the data analysis; FBH: contributed to the discussion of results and reviewed and edited the manuscript; FMS: directed the study and reviewed the manuscript; LQ: designed the research question, contributed to discussion, and reviewed and edited the manuscript; and all authors: reviewed and approved the final manuscript. None of the authors had a conflict of interest.
Footnotes
Abbreviations used: DXA, dual-energy X-ray absorptiometry; LD, linkage disequilibrium; Pounds Lost, Preventing Overweight Using Novel Dietary Strategies; SNP, single-nucleotide polymorphism.
REFERENCES
- 1.Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, Helgason A, Stefansson H, Emilsson V, Helgadottir A, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet 2006;38:320–3 [DOI] [PubMed] [Google Scholar]
- 2.Florez JC, Jablonski KA, Bayley N, Pollin TI, de Bakker PI, Shuldiner AR, Knowler WC, Nathan DM, Altshuler D. TCF7L2 polymorphisms and progression to diabetes in the Diabetes Prevention Program. N Engl J Med 2006;355:241–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang C, Qi L, Hunter DJ, Meigs JB, Manson JE, van Dam RM, Hu FB. Variant of transcription factor 7-like 2 (TCF7L2) gene and the risk of type 2 diabetes in large cohorts of U.S. women and men. Diabetes 2006;55:2645–8 [DOI] [PubMed] [Google Scholar]
- 4.Waters KM, Stram DO, Hassanein MT, Le Marchand L, Wilkens LR, Maskarinec G, Monroe KR, Kolonel LN, Altshuler D, Henderson BE, et al. Consistent association of type 2 diabetes risk variants found in Europeans in diverse racial and ethnic groups. PLoS Genet 2010;6:e1001078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Timpson NJ, Lindgren CM, Weedon MN, Randall J, Ouwehand WH, Strachan DP, Rayner NW, Walker M, Hitman GA, Doney AS, et al. Adiposity-related heterogeneity in patterns of type 2 diabetes susceptibility observed in genome-wide association data. Diabetes 2009;58:505–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jin T, Liu L. The Wnt signaling pathway effector TCF7L2 and type 2 diabetes mellitus. Mol Endocrinol 2008;22:2383–92 [DOI] [PubMed] [Google Scholar]
- 7.Cauchi S, Choquet H, Gutierrez-Aguilar R, Capel F, Grau K, Proenca C, Dina C, Duval A, Balkau B, Marre M, et al. Effects of TCF7L2 polymorphisms on obesity in European populations. Obesity (Silver Spring) 2008;16:476–82 [DOI] [PubMed] [Google Scholar]
- 8.Wang J, Kuusisto J, Vanttinen M, Kuulasmaa T, Lindstrom J, Tuomilehto J, Uusitupa M, Laakso M. Variants of transcription factor 7-like 2 (TCF7L2) gene predict conversion to type 2 diabetes in the Finnish Diabetes Prevention Study and are associated with impaired glucose regulation and impaired insulin secretion. Diabetologia 2007;50:1192–200 [DOI] [PubMed] [Google Scholar]
- 9.Helgason A, Palsson S, Thorleifsson G, Grant SF, Emilsson V, Gunnarsdottir S, Adeyemo A, Chen Y, Chen G, Reynisdottir I, et al. Refining the impact of TCF7L2 gene variants on type 2 diabetes and adaptive evolution. Nat Genet 2007;39:218–25 [DOI] [PubMed] [Google Scholar]
- 10.Haupt A, Thamer C, Heni M, Ketterer C, Machann J, Schick F, Machicao F, Stefan N, Claussen CD, Haring HU, et al. Gene variants of TCF7L2 influence weight loss and body composition during lifestyle intervention in a population at risk for type 2 diabetes. Diabetes 2010;59:747–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Phillips CM, Goumidi L, Bertrais S, Field MR, McManus R, Hercberg S, Lairon D, Planells R, Roche HM. Dietary saturated fat, gender and genetic variation at the TCF7L2 locus predict the development of metabolic syndrome. J Nutr Biochem 2012;23:239–44 [DOI] [PubMed] [Google Scholar]
- 12.Delgado-Lista J, Perez-Martinez P, Garcia-Rios A, Phillips CM, Williams CM, Gulseth HL, Helal O, Blaak EE, Kiec-Wilk B, Basu S, et al. Pleiotropic effects of TCF7L2 gene variants and its modulation in the metabolic syndrome: from the LIPGENE study. Atherosclerosis 2011;214:110–6 [DOI] [PubMed] [Google Scholar]
- 13.Ruchat SM, Elks CE, Loos RJ, Vohl MC, Weisnagel SJ, Rankinen T, Bouchard C, Perusse L. Evidence of interaction between type 2 diabetes susceptibility genes and dietary fat intake for adiposity and glucose homeostasis-related phenotypes. J Nutrigenet Nutrigenomics 2009;2:225–34 [DOI] [PubMed] [Google Scholar]
- 14.Fisher E, Boeing H, Fritsche A, Doering F, Joost HG, Schulze MB. Whole-grain consumption and transcription factor-7-like 2 (TCF7L2) rs7903146: gene-diet interaction in modulating type 2 diabetes risk. Br J Nutr 2009;101:478–81 [DOI] [PubMed] [Google Scholar]
- 15.Cornelis MC, Qi L, Kraft P, Hu FB. TCF7L2, dietary carbohydrate, and risk of type 2 diabetes in US women. Am J Clin Nutr 2009;89:1256–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fisher E, Meidtner K, Angquist L, Holst C, Hansen RD, Halkjaer J, Masala G, Ostergaard JN, Overvad K, Palli D, et al. Influence of dietary protein intake and glycemic index on the association between TCF7L2 HapA and weight gain. Am J Clin Nutr 2012;95:1468–76 [DOI] [PubMed] [Google Scholar]
- 17.Grau K, Cauchi S, Holst C, Astrup A, Martinez JA, Saris WH, Blaak EE, Oppert JM, Arner P, Rossner S, et al. TCF7L2 rs7903146-macronutrient interaction in obese individuals’ responses to a 10-wk randomized hypoenergetic diet. Am J Clin Nutr 2010;91:472–9 [DOI] [PubMed] [Google Scholar]
- 18.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O'Donnell CJ, de Bakker PI. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics 2008;24:2938–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sacks FM, Bray GA, Carey VJ, Smith SR, Ryan DH, Anton SD, McManus K, Champagne CM, Bishop LM, Laranjo N, et al. Comparison of weight-loss diets with different compositions of fat, protein, and carbohydrates. N Engl J Med 2009;360:859–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Souza RJ, Bray GA, Carey VJ, Hall KD, LeBoff MS, Loria CM, Laranjo NM, Sacks FM, Smith SR. Effects of 4 weight-loss diets differing in fat, protein, and carbohydrate on fat mass, lean mass, visceral adipose tissue, and hepatic fat: results from the POUNDS LOST trial. Am J Clin Nutr 2012;95:614–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qi Q, Bray GA, Smith SR, Hu FB, Sacks FM, Qi L. Insulin receptor substrate 1 gene variation modifies insulin resistance response to weight-loss diets in a 2-year randomized trial: the Preventing Overweight Using Novel Dietary Strategies (POUNDS LOST) trial. Circulation 2011;124:563–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu FB. Measurements of adiposity and body composition. In: Obesity epidemiology. New York, NY: Oxford University Press, 2008:53–83.
- 23.Liu K, Muse SV. PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 2005;21:2128–9 [DOI] [PubMed] [Google Scholar]
- 24.Storey JD. The positive false discovery rate: a Bayesian interpretation and the q-value. Ann Stat 2003;31:2013–35 [Google Scholar]
- 25.Astrup A, Pedersen SD. Is a protein calorie better for weight control? Am J Clin Nutr 2012;95:535–6 [DOI] [PubMed] [Google Scholar]
- 26.McCaffery JM, Jablonski KA, Franks PW, Dagogo-Jack S, Wing RR, Knowler WC, Delahanty L, Dabelea D, Hamman R, Shuldiner AR, et al. TCF7L2 polymorphism, weight loss and proinsulin:insulin ratio in the diabetes prevention program. PLoS ONE 2011;6:e21518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Borel AL, Nazare JA, Smith J, Almeras N, Tremblay A, Bergeron J, Poirier P, Despres JP. Improvement in insulin sensitivity following a 1-year lifestyle intervention program in viscerally obese men: contribution of abdominal adiposity. Metabolism 2012;61:262–72 [DOI] [PubMed] [Google Scholar]
- 28.Elisha B, Rabasa-Lhoret R, Messier V, Abdulnour J, Karelis AD. Relationship between the body adiposity index and cardiometabolic risk factors in obese postmenopausal women. Eur J Nutr 2012; (in press). [DOI] [PubMed] [Google Scholar]
- 29.Malo N, Libiger O, Schork NJ. Accommodating linkage disequilibrium in genetic-association analyses via ridge regression. Am J Hum Genet 2008;82:375–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prokunina-Olsson L, Kaplan LM, Schadt EE, Collins FS. Alternative splicing of TCF7L2 gene in omental and subcutaneous adipose tissue and risk of type 2 diabetes. PLoS ONE 2009;4:e7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pang DX, Smith AJ, Humphries SE. Functional analysis of TCF7L2 genetic variants associated with type 2 diabetes. Nutr Metab Cardiovasc Dis (Epub ahead of print 6 March 2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palmer ND, Lehtinen AB, Langefeld CD, Campbell JK, Haffner SM, Norris JM, Bergman RN, Goodarzi MO, Rotter JI, Bowden DW. Association of TCF7L2 gene polymorphisms with reduced acute insulin response in Hispanic Americans. J Clin Endocrinol Metab 2008;93:304–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palmer ND, Hester JM, An SS, Adeyemo A, Rotimi C, Langefeld CD, Freedman BI, Ng MC, Bowden DW. Resequencing and analysis of variation in the TCF7L2 gene in African Americans suggests that SNP rs7903146 is the causal diabetes susceptibility variant. Diabetes 2011;60:662–8 [DOI] [PMC free article] [PubMed] [Google Scholar]