Abstract
Take a look at a textbook illustration of a cell and you will immediately be able to locate the nucleus, which is often drawn as a spherical or ovoid shaped structure. But not all cells have such nuclei. In fact, some disease states are diagnosed by the presence of nuclei that have an abnormal shape or size. What defines nuclear shape and nuclear size, and how does nuclear geometry affect nuclear function? While the answer to the latter question remains largely unknown, significant progress has been made towards understanding the former. In this review, we provide an overview of the factors and forces that affect nuclear shape and size, discuss the relationship between ER structure and nuclear morphology, and speculate on the possible connection between nuclear size and its shape. We also note the many interesting questions that remain to be explored.
Keywords: nuclear shape, endoplasmic reticulum, mitosis, nuclear envelope, laminopathies
The defining feature of the eukaryotic cell is the nucleus, a double membrane-bound compartment that contains the cell’s chromosomes [Pederson, 2011]. The nucleus is enclosed by the nuclear envelope (NE), which is comprised of two lipid bilayers, the inner nuclear membrane (INM) and the outer nuclear membrane (ONM) [Wilson and Berk, 2010] (Fig. 1a). In metazoans, underlying the INM is the nuclear lamina, a network of lamin filaments and associated proteins [Wilson and Berk, 2010]. The inner and outer nuclear membranes are connected at sites of nuclear pore complexes (NPCs) (Fig. 1b). NPCs comprise multiple copies of ~30 nucleoporin (Nup) proteins, which form a ~60 MDa structure that spans the NE and allows transport of molecules between the cytoplasm and the nucleus [Wente, 2000; Hetzer et al., 2005]. The central pore of the NPC has a passive diffusion limit of around 40 kDa, with larger molecules requiring facilitated transport mechanisms [Wente and Rout, 2010]. The density of NPCs on the NE varies between organisms and cell types, with a calculated density of 14.6 NPCs/μm2 in S. cerevisiae [Winey et al., 1997] and 11 NPCs/μm2 in cultured HeLa cells [Maul and Deaven, 1977]. Despite their connection at NPCs, the ONM and the INM differ in their protein composition: The ONM is continuous with the endoplasmic reticulum (ER), is studded with ribosomes, and contains many of the same proteins that reside in the ER, and which do not move into the INM [Hetzer et al., 2005; English et al., 2009]. The INM, on the other hand, contains proteins that interact with the nuclear lamina, chromatin-associated proteins, and other nuclear proteins [Zuleger et al., 2011]. Interestingly, the distance between the INM and ONM is constant, likely due to a protein complex, the LINC complex, which spans both membranes (see below).
Fig. 1.
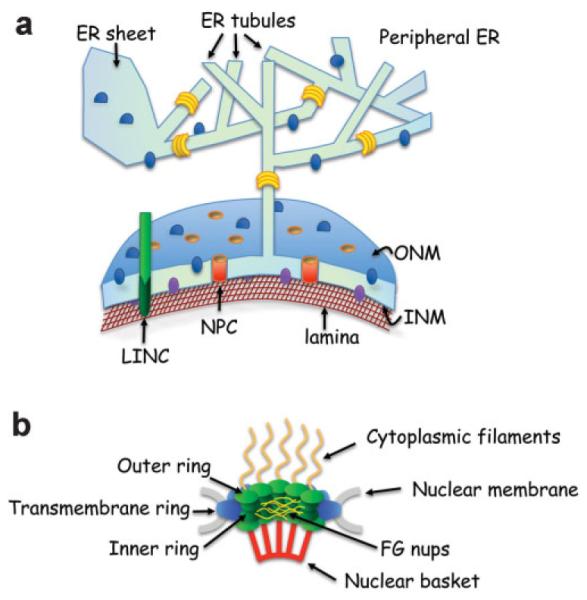
The nuclear envelope. a: The nuclear envelope (NE) is composed of an outer nuclear membrane (ONM) and an inner nuclear membrane (INM), which meet at sites where nuclear pore complexes (NPCs, in orange, see panel b for details) are embedded. The ONM is continuous with the ER membrane, and the lumen between the ONM and INM is continuous with the ER lumen (in light green). Most proteins (blue spheres) and ribosomes (not shown) that reside in the peripheral ER can diffuse to the ONM. However, some proteins, such as the reticulons (in yellow), are confined to highly curved membrane; they are enriched in the peripheral ER that is in the form of tubules, and are present in the curved memrbane that is around the NPCs (not shown). The protein composition of the INM (purple spheres) is distinct from that of the ONM. Underlying the INM is the nuclear lamina (brown meshwork). The LINC complex (in green) spans the ONM and INM (see Fig. 2). b: A cross section through the NPC, with its main parts highlighted. Note that each of the structures shown is made up of various proteins.
In metazoans, the nuclear lamina contributes to the rigidity of the nucleus, allowing it to withstand pushing/pulling forces and enabling nuclear migration during development [Dahl et al., 2008; Aoki et al., 2011]. Moreover, connections between the nuclear lamina and chromatin likely play a role in genome organization, for example by facilitating the positioning of chromosomes into defined regions, or “territories”, within the nucleus [Cremer et al., 2006]. In all organisms, interactions between nuclear periphery proteins and chromatin also play a complex role in transcriptional control [Brickner and Walter, 2004; Casolari et al., 2004; Akhtar and Gasser, 2007; Egecioglu and Brickner, 2011]: In some cases, positioning of chromatin at the nuclear periphery is associated with transcriptional silencing, while in others it leads to transcriptional activation. In general, however, heterochromatin is positioned near the nuclear periphery, and this localization is altered in cells of patients with mutations in genes coding for components of the nuclear lamina [Zuleger et al., 2011]. Finally, tethering of chromatin to the NE plays a role in DNA repair and recombination [Mekhail and Moazed, 2010; Oza and Peterson, 2010]. Thus, the NE is much more that just a diffusion barrier between the nucleus and cytoplasm.
The nucleus is not a static structure. The most dramatic changes to nuclear structure occur during mitosis, when chromosome segregation takes place. Different cell types undergo mitosis via different strategies. Most eukaryotic cells undergo “open” mitosis, where the NE breaks down completely at the onset of mitosis; the lamina is disassembled, NPCs dismantle into sub-complexes, and NE proteins and lipids are absorbed into the peripheral ER [Ellenberg et al., 1997; Yang et al., 1997; Daigle et al., 2001; Mattaj, 2004]. NE breakdown allows the centrosome-bound microtubules in the cytoplasm to access the chromosomes and instigate chromosome segregation. Once mitosis is complete, the NE reforms around the chromosomes of each daughter cell and expands to form the two daughter nuclei. In contrast, some fungal species undergo “closed” mitosis, during which the NE remains intact, and chromosomes are segregated by microtubules that emanate from NE-embedded structures called spindle pole bodies [Jaspersen and Winey, 2004]. In this case, the nucleus elongates to accommodate the movement of the segregating chromosomes, and this is followed by nuclear division and rounding of the two daughter nuclei. There are also variations on the open or closed mechanisms of mitosis. For example, in Aspergillus nidulans, large holes form in the NE during mitosis due to partial NPC disassembly [De Souza and Osmani, 2007]. In Schizosaccharomyces japonicas, the nuclear memrbane ruptures during anaphase as the spindle elongates in a nucleus that does not sufficiently increase in surface area [Aoki et al., 2011; Yam et al., 2011]. If fatty acid synthesis is inhibited in S. pombe, the NE is unable to grow and, as in S. japonicus, nuclei become temporarily fusiform in shape and the mitotic spindle buckles [Yam et al., 2011]. However, under these conditions the S. pombe nuclear membrane does not rupture, suggesting that S. japonicus probably has a mechanism that enables nuclear membrane rupture. Regardless of the flavor of mitosis used by different cell types, in all cases the daughter cell nuclei somehow acquire a shape and size that was characteristic of the interphase nucleus of the parent cell.
Our understanding of how nuclear shape and size are determined is rather poor. Although we know that altered nuclear shape and size are associated with aging and certain disease states, such as cancer [Zink et al., 2004; Capell and Collins, 2006; Webster et al., 2010; Chow et al., 2012], our knowledge of the role that nuclear shape plays in nuclear function is very limited. In this review we discuss the cellular components and processes that play a role in determining nuclear shape and go on to consider what determines nuclear size. We speculate on how nuclear shape could be affected by nuclear size, and specifically by the requirement for a constant nuclear/cytoplasmic volume ratio. The internal organization of the nucleus is not discussed here in detail; it has been the subject of many excellent reviews, and those are cited throughout this article.
CELLULAR COMPONENTS THAT AFFECT NUCLEAR SHAPE
THE NUCLEAR PERIPHERY
Based on the extensive association between the NE and the nuclear lamina, it stands to reason that the nuclear lamina can affect nuclear shape. Lamins, the major component of the nuclear lamina, are present in all the metazoans but absent from plants and unicellular eukaryotes [Dittmer and Misteli, 2011; Simon and Wilson, 2011], although a protein similar to the metazoan lamin that affects nuclear morphology was recently discovered in Dictyostelium [Kruger et al., 2011]. There are two major types of nuclear lamins: A-type and B-type. In mammals A-type lamins are encoded by LMNA, and B-type lamins are encoded by LMNB1 and LMNB2. Lamin genes have multiple splice variants (for example, Lamin C is a splice variant of the LMNA gene). Most invertebrates express only one lamin isoform, similar to the mammalian B-type lamin. Lamin gene expression is developmentally regulated: B-type lamins are expressed throughout development, while A-type lamins are expressed in differentiated cells [Dechat et al., 2008].
Certain mutations in genes coding for lamins, lamin processing enzymes or lamin associated proteins result in heritable diseases known as laminopathies [Capell and Collins, 2006]. These diseases can affect skeletal muscle, cardiac muscle, adipose tissue, or bone, and can lead to progeria-like (premature aging) symptoms. One of the most striking cellular features observed in these laminopathies is abnormal nuclear morphology, underscoring the central role that the nuclear lamina plays in controlling nuclear shape. For example, Hutchinson-Gilford progeria syndrome (HGPS) is caused by a mutation in LMNA that leads to an alternatively spliced product known as progerin. Cells from HGPS patients exhibit misshaped nuclei and nuclear blebbing [Worman, 2012]. Interestingly, normal aging cells exhibit similar phenotypes, including the accumulation of progerin and the deformation of nuclear shape [Scaffidi and Misteli, 2006]. Given the link between the NE, the nuclear lamina and chromatin, it is tempting to speculate that changes in nuclear morphology can have deleterious affects on gene expression and DNA repair, thereby contributing to aging or disease process.
Under certain conditions, parts of the NE extend into the nuclear interior. These structures, known as the nucleoplasmic reticulum [Malhas et al., 2011], are comprised of invaginations into the nucleus of either the INM alone or both the INM and ONM. In the latter case the invaginations also contain NPCs and nuclear lamina. While the nucleoplasmic reticulum can be seen in cultured cells and can be induced by over expression of certain NE proteins or alterations in lipid composition, it is also clear that it exists in certain cell types under physiological conditions and in certain disease states, most notably cancer. The function(s) of the nucleoplasmic reticulum, its prevalence and its contribution to overall nuclear structure remain to be explored.
Yeast lack lamins, but other components of the NE have been shown to affect nuclear morphology. For example, altered levels of certain NPC components, such as depletion of Nup1 (an FG nucleoporin, Fig. 1b) or Nup85 (in the outer ring, Fig. 1b), leads to the formation of projections from the nucleus that protrude into the cytoplasm [Bogerd et al., 1994; Goldstein et al., 1996]. Depletion of Nup170 (inner ring, Fig. 1b) in a strain that is lacking the transmembrane NPC protein, Pom152 (lumenal ring, Fig. 1b), leads to nuclear deformations [Aitchison et al., 1995]. One potential mechanism by which changes in NPC components could affect nuclear shape is through altered interactions between the NE and the nuclear interior. It is conceivable that yeast have a structure that is functionally analogous to the nuclear lamina of higher eukaryotes and that contributes to nuclear integrity. If altered NPC composition disrupts connections between the NE and structural proteins within the nucleus, the NE could be more easily pulled away from the main body of the nucleus, for example during nuclear migration. Alternatively, NPCs may affect their surrounding membrane environment [Witkin et al., 2010]. The Mps3 protein, although not part of the NPC, is an integral INM protein that was recently shown to affect membrane composition (see below) [Friederichs et al., 2011]. Therefore, altering NPCs, which are abundant in the NE, could lead to uncontrolled proliferation of the NE and changes in nuclear shape.
In both lower and higher eukaryotes the NE contains proteins, known as the KASH- and SUN-domain proteins, which physically link the cytoskeleton with either the nuclear lamina (in higher eukaryotes) or chromatin (in yeast). KASH (Klarsicht-Anc1-syne1 homology) domain proteins are anchored in the ONM, while SUN (Sad1/UNC-84) domain proteins are anchored in the INM [Fridkin et al., 2009; Razafsky and Hodzic, 2009] (Fig. 2). The C-termini of the KASH-domain and SUN-domain proteins interact with each other in the lumenal space of the NE, forming trans-nuclear membrane bridges called LINC (for linkers of the nucleoskeleton to the cytoskeleton) complexes [Crisp et al., 2006]. The N-termini of the KASH and SUN domain proteins are located in the cytoplasm and nucleoplasm, respectively. The cytoplasmic portion of the KASH-domain proteins interacts with various components of the cytoskeleton, including actin filaments and microtubules. In higher eukaryotes the nucleoplasmic domains of the SUN-domain proteins interact with nuclear lamina proteins, while in yeast they interact with chromatin [Bupp et al., 2007; Oza et al., 2009]. The LINC complex plays a key role in nuclear positioning, chromosome movement, and mechanotransduction [Dahl et al., 2008]. It also appears to affect both the integrity of the NE and nuclear shape. For example, in HeLa cell lines, down regulation of the SUN-domain proteins SUN1 and SUN2 increases the space between the INM and ONM [Crisp et al., 2006]. Furthermore, in granulocytes (neutrophils), which are a cell type that normally has multilobed nuclei, the levels of certain KASH and SUN domain proteins are reduced [Olins et al., 2009]. This suggests that the expression levels of LINC complex proteins might be regulated in order to control the shape and architecture of the nucleus. Consistent with this possibility, reducing the levels of a particular isoform of nesprin (a KASH-domain protein) leads to a range of nuclear shape changes, from minor NE blebbing to severely misshapen and giant nuclei [Luke et al., 2008]. Similarly, in Dictyostelium, expression of a mutant form of the Sun-1 protein, which lacks a chromatin-binding domain, leads to severe nuclear deformities [Xiong et al., 2008]. Thus, the LINC complex, along with other proteins at the nuclear periphery, plays a central role in defining nuclear shape.
Fig. 2.
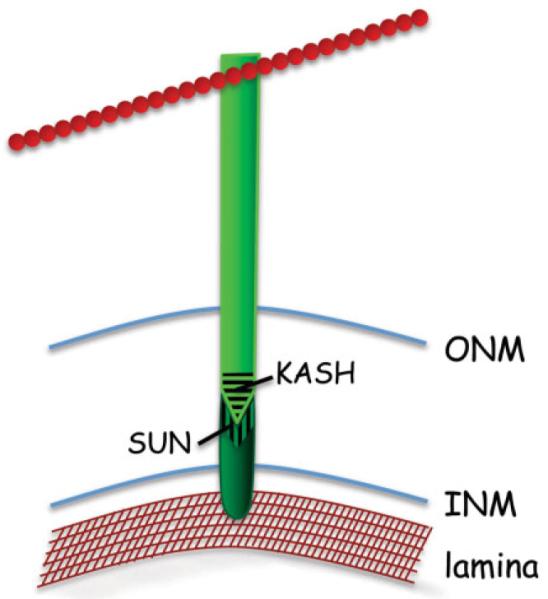
The LINC complex. The LINC complex is composed of two protein types: a KASH-domain protein (light green), which spans the ONM, and a SUN-domain protein (dark green), which spans the INM. The KASH and SUN domain proteins interact in the NE lumen via their respective domains (striated regions). KASH-domain proteins interact in the cytoplasm with elements of the cytoskeleton (red), such as microtubules and actin cable. SUN-domain proteins interact in the nucleoplasm with the nuclear lamina (brown mesh-work).
MEMBRANES OF THE ENDOPLASMIC RETICULUM
Since the nucleus is bound by a NE, and the NE is part of the ER, it is conceivable that maintaining correct nuclear size and shape during interphase relies on mechanisms that partition lipids and proteins between the peripheral ER and NE. This could be accomplished by the affinity of peripheral ER proteins to cytoskeletal elements and NE proteins to nuclear proteins, or by diffusion barriers between the different ER membranes. For example, NPCs likely play an important role in controlling the movement of proteins between the ONM and INM [Ohba, 2004]. INM proteins are synthesized in the peripheral ER and diffuse to the ONM before being transported to the INM via NPCs [Mattaj, 2004]. A recent study using Xenopus extracts showed that depletion of the NPC protein Nup188 (inner ring, Fig. 1b) led to increased rate of transport of proteins into the INM and a subsequent increase in nuclear size [Theerthagiri et al., 2010]. Notably, Nup188 depletion only affected rate of transport and did not compromise the size stringency of protein transport into the INM, which is normally restricted to 40 kDa [Ohba, 2004], nor did it allow entry of ONM or ER proteins into the INM [Theerthagiri et al., 2010]. Thus, Nup188 and other NPC-associated proteins may act as gatekeepers that control protein flux to the INM, thereby establishing the different domains of the ER and helping to maintain nuclear size and shape.
Our understanding of how lipids are added to the NE is rather limited. Where lipids are added to the NE and whether or not lipid can flow freely between the peripheral ER and the NE are open questions. If lipid is freely transferred between the peripheral ER and NE, then expansion of the ER membrane could lead to the expansion of the NE, potentially dragging the chromatin with it. There are several mutants that have been identified where peripheral ER and NE expansion occur concomitantly. For example, in yeast, deletion of certain genes affecting lipid biosynthesis (e.g. SPO7, NEM1, or PAH1, see below) or early protein secretion pathways (SEC31, SEC53, SAR1), leads to both peripheral ER and NE expansion [Siniossoglou et al., 1998; Matynia et al., 2002; Santos-Rosa et al., 2005; Campbell et al., 2006]. In C. elegans, inactivating the pathway controlled by the Pah1 homolog, known as lipin, leads to expansion of the ER and defects in NE breakdown and reassembly, which, in turn, affect nuclear shape [Golden et al., 2009; Gorjánácz and Mattaj, 2009]. However, there are also examples where ER expansion occurs without any effect on the NE, such as in S. cerevisiae opi1Δ mutants, [Wright et al., 1988; O’Hara et al., 2006] and vice versa [Friederichs et al., 2011], indicating that there is likely a barrier to membrane expansion between the peripheral ER and NE.
The membrane of the peripheral ER exists in two major forms: Membrane tubules and flat, or sheet-like, membrane, often in the shape of cisternae [English et al., 2009]. In contrast, the NE is a large membrane sheet. Thus, the partitioning of ER membrane between sheets and tubules could affect nuclear shape, NE expansion, and NE reformation after open mitosis. For example, it is conceivable that cells can control the amount of membrane available to the nucleus by regulating the amount of ER membrane that is captured into tubules. Several classes of proteins contribute to ER shape. The conserved reticulons and DP1/Yop1 are involved in stabilizing ER tubules [Voeltz et al., 2006; Hu et al., 2008; West et al., 2011]. The mammalian atlastins (Sey1 in yeast), which are dynamin-like GTPases, form connections between ER tubules [Hu et al., 2009; Orso et al., 2009]. In Xenopus, one of the reticulons, Rtn4a, is found at ER/ONM junctions and could play a role in facilitating nuclear growth by stabilizing regions of high curvature where membrane is added to the NE [Kiseleva et al., 2007]. Studies in C. elegans indicate that reticulons play an important role in NE breakdown prior to mitosis [Audhya et al., 2007]: Depletion of YOP1/RET1 in C. elegans blocked the release of INM proteins during mitosis and prevented NE breakdown [Audhya et al., 2007]. It was hypothesized that this blockage is caused by the altered ER morphology [Audhya et al., 2007]. Likewise, alteration of ER structure in C. elegans embryos by interfering with lipid synthesis also coincided with defects in NE breakdown, and resulted in abnormal nuclei in the subsequent cell cycle [Golden et al., 2009; Gorjánácz and Mattaj, 2009]. This link between NE disassembly and peripheral ER morphology demonstrates that the partitioning of proteins and membrane between different domains of the ER plays an important physiological role, and that the dynamics of the NE are dependent on the structure of the peripheral ER.
The balance between sheets and tubules in the peripheral ER is important not only for NE breakdown but also for NE reformation after open mitosis. Two different models have been proposed for the structure of the ER during open mitosis. Evidence from mammalian cells suggests that during mitosis the peripheral ER is almost entirely tubular [Puhka et al., 2007], and studies by Anderson and Hetzer [2007, 2008] have suggested that tubules are required for NE reformation. In contrast, Lu et al. [2009, 2011] found that during mitosis in mammalian cells the ER is almost entirely composed of flat membrane, and that the nucleus reforms from these membrane sheets. It is noteworthy, however, that in both cases, the ER remains as one continuous structure throughout the cell cycle. Regardless of whether the ER is in sheet or tubule form during mitosis, it is clear that dynamic changes that occur in the ER during the cell cycle can influence the NE. For example, over-expression of reticulons, which stabilize tubular ER, leads to slower NE reformation, whereas depletion of reticulons leads to an increased rate of NE reformation [Anderson and Hetzer, 2008]. The driving forces behind the changes in peripheral ER structure, how these changes are regulated throughout the cell cycle, and how peripheral ER dynamics impact nuclear shape are interesting areas for future study.
In cells that undergo closed mitosis, such as yeast, the nucleus must increase in size to allow chromosome segregation to occur. This is indeed the case in S. pombe and S. cerevisiae, where NE surface area increases during mitosis [Jorgensen et al., 2007; Neumann and Nurse, 2007]. During NE expansion, there is a requirement for coordinated growth between the INM and ONM in order to maintain even lumenal spacing and normal nuclear shape. How this co-ordination is achieved is not known, but the movement of lipids and proteins from the expanding ONM into the INM across the inner face of NPCs is likely to be important for this process. It has been demonstrated that lipid synthesis plays a role in NE expansion during closed mitosis. In S. cerevisiae, constitutive dephosphorylation of Pah1, which regulates phospholipid synthesis, prevents NE expansion and mitosis cannot be completed [Santos-Rosa et al., 2005]. How the addition of lipid to the NE is regulated is not known.
LIPID METABOLISM AND TRAFFICKING
Since phospholipids are a major component of the NE, lipid synthesis likely plays a role in determining nuclear shape and size. Direct evidence of a role for lipid synthesis in determining nuclear shape comes from studies of yeast with altered lipid metabolism, including the spo7Δ, nem1Δ, and pah1Δ mutants, mentioned earlier in this review [Siniossoglou, 2009]. The S. cerevisiae proteins Spo7 and Nem1 form an ER-associated phosphatase complex that activates the Pah1 protein through dephosphorylation [Santos-Rosa et al., 2005]. Pah1 is a key regulator of lipid metabolism; it converts phosphatidic acid (PA) to diacylglycerol (DAG) and it regulates the expression of lipid biosynthesis genes [Santos-Rosa et al., 2005; O’Hara et al., 2006]. Deletion of SPO7, NEM1, or PAH1 in S. cerevisiae causes ER expansion and the formation of an abnormally shaped nucleus [Siniossoglou et al., 1998; Santos-Rosa et al., 2005; Campbell et al., 2006]. S. cerevisiae nuclei are normally round and the nucleolus forms a cap-like structure close to the NE (Fig. 3). In spo7Δ, nem1Δ, or pah1Δ cells, the NE expands at the site of the nucleolus to form a “flare”, but the NE surrounding the bulk of the DNA maintains its normal round shape [Campbell et al., 2006] (Fig. 3). There are several possible explanations as to why NE expansion is limited to the nucleolar region: The NE adjacent to the nucleolus could be the site of lipid addition, or lipid could be added at other sites in the NE, but the region of membrane around the nucleolus is the most pliable and thus accumulates the extra membrane. The expansion of the NE only in the nucleolar region raises the possibility that there are different lipid domains within the NE, which have different physical properties. These domains could be established by the interactions between the NE and the underlying chromatin, which could affect membrane plasticity, or by altered membrane composition in different regions of the NE.
Fig. 3.

Uneven expansion of the budding yeast nucleus. In wild type budding yeast, the nucleolus (in red) forms a crescent shaped structure apposing the NE (in green) that caps the chromatin (in blue). In spo7Δ, nem1Δ, or pah1Δ mutant cells, the NE expands only in the region that is adjacent to the nucleolus, forming a “flare”, while the NE adjacent to the chromatin retains it original shape.
Recent studies in yeast have shown that vesicle trafficking plays an important role in regulating nuclear shape [Webster et al., 2010]. If a conditional spo7 mutant is combined with mutations in a subset of vesicle trafficking genes, NE expansion is no longer confined to the nucleolar region, and nuclei have a multi-flare morphology, with NE protrusions occurring around the entire nucleus [Webster et al., 2010]. These observations indicate that in the face of excess lipid, the confinement of NE expansion to the nucleolar region is dependent on vesicle trafficking [Webster et al., 2010]. The mechanism by which vesicle trafficking is involved in controlling nuclear shape in yeast, and whether vesicle trafficking affects nuclear shape in other organisms, remains to be elucidated.
Several lines of evidence suggest that the physical properties of the NE, determined by the types of lipids and proteins present, have an effect on nuclear shape. Some yeast mutants that have abnormal nuclear shape can be restored to wild type nuclear morphology by altering membrane fluidity. For example, in mutants where components of the chromatin-remodeling complex, RSC, are disrupted, nuclei become abnormally shaped due to significant membrane proliferation at the NE, with extensive sheets of membrane protruding into the cytoplasm [Titus et al., 2010]. Normal nuclear shape in these mutants can be restored by the addition of benzyl alcohol, which increases membrane fluidity [Titus et al., 2010]. Similarly, yeast mutants that lack the integral ER protein Apq12 have alterations in both NE and ER shape that can be restored to normal by the addition of benzyl alcohol [Scarcelli et al., 2007]. In human cell lines, expression of mutant lamin B receptor (LBR) protein, an INM protein with a conserved C-terminal sterol reductase domain, caused expansion of the ER lumen and the space between the INM and ONM [Zwerger et al., 2010]. A recent study, also in yeast, has not only shown that lipid composition of the NE affects nuclear shape, but that the INM protein, Mps3, may play a role in lipid homeostasis [Friederichs et al., 2011]. Mps3 is a SUN domain protein that localizes to the spindle pole body (SPB, the yeast’s centrosome equivalent) and INM [Jaspersen et al., 2006]. Over-expression of a dominant lethal allele of MPS3 caused excessive NE proliferation and abnormal nuclear morphology, with several stacked layers of nuclear membrane and multiple nuclear extensions and lobes [Friederichs et al., 2011]. The ER of these mutants was unaffected. The growth defect of these mutants could be rescued by deletion of the FAA3 or DEP1 genes, which are involved in lipid metabolism, or by the addition of chemicals that affect membrane properties, such as benzyl alcohol and oleic acid. The mechanism by which Mps3 affects lipid homeostasis is not known, but it is clear that INM proteins and lipid metabolism can have a substantial effect on nuclear shape.
CHROMOSOMES
It has long been known that chromatin is arranged into higher order chromosomal territories (CTs). Using chromosome-specific probes and techniques that minimally disturb the 3-dimensional structure of the nucleus, Bolzer et al. [2005] have shown that there exists a probable order of prometaphase chromosomes and CTs in nuclei of quiescent and cycling human fibroblasts. In these cells, which have flat-ellipsoidal nuclei, chromosomes were arranged according to size, with smaller chromosomes more likely to be toward the center of the nucleus, and larger chromosomes more likely to be near the nuclear periphery. This is in contrast to the organization of chromosomes in lymphocytes and other human cell types that have spherical nuclei, where the location of chromosomes correlated not with size but with gene density: Chromosomes with high gene density tended to be closer to the center of the nucleus, while gene poor chromosomes had a preference for the nuclear periphery [Boyle et al., 2001]. These observations raise the question of whether nuclear shape dictates the arrangement of CTs or whether it is the other way round.
Abnormal nuclear shape is a hallmark of cancerous cells, but the relationship between cellular transformation and nuclear morphology is poorly understood: Do alterations in nuclear shape contribute to cellular transformation, or are they merely a consequence of it? Gisselsson et al. [2001] observed that there is a linear correlation between abnormal nuclear shape and chromosome instability. Indeed, lagging chromosomes tend to lead to the formation of nuclear blebs. For example, a genome-wide RNAi screen in human cells identified many genes whose inactivation led to abnormal nuclear shapes, including nuclear blebs, polylobed, and grape-shaped nuclei [Neumann et al., 2010]. These genes were often required for proper chromosomal segregation, underscoring the relationship between chromosome organization and nuclear shape. It is tempting to speculate that changes in nuclear shape caused by aberrant chromosome segregation can, in turn, lead to changes in gene expression that further alter nuclear function, potentially contributing to cellular transformation.
Finally, chromatin can exist as euchromatin or heterochromatin, with the latter often positioned at the nuclear periphery. Whether the state of the chromatin, and specifically its degree of compaction, affects nuclear shape or size remains to be determined.
WHAT DETERMINES NUCLEAR SIZE?
Not only do nuclei in a given tissue have a characteristic nuclear shape, they also have a typical size. It has been demonstrated that there is a direct relationship between cell size and nuclear size, and in a given cell type there is a constant nuclear/cell (N/C) volume ratio [Gregory, 2005]. The N/C ratio is often disturbed in cancer cells, but the relationship between nuclear size and cell function is unclear [Zink et al., 2004; Chow et al., 2012]. Why the N/C ratio is so crucial, and the mechanisms by which nuclear and cell volumes are coordinated are largely unknown.
The regulation of nuclear size is likely to be important for optimal nuclear function [Webster et al., 2009]. Because the NE is a selective diffusion barrier, changes in nuclear volume can affect the concentration of nuclear proteins, DNA, and RNA. Consequently, the enzymatic activities of RNA and DNA polymerases, the ability of different chromosomal domains to associate with each other, and the assembly of key structures such as the nucleolus could all be affected by changes in nuclear volume. It is unlikely that DNA content determines nuclear size since different cell types within the same organism have the same DNA content but can have different nuclear sizes. Moreover, there is not a sharp increase in nuclear size when DNA content doubles during S-phase in yeast; instead, cell volume increases gradually throughout the cell cycle, and nuclear volume scales proportionately to cell volume rather than DNA content [Jorgensen et al., 2007; Neumann and Nurse, 2007]. If it is not DNA content, what is responsible for determining nuclear size?
When searching for mechanisms that could regulate nuclear size, two possibilities come to mind: (i) That nuclear size is determined autonomously (in which case cell size would scale appropriately); or (ii) that nuclear size scales proportionately with something else in the cell. Evidence from studies in yeast and frogs strongly supports the latter: When multi-nucleated S.pombe cells were generated such that individual nuclei were unevenly distributed within the cell, nuclei surrounded by a larger cytoplasmic volume grew faster and were larger than nuclei surrounded by less cytoplasm [Neumann and Nurse, 2007]. If indeed cell volume determines nuclear volume, one can envision two scenarios, which are not mutually exclusive: (i) A cellular structure, such as the ER, determines to what size the nucleus can grow. This, for example, could explain the aforementioned yeast experiment, if we assume that rather than being proportional to the amount of surrounding cytoplasm, nuclear volume is actually proportional to the amount of surrounding ER; and (ii) nuclear size is determined by a limiting soluble component that originates in the cytoplasm and is transported to the nucleus. If the second possibility were true, then nuclear import/export would be expected to play a role in determining nuclear size. Yeast studies differed in their observations regarding the influence of nuclear import/export on nuclear size. Inhibition of nuclear export had no effect on nuclear volume in S. cerevisiae [Jorgensen et al., 2007], but when nuclear export was blocked for a longer time period in S. pombe, nuclear volume increased by 50%, indicating that transport between the nucleus and cytoplasm could play a role in regulating nuclear size [Neumann and Nurse, 2007]. Consistent with this observation, nuclear import is required for NE expansion after open mitosis in Xenopus [D’Angelo et al., 2006].
Further evidence that nuclear size is dependent on a cytoplasmic factor came from a cell-free study using extracts from two frog species [Levy and Heald, 2010]. Levy and Heald [2010] exploited the difference in cell and nuclear size between the larger pseudotetraploid Xenopus laevis and smaller diploid Xenopus tropicalis. Regardless of the source of sperm DNA used (i.e., from X. tropicalis or X. laevis), nuclei formed in the X. laevis extract had a larger NE surface area than nuclei formed in the X. tropicalis extract. Furthermore, formation of nuclei in mixtures of X. laevis and X. tropicalis extracts produced nuclei of intermediate sizes, indicating that titratable cytoplasmic factors are responsible for nuclear scaling in Xenopus. In particular, Levy and Heald [2010] found that the rate of import of lamin B3, regulated by transport factors importin α and Nft2, affects nuclear size in Xenopus extracts. For example, increased amounts of importin α led to higher rates of lamin B3 import and larger nuclei [Levy and Heald, 2010], consistent with previous studies where over-expression of lamins in Xenopus cells promoted growth of the NE [Prufert, 2004].
While the above mentioned studies suggest that lamins can be limiting for nuclear size, it seems unlikely that this is the sole mechanism by which scaling is achieved, for several reasons. First, yeast lack lamins but still maintain a constant N/C ratio. Second, although the studies in Xenopus extracts show that lamin import can influence nuclear size [Levy and Heald, 2010], this study does not address the question of nuclear scaling because the cell free system has no defined cell volume. In addition, there are several studies that implicate other NE-associated proteins beside lamins in determining nuclear size. For example, as noted earlier, alteration of the NPC in Xenopus extracts by depletion of the Nup188 nucleoporin led to the formation of enlarged nuclei that correlated with an increased rate of integral INM protein accumulation [Theerthagiri et al., 2010]. INM-associated proteins from Arabidopsis and Drosophila have also been shown to influence nuclear size [Brandt et al., 2006; Dittmer et al., 2007]. The fly protein, Kugelkern, is targeted to the INM through its carboxyterminal farnesylation, much like lamins [Brandt et al., 2006]. In the absence of Kugelkern, nuclei fail to fully elongate during Drosophila embryo cellularization [Brandt et al., 2006]. In Arabidopsis, LINC1 and LINC2 genes encode nuclear periphery-associated proteins with extensive coiled coil domains, which have structural similarity to lamins [Rose et al., 2005]. Disruption of either of these genes leads to decreased nuclear size [Dittmer et al., 2007]. Finally, it remains to be determined whether cells can regulate nuclear size by controlling the amount of membrane that is added to the NE. Any one of these cellular components could contribute to the mechanism that dictates constant nuclear/cell volume ratio.
COULD THE REQUIREMENT FOR A CONSTANT NUCLEAR/CELL VOLUME RATIO LEAD TO ALTERATIONS IN NUCLEAR SHAPE?
Since an increase in nuclear volume requires additional nuclear membrane, it is likely that lipid biosynthesis rates affect nuclear growth [Siniossoglou, 2009]. As mentioned earlier, spo7Δ mutants, in which lipid biosynthesis is altered, exhibit a single nuclear “flare” [Siniossoglou et al., 1998; Campbell et al., 2006], and when vesicle trafficking genes are mutated in cells depleted of Spo7 activity, multiple flares form around the entire nucleus [Webster et al., 2010]. Strikingly, in both single and multi-flared nuclei, the surface area of the NE increased but the nuclear/cell volume was unchanged compared to wild type cells [Webster et al., 2010]. These observations are consistent with a model where the maintenance of the nuclear/cell volume ratio takes precedence over maintenance of normal nuclear shape, at least in these particular circumstances [Webster et al., 2010]. In other words, under conditions of increased NE surface area, nuclear shape can change in one of two ways: The nucleus can expand isometrically, creating a larger spherical or ellipsoid structure with a larger volume, or the NE can form protrusions and invaginations, such that NE surface increases but nuclear volume does not (Fig. 4). The observations thus far in yeast [Webster et al., 2010] are consistent with the latter, and it will be interesting to see if nuclear invaginations observed in higher eukaryotes, such as the nucleoplasmic reticulum, are a consequence of the requirement to adhere to a constant nuclear/cell volume ratio.
Fig. 4.

Changes in nuclear shape in response to increase in NE surface area. In a hypothetical situation where the NE surface area (SA = x) of a nucleus with a given volume (volume = y) increases by 20%, the nucleus can change in shape in one of two ways: a: The NE can expand uniformly. In this case, a 20% increase in SA will result in a 30% increase in nuclear volume. b: The NE can form invaginations and/or protrusions such that nuclear volume remains unchanged. The dashed red line shows the circumference of the original nucleus, before NE expansion.
CONCLUDING REMARKS
Significant progress has been made in recent years in our understanding of the structural components of the nucleus, and in particular the NE. However, much remains to be uncovered regarding how the NE and other cellular components contribute to nuclear shape and size. The effect of ER dynamics on nuclear shape is an intriguing area of study. The balance between sheet-like and tubular forms of the peripheral ER at different stages of the cell cycle could play a role in determining nuclear shape and reformation after open mitosis. Likewise, an increased understanding of what controls the flow of lipids and proteins between different ER domains, and the regulation of lipid homeostasis in the NE, are needed in order to understand the regulation of nuclear size and shape. Observations in yeast are consistent with the possibility that nuclear shape could be affected by the requirement for a constant nuclear/cell volume ratio. Whether the maintenance of this ratio takes precedence over maintaining normal nuclear shape in other systems is not known. Finally, aging and certain disease states are associated with changes in nuclear shape and size, but it is not clear whether altered nuclear shape or size is the cause or consequence of cell malfunction. Increasing our understanding of the mechanisms by which nuclear shape and size are determined will not only address a fundamental unanswered question in cell biology, but may also provide novel therapeutic strategies.
ACKNOWLEDGMENTS
We thank Will Prinz, Andy Golden, Daphna Joseph-Strauss, Brandon Lee, and Christopher May for comments on the manuscript. Our apologies to colleagues whose work we have been unable to cite due to space limitations. A.D.W., A.B., and O.C.F. are funded by an intramural grant from the National Institutes of Diabetes and Digestive and Kidney Diseases.
Grant sponsor: National Institutes of Diabetes and Digestive and Kidney Diseases.
Abbreviations
- NE
nuclear envelope
- INM
inner nuclear membrane
- ONM
outer nuclear membrane
- ER
endoplasmic reticulum
- NPC
nuclear pore complex
- SPB
spindle pole body
REFERENCES
- Aitchison JD, Rout MP, Marelli M, Blobel G, Wozniak RW. Two novel related yeast nucleoporins Nup170p and Nup157p: Complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. J Cell Biol. 1995;131:1133–1148. doi: 10.1083/jcb.131.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar A, Gasser SM. The nuclear envelope and transcriptional control. Nat Rev Genet. 2007;8:507–517. doi: 10.1038/nrg2122. [DOI] [PubMed] [Google Scholar]
- Anderson DJ, Hetzer MW. Nuclear envelope formation by chromatin-mediated reorganization of the endoplasmic reticulum. Nat Cell Biol. 2007;9:1160–1166. doi: 10.1038/ncb1636. [DOI] [PubMed] [Google Scholar]
- Anderson DJ, Hetzer MW. Reshaping of the endoplasmic reticulum limits the rate for nuclear envelope formation. J Cell Biol. 2008;182:911–924. doi: 10.1083/jcb.200805140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki K, Hayashi H, Furuya K, Sato M, Takagi T, Osumi M, Kimura A, Niki H. Breakage of the nuclear envelope by an extending mitotic nucleus occurs during anaphase in Schizosaccharomyces japonicus. Genes Cells. 2011;16:911–926. doi: 10.1111/j.1365-2443.2011.01540.x. [DOI] [PubMed] [Google Scholar]
- Audhya A, Desai A, Oegema K. A role for Rab5 in structuring the endoplasmic reticulum. J Cell Biol. 2007;178:43–56. doi: 10.1083/jcb.200701139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd AM, Hoffman JA, Amberg DC, Fink GR, Davis LI. nup1 mutants exhibit pleiotropic defects in nuclear pore complex function. J Cell Biol. 1994;127:319–332. doi: 10.1083/jcb.127.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolzer A, Kreth G, Solovei I, Koehler D, Saracoglu K, Fauth C, Muller S, Eils R, Cremer C, Speicher MR, Cremer T. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol. 2005;3:e157. doi: 10.1371/journal.pbio.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle S, Gilchrist S, Bridger JM, Mahy NL, Ellis JA, Bickmore WA. The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum Mol Genet. 2001;10:211–219. doi: 10.1093/hmg/10.3.211. [DOI] [PubMed] [Google Scholar]
- Brandt A, Papagiannouli F, Wagner N, Wilsch-Brauninger M, Braun M, Furlong EE, Loserth S, Wenzl C, Pilot F, Vogt N, Lecuit T, Krohne G, Grosshans J. Developmental control of nuclear size and shape by Kugelkern and Kurzkern. Curr Biol. 2006;16:543–552. doi: 10.1016/j.cub.2006.01.051. [DOI] [PubMed] [Google Scholar]
- Brickner JH, Walter P. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. 2004;2:e342. doi: 10.1371/journal.pbio.0020342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bupp JM, Martin AE, Stensrud ES, Jaspersen SL. Telomere anchoring at the nuclear periphery requires the budding yeast Sad1-UNC-84 domain protein Mps3. J Cell Biol. 2007;179:845–854. doi: 10.1083/jcb.200706040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JL, Lorenz A, Witkin KL, Hays T, Loidl J, Fix O. Yeast nuclear envelope subdomains with distinct abilities to resist membrane expansion. Mol Biol Cell. 2006;17:1768–1778. doi: 10.1091/mbc.E05-09-0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capell BC, Collins FS. Human laminopathies: Nuclei gone genetically awry. Nat Rev Genet. 2006;7:940–952. doi: 10.1038/nrg1906. [DOI] [PubMed] [Google Scholar]
- Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 2004;117:427–439. doi: 10.1016/s0092-8674(04)00448-9. [DOI] [PubMed] [Google Scholar]
- Chow KH, Factor RE, Ullman KS. The nuclear envelope environment and its cancer connections. Nat Rev Cancer. 2012;12:196–209. doi: 10.1038/nrc3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T, Cremer M, Dietzel S, Müller S, Solovei I, Fakan S. Chromosome territories-a functional nuclear landscape. Curr Opin Cell Biol. 2006;18:307–316. doi: 10.1016/j.ceb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, Stahl PD, Hodzic D. Coupling of the nucleus and cytoplasm: Role of the LINC complex. J Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo MA, Anderson DJ, Richard E, Hetzer MW. Nuclear pores form de novo from both sides of the nuclear envelope. Science. 2006;312:440–443. doi: 10.1126/science.1124196. [DOI] [PubMed] [Google Scholar]
- Dahl KN, Ribeiro AJ, Lammerding J. Nuclear shape, mechanics, and mechanotransduction. Circ Res. 2008;102:1307–1318. doi: 10.1161/CIRCRESAHA.108.173989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle N, Beaudouin J, Hartnell L, Imreh G, Hallberg E, Lippincott-Schwartz J, Ellenberg J. Nuclear pore complexes form immobile networks and have a very low turnover in live mammalian cells. J Cell Biol. 2001;154:71–84. doi: 10.1083/jcb.200101089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza CP, Osmani SA. Mitosis, not just open or closed. Eukaryot Cell. 2007;6:1521–1527. doi: 10.1128/EC.00178-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechat T, Pfleghaar K, Sengupta K, Shimi T, Shumaker DK, Solimando L, Goldman RD. Nuclear lamins: Major factors in the structural organization and function of the nucleus and chromatin. Genes Dev. 2008;22:832–853. doi: 10.1101/gad.1652708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmer TA, Misteli T. The lamin protein family. Genome Biol. 2011;12:222. doi: 10.1186/gb-2011-12-5-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmer TA, Stacey NJ, Sugimoto-Shirasu K, Richards EJ. LITTLE NUCLEI genes affecting nuclear morphology in Arabidopsis thaliana. Plant Cell. 2007;19:2793–2803. doi: 10.1105/tpc.107.053231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egecioglu D, Brickner JH. Gene positioning and expression. Curr Opin Cell Biol. 2011;23:338–345. doi: 10.1016/j.ceb.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenberg J, Siggia ED, Moreira JE, Smith CL, Presley JF, Worman HJ, Lippincott-Schwartz J. Nuclear membrane dynamics and reassembly in living cells: Targeting of an inner nuclear membrane protein in interphase and mitosis. J Cell Biol. 1997;138:1193–1206. doi: 10.1083/jcb.138.6.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English AR, Zurek N, Voeltz GK. Peripheral ER structure and function. Curr Opin Cell Biol. 2009;21:596–602. doi: 10.1016/j.ceb.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridkin A, Penkner A, Jantsch V, Gruenbaum Y. SUN-domain and KASH-domain proteins during development, meiosis and disease. Cell Mol Life Sci. 2009;66:1518–1533. doi: 10.1007/s00018-008-8713-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friederichs JM, Ghosh S, Smoyer CJ, McCroskey S, Miller BD, Weaver KJ, Delventhal KM, Unruh J, Slaughter BD, Jaspersen SL. The SUN protein Mps3 is required for spindle pole body insertion into the nuclear membrane and nuclear envelope homeostasis. PLoS Genet. 2011;7:e1002365. doi: 10.1371/journal.pgen.1002365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gisselsson D, Bjork J, Hoglund M, Mertens F, Dal Cin P, Akerman M, Mandahl N. Abnormal nuclear shape in solid tumors reflects mitotic instability. Am J Pathol. 2001;158:199–206. doi: 10.1016/S0002-9440(10)63958-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden A, Liu J, Cohen-Fix O. Inactivation of the C. elegans lipin homolog leads to ER disorganization and to defects in the breakdown and reassembly of the nuclear envelope. J Cell Sci. 2009;122:1970–1978. doi: 10.1242/jcs.044743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein AL, Snay CA, Heath CV, Cole CN. Pleiotropic nuclear defects associated with a conditional allele of the novel nucleoporin Rat9p/Nup85p. Mol Biol Cell. 1996;7:917–934. doi: 10.1091/mbc.7.6.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorjánácz M, Mattaj IW. Lipin is required for efficient breakdown of the nuclear envelope in Caenorhabditis elegans. J Cell Sci. 2009;122:1963–1969. doi: 10.1242/jcs.044750. [DOI] [PubMed] [Google Scholar]
- Gregory T. Genome size evolution in animals. Elsevier Academic Press; London: 2005. pp. 4–87. [Google Scholar]
- Hetzer MW, Walther TC, Mattaj IW. Pushing the envelope: Structure, function, and dynamics of the nuclear periphery. Annu Rev Cell Dev Biol. 2005;21:347–380. doi: 10.1146/annurev.cellbio.21.090704.151152. [DOI] [PubMed] [Google Scholar]
- Hu J, Shibata Y, Voss C, Shemesh T, Li Z, Coughlin M, Kozlov MM, Rapoport TA, Prinz WA. Membrane proteins of the endoplasmic reticulum induce high-curvature tubules. Science. 2008;319:1247–1250. doi: 10.1126/science.1153634. [DOI] [PubMed] [Google Scholar]
- Hu J, Shibata Y, Zhu P-P, Voss C, Rismanchi N, Prinz WA, Rapoport TA, Blackstone C. A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell. 2009;138:549–561. doi: 10.1016/j.cell.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen SL, Martin AE, Glazko G, Giddings TH, Morgan G, Mushegian A, Winey M. The Sad1-UNC-84 homology domain in Mps3 interacts with Mps2 to connect the spindle pole body with the nuclear envelope. J Cell Biol. 2006;174:665–675. doi: 10.1083/jcb.200601062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen SL, Winey M. The budding yeast spindle pole body: Structure, duplication, and function. Annu Rev Cell Dev Biol. 2004;20:1–28. doi: 10.1146/annurev.cellbio.20.022003.114106. [DOI] [PubMed] [Google Scholar]
- Jorgensen P, Edgington NP, Schneider BL, Rupes I, Tyers M, Futcher B. The size of the nucleus increases as yeast cells grow. Mol Biol Cell. 2007;18:3523–3532. doi: 10.1091/mbc.E06-10-0973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiseleva E, Morozova KN, Voeltz GK, Allen TD, Goldberg MW. Reticulon 4a/NogoA locates to regions of high membrane curvature and may have a role in nuclear envelope growth. J Struct Biol. 2007;160:224–235. doi: 10.1016/j.jsb.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger A, Batsios P, Baumann O, Luckert E, Schwarz H, Stick R, Meyer I, Graf R. Characterization of NE81, the first lamin-like nucleoskeleton protein in a unicellular organism. Mol Biol Cell. 2011;23:360–370. doi: 10.1091/mbc.E11-07-0595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy DL, Heald R. Nuclear size is regulated by importin α and Ntf2 in xenopus. Cell. 2010;143:288–298. doi: 10.1016/j.cell.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Ladinsky MS, Kirchhausen T. Cisternal organization of the endoplasmic reticulum during mitosis. Mol Biol Cell. 2009;20:3471–3480. doi: 10.1091/mbc.E09-04-0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Ladinsky MS, Kirchhausen T. Formation of the postmitotic nuclear envelope from extended ER cisternae precedes nuclear pore assembly. J Cell Biol. 2011;194:425–440. doi: 10.1083/jcb.201012063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke Y, Zaim H, Karakesisoglou I, Jaeger VM, Sellin L, Lu W, Schneider M, Neumann S, Beijer A, Munck M, Padmakumar VC, Gloy J, Walz G, Noegel AA. Nesprin-2 Giant (NUANCE) maintains nuclear envelope architecture and composition in skin. J Cell Sci. 2008;121:1887–1898. doi: 10.1242/jcs.019075. [DOI] [PubMed] [Google Scholar]
- Malhas A, Goulbourne C, Vaux DJ. The nucleoplasmic reticulum: Form and function. Trends Cell Biol. 2011;21:362–373. doi: 10.1016/j.tcb.2011.03.008. [DOI] [PubMed] [Google Scholar]
- Mattaj IW. Sorting out the nuclear envelope from the endoplasmic reticulum. Nat Rev Mol Cell Biol. 2004;5:65–69. doi: 10.1038/nrm1263. [DOI] [PubMed] [Google Scholar]
- Matynia A, Salus SS, Sazer S. Three proteins required for early steps in the protein secretory pathway also affect nuclear envelope structure and cell cycle progression in fission yeast. J Cell Sci. 2002;115:421–431. doi: 10.1242/jcs.115.2.421. [DOI] [PubMed] [Google Scholar]
- Maul GG, Deaven L. Quantitative determination of nuclear pore complexes in cycling cells with differing DNA content. J Cell Biol. 1977;73:748–760. doi: 10.1083/jcb.73.3.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekhail K, Moazed D. The nuclear envelope in genome organization, expression and stability. Nat Rev Mol Cell Biol. 2010;11:317–328. doi: 10.1038/nrm2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann B, Walter T, Heriche JK, Bulkescher J, Erfle H, Conrad C, Rogers P, Poser I, Held M, Liebel U, Cetin C, Sieckmann F, Pau G, Kabbe R, Wunsche A, Satagopam V, Schmitz MH, Chapuis C, Gerlich DW, Schneider R, Eils R, Huber W, Peters JM, Hyman AA, Durbin R, Pepperkok R, Ellenberg J. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature. 2010;464:721–727. doi: 10.1038/nature08869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann FR, Nurse P. Nuclear size control in fission yeast. J Cell Biol. 2007;179:593–600. doi: 10.1083/jcb.200708054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hara L, Han GS, Peak-Chew S, Grimsey N, Carman GM, Siniossoglou S. Control of phospholipid synthesis by phosphorylation of the yeast lipin Pah1p/Smp2p Mg2+-dependent phosphatidate phosphatase. J Biol Chem. 2006;281:34537–34548. doi: 10.1074/jbc.M606654200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba T. Energy- and temperature-dependent transport of integral proteins to the inner nuclear membrane via the nuclear pore. J Cell Biol. 2004;167:1051–1062. doi: 10.1083/jcb.200409149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olins AL, Hoang TV, Zwerger M, Herrmann H, Zentgraf H, Noegel AA, Karakesisoglou I, Hodzic D, Olins DE. The LINC-less granulocyte nucleus. Eur J Cell Biol. 2009;88:203–214. doi: 10.1016/j.ejcb.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orso G, Pendin D, Liu S, Tosetto J, Moss TJ, Faust JE, Micaroni M, Egorova A, Martinuzzi A, McNew JA, Daga A. Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin. Nature. 2009;460:978–983. doi: 10.1038/nature08280. [DOI] [PubMed] [Google Scholar]
- Oza P, Jaspersen SL, Miele A, Dekker J, Peterson CL. Mechanisms that regulate localization of a DNA double-strand break to the nuclear periphery. Genes Dev. 2009;23:912–927. doi: 10.1101/gad.1782209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oza P, Peterson CL. Opening the DNA repair toolbox: Localization of DNA double strand breaks to the nuclear periphery. Cell Cycle. 2010;9:43–49. doi: 10.4161/cc.9.1.10317. [DOI] [PubMed] [Google Scholar]
- Pederson T. The nucleus introduced. In: Misteli T, Spector DL, editors. The nucleus. Cold Spring Harbor Press; New York: 2011. pp. 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prufert K. The lamin CxxM motif promotes nuclear membrane growth. J Cell Sci. 2004;117:6105–6116. doi: 10.1242/jcs.01532. [DOI] [PubMed] [Google Scholar]
- Puhka M, Vihinen H, Joensuu M, Jokitalo E. Endoplasmic reticulum remains continuous and undergoes sheet-to-tubule transformation during cell division in mammalian cells. J Cell Biol. 2007;179:895–909. doi: 10.1083/jcb.200705112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razafsky D, Hodzic D. Bringing KASH under the SUN: The many faces of nucleo-cytoskeletal connections. J Cell Biol. 2009;186:461–472. doi: 10.1083/jcb.200906068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose A, Schraegle SJ, Stahlberg EA, Meier I. Coiled-coil protein composition of 22 proteomes-differences and common themes in subcellular infrastructure and traffic control. BMC Evol Biol. 2005;5:66. doi: 10.1186/1471-2148-5-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H, Leung J, Grimsey N, Peak-Chew S, Siniossoglou S. The yeast lipin Smp2 couples phospholipid biosynthesis to nuclear membrane growth. EMBO J. 2005;24:1931–1941. doi: 10.1038/sj.emboj.7600672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi P, Misteli T. Lamin A-dependent nuclear defects in human aging. Science. 2006;312:1059–1063. doi: 10.1126/science.1127168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarcelli JJ, Hodge CA, Cole CN. The yeast integral membrane protein Apq12 potentially links membrane dynamics to assembly of nuclear pore complexes. J Cell Biol. 2007;178:799–812. doi: 10.1083/jcb.200702120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon DN, Wilson KL. The nucleoskeleton as a genome-associated dynamic ‘network of networks’. Nat Rev Mol Cell Biol. 2011;12:695–708. doi: 10.1038/nrm3207. [DOI] [PubMed] [Google Scholar]
- Siniossoglou S. Lipins, lipids and nuclear envelope structure. Traffic. 2009;10:1181–1187. doi: 10.1111/j.1600-0854.2009.00923.x. [DOI] [PubMed] [Google Scholar]
- Siniossoglou S, Santos-Rosa H, Rappsilber J, Mann M, Hurt E. A novel complex of membrane proteins required for formation of a spherical nucleus. EMBO J. 1998;17:6449–6464. doi: 10.1093/emboj/17.22.6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theerthagiri G, Eisenhardt N, Schwarz H, Antonin W. The nucleoporin Nup188 controls passage of membrane proteins across the nuclear pore complex. J Cell Biol. 2010;189:1129–1142. doi: 10.1083/jcb.200912045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titus LC, Dawson TR, Rexer DJ, Ryan KJ, Wente SR. Members of the RSC chromatin-remodeling complex are required for maintaining proper nuclear envelope structure and pore complex localization. Mol Biol Cell. 2010;21:1072–1087. doi: 10.1091/mbc.E09-07-0615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124:573–586. doi: 10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]
- Webster M, Witkin KL, Cohen-Fix O. Sizing up the nucleus: Nuclear shape, size and nuclear-envelope assembly. J Cell Sci. 2009;122:1477–1486. doi: 10.1242/jcs.037333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster MT, McCaffery JM, Cohen-Fix O. Vesicle trafficking maintains nuclear shape in Saccharomyces cerevisiae during membrane proliferation. J Cell Biol. 2010;191:1079–1088. doi: 10.1083/jcb.201006083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente SR. Gatekeepers of the nucleus. Science. 2000;288:1374–1377. doi: 10.1126/science.288.5470.1374. [DOI] [PubMed] [Google Scholar]
- Wente SR, Rout MP. The nuclear pore complex and nuclear transport. Cold Spring Harb Perspect Biol. 2010;2:a000562. doi: 10.1101/cshperspect.a000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West M, Zurek N, Hoenger A, Voeltz GK. A 3D analysis of yeast ER structure reveals how ER domains are organized by membrane curvature. J Cell Biol. 2011;193:333–346. doi: 10.1083/jcb.201011039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson KL, Berk JM. The nuclear envelope at a glance. J Cell Sci. 2010;123:1973–1978. doi: 10.1242/jcs.019042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Yarar D, Giddings TH, Jr, Mastronarde DN. Nuclear pore complex number and distribution throughout the Saccharomyces cerevisiae cell cycle by three-dimensional reconstruction from electron micrographs of nuclear envelopes. Mol Biol Cell. 1997;8:2119–2132. doi: 10.1091/mbc.8.11.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkin KL, Friederichs JM, Cohen-Fix O, Jaspersen SL. Changes in the nuclear envelope environment affect spindle pole body duplication in Saccharomyces cerevisiae. Genetics. 2010;186:867–883. doi: 10.1534/genetics.110.119149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worman HJ. Nuclear lamins and laminopathies. J Pathol. 2012;226:316–325. doi: 10.1002/path.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright R, Basson M, D’Ari L, Rine J. Increased amounts of HMG-CoA reductase induce “karmellae”: A proliferation of stacked membrane pairs surrounding the yeast nucleus. J Cell Biol. 1988;107:101–114. doi: 10.1083/jcb.107.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong H, Rivero F, Euteneuer U, Mondal S, Mana-Capelli S, Larochelle D, Vogel A, Gassen B, Noegel AA. Dictyostelium Sun-1 connects the centrosome to chromatin and ensures genome stability. Traffic. 2008;9:708–724. doi: 10.1111/j.1600-0854.2008.00721.x. [DOI] [PubMed] [Google Scholar]
- Yam C, He Y, Zhang D, Chiam K-H, Oliferenko S. Divergent strategies for controlling the nuclear membrane satisfy geometric constraints during nuclear division. Curr Biol. 2011;21:1314–1319. doi: 10.1016/j.cub.2011.06.052. [DOI] [PubMed] [Google Scholar]
- Yang L, Guan T, Gerace L. Integral membrane proteins of the nuclear envelope are dispersed throughout the endoplasmic reticulum during mitosis. J Cell Biol. 1997;137:1199–1210. doi: 10.1083/jcb.137.6.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink D, Fischer AH, Nickerson JA. Nuclear structure in cancer cells. Nat Rev Cancer. 2004;4:677–687. doi: 10.1038/nrc1430. [DOI] [PubMed] [Google Scholar]
- Zuleger N, Robson MI, Schirmer EC. The nuclear envelope as a chromatin organizer. Nucleus. 2011;2:339–349. doi: 10.4161/nucl.2.5.17846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwerger M, Kolb T, Richter K, Karakesisoglou I, Herrmann H. Induction of a massive endoplasmic reticulum and perinuclear space expansion by expression of lamin B receptor mutants and the related sterol reductases TM7SF2 and DHCR7. Mol Biol Cell. 2010;21:354–368. doi: 10.1091/mbc.E09-08-0739. [DOI] [PMC free article] [PubMed] [Google Scholar]


