Abstract
Although many drugs and environmental chemicals are teratogenic, the mechanisms by which most toxicants disrupt embryonic development are not well understood. MicroRNAs, single-stranded RNA molecules of ~22 nt that regulate protein expression by inhibiting mRNA translation and promoting mRNA sequestration or degradation, are important regulators of a variety of cellular processes including embryonic development and cellular differentiation. Recent studies have demonstrated that exposure to xenobiotics can alter microRNA expression and contribute to the mechanisms by which environmental chemicals disrupt embryonic development. In this study we tested the hypothesis that developmental exposure to 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD), a well-known teratogen, alters microRNA expression during zebrafish development. We exposed zebrafish embryos to DMSO (0.1%) or TCDD (5 nM) for 1 hr at 30 hours post fertilization (hpf) and measured microRNA expression using several methods at 36 and 60 hpf. TCDD caused strong induction of CYP1A at 36 hpf (62-fold) and 60 hpf (135-fold) as determined by real-time RT-PCR, verifying the effectiveness of the exposure. MicroRNA expression profiles were determined using microarrays (Agilent and Exiqon), next-generation sequencing (SOLiD), and real-time RT-PCR. The two microarray platforms yielded results that were similar but not identical; both showed significant changes in expression of miR-451, 23a, 23b, 24 and 27e at 60 hpf. Multiple analyses were performed on the SOLiD sequences yielding a total of 16 microRNAs as potentially differentially expressed by TCDD in zebrafish embryos. However, miR-27e was the only microRNA to be identified as differentially expressed by all three methods (both microarrays, SOLiD sequencing, and real-time RT-PCR). These results suggest that TCDD exposure causes modest changes in expression of microRNAs, including some (miR-451, 23a, 23b, 24 and 27e) that are critical for hematopoiesis and cardiovascular development.
Keywords: dioxin, microRNA, zebrafish, development
INTRODUCTION
Ribonucleic acids (RNA) have historically well-known cellular functions: encoding proteins (mRNA), serving as structural components of ribosomes (rRNA), and translating mRNA codons into amino acids (tRNA). Within the last two decades, a new type of RNA—small noncoding RNAs (e.g., microRNAs)—has emerged as an important regulator of a variety of cellular processes. MicroRNAs were first identified in 1993 in the nematode Caenorhabditis elegans (Lee et al., 1993; Wightman et al., 1993) but have more recently been recognized as having broader significance (Lagos-Quintana et al., 2001; Hobert 2008; Makeyev and Maniatis 2008).
MicroRNAs are single-stranded RNA molecules of ~21-22 nucleotides that function to regulate the expression of mRNAs by inhibiting their translation into proteins through promotion of sequestration and degradation. MicroRNA genes are transcribed by RNA polymerase II as longer, primary microRNAs (pri-microRNAs), which are processed in the nucleus by Drosha, an RNase III enzyme, into 70-80-nucleotide precursor microRNAs (pre-microRNAs). After transport out of the nucleus, the pre-microRNA is cleaved by the enzyme Dicer into the mature microRNA (Wienholds and Plasterk 2005). MicroRNAs function to inhibit translation as a complex with Dicer and an Argonaute protein, which together form the RNA-induced silencing complex (RISC). The microRNA in the RISC complex binds by contiguous Watson-Crick base pairing between specific sites on the 3′-UTR of its target mRNA and the ‘seed region’ on the 5′- end of the microRNA. Outside of this region, base-pairing between the microRNA and its target form a duplex that is not completely complementary and serves as the cleavage site of the mRNA. Some microRNAs may target hundreds of different messenger RNAs (Lim et al., 2005), although there are also cases of much greater specificity between microRNA and target. Many genes are targets of regulation by microRNAs; it has been estimated that 30-60% of the genes in a given animal genome are regulated in this way (Wienholds and Plasterk 2005; Bushati and Cohen 2007; Friedman et al., 2009).
MicroRNAs are abundant and evolutionarily conserved in multicellular animals (Sempere et al., 2006). The human genome encodes hundreds of microRNAs (Bentwich et al., 2005), and large numbers of microRNA sequences occur in the genomes of other animals, as demonstrated by studies in C. elegans, Drosophila melanogaster, and the zebrafish Danio rerio (Lee and Ambros 2001; Lim et al., 2001; Aravin et al., 2003; Lai et al., 2003; Weinholds et al., 2005; Kloosterman et al., 2006). While much still remains to be learned about the various functions of microRNAs, it is clear that they play an essential role in embryonic development by serving as a regulatory mechanism for the spatial and temporal expression of mRNA transcripts (Carrington and Ambros 2003; Mishima 2012). Furthermore, microRNAs are involved in regulating cellular responses to a variety of environmental or cellular stressors and are misexpressed in certain human diseases (Bhattacharyya et al., 2006; Kulshreshtha et al., 2007; Hudder and Novak 2008; Stern-Ginossar et al., 2008; Simone et al., 2009; Yokoi and Nakajima 2011; Tal et al., 2012).
With a fundamental role in development and potential to be misregulated by cellular stress, microRNAs have become of major interest to the fields of toxicology and teratology (Jardim et al., 2009; Pogribny 2009; Tryndyak et al., 2009; Wang et al., 2009; Gueta et al., 2010; Halappanavar et al., 2011). The primary objective of this study was to investigate the potential for microRNAs to be differentially expressed during development in response to exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD). Although TCDD is well known to be teratogenic in a variety of vertebrate organisms (Smith et al., 1976; Cheung et al., 1981; Couture et al., 1990; Peters et al., 1999; Mehta et al., 2008), the exact mechanisms by which these developmental defects arise still remains to be elucidated. We utilized the zebrafish (Danio rerio) which has emerged as an excellent model organism for investigations into both development and developmental toxicology. The teratogenic effects of TCDD in zebrafish embryos may manifest as several phenotypes, including lower jaw malformations, cardiac malformations, neurodevelopmental defects and disruption of normal erythropoiesis (Belair et al., 2001; Dong et al., 2001; Teraoka et al., 2002; Hill et al., 2003; Antkiewicz et al., 2005). Although the toxicity of TCDD is known to be primarily regulated by the aryl hydrocarbon receptor 2 (AHR2) during zebrafish development (Prasch et al., 2003; Carney et al., 2004; Antkiewicz et al., 2006), many of the specific target genes responsible for the phenotypes have yet to be identified. Given the important role of microRNAs during embryonic development, we sought to determine if any microRNAs are differentially expressed in zebrafish embryos in response to TCDD treatment as an avenue for future mechanistic studies.
MicroRNA quantification is currently done using methods similar to those used for quantifying mRNA transcript levels. These methods include real-time RT-PCR for measuring individual microRNAs using microRNA-specific primers (Chen et al., 2005), microRNA microarrays for high-throughput quantification of all or most of the known microRNAs (Ach et al., 2008) and small RNA sequencing to measure all known and unknown small RNA species (Soares et al., 2009; Cifuentes et al., 2010). Previous comparisons have suggested that these methods vary in their ability to detect differential expression of microRNAs in vitro (Ach et al., 2008; Git et al., 2010; Yauk et al., 2010). Hence, a secondary objective of this study was to compare the three methods in their ability to detect differential expression of microRNAs in an in vivo model system. We quantified microRNA expression using 1) two widely used microRNA microarray platforms (Exiqon and Agilent microarrays), 2) a deep sequencing approach (SOLiD sequencing), and 3) real-time RT-PCR using a stem-loop primer method to confirm some of the changes observed with the high-throughput methods.
MATERIALS AND METHODS
Fish husbandry
The TL (Tupfel/Long fin mutations) wild-type strain of zebrafish was used for all experiments. Fertilized eggs were obtained from multiple group breedings from tanks of 30 female and 15 male fish. Procedures used in these experiments were approved by the Animal Care and Use Committee of the Woods Hole Oceanographic Institution.
Exposure of zebrafish embryos to TCDD and Isolation of Total RNA
Groups of embryos were placed in large glass petri dishes at a density of 3 embryos per mL of 0.3× Danieau’s, and then exposed to carrier (0.1% DMSO) or 5 nM TCDD (dissolved in DMSO) for 1 hour, starting at 30 hours post fertilization (hpf). Each treatment consisted of three biological replicates of 200 pooled embryos. After the 1 hour TCDD exposure the embryos were washed three times with 0.3x Danieau’s and maintained at 28.5 °C at a density of 3 embryos per mL. At 36 and 60 hpf embryos were euthanized, preserved with liquid nitrogen and stored at −80°C.
For isolation of total RNA, we evaluated two different isolation methods (QIAGEN miRNeasy kit and STAT-60). Similar to previous studies (Ach et al., 2008), we did not see any differences in microRNA quantification between the two RNA isolation methods using real-time RT-PCR (data not shown). For subsequent microRNA profiling, we used total RNA isolated using the QIAGEN miRNeasy protocol. Total RNA quality was assessed using an Agilent 2100 Bioanalyzer which revealed that all samples were of extremely high quality (RNA integrity numbers of 9.8-10). CYP1A gene expression and microRNA quantification using different approaches (microarrays, deep sequencing and real-time RT-PCR) were conducted on the same samples of total RNA.
Measurement of CYP1A Gene expression by real-time RT-PCR
Complementary DNA (cDNA) was synthesized from 2 μg total RNA using random hexamers and the Omniscript cDNA Synthesis Kit (Qiagen). Quantitative PCR was performed using the iQ SYBR Green Supermix (Bio-Rad, Hercules, CA) in a MyiQ Single-Color Real-Time PCR Detection system (Bio-Rad). At the end of each PCR run, a melt curve analysis was performed to ensure that only a single product was amplified. Three technical replicates were used for each sample. A standard curve for each gene was generated by serially diluting plasmids containing a full-length copy of either CYP1A or β-actin. Total molecule numbers were calculated for each sample and normalized by a β-actin correction factor. Changes in expression are reported as changes in fold induction by normalizing molecule numbers to the appropriate control. Real-time RT-PCR primers and the thermocycler protocols for CYP1A and β-actin are provided elsewhere (Evans et al., 2005).
Exiqon microarray analysis
MicroRNA profiling was performed using custom designed locked-nucleic acid (LNA)-probe microarray (miRCURY LNA array v.9.2 profiling service, Exiqon A/S, Denmark). The microRNA probes were based on zebrafish microRNA sequences from miRbase v. 12. Each sample was labeled with Hy3 and hybridized against a Hy5-labeled universal RNA reference made from equal amounts of all RNAs from this experiment. Signals were background corrected and normalized using global LOWESS. A student’s t-test was used to determine changes in microRNA expression data between DMSO and TCDD treatments for each time point, or between 36 and 60 hpf DMSO timepoints. A Bonferroni corrected p-value of 0.000196 (0.05/255 microRNAs) was used to determine statistical significance.
Agilent microarray analysis
Agilent microRNA microarrays contain 8 individual microarrays with approximately 15,000 features per array (8X15K). Custom-made Agilent oligonucleotide microRNA microarrays were designed based on zebrafish mature microRNA sequences from miRbase v.16. Each individual array contained a total of 548 unique probes representing 259 mature microRNAs from zebrafish (245), Fugu rubripes (11) and Tetraodon nigriviridis (3). Total RNA labeling and hybridization were carried out using Agilent’s microRNA complete labeling and hybridization kit (Agilent Technologies). Total RNA (100 ng) was dephosphorylated by treating with calf intestinal alkaline phosphatase and labeled with cyanine3-pCp by incubating with T4 RNA ligase for 2 hr at 16°C. Any unlabeled dye was removed by running the labeled RNA through Micro Biospin 6 columns (BioRad). Labeled and purified RNA was vacuum dried and resuspended in nuclease free water. Prior to hybridization, blocking agent and hybridization buffer were added to the samples. Microarrays were hybridized in Agilent SureHyb chambers (Agilent Technologies) for 20 hr at 55°C in a rotating hybridization oven at 20 rotations per minute (rpm). At the end of the hybridization, the hybridized arrays were washed using Gene Expression wash buffers following manufacturer’s instructions. Immediately after washing, the microarrays were scanned using GenePix 4000B scanner (Molecular Devices, CA). Each slide was initially scanned in preview mode under low resolution to determine the appropriate photo multiplier tube (PMT) settings. PMT settings were manually adjusted to have the majority of the features fall between 15,000 to 50,000 intensity units. Full scans were done at PMT settings between 650-700 for all the slides used in this study. Agilent Feature Extraction (AFE) software 9.5.3 using default settings was used to extract data.
Data analysis was carried using AgiMicroRna, a Bioconductor package (Lopez-Romero 2011). Total Gene Signal (TGS), a single intensity measure for each microRNA computed by the AFE algorithm was used in statistical analysis. TGS was background-subtracted and quantile-normalized using the robust multiarray average (RMA) algorithm (Irizarry et al., 2003; Lopez-Romero 2011). Differential expression of microRNAs was determined by fitting a linear model using an empirical Bayes approach (Smyth 2004). We controlled for multiple testing by estimating false discovery rate (FDR) using the Benjamini and Hochberg method (Benjamini and Hochberg 1995). Adjusted p-values less than 0.05 were considered to be significant.
Next-generation SOLiD sequencing
The same total RNA used for the array hybridizations was also used for high-throughput sequencing using the SOLiD technology. Prior to sequencing, the quality of the RNA and presence of the microRNA fraction was confirmed with the Agilent Small RNA kit for the 2100 Bioanalyzer which revealed that the microRNA fraction represented 3-4% of the total RNA pool for each sample. Small RNA library construction and microRNA sequencing were performed by the Genomic Services Lab at the HudsonAlpha Institute for Biotechnology in Huntsville, AL, USA. Small RNA libraries were prepared using the small RNA expression kit (SREK) protocol (Applied Biosystems, Foster City, CA). Sequencing was done on a SOLiD V3 system (Applied Biosystems).
Small RNA sequence analysis
Three different methods were used to analyze the SOLiD sequences – CLC Genomics Workbench 4.7.2, Genome Mapping, and DNASTAR ArrayStar.
CLC Genomics Workbench
The analysis work flow for CLC Genomics involved adaptor trimming, counting different unique small RNA sequences and mapping them to zebrafish precursor microRNA sequences from miRBase version 17 and other noncoding RNAs from zebrafish ensembl version 63 (ftp://ftp.ensembl.org/pub/release-63/fasta/danio_rerio/ncrna/). Adaptor trimming was done in color space using default parameters – trimming the adaptor sequence, deleting the reads without the adaptor sequence, discarding any reads that were less than 15 nucleotides in length, and deleting any reads with more than 5 consecutive ambiguous nucleotides. Reads were aligned to precursor microRNA and other non-coding RNA sequences allowing two mismatches. The criteria used for determining isomiRs were the presence or absence of up to 5 additional bases on either 5′ or 3′ end of the reads. The output from this analysis is grouped into the total number of mature microRNA and mature star sequence counts. These counts are further classified into isomiRs – both length and sequence variants.
Statistical analysis of the count data was done using edgeR, a bioconductor package for differential expression analysis of deep sequencing data (Robinson et al., 2010). Count data were normalized using trimmed mean of M values (TMM) method and a tagwise dispersion model was fitted to obtain significant differences in microRNA expression between different treatment groups (Robinson and Oshlack 2010). Multiple testing correction was done using Benjamini-Hochberg method (Benjamini and Hochberg 1995).
Genome mapping method
This is the most widely used method for annotating the sequence reads. Prior to mapping the reads to the zebrafish genome, adaptor sequences were trimmed in color space using cutadapt software (Schulte et al., 2010). Trimmed reads were mapped to the reference zebrafish genome (Zv7) using BWA algorithm (Li and Durbin 2009), with default settings. Read counts for annotated microRNAs and other non-coding RNAs were obtained from the genomic co-ordinates using HTSeq (http://www-huber.embl.de/users/anders/HTSeq/doc/overview.html) software. Statistical testing was done using edgeR as described in the previous section.
QSeq/ArrayStar software analysis
Analysis of changes in microRNA expression was determined with the QSeq/ArrayStar software package (DNASTAR, Inc.) by comparing the individual sequence reads to a template database consisting of mature and star forms for annotated microRNAs (miRBase v.17). Statistical significance was calculated using a student’s t-test with a Benjamini Hochberg False Discovery Rate (FDR=0.05) to compare DMSO and TCDD treatments (Benjamini and Hochberg 1995).
Mature microRNA cDNA synthesis and real-time RT-PCR
cDNA synthesis from mature microRNAs was done using a stem-loop reverse transcription (RT) primer method (Chen et al., 2005). Briefly, separate stem-loop RT primers were designed for each mature microRNA in such a way that the RT primer binds to the 3′ end of the microRNA (Table 1). Polyacrylamide gel electrophoresis (PAGE)-purified RT primers were synthesized by MWG Operon (Huntsville, AL). The reverse transcription reaction was carried out using TaqMan microRNA reverse transcription kit (Applied Biosystems, CA) following manufacturer’s instructions. Each reverse transcription reaction contained 100 ng of total RNA, 50 nM stem-loop primer, 1x RT buffer, 0.25 mM each of dNTPs, 0.25 U/μL RNase inhibitor and 3.33 U/μL of multi-scribe reverse transcriptase in a total volume of 15 μL. The amount of total RNA and the concentration of RT primer used were determined empirically. Briefly, preliminary experiments were conducted with varying concentrations of input RNA (50, 100, 200, 300 and 500 ng total RNA) and RT primer (50, 100, 667, 2,000 nM) for cDNA synthesis prior to PCR. High input RNA concentrations and RT primer caused increased appearance of non-specific products.
Table 1.
MicroRNA Stem-loop and qRT-PCR primers used in this study.
| Small RNA | Tm (°C) | Primer (5′ – 3′) | |
|---|---|---|---|
| U6 | For | 60 | TCGCTACGGTGGCACATA |
| Rev | TATGGAGCGCTTCACGG | ||
| miR-9 | For | 62 | CGCGGCGGTCTTTGGTTATCTA |
| miR-141 | For | 60 | GCCGCTAACACTGTCTGGTA |
| miR-144 | For | 60 | GCCCGGCCTACAGTATAGATG |
| miR-451 | For | 60 | GGCCCAAACCGTTACCA |
| miR-27e | For | 65 | CGCGCTTCACAGTGGCT |
| miR | Rev | GTGCAGGGTCCGAGGTATTC | |
| miR-9 | RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCATAC | |
| miR-141 | RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGCATCG | |
| miR-144 | RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAGTACA | |
| miR-451 | RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAACTCA | |
| miR-27e | RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCACTGA |
Real-time RT-PCR was performed using iQ SYBR Green Supermix (Bio-Rad, Hercules, CA) as described in the previous section. Each 25 μL PCR reaction consisted of 1 μL of RT product, 1X PCR master mix, 1.5 μM microRNA specific forward primer and 1.5 μM common reverse primer. The forward primers contained 5-8 extra nucleotides at the 5′ end in order to increase the melting temperature. The reverse primer was designed based on the stem-loop primer and was common for all microRNAs. The PCR primer sequences and their melting temperatures are given in Table 1. The optimum annealing/extension temperature (Tm) of all the primer pairs was established by running a gradient PCR. The PCR reaction conditions used were 95 °C for 3 min followed by 40 cycles of 95 °C for 15 sec and Tm for 1 min. At the end of the PCR run, melt curve analysis were performed to ensure that only one product was amplified. All the samples were run in triplicate. U6 snRNA was used as a reference gene. To verify the amplification of a single product the real-time RT-PCR reactions were electrophoresed on a 15% polyacrylamide gel and stained with ethidium bromide to visualize the amplified PCR products. Some of these products were also cloned into pGEM-Teasy vector and sequenced.
RESULTS
Induction of CYP1A as determined by real-time RT-PCR
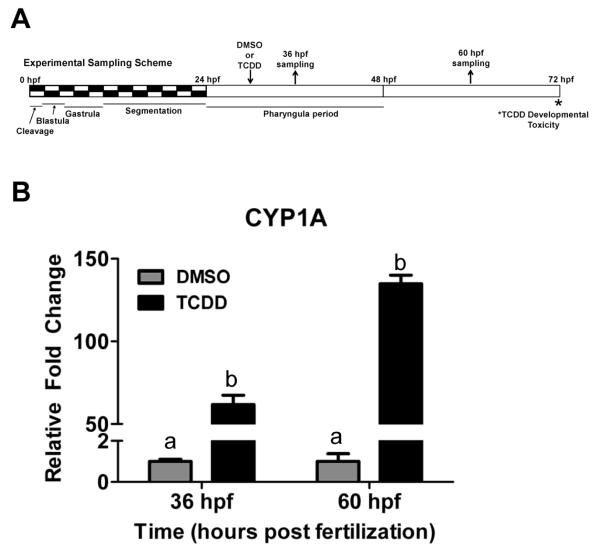
Zebrafish were exposed for one hour to 5 μM TCDD starting at 30 hpf to minimize the chances of developmental delay associated with earlier exposure, which might indirectly affect microRNA expression (Figure 1A). Dosing zebrafish embryos at any time prior to 66 hpf is sufficient to induce the teratogenic effects of TCDD which begin to significantly manifest at 72 hpf (Belair et al., 2001). To confirm the level of AHR activation by the one hour exposure to TCDD, the induction of cytochrome P450 1A1 was assessed by real-time RT-PCR at 36 hpf and 60 hpf. TCDD caused strong induction of CYP1A at 36 hpf (62-fold) and 60 hpf (135-fold) (Figure 1B).
Figure 1.
(A) Exposure regime and sampling times in the study. Zebrafish embryos at 30 hpf were exposed to 5 nM TCDD or DMSO for 1 h and sampled at 36 and 60 hpf. (B) Induction of CYP1A transcripts in response to TCDD exposure. CYP1A expression is expressed as fold change from DMSO-treated group. Different letters represent significant differences from DMSO control (Two-way ANOVA; p < 0.01).
Changes in microRNA expression as determined by Exiqon microarray profiling
Exiqon microarray data generated in this study have been deposited in the NCBI Gene Expression Omnibus (accession number GSE39809). Comparison of the two time points (36 and 60 hpf DMSO groups) revealed developmental changes in the expression of multiple microRNAs (Supplementary Fig. 1). A total of 82 out of 255 microRNAs on the array (32%) were differentially expressed (Bonferroni corrected p-value; p<0.000196). Sixty of the microRNAs increased between 36 and 60 hpf (1.2- to 5.4-fold), consistent with previous reports showing increased microRNA expression between 24 and 72 hpf (Weinholds et al., 2005; Thatcher et al., 2007; Wei et al., 2012). Twenty microRNAs were down-regulated between 36 and 60 hpf, including several members of the miR-430 family, which have been shown to be expressed early in development (3- to 24-hpf) and then decrease rapidly after 24-hpf (Kloosterman et al., 2006). Thus, we were able to reproduce and extend results of previous studies of changing microRNA expression during development.
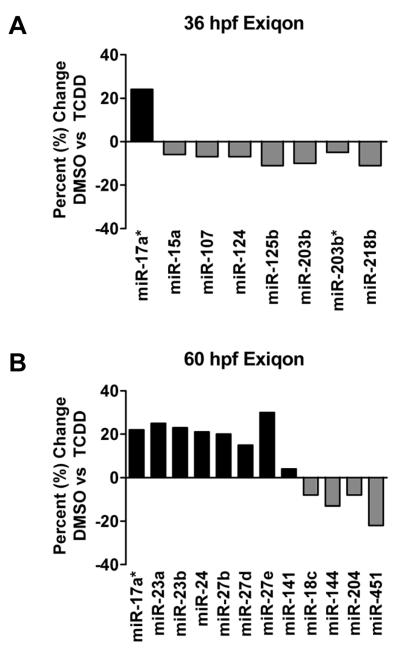
A total of 19 microRNAs from both the 36 and 60 hpf timepoints were determined to be differentially expressed as a result of TCDD exposure (Figure 2). Overall the changes in expression were modest (−1.22 to 1.30-fold change) but statistically significant at a p-value of 0.01. Embryos sampled at 36-hpf had eight microRNAs with differential expression between DMSO and TCDD treatments. Among them, only one microRNA (miR-17a*) was up-regulated, while seven microRNAs were down-regulated (miR-15a, 107, 124, 125b, 203b, 203b*, and 218b). Embryos sampled at 60-hpf had twelve microRNAs with differential expression between treatments; eight were up-regulated (miR-17a*, 23a, 23b, 24, 27b, 27d, 27e, and 141) and four were down-regulated (18c, 144, 204, and 451). Of the microRNAs differentially expressed in response to TCDD at these time points, only miR-218b was statistically significant at a Bonferroni corrected p-value of 0.000196.
Figure 2.
TCDD-induced changes in microRNA expression observed with Exiqon LNA microarrays at 36 hpf (A) and 60 hpf (B). Data are expressed as percent change of expression normalized to the DMSO carrier control treatment. All of the changes in microRNAs plotted in this graph were statistically significant based on p-value <0.01. However only miR-218b was significant when using the Bonferroni correction (0.000196) to safeguard against false positives.
Changes in microRNA expression as determined by Agilent microarray profiling
Agilent microarray data generated in this study have been deposited in the NCBI Gene Expression Omnibus (accession number GSE39809). Similar to results obtained with Exiqon microarrays, we observed developmental changes in microRNA expression using a custom Agilent microRNA microarray. Between 36 and 60 hpf, 137 microRNAs were differentially expressed (adjusted p<0.05) with fold changes ranging from −2.0 to 2.3 (Supplementary Fig. 2).
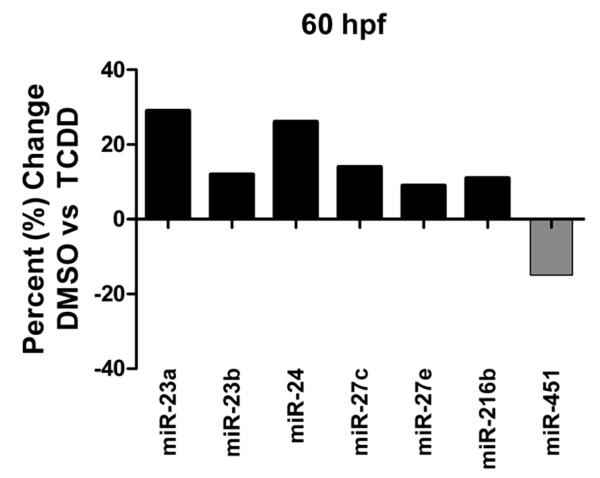
No significant changes in microRNA expression were detected using the Agilent microarray in 36 hpf zebrafish embryos exposed to TCDD. However at 60 hpf there were modest but statistically significant changes in expression of seven microRNAs in response to TCDD (up-regulation of miR-23a, 23b, 24, 27c, 27e and 216b; down-regulation of miR-451) (Figure 3). Two of the seven differentially expressed microRNAs (miR-216b and miR-27c) were detected only using the Agilent array, whereas the other five microRNAs (miR-23a, miR-23b, miR-24, miR-27e, and miR-451) were also detected by the Exiqon array.
Figure 3.
TCDD-induced changes in microRNA expression observed with Agilent microarrays at 60 hpf. Data are expressed as percent change of expression normalized to the DMSO carrier control treatment. No significant changes in microRNA expression were observed at 36 hpf. Seven differentially expressed miRNAs were observed at 60 hpf. All seven of the miRNAs plotted in this graph were determined to be statistically significant based on an adjusted p-value <0.05 (Benjamini-Hochberg method).
Changes in microRNA expression as determined by SOLiD sequencing
A total of 210 million raw reads from 12 samples at an average of 17.5 million reads per sample was obtained from the SOLiD sequencing. Raw sequencing data generated in this study have been deposited in the NCBI Gene Expression Omnibus (accession number GSE39809). Sequencing results were analyzed using CLC Genomics Workbench 4.7.2. After removing the low quality reads and removing the adaptor sequences, unique small RNAs were mapped to known zebrafish precursor microRNAs (miRbase v.17) and other noncoding RNAs (Ensembl v.63). At 36 hpf, an average of 24% of all the trimmed reads were annotated as small RNAs, whereas at 60 hpf, 46% of the reads mapped to known small RNA sequences (Table 2). Among these reads, 88-92% of them were annotated as mature microRNAs, 6.5-10% were annotated as star sequences and the remaining (1.1-2.5%) matched to other non-coding RNAs (Table 2). A detailed list of read counts of mature microRNAs, star sequences and ncRNAs is given in Supplementary Table 1.
Table 2.
Summary of miRNA and ncRNA abundance from SOLiD sequencing using CLC Genomics analysis.
| Sample | Total raw reads |
Raw reads after trimming |
Annotated small RNAs |
miRNAs | Other ncRNAs | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| reads | % | Mature | % | Star | % | reads | % | |||
| DMSO 36hpf-1 |
8,846,676 | 4,227,737 | 821,505 | 19.4 | 738,954 | 90.0 | 62,177 | 7.6 | 20,374 | 2.5 |
| DMSO 36hpf-2 |
10,392,462 | 5,789,720 | 1,774,526 | 30.6 | 1,624,074 | 91.5 | 120,123 | 6.8 | 30,329 | 1.7 |
| DMSO 36hpf-3 |
60,245,391 | 18,498,582 | 4,543,617 | 24.6 | 4,067,982 | 89.5 | 392,972 | 8.6 | 82,663 | 1.8 |
| TCDD 36hpf-1 |
10,467,422 | 5,663,848 | 1,547,597 | 27.3 | 1,394,241 | 90.1 | 125,150 | 8.1 | 28,206 | 1.8 |
| TCDD 36hpf-2 |
11,098,349 | 4,707,922 | 945,175 | 20.1 | 856,991 | 90.7 | 66,672 | 7.1 | 21,512 | 2.3 |
| TCDD 36hpf-3 |
6,115,523 | 2,877,262 | 583,910 | 20.3 | 523,013 | 89.6 | 47,019 | 8.1 | 13,878 | 2.4 |
| DMSO 60hpf-1 |
16,678,613 | 9,805,841 | 4,870,267 | 49.7 | 4,493,580 | 92.3 | 320,732 | 6.6 | 55,955 | 1.1 |
| DMSO 60hpf-2 |
12,582,035 | 6,644,382 | 3,078,161 | 46.3 | 2,829,596 | 91.9 | 211,001 | 6.9 | 37,564 | 1.2 |
| DMSO 60hpf-3 |
14,142,261 | 8,489,655 | 4,323,169 | 50.9 | 3,991,202 | 92.3 | 280,772 | 6.5 | 51,195 | 1.2 |
| TCDD 60hpf-1 |
19,450,897 | 8,833,248 | 4,308,553 | 48.8 | 3,852,539 | 89.4 | 405,765 | 9.4 | 50,249 | 1.2 |
| TCDD 60hpf-2 |
11,637,590 | 6,094,678 | 2,684,617 | 44.0 | 2,401,231 | 89.4 | 252,139 | 9.4 | 31,247 | 1.2 |
| TCDD 60hpf-3 |
28,775,825 | 15,768,488 | 5,545,552 | 35.2 | 4,911,952 | 88.6 | 553,578 | 10.0 | 80,022 | 1.4 |
We observed a significant increase in the number of annotated small RNA reads at 60 hpf compared to 36 hpf, suggesting increased abundance of small RNAs later in development. However, there are no major differences in the diversity of small RNA between the two time points investigated. Out of 358 known microRNAs in miRBase, 97% were found in our samples, whereas only 33% of the other non-coding RNAs (1483 out of 4431 known ncRNAs) were found. Statistical comparison of small RNAs between the two developmental time points (36 hpf DMSO vs 60 hpf DMSO) revealed 336 small RNAs to be differentially expressed (adjusted p.value < 0.05). Of these, 252 were microRNAs and 84 are other ncRNAs. Out of 336 differentially expressed small RNAs, 186 were up-regulated and 150 were down-regulated. The magnitude of change ranged from 1.2 to 31-fold for up-regulated small RNAs and −1.32 to −4.34 fold change for down-regulated small RNAs. A heat map representation of these developmental changes is shown in Supplementary Figure 3.
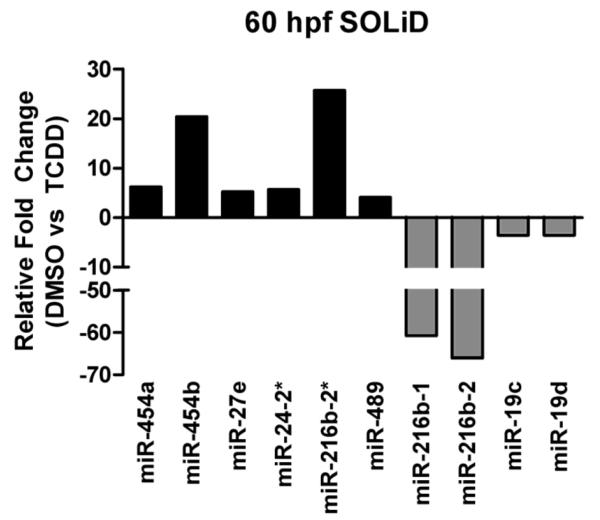
There were no statistically significant changes in microRNA expression between DMSO and TCDD treatments at 36 hpf as determined by SOLiD sequencing. However, TCDD exposure resulted in differential expression of 10 microRNAs and 14 other ncRNAs at 60 hpf. The results of statistical analysis for all 24 of these small RNAs are shown in Table 3. Of the microRNAs differentially expressed in response to TCDD at 60 hpf, six were up-regulated (miR-454a, miR-454b, miR-27e, miR-24-2*, miR-216b-2* and miR-489) and four were down-regulated (miR-216b-1, miR-216b-2, miR-19c and miR-19d) (Figure 4).
Table 3.
Statistical analysis results showing TCDD-induced differential expression of small RNAs at 60 hours post-fertilization, ranked by p-value.
| Read counts | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Small RNA | Fold Change |
p.value | Adj. p.value | DMSO #1 |
DMSO #2 |
DMSO #3 |
TCDD #1 |
TCDD #2 |
TCDD #3 |
| mir-216b-2 | −66.0 | 2.89E-18 | 3.84E-15 | 1285 | 106 | 648 | 13 | 5 | 15 |
| mir-216b-1 | −60.8 | 9.21E-18 | 6.12E-15 | 1284 | 108 | 642 | 13 | 7 | 15 |
| mir-454b | 20.4 | 2.00E-12 | 8.84E-10 | 89 | 341 | 71 | 549 | 1697 | 14663 |
| 5S ribosomal RNA |
13.3 | 5.96E-07 | 1.98E-04 | 3 | 3 | 2 | 36 | 28 | 61 |
| mir-454a | 6.2 | 5.05E-06 | 1.34E-03 | 45 | 158 | 72 | 60 | 188 | 2615 |
| U4 | 15.8 | 1.93E-05 | 4.07E-03 | 1 | 2 | 0 | 23 | 14 | 15 |
| U5 | −5.3 | 2.14E-05 | 4.07E-03 | 154 | 4352 | 900 | 159 | 329 | 1045 |
| SNORD37 | −6.5 | 2.79E-05 | 4.50E-03 | 50 | 64 | 18 | 3 | 6 | 17 |
| mir-27e | 5.2 | 3.05E-05 | 4.50E-03 | 97 | 76 | 83 | 582 | 338 | 601 |
| U1.68-201 | −6.7 | 6.49E-05 | 8.62E-03 | 102 | 3 | 32 | 1 | 13 | 4 |
| mir-24-2 * | 5.7 | 9.36E-05 | 1.13E-02 | 5 | 19 | 4 | 109 | 31 | 55 |
| SNORD30 | −5.3 | 2.12E-04 | 2.24E-02 | 46 | 66 | 12 | 5 | 11 | 11 |
| SNORD42 | −4.8 | 2.21E-04 | 2.24E-02 | 90 | 50 | 25 | 8 | 7 | 27 |
| AC024175.7 | −4.5 | 2.36E-04 | 2.24E-02 | 122 | 90 | 105 | 8 | 26 | 47 |
| mir-216b-2 * | 25.7 | 2.76E-04 | 2.28E-02 | 0 | 1 | 0 | 9 | 10 | 9 |
| AC024175.8 | −4.2 | 2.76E-04 | 2.28E-02 | 393 | 366 | 84 | 64 | 95 | 38 |
| SNORD12/106 | −4.4 | 2.92E-04 | 2.28E-02 | 195 | 67 | 38 | 11 | 22 | 42 |
| mir-489 | 4.1 | 3.44E-04 | 2.54E-02 | 22 | 63 | 84 | 406 | 70 | 441 |
| SNORD29 | −3.9 | 4.24E-04 | 2.96E-02 | 433 | 282 | 240 | 56 | 43 | 214 |
| U5 | −4.8 | 4.94E-04 | 3.16E-02 | 14 | 23 | 21 | 4 | 0 | 12 |
| SNORD75 | −4.3 | 5.00E-04 | 3.16E-02 | 65 | 55 | 26 | 12 | 7 | 22 |
| SNORD59 | −4.8 | 7.73E-04 | 4.67E-02 | 21 | 32 | 22 | 0 | 5 | 15 |
| mir-19c | −3.6 | 8.29E-04 | 4.71E-02 | 64686 | 36308 | 35656 | 18277 | 10682 | 10339 |
| mir-19d | −3.6 | 8.51E-04 | 4.71E-02 | 27978 | 16884 | 21172 | 10894 | 5704 | 825 |
Figure 4.
TCDD-induced changes in microRNA expression observed with the CLC Genomics analysis of SOLiD microRNA sequencing. Data are expressed as fold change in expression normalized to the DMSO carrier control treatment. No significant changes in microRNA expression were observed at 36 hpf. However at 60 hpf, ten miRNAs were differentially expressed in a statistically significant manner based on an adjusted p-value <0.05 (Benjamini-Hochberg method).
Comparison between microarrays and next-generation sequencing
All three high-throughput methods for analyzing global changes in microRNA expression (two microarray platforms and SOLiD sequencing) successfully detected differential expression between our two developmental timepoints, consistent with previous reports characterizing microRNA expression between 24 and 72 hpf (Weinholds et al., 2005; Thatcher et al., 2007; Wei et al., 2012). Our ability to consistently reproduce these results with all three high-throughput methods, with additional confirmation by quantitative RT-PCR, validates the utility of the different methods even though there are apparent differences in specific results among the different approaches.
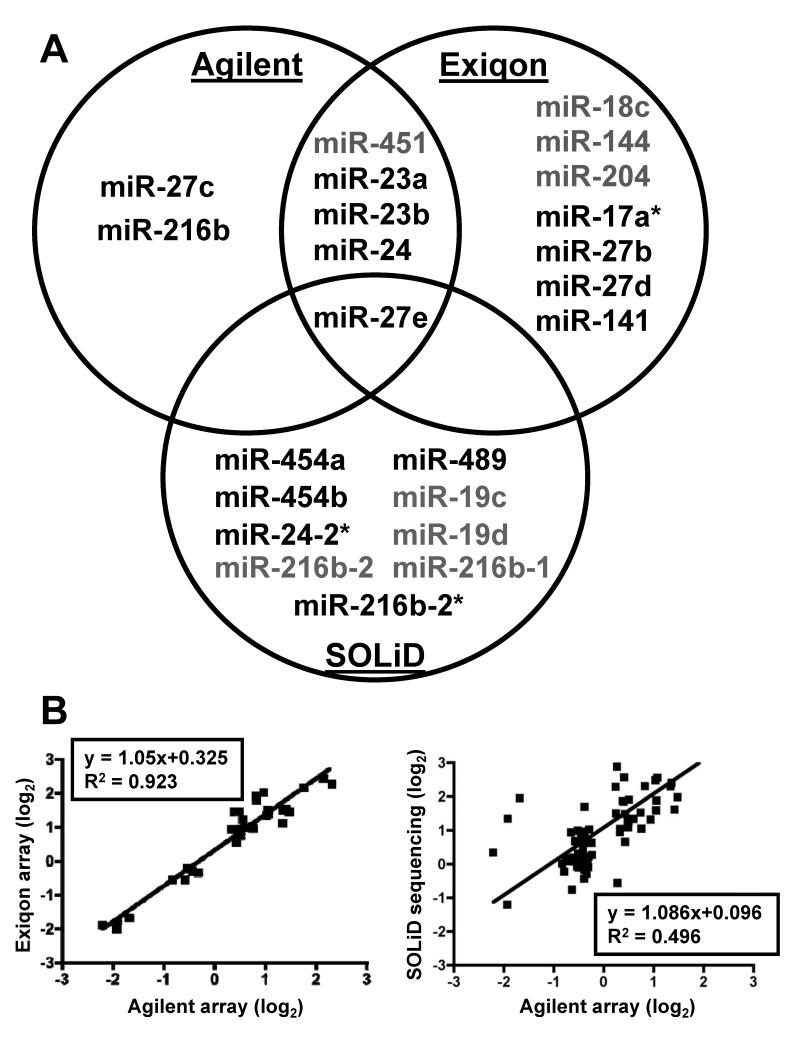
The three high-throughput methods produced unique but partially overlapping signatures of microRNAs differentially expressed in response to TCDD (Figure 5A). The two microarray platforms displayed the greatest level of congruence, with five microRNAs detected as differentially expressed between DMSO and TCDD treatments, including one down-regulated (miR-451) and four up-regulated microRNAs (miR-23a, 23b, 24, and 27e). We also compared the developmental changes (36 hpf vs 60 hpf DMSO) between the two microarray platforms and observed a very strong correlation (R2 = 0.92; Figure 5B). Only one microRNA (miR-27e) was detected by all three methods as differentially expressed in response to TCDD. The only disagreement between the methods in terms of direction of change was in the expression of miR-216b-1, which was considered slightly up-regulated as determined by the Agilent array and down-regulated as determined by SOLiD sequencing.
Figure 5.
A) Venn diagram showing the significant TCDD-induced changes in microRNA expression measured by microarray or SOLiD sequencing analysis at 60 hpf. Each quantification method resulted in some unique changes in microRNAs but some changes were common to different platforms. The microRNA changes that were common to different methods are shown in the intersections of the circles. MicroRNAs shown in green were downregulated and those in red were upregulated. B) Left: Scattor plot showing the correlation between the developmental changes in microRNA expression (60 hpf vs 36 hpf) as measured by the two microarray platforms. Right: Scattor plot showing the correlation between the developmental changes in microRNA expression (60 hpf vs 36 hpf) as measured by SOLiD sequencing as compared to Agilent arrays. Fold change values of microRNAs that were common to both the platforms were plotted.
The average coverage of known zebrafish microRNAs detected using the Exiqon and Agilent microarrays was 58% and 65%, respectively. In contrast, next-generation sequencing allowed for an average detection of 95% of all known mature microRNAs and 87% of star microRNA sequences in all six samples at each time point (36 and 60 hpf), including the ability to differentiate between individual microRNA isomirs. This additional coverage allowed for the detection by SOLiD sequencing of expression changes for miR-454a, 454b, miR-216b-1, miR-216b-2, miR-489, miR-19c, and miR-19d, which had probes on the arrays but were difficult to detect in our samples due to lower sensitivity of the arrays, and miR-216b-2* and miR-24-2*, which were not represented on the arrays. Comparison of developmental changes (36 hpf vs 60 hpf DMSO) in microRNA expression between the Agilent array and SOLiD sequencing (CLC analysis) resulted in a weaker correlation (R2 = 0.496) than seen for the microarray comparisons (R2 = 0.92) (Figure 5B) but the overall trend was consistent with the expected changes in microRNA expression that occur during development.
Two additional approaches to analysis of the SOLiD data (Genome Mapping and DNASTAR QSeq) were used to determine their consistency with results obtained by CLC Genomics workbench. In contrast to CLC Genomics workbench, which matches SOLiD reads to microRNA precursors in miRBase, DNASTAR QSeq was used to compare the sequences to mature microRNAs in miRBase and Genome Mapping maps the sequences to the zebrafish genome and compares the map positions to the coordinates of known microRNA precursors. Sixteen different microRNAs (mature or star sequences) were identified as differentially expressed between DMSO and TCDD treatments at 60 hpf by at least one of the three analysis methods (Table 4). Consistent with the microarray analyses, all three analyses of the SOLiD data identified miR-27e as significantly up-regulated at 60 hpf in response to TCDD exposure. The CLC Genomics and Genome Mapping approaches identified six microRNAs in common, whereas Genome Mapping and QSeq had two microRNAs in common and CLC Genomics and QSeq had only one microRNA showing a statistically significant change by both methods. Although the statistical significance may have differed, the calculated difference for 15 of the 16 microRNAs was consistent in direction of fold change among these approaches. The only discrepancy was in the detection of miR-23a, which was determined to be significantly up-regulated by the QSeq approach, but down-regulated by both the CLC Genomics and Genome Mapping approaches. Interestingly, both the Agilent and Exiqon arrays determined miR-23a to be significantly up-regulated in response to TCDD exposure at 60 hpf. Overall, the microarrays appeared to have less variation among samples, allowing for greater statistical power, while the high-throughput sequencing allowed for much deeper coverage of the entire microRNAome.
Table 4.
Comparison of differentially expressed microRNAs (TCDD vs. DMSO, 60 hpf) identified by three SOLiD sequencing analyses. The highlighted values are statistically significant. For comparison, results from both microarray platforms were included for all microRNAs identified as significant in the SOLiD sequencing data.
| CLC Workbench | Genome Mapping1 |
DNASTAR QSeq | Exiqon Arrays | Agilent Arrays | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| miRNA | Fold Diff. |
p-value | Fold Diff. |
p-value | Fold Diff. |
p-value | Fold Diff. |
p-value | Fold Diff. |
pvalue |
| mir- 216b-2 |
−66.0 | 3.84E- 15 |
NA | NA | NA | NA | NA | NA | NA | NA |
| mir- 216b-1 |
−60.8 | 6.12E- 15 |
−29.67 | 1.28E- 20 |
NA | NA | NA | NA | NA | NA |
| mir-216b | NA | NA | NA | NA | −5.2 | 0.289 | ND | ND | 1.109 | 0.03 |
| mir-454b | 20.4 | 2.00E- 12 |
8.27 | 1.81E- 09 |
6.2 | 0.32 | ND | ND | 1.07 | 0.88 |
| mir-454a | 6.2 | 5.05E- 07 |
2.57 | 0.049 | 1.3 | 0.857 | ND | ND | 1.022 | 0.93 |
| mir-27e | 5.2 | 3.05E- 05 |
4.87 | 2.5E-05 | 6.0 | 0.00467 | 1.3 | 0.00555 | 1.08 | 0.03 |
| mir-24- 2* |
5.7 | 9.36E- 05 |
NA | NA | NA | NA | NA | NA | NA | NA |
| mir- 216b-2* |
25.7 | 0.0228 | NA | NA | NA | NA | NA | NA | NA | NA |
| mir-489 | 4.1 | 0.0254 | 3.98 | 0.001 | 3.0 | 0.138 | ND | ND | −1.038 | 0.88 |
| mir-19c | −3.6 | 0.0471 | −2.36 | 0.095 | −1.4 | 0.791 | −1.07 | 0.0805 | 1.018 | 0.92 |
| mir-19d | −3.6 | 0.0471 | −3.13 | 0.006 | −1.8 | 0.152 | −1.04 | 0.144 | −1.016 | 0.93 |
| mir-23a | −0.63 | 1.00 | −0.63 | 0.877 | 1.5 | 0.00467 | 1.25 | 8.17E-04 | 1.288 | 1.0E-5 |
| mir-738 | NA | NA | NA | NA | −3.2 | 0.017 | ND | ND | 1.024 | 0.92 |
| mir-1388 | 3.2 | 0.157 | 2.93 | 0.032 | 3.1 | 0.0419 | NA | NA | NA | NA |
| mir-153b | −2.4 | 0.369 | −1.97 | 0.406 | −2.3 | 0.0458 | 1.03 | 0.577 | 1.064 | 0.88 |
| mir-93 | −2.08 | 0.601 | −3.28 | 0.004 | −12.8 | 0.434 | ND | ND | 1.024 | 0.92 |
| mir-19b | −2.77 | 0.216 | −2.67 | 0.032 | −2.1 | 0.184 | −1.07 | 0.048 | 1.002 | 0.97 |
NA = Not applicable due to inability of method to detect that specific miRNA isomer
ND = Not detected, probe was present on the array but hybridization intensity was not sufficient for accurate quantification
precursor miRNA reads
Assessment of microRNA expression as determined by real-time RT-PCR
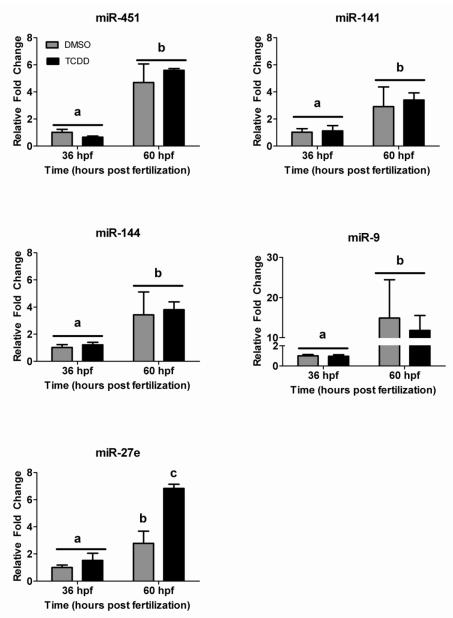
In an effort to confirm observed differences in microRNA expression, real-time RT-PCR was used to measure changes between treatments and timepoints for four microRNAs identified by one or more of the three high-throughput methods (mir-27e, miR-451, miR-141, and miR-144) and one microRNA (miR-9) known to be significantly differentially expressed between the two timepoints (Weinholds et al., 2005). Real-time RT-PCR analysis confirmed that all five microRNAs were significantly up-regulated in expression between 36 and 60 hpf as a result of increased basal expression during development, independent of TCDD exposure (Figure 6). However, of the four suspect microRNAs, only miR-27e was differentially expressed in response to TCDD exposure in a statistically significant manner as determined by real-time RT-PCR.
Figure 6.
Confirmation of microarray results with quantitative RT-PCR. We selected five microRNAs that were significantly altered between the two time points and showed TCDD-induced change in microRNA expression (miR-9, 451, 141, 144 and 27e) to confirm by qRT-PCR. Results are expressed as fold change from the DMSO control at 36 hpf, normalized to U6 – small nucleolar RNA. Different letters denote significant changes in expression over time or with TCDD treatment (Two-way ANOVA, p<0.05).
DISCUSSION
TCDD-induced changes in microRNA expression in developing embryos
TCDD is an environmental pollutant of global concern that is capable of inducing several abnormal developmental phenotypes in animals. The toxicity of TCDD occurs in an AHR-dependent manner with inappropriate activation of the AHR signaling pathway causing misregulation of target genes involved in basic biological processes. Previous studies in rodents have shown that while activation of the AHR pathway with known agonists (TCDD and benzo(a)pyrene) results in robust changes in mRNA expression, dramatic changes in microRNA expression do not occur, suggesting that it is unlikely that microRNAs play any significant role in mediating TCDD toxicity in adult mice and rats (Moffat et al., 2007; Yauk et al., 2011). However, these observations do not preclude a potential role of microRNAs in TCDD-mediated developmental defects. Exposure of zebrafish embryos to TCDD results in several severe abnormal developmental phenotypes, such as lower jaw malformations, cardiac malformations, neurodevelopmental defects and disruption of normal erythropoiesis (Belair et al., 2001; Dong et al., 2001; Teraoka et al., 2002; Hill et al., 2003; Antkiewicz et al., 2005). Interestingly, exposure of zebrafish embryos to TCDD prior to 72 hpf is capable of producing the observed phenotypes that begin to manifest at 72 hpf, where as exposure after 96 hpf produces no developmental phenotypes (Belair et al., 2001). This suggests that there are specific developmental regulatory pathways affected by TCDD in a time-dependent manner. To date, the only specific genes directly linked to a TCDD-induced phenotype in zebrafish embryos are sox9b, which plays a role in mediating the lower jaw malformations (Xiong et al., 2008), and R-Spondin1 (rspo1), which is involved in defective fin regeneration (Mathew et al., 2008). We report here our initial investigation into the potential for TCDD exposure to result in misexpression of microRNAs during zebrafish development.
Through a combination of microarray profiling and high-throughput sequencing, we identified numerous microRNAs that are potentially misexpressed as a result of TCDD exposure. Although the magnitude of the observed changes in microRNA expression was modest, several of the microRNAs may be of biological significance with respect to TCDD developmental toxicity. For example, some of the microRNAs identified in this study have been implicated in the direct regulation of various AHR pathway genes. MiR-24 has been shown to negatively regulate the aryl hydrocarbon receptor nuclear translocator (ARNT), the dimerization partner of AHR, in human liver (Oda et al., 2012). MiR-27b has been shown to negatively regulate CYP1B1 (Tsuchiya et al., 2006), with decreased expression of miR-27b as a possible explanation for the high level of CYP1B1 protein expression in human cancerous tissue (Devlin et al., 2010).
When considering the developmental phenotypes associated with TCDD exposure, the possible down-regulation of miR-144 and miR-451 by TCDD at 60 hpf is intriguing because these microRNAs have been shown to have a conserved role in the regulation of vertebrate erythropoiesis (Dore et al., 2008; Pase et al., 2009; Rasmussen et al., 2010). In zebrafish, miR-144 and miR-451 are processed from a single precursor transcript (Pase et al., 2009) and loss of these microRNAs results in inhibition of erythrocyte maturation (Dore et al., 2008; Pase et al., 2009), a phenotype consistent with TCDD-dependent inhibition of definitive erythropoiesis in zebrafish embryos (Belair et al., 2001). Related to the cardiac malformations caused by TCDD, up-regulation of miR-23a has been shown to positively regulate cardiac hypertrophy (Lin et al., 2009; Wang et al., 2012) and miR-24 has been shown to inhibit cardiomyocyte apoptosis and regulate cardiac fibrosis (Qian et al., 2011; Wang et al., 2012). TCDD has been demonstrated to induce cardiac hypertrophy in mice after in utero exposure (Kopf et al., 2008). However, during embryonic development TCDD exposure usually results in a decrease in myocyte proliferation (Ivnitski et al., 2001; Carney et al., 2006), which is inconsistent with the up-regulation of miR-23a and miR-24. However, TCDD exposure does result in failure of the development of heart valves and the bulbus arteriosus during zebrafish development (Mehta et al., 2008). It is possible that changes in miR-23a and miR-24 expression result in spatially restricted alterations in cardiac morphology during zebrafish development.
An additional phenotype associated with TCDD exposure in zebrafish embryos is the development of severe edema, most noticeable in the pericardial region and yolk sac (Henry et al., 1997; Hill et al., 2004). Interestingly, miR-204 down-regulation has been associated with the reduced expression of various claudin mRNA/proteins (Wang et al., 2010). Claudins are small transmembrane proteins that serve as a major component of tight junctions, protein barriers that control the flow of molecules between cells of epithelial tissues. Disruption of tight junction formation during zebrafish development by morpholino targeting of tjp3/zo-3, which encodes for a tight junction scaffold protein, resulted in edema, loss of blood circulation and tail fin malformations (Kiener et al., 2008), all phenotypes consistent with TCDD exposure during zebrafish development (Henry et al., 1997).
Although some of the changes in microRNA expression described above have potential links to TCDD effects, only one microRNA—miR-27e—was demonstrated consistently to be significantly altered (up-regulated), regardless of the method used (both array platforms, all three SOLiD analyses, and real-time RT-PCR). The miR-27 family in zebrafish consists of five members, three others of which (miR-27b, miR-27c, and miR-27d) may also have been up-regulated, as suggested by the microarray results. The functions of miR-27 in zebrafish are not well understood, but recent studies suggest an important role in regulating vascular development (Biyashev et al., 2012; Urbich et al., 2012). Zebrafish miR-27 forms share high sequence identity (including identical seed sequences) with mammalian miR-27 members (e.g. miR-27a and miR-27b in humans) (Supplementary Fig. 4). Zebrafish miR-27e occurs in a cluster with mir-23a-1 and mir-24-4, and in mammals there is a mir-23a ~ mir-27a ~ mir-24-2 cluster that is transcribed as a single primary transcript (Chhabra et al., 2010). Whether members of the zebrafish mir-23a-1 ~ mir-27e ~ mir-24-4 cluster are co-regulated or co-transcribed is not known, but mir-23a and mir-24 both were suggested by array data to be up-regulated by TCDD (Fig. 5A), consistent with that possibility. In addition, DNA sequence analysis of the 10kb region upstream of mir-23a-1~ mir-27e ~ mir-24-4 cluster revealed the presence of three predicted xenobiotic response elements (XREs; core sequence 5′-GCGTG-3′; −9104, −9180 and −9215). Additional studies will be needed to determine whether these XREs are involved in regulating expression of this cluster in response to TCDD. Together, these results point to mir-27e and related microRNAs as promising subjects for further research to investigate the role of altered microRNA expression in the mechanism of TCDD developmental toxicity.
Small RNA expression profiling
The secondary objective of the present investigation was to compare different methods for measuring expression of microRNAs in zebrafish embryos exposed to TCDD. Several previous studies have analyzed global microRNA expression in zebrafish embryos using microarrays (Thatcher et al., 2007; Zhang et al., 2011; Tal et al., 2012; Wu et al., 2012) and deep sequencing (Soares et al., 2009; Cifuentes et al., 2010; Yang et al., 2011; Wei et al., 2012). Some of these studies have also used real-time RT-PCR to confirm their results. Although the microarray-based methods allow global quantification of all the known microRNAs, there are differences between microarray platforms such as Exiqon and Agilent in the probe design and hybridization chemistry. Similarly, there are differences in sequencing chemistry between different deep sequencing platforms such as Roche 454 sequencing, Illumina Genome Analyzer, and ABI SOLiD. These high-throughput approaches have been used to determine the expression of microRNAs and other non-coding RNAs during zebrafish development but the specificity and accuracy of these methods in quantifying microRNA expression are not fully understood. This is particularly important given the fact that subtle changes in microRNA expression can have a major impact on gene expression patterns. Thus, it is essential to do a systematic comparison of different methods and identify the best approach for microRNA expression profiling. Such comparisons have been done previously in cancer cell lines (Git et al., 2010) and in human tissues and cell lines (Ach et al., 2008) and using reference samples (Yauk et al., 2010), but similar studies have not been reported in any in vivo vertebrate developmental model system.
Using both Exiqon and Agilent microarrays, we were able to detect the majority of the microRNA changes that occur during development. For the microRNAs that were represented in both arrays there was a significant correlation in their fold changes. These results suggest that, despite the differences in probe design, both microarray platforms are able to capture differentially expressed microRNAs. A previous study comparing the performance of four different commercial microRNA microarrays using a dilution series of spike-in controls concluded that Agilent microarrays have high accuracy as determined by overall changes in signal intensity with corresponding changes in spike-in concentration, while Exiqon microarrays have high specificity as determined by the increase in the signal intensity of only the probes that were specifically altered in the spike-in controls (Sah et al., 2010). We are unable to make a similar analysis to measure accuracy and precision because we did not incorporate spiked microRNAs into our hybridization reactions. However, based on the similarity in relative fold change values of the biological replicates between the two array platforms (Figure 5B), we can conclude that both platforms demonstrated similar detection capabilities.
There was strong congruence between the real-time RT-PCR results and small RNA sequencing results, as determined by the similar fold differences measured by the two methods for the developmental changes in expression of microRNAs or the changes in miR-27e expression in response to TCDD. However, one unexpected result was the inability of real-time RT-PCR to confirm some of the more modest TCDD-induced changes measured by the microarray platforms. Although real-time RT-PCR is considered the ‘gold standard’ for verifying changes in expression of mRNA, the short length of microRNAs presents additional challenges in the use of RT-PCR, particularly at smaller levels of change (~2-fold differences in microRNA expression). The stem-loop method used here, although considered robust for microRNAs (Chen et al., 2005), may have been limited by the modest differences in microRNA expression caused by TCDD and by the relatively small number of samples, reducing the power of this method to confirm the changes observed by microarray or SOLiD sequencing.
Interestingly, we found substantial variation in the detection of differentially expressed microRNAs by three different approaches to analysis of the next-generation sequencing data (CLC Genomics, Genome Mapping, and QSeq). One of the main reasons for this variation could be the differences in the mapping algorithms used in these methods. For instance, CLC genomics software maps the adaptor trimmed reads to the precursor microRNA sequences and other non-coding RNAs, whereas in the genome mapping method trimmed reads were mapped to the genome, and in the Qseq method reads were mapped to the mature microRNA and star sequences separately (Flow charts summarizing the three analysis pipelines can be found in Supplementary Figure 5). In addition, there are also differences in the counting of the aligned reads between commercial software (CLC Bio and QSeq) and the genome mapping method. Commercial software gives the read counts for mature, star and hairpin sequences separately whereas with genome mapping, we were only able to obtain the read counts for precursor microRNA sequences, which include the reads aligned to mature, star and hairpin regions of the precursor microRNA. Furthermore, there are several microRNAs for which genome coordinates are unknown; their read counts are not obtained from the genome mapping method. Despite these differences, all three approaches were fairly consistent in their calculations for changes in fold expression of microRNAs between DMSO and TCDD exposures. Differences in the method of identification of mature and/or star sequences and the inclusion or exclusion of other ncRNAs may have led to a difference in the observed abundance of specific microRNA sequences, as well as the number of observations used to normalize between samples. These differences in normalized abundance and number of observations could result in differences in sample-to-sample variability that affect the p-value calculations and result in different observations of statistical significance between samples. It is likely that all three methods used to analyze the SOLiD sequences yielded valuable data. However, only additional functional studies on individual microRNAs will fully confirm their role in TCDD toxicity. Our observations highlight the strengths, as well as some of the challenges that still exist, with analyzing high throughput sequencing datasets.
With the advent of deep sequencing technologies, the limitations associated with probe-based methods such as microarrays and real-time RT-PCR have been overcome. Using SOLiD sequencing we were able to quantify not only all the known microRNAs at single copy resolution but also other non-coding RNAs such as snRNA, snoRNA, tRNAs, rRNAs and vault RNAs. In addition, we detected multiple mature microRNA variants (isomiRs) and 3′ terminal addition of single nucleotides as shown previously in deep sequencing datasets (Cifuentes et al., 2010; Schulte et al., 2010). SOLiD sequencing identified twice the number of developmentally regulated microRNAs compared to microarrays and most of these changes were comparable to previously identified developmental changes (Soares et al., 2009; Wei et al., 2012). Overall these results suggest that deep sequencing may provide a more comprehensive assessment of microRNA expression in developing embryos.
CONCLUSION
The results presented here provide new information concerning the potential role of microRNAs in mediating abnormal developmental phenotypes associated with TCDD exposure. We tested two different methods for assessing global changes in microRNA expression, a microarray-based approach (two platforms) and a high-throughput sequencing approach. Both methods had strengths and weaknesses. The microarray approach produced less variation among samples allowing for greater precision and thus greater statistical power, but lacked the depth and coverage provided by high-throughput sequencing. Although the changes in microRNA expression in TCDD-exposed embryos were not dramatic, many of the identified microRNAs are known to participate in the regulation of signaling pathways related to the different developmental phenotypes observed with TCDD exposure. These changes warrant further study, including assessment of possible tissue-specific changes that may have been obscured by the whole-embryo approach used in this study.
Supplementary Material
Highlights.
Zebrafish embryos were exposed to TCDD at two different developmental timepoints.
Compared different methods in detecting global changes in microRNA expression.
TCDD caused significant changes in microRNA expression in zebrafish embryos.
Differentially expressed microRNAs have roles related to TCDD-induced phenotypes.
ACKNOWLEDGEMENTS
This work was supported in part by National Institutes of Health grants [R21ES017304 to MEH and MJJ, R00ES017044 to MJJ]; additional support was provided by a Woods Hole Oceanographic Institution (WHOI) Independent Study Award (funded by the Andrew W. Mellon Foundation Endowed Fund for Innovative Research) and by the Walter A. and Hope Noyes Smith Senior Scientist Chair to MEH. The U.S. Government is authorized to produce and distribute reprints for governmental purposes notwithstanding any copyright notation that may appear hereon. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ach RA, Wang H, Curry B. Measuring microRNAs: comparisons of microarray and quantitative PCR measurements, and of different total RNA prep methods. BMC Biotechnol. 2008;8:69. doi: 10.1186/1472-6750-8-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antkiewicz DS, Burns CG, Carney SA, Peterson RE, Heideman W. Heart malformation is an early response to TCDD in embryonic zebrafish. Toxicol. Sci. 2005;84:368–377. doi: 10.1093/toxsci/kfi073. [DOI] [PubMed] [Google Scholar]
- Antkiewicz DS, Peterson RE, Heideman W. Blocking expression of AHR2 and ARNT1 in zebrafish larvae protects against cardiac toxicity of 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol. Sci. 2006;94:175–182. doi: 10.1093/toxsci/kfl093. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T. The small RNA profile during Drosophila melanogaster development. Dev. Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- Belair CD, Peterson RE, Heideman W. Disruption of erythropoiesis by dioxin in the zebrafish. Dev. Dyn. 2001;222:581–594. doi: 10.1002/dvdy.1213. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a pratical and powerful approach to multiple testing. J. R. Stat. Soc. Ser B. 1995;57:289–300. [Google Scholar]
- Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, Sharon E, Spector Y, Bentwich Z. Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- Biyashev D, Veliceasa D, Topczewski J, Topczewska JM, Mizgirez I, Vinokour E, Reddi AL, Licht JD, Revskoy SY, Volpert OV. MiR-27b controls venous specification and tip cell fate. Blood. 2012;119:2679–2687. doi: 10.1182/blood-2011-07-370635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushati N, Cohen SM. microRNA functions. Annu. Rev. Cell Dev. Biol. 2007;32:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Carney SA, Peterson RE, Heideman W. 2,3,7,8-tetrachlorodibenzo-p-dioxin activation of the aryl hydrocarbon receptor/aryl hydrocarbon receptor nuclear translocator pathway causes developmental toxicity through a CYP1A-independent mechanism in zebrafish. Mol. Pharmacol. 2004;66:512–522. doi: 10.1124/mol.66.3.. [DOI] [PubMed] [Google Scholar]
- Carney SA, Chen J, Burns CG, Xiong KM, Peterson RE, Heideman W. Aryl hydrocarbon receptor activation produces heart-specific transcriptional and toxic responses in developing zebrafish. Mol. Pharmacol. 2006;70:549–561. doi: 10.1124/mol.106.025304. [DOI] [PubMed] [Google Scholar]
- Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung MO, Gilbert EF, Peterson RE. Cardiovascular teratogenicity of 2,3,7,8-tetrachlorodibenzo-p-dioxin in the chick embryo. Toxicol. Appl. Pharmacol. 1981;61:197–204. doi: 10.1016/0041-008x(81)90409-9. [DOI] [PubMed] [Google Scholar]
- Chhabra R, Dubey R, Saini N. Cooperative and individualistic functions of the microRNAs in the miR-23a~27a~24-2 cluster and its implication in human diseases. Mol. Cancer. 2010;9:232. doi: 10.1186/1476-4598-9-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, Ma E, Mane S, Hannon GJ, Lawson ND, Wolfe SA, Giraldez AJ. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couture LA, Harris MW, Birnbaum LS. Characterization of the peak period of sensitivity for the induction of hydronephrosis in C57BL/6N mice following exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Fundam. Appl Toxicol. 1990;15:142–150. doi: 10.1016/0272-0590(90)90171-f. [DOI] [PubMed] [Google Scholar]
- Devlin AH, Thompson P, Robson T, McKeown SR. Cytochrome P450 1B1 mRNA untranslated regions interact to inhibit protein translation. Mol. Carcinog. 2010;49:190–199. doi: 10.1002/mc.20589. [DOI] [PubMed] [Google Scholar]
- Dong W, Teraoka H, Kondo S, Hiraga T. 2,3,7,8-tetrachlorodibenzo-p-dioxin induces apoptosis in the dorsal midbrain of zebrafish embryos by activation of aryl hydrocarbon receptor. Neurosci. Lett. 2001;303:169–172. doi: 10.1016/s0304-3940(01)01743-8. [DOI] [PubMed] [Google Scholar]
- Dore LC, Amigo JD, Santos COD, Zhang Z, Gai X, Tobias JW, Yu D, Klein AM, Dorman C, Wu W, Hardison RC, Paw BH, Weiss MJ. A GATA-1-regulated microRNA locus essential for erythropoiesis. Proc. Natl. Acad. Sci. U S A. 2008;105:3333–3338. doi: 10.1073/pnas.0712312105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans BR, Karchner SI, Franks DG, Hahn ME. Duplicate aryl hydrocarbon receptor repressor genes (ahrr1 and ahrr2) in the zebrafish Danio rerio: structure function, evolution, and AHR-dependent regulation in vivo. Arch. Biochem. Biophys. 2005;441:151–167. doi: 10.1016/j.abb.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Git A, Dvinge H, Salmon-Divon M, Osborne M, Kutter C, Hadfield J, Bertone P, Caldas C. Systematic comparison of microarray profiling, real-time PCR, and next-generation sequencing technologies for measuring differential microRNA expression. RNA. 2010;16:991–1006. doi: 10.1261/rna.1947110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueta K, Molotski N, Gerchikov N, Mor E, Savion S, Fein A, Toder V, Shomron N, Torchinsky A. Teratogen-induced alterations in microRNA-34, microRNA-125b and microRNA-155 expression: correlation with embryonic p53 genotype and limb phenotype. BMC Dev. Biol. 2010;10:20. doi: 10.1186/1471-213X-10-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halappanavar S, Jackson P, Williams A, Jensen KA, Hougaard KS, Vogel U, Yauk CL, Wallin H. Pulmonary responses to surface-coated nanotitanium dioxide particles includes induction of acute phase response genes, inflammatory cascades, and changes in microRNAs: a toxicogenomic study. Environ. Mol. Mutagen. 2011;52:425–439. doi: 10.1002/em.20639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry TR, Spitsbergen JM, Hornung MW, Abnet CC, Peterson RE. Early life stage toxicity of 2,3,7,8-tetrachlorodibenzo-p-dioxin in zebrafish (Danio rerio) Toxicol. Appl. Pharmacol. 1997;142:56–68. doi: 10.1006/taap.1996.8024. [DOI] [PubMed] [Google Scholar]
- Hill A, Howard CV, Stahle U, Cossins A. Neurodevelopmental defects in zebrafish (Danio rerio) at environmentally relevant dioxin (TCDD) concentrations. Toxicol. Sci. 2003;76:392–399. doi: 10.1093/toxsci/kfg241. [DOI] [PubMed] [Google Scholar]
- Hill AJ, Bello SM, Prasch AL, Peterson RE, Heideman W. Water permeability and TCDD-induced edema in zebrafish early-life stages. Toxicol. Sci. 2004;78:78–87. doi: 10.1093/toxsci/kfh056. [DOI] [PubMed] [Google Scholar]
- Hobert O. Gene regulation by transcription factors and microRNAs. Science. 2008;319:1785–1786. doi: 10.1126/science.1151651. [DOI] [PubMed] [Google Scholar]
- Hudder A, Novak RF. miRNAs: effectors of environmental influences on gene expression and disease. Toxicol. Sci. 2008;103:228–240. doi: 10.1093/toxsci/kfn033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatitics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Ivnitski I, Elmaoued R, Walker MK. 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) inhibition of coronary development is preceded by a decrease in myocyte proliferation and an increase in cardiac apoptosis. Teratology. 2001;64:201–212. doi: 10.1002/tera.1065. [DOI] [PubMed] [Google Scholar]
- Jardim MJ, Fry RC, Jaspers I, Dailey L, Diaz-Sanchez D. Disruption of microRNA expression in human airway cells by diesel exhaust particles is linked to tumorigenesis-associated pathways. Environ. Health Perspect. 2009;117:1745–1751. doi: 10.1289/ehp.0900756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiener TK, Selptsova I, Hunziker W. Tjp3/zo-3 is critical for epidermal barrier function in zebrafish embryos. Dev. Biol. 2008;316:36–49. doi: 10.1016/j.ydbio.2007.12.047. [DOI] [PubMed] [Google Scholar]
- Kloosterman WP, Steiner FA, Berezikov E, Bruijn E. d., Belt J. v. d., Verheul M, Cuppen E, Plasterk RH. Cloning and expression of new microRNAs from zebrafish. Nucleic Acids Res. 2006;34:2558–2569. doi: 10.1093/nar/gkl278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopf PG, Huwe JK, Walker MK. Hypertension, cardiac hypertrophy, and impaired vascular relaxation induced by 2,3,7,8-tetrachlorodibenzo-p-dioxin are associated with increased superoxide. Cardiovasc. Toxicol. 2008;8:181–193. doi: 10.1007/s12012-008-9027-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulshreshtha R, Ferracin M, Negrini M, Calin GA, Davuluri RV, Ivan M. Regulation of microRNA Expression: The hypoxic component. Cell Cycle. 2007;6:1426–1431. [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Lai EC, Tomancak P, Williams RW, Rubin GM. Computational identification of Drosophila microRNA genes. Genome Biol. 2003;4:R42. doi: 10.1186/gb-2003-4-7-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(7):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray anaysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Lim NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lin Z, Murtaza I, Wang K, Jiao J, Gao J, Li PF. miR-23a functions downstream of NFATc3 to regulate cardiac hypertrophy. Proc. Natl. Acad. Sci. U S A. 2009;106:12103–12108. doi: 10.1073/pnas.0811371106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Romero P. Pre-processing and differential expression analysis of Agilent microRNA arrays using the AgiMicroRna Bioconductor library. BMC Genomics. 2011;12:64. doi: 10.1186/1471-2164-12-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev EV, Maniatis T. Multilevel regulation of gene expression by microRNAs. Science. 2008;319:1789–1790. doi: 10.1126/science.1152326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew LK, Sengupta SS, Ladu J, Andreasen EA, Tanguy RL. Crosstalk between AHR and Wnt signaling through R-Spondin1 impairs tissue regeneration in zebrafish. FASEB J. 2008;22:3087–3096. doi: 10.1096/fj.08-109009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta V, Peterson RE, Heideman W. 2,3,7,8-tetrachlorodibenzo-p-dioxin exposure prevents cardiac valve formation in developing zebrafish. Toxicol. Sci. 2008;104(2):303–311. doi: 10.1093/toxsci/kfn095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima Y. Widespread roles of microRNAs during zebrafish development and beyond. Dev. Growth Differ. 2012;54:55–65. doi: 10.1111/j.1440-169X.2011.01306.x. [DOI] [PubMed] [Google Scholar]
- Moffat ID, Boutros PC, Celius T, Linden J, Pohjanvirta R, Okey AB. microRNA in adult rodent liver are refractory to dioxin treatment. Toxicol. Sci. 2007;99:470–487. doi: 10.1093/toxsci/kfm189. [DOI] [PubMed] [Google Scholar]
- Oda Y, Nakajima M, Mohri T, Takaiya M, Aoki Y, Fukami T, Yokoi T. Aryl hydrocarbon receptor nuclear translocator in human liver is regulated by miR-24. Toxicol. Appl. Pharmacol. 2012;260:222–231. doi: 10.1016/j.taap.2012.02.012. [DOI] [PubMed] [Google Scholar]
- Pase L, Layton JE, Kloosterman WP, Carradice D, Waterhouse PM, Lieschke GJ. miR-451 regulates zebrafish erythroid maturation in vivo via its target gata2. Blood. 2009;113:1794–1804. doi: 10.1182/blood-2008-05-155812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM, Narotsky MG, Elizondo G, Fernandez-Salguero PM, Gonzalez FJ, Abbott BD. Amelioration of TCDD-induced teratogenesis in aryl hydrocarbon receptor (AhR)-null mice. Toxicol. Sci. 1999;47:86–92. doi: 10.1093/toxsci/47.1.86. [DOI] [PubMed] [Google Scholar]
- Pogribny IP. MicroRNA dysregulation during chemical carcinogenesis. Epigenomics. 2009;1:281–290. doi: 10.2217/epi.09.17. [DOI] [PubMed] [Google Scholar]
- Prasch AL, Teraoka H, Carney SA, Dong W, Hiraga T, Stegeman JJ, Heideman W, Peterson RE. Aryl hydrocarbon receptor 2 mediates 2,3,7,8-tetrachlorodibenzo-p-dioxin developmental toxicity in zebrafish. Toxicol. Sci. 2003;76:138–150. doi: 10.1093/toxsci/kfg202. [DOI] [PubMed] [Google Scholar]
- Qian L, Laake LWV, Huang Y, Liu S, Wendland MF, Srivastava D. miR-24 inhibits apoptosis and represses Bim in mouse cardiomyocytes. J. Exp. Med. 2011;208:549–560. doi: 10.1084/jem.20101547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen KD, Simmini S, Abreu-Goodger C, Bartonicek N, Giacomo MD, Bilbao-Cortes D, Horos R, Linderm MV, Enright AJ, O’Carroll D. The miR-144/451 locus is required for erythroid homeostasis. J. Exp. Med. 2010;207:1351–1358. doi: 10.1084/jem.20100458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sah S, McCall MN, Eveleigh D, Wilson M, Irizarry RA. Performance evaluation of commercial miRNA expression array platforms. BMC Res. Notes. 2010;3:80. doi: 10.1186/1756-0500-3-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte JH, Marschall T, Martin M, Rosenstiel P, Mestdagh P, Schlierf S, Thor T, Vandesompele J, Eggert A, Schreiber S, Rahmann S, Schramm A. Deep sequencing reveals differential expression of microRNAs in favorable versus unfavorable neuroblastoma. Nucleic Acids Res. 2010;38:5919–5928. doi: 10.1093/nar/gkq342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sempere LF, Cole CN, McPeek MA, Peterson KJ. The phylogenetic distribution of metazoan microRNAs: insights into evolutionary complexity and constraint. J. Exp. Zool. B Mol. Dev. Evol. 2006;306:575–588. doi: 10.1002/jez.b.21118. [DOI] [PubMed] [Google Scholar]
- Simone NL, Soule BP, Saleh AD, Savage JE, Degraff W, Cook J, Harris CC, Guis D, Mitchell JB. Ionizing radiation-induced oxidative stress alters miRNA expression. PLos ONE. 2009;4:e6377. doi: 10.1371/journal.pone.0006377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith FA, Schwetz BA, Nitschke KD. Teratogenicity of 2,3,7,8-tetrachlorodibenzo-p-dioxin in CF-1 mice. Toxicol. Appl. Pharmacol. 1976;38:517–523. doi: 10.1016/0041-008x(76)90183-6. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;22:897–899. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Soares AR, Pereira PM, Santos B, Egas C, Gomes AC, Arrais J, Oliveira JL, Moura GR, Santos MA. Parallel DNA pyrosequencing unveils new zebrafish microRNAs. BMC Genomics. 2009;10:195. doi: 10.1186/1471-2164-10-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern-Ginossar N, Gur C, Biton M, Horwitz E, Elboim M, Stanietsky N, Mandelboim M, Mandelboim O. Human microRNAs regulate stress-induced immune responses mediated by the receptor NKGD2. Nat. Immunol. 2008;9:1065–1073. doi: 10.1038/ni.1642. [DOI] [PubMed] [Google Scholar]
- Tal TL, Franzosa JA, Tilton SC, Philbrick KA, Iwaniec UT, Turner RT, Waters KM, Tanguay RL. MicroRNAs control neurobehavioral development and function in zebrafish. FASEB J. 2012;26:1452–1461. doi: 10.1096/fj.11-194464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teraoka H, Dong W, Ogawa S, Tsukiyama S, Okuhara Y, Niiyama M, Ueno N, Peterson RE, Hiraga T. 2,3,7,8-tetrachlorodibenzon-p-dioxin toxicity n the zebrafish embryo: altered regional blood flow and impaired lower jaw development. Toxicol. Sci. 2002;65:192–199. doi: 10.1093/toxsci/65.2.192. [DOI] [PubMed] [Google Scholar]
- Thatcher EJ, Flynt AS, Li N, Patton JR, Patton JG. miRNA expresison analysis during normal zebrafish development and following inhibition of the Hedgehog and Notch signaling pathways. Dev. Dyn. 2007;236:2172–2180. doi: 10.1002/dvdy.21211. [DOI] [PubMed] [Google Scholar]
- Tryndyak VP, Ross SA, Beland FA, Pogribny IP. Down-regulation of the microRNAs miR-34a, miR-127, and miR-200b in rat liver during hepatocarcinogenesis induced by a methyl-deficient diet. Mol. Carcinog. 2009;48:479–487. doi: 10.1002/mc.20484. [DOI] [PubMed] [Google Scholar]
- Tsuchiya Y, Nakajima M, Takagi S, Taniya T, Yokoi T. MicroRNA regulates the expression of human cytochrome P450 1B1. Cancer Res. 2006;66:9090–9098. doi: 10.1158/0008-5472.CAN-06-1403. [DOI] [PubMed] [Google Scholar]
- Urbich C, Kaluza D, Fromel T, Knau A, Bennewitz K, Boon RA, Bonauer A, Doebele C, Boeckel J-N, Herenreider E, Zeiher AM, Kroll J, Fleming I, Dimmeler S. MicroRNa-27a/b controls endothelial cell repulsion and angiogenesis by targeting semaphorin 6A. Blood. 2012;119:1607–1616. doi: 10.1182/blood-2011-08-373886. [DOI] [PubMed] [Google Scholar]
- Wang FE, Zhang C, Maminishkis A, Dong L, Zhi C, Li R, Zhao J, Majerciak V, Gaur AB, Chen S, Miller SS. MicroRNA-204/211 alters epithelial physiology. FASEB J. 2010;24:1552–1571. doi: 10.1096/fj.08-125856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Huang W, Xu R, Nie Y, Cao X, Meng J, Xu X, Hu S, Zheng Z. MicroRNA-24 regulates cardiac fibrosis after myocardial infarction. J. Cell. Mol. Med. 2012 doi: 10.1111/j.1582-4934.2012.01523.x. ePub ahead of print, PMID 22260784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Lin ZQ, Long B, Li JH, Zhou J, Li PF. Cardiac hypertrophy is positively regulated by microRNA miR-23a. J. Biol. Chem. 2012;287:589–599. doi: 10.1074/jbc.M111.266940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LL, Zhang Z, Li Q, Yang R, Pei X, Xu Y, Wang J, Zhou SF, Li Y. Ethanol exposure induces differential microRNA and target gene expression and teratogenic effects which can be suppressed by folic acid supplementation. Hum. Reprod. 2009;24:562–579. doi: 10.1093/humrep/den439. [DOI] [PubMed] [Google Scholar]
- Wei C, Salichos L, Wittgrove CM, Rokas A, Patton JG. Transcriptome-wide analysis of small RNA expression in early zebrafish development. RNA. 2012;18:915–929. doi: 10.1261/rna.029090.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinholds E, Kloosterman WP, Mishka E, Alvarez-Saavedra E, Berezikov E, Bruijn E. d., Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- Wienholds E, Plasterk RH. MicroRNA function in animal development. FEBS Lett. 2005;579:5911–5922. doi: 10.1016/j.febslet.2005.07.070. [DOI] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Wu TH, Pan CY, Lin MC, Hsieh JC, Hui CF, Chen JY. In vivo screening of zebrafish microRNA responses to bacterial infection and their possible roles in regulating immune response genes after lipopolysaccharide stimulation. Fish Physiol. Biochem. 2012 doi: 10.1007/s10695-012-9617-1. ePub ahead of print, PMID: 22419229. [DOI] [PubMed] [Google Scholar]
- Xiong KM, Peterson RE, Heideman W. Aryl hydrocarbon receptor-meidated down-regulation of sox9b causes jaw malformation in zebrafish embryos. Mol. Pharmacol. 2008;74:1544–1553. doi: 10.1124/mol.108.050435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R, Dai Z, Chen S, Chen L. microRNA-mediated gene regulation plays a minor role in the transcriptomic plasticity of cold-acclimated zebrafish brain tissue. BMC Genomics. 2011;12:605. doi: 10.1186/1471-2164-12-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yauk CL, Rowan-Carroll A, Stead JD, Williams A. Cross-platform analysis of global microRNA expression technologies. BMC Genomics. 2010;11:330. doi: 10.1186/1471-2164-11-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yauk CL, Jackson K, Malowany M, Williams A. Lack of change in microRNA expression in adult mouse liver following treatment with benzo(a)pyrene despite robust mRNA transcriptional response. Mutat. Res. 2011;722:131–139. doi: 10.1016/j.mrgentox.2010.02.012. [DOI] [PubMed] [Google Scholar]
- Yokoi T, Nakajima M. Toxicological implicaitons of modulation of gene expression by microRNAs. Toxicol. Sci. 2011;123:1–14. doi: 10.1093/toxsci/kfr168. [DOI] [PubMed] [Google Scholar]
- Zhang L, Li Y-Y, Zeng H.-c., Wei J, Wan Y.-j., Chen J, Xu S.-q. MicroRNA expression changes during zebrafish development induced by perfluorooctane sulfonate. J. Appl. Toxicol. 2011;31:210–222. doi: 10.1002/jat.1583. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.