A chemical biology study characterizes the role of Haspin kinase in centromere recruitment of the chromosome passenger complex and in spindle assembly checkpoint function.
Abstract
By phosphorylating Thr3 of histone H3, Haspin promotes centromeric recruitment of the chromosome passenger complex (CPC) during mitosis. Aurora B kinase, a CPC subunit, sustains chromosome bi-orientation and the spindle assembly checkpoint (SAC). Here, we characterize the small molecule 5-iodotubercidin (5-ITu) as a potent Haspin inhibitor. In vitro, 5-ITu potently inhibited Haspin but not Aurora B. Consistently, 5-ITu counteracted the centromeric localization of the CPC without affecting the bulk of Aurora B activity in HeLa cells. Mislocalization of Aurora B correlated with dephosphorylation of CENP-A and Hec1 and SAC override at high nocodazole concentrations. 5-ITu also impaired kinetochore recruitment of Bub1 and BubR1 kinases, and this effect was reversed by concomitant inhibition of phosphatase activity. Forcing localization of Aurora B to centromeres in 5-ITu also restored Bub1 and BubR1 localization but failed to rescue the SAC override. This result suggests that a target of 5-ITu, possibly Haspin itself, may further contribute to SAC signaling downstream of Aurora B.
Introduction
Haspin (also known as germ cell–specific gene 2 protein/GSG2 or haploid germ cell–specific nuclear protein kinase) is a serine/threonine kinase. Its overall fold conforms to the eukaryotic protein kinase (ePK) domain, but diverges in crucial ways from typical ePK members. Accordingly, Haspin is often classified as an atypical ePK family member (Tanaka et al., 1999; Higgins, 2001; Eswaran et al., 2009; Villa et al., 2009). Haspin’s best-characterized and conserved function to date is exercised at centromeres (Higgins, 2010). Centromeres are genetic loci that mark the site of construction of kinetochores, structures that mediate the interaction of chromosomes with spindle microtubules during mitosis (Tanaka et al., 1999; Higgins, 2001; Eswaran et al., 2009; Santaguida and Musacchio, 2009; Villa et al., 2009). Haspin phosphorylates histone H3 on threonine 3 (P-T3-H3), a phosphomark that accumulates specifically at centromeres during prometaphase (Polioudaki et al., 2004; Dai et al., 2005; Markaki et al., 2009; Higgins, 2010). P-T3-H3 is dephosphorylated after anaphase by the PP1 phosphatase with the contribution of the PP1 regulator Repo-Man (Qian et al., 2011).
Ablation of Haspin by RNAi perturbs chromosome bi-orientation and sister chromatid cohesion (Dai et al., 2005, 2006; Markaki et al., 2009; Higgins, 2010). These consequences of Haspin inhibition might reflect impaired centromeric localization of the chromosome passenger complex (CPC; Kelly et al., 2010; Wang et al., 2010; Yamagishi et al., 2010). The CPC is a complex of the four subunits Survivin, Borealin, INCENP, and Aurora B. The latter, also a serine/threonine kinase, regulates aspects of mitotic progression including chromosome condensation and bi-orientation, the spindle assembly checkpoint (SAC), and cytokinesis (Ruchaud et al., 2007).
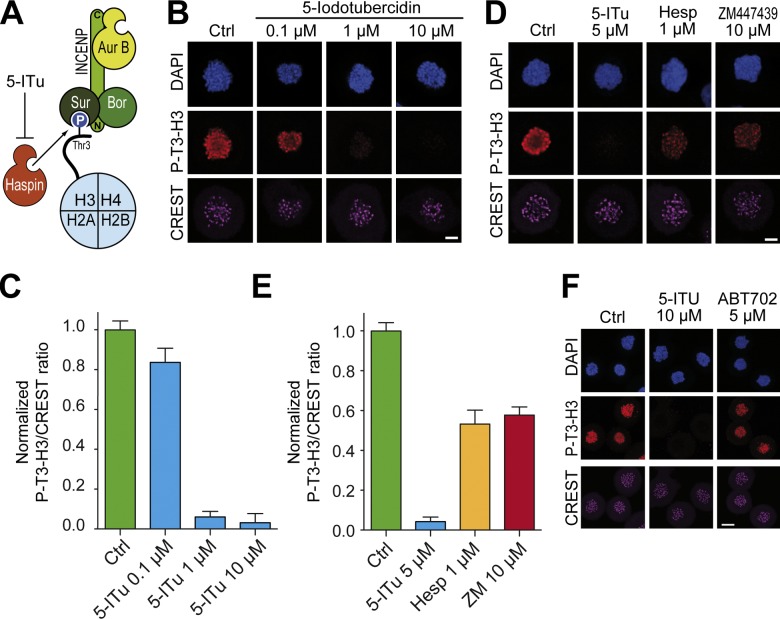
Two related pathways mediate centromere recruitment of the CPC. First, Bub1-dependent phosphorylation of histone H2A at Thr120 is proposed to create a binding site for Borealin through Shugoshin (Kawashima et al., 2010; Tsukahara et al., 2010; Storchová et al., 2011; van der Waal et al., 2012a). Second, Survivin binds directly to P-T3-H3 in the context of the N-terminal tail of histone H3 (Tanaka et al., 1999; Higgins, 2001; Eswaran et al., 2009; Villa et al., 2009; Kelly et al., 2010; Wang et al., 2010; Yamagishi et al., 2010; Jeyaprakash et al., 2011; Qian et al., 2011; F. Wang et al., 2011; Du et al., 2012; Niedzialkowska et al., 2012). Thus, by phosphorylating T3-H3, Haspin promotes phosphorylation-dependent centromeric recruitment of the CPC (Fig. 1 A).
Figure 1.
5-ITu inhibits histone 3 Thr3 phosphorylation. (A) Schematic representation of CPC recruitment to centromeres. Haspin phosphorylates T3-H3 at centromeres. Survivin (Sur), a CPC subunit, recognizes P-T3-H3, promoting recruitment of other CPC subunits. 5-ITu has been shown to inhibit Haspin in vitro (Villa et al., 2009; Eswaran et al., 2009). (B) HeLa cells were released from a double-thymidine arrest. After 5 h, nocodazole (3.3 µM) and the indicated concentrations of 5-ITu were added. After 2 h, 10 µM MG132 was added to prevent mitotic exit. After 90 min, cells were harvested, processed for immunofluorescence, and stained with DAPI (DNA), CREST sera (kinetochores), and an antibody against phosphorylated threonine 3 of histone H3 (P-T3-H3). Bar, 5 µm. (C) Quantitation of data from the experiment illustrated in B. Graphs report mean ± SEM. Data were normalized to the indicated control ratio. Details of quantitation are to be found in Materials and methods. (D) HeLa cells were treated as in B. Two Aurora B inhibitors (Hesperadin and ZM447439) were tested and their effect on the levels of P-T3-H3 compared. Bar, 5 µm. (E) Quantitation of data from the experiment illustrated in D. Graphs report mean ± SEM. Data were normalized to the indicated control ratio. (F) HeLa cells were treated with 3.3 µM nocodazole, and after 6 h mitotic cells were isolated by mitotic shake-off. Cells were replated in 10 µM MG132, and after 30 min the indicated inhibitors were added. After 90 min, cells were harvested and processed for immunofluorescence. Bar, 10 µm.
Aurora B generates additional mitotic phosphomarks on histones or histone variants by targeting Ser10 on histone H3 (P-S10-H3) along chromosome arms (Hsu et al., 2000; Giet and Glover, 2001; Higgins, 2010) and Ser7 of CENP-A (P-S7-CENP-A) Zeitlin et al., 2001b; Santaguida and Musacchio, 2009), the centromeric variant of histone H3 (Polioudaki et al., 2004; Dai et al., 2005; Markaki et al., 2009; Santaguida and Musacchio, 2009). Aurora B phosphorylates additional substrates at the centromere and kinetochores, including the kinetochore subunits Hec1/Ndc80, Dsn1, and Knl1/Spc105, and the microtubule regulators MCAK and CENP-E (Ruchaud et al., 2007; Qian et al., 2011; van der Waal et al., 2012b). Aurora B is also required for kinetochore recruitment and optimal activity of several SAC proteins (Ditchfield et al., 2003; Hauf et al., 2003; Vigneron et al., 2004; Dai et al., 2005, 2006; Markaki et al., 2009; Becker et al., 2010; Higgins, 2010; Santaguida et al., 2011; Saurin et al., 2011; Matson et al., 2012). Furthermore, Aurora B activity might counteract the kinetochore localization of the PP1 phosphatase, which opposes SAC signaling (Lampson and Cheeseman, 2011; Lesage et al., 2011).
Depletion of Haspin prevented CPC recruitment to centromeres (Kelly et al., 2010; Wang et al., 2010; Yamagishi et al., 2010). This in turn correlated with chromosome bi-orientation defects, possibly through deregulation of mitotic centromere associated kinesin (MCAK; Wang et al., 2010). However, other landmarks of centromeric localization of Aurora B, such as P-S7-CENP-A, were only modestly impaired (Wang et al., 2010). Similarly, SAC function was also modestly impaired in cells in which Haspin function was inhibited by RNAi (Wang et al., 2010). It is unclear whether the relatively modest consequences of repressing Haspin by RNAi are due to limited penetrance of the targeting approach, or alternatively to the ability of the CPC to control kinetochore and centromere function through a soluble pool that is not affected by inhibition of Haspin. Arguing against the second alternative, previous analyses have suggested that Aurora B concentration at centromeres or kinetochores might be important for its activation (Kelly et al., 2007; Rosasco-Nitcher et al., 2008; DeLuca et al., 2011; E. Wang et al., 2011).
Here, we further investigated the role of Haspin in the regulation of the CPC by means of a small-molecule ATP-competitive Haspin inhibitor, 5-iodotubercidin (5-Iodo-7-β-d-ribofuranosyl-7H-pyrrolo[2,3-d]pyrimidin-4-amine, abbreviated as 5-ITu). 5-ITu was originally characterized as an inhibitor of adenosine kinase (Wotring and Townsend, 1979; Newby, 1981), but was later shown to target protein kinases as well (Massillon et al., 1994). We and others recently demonstrated that 5-ITu is a potent and selective Haspin inhibitor in vitro (Fedorov et al., 2007; Eswaran et al., 2009; Balzano et al., 2011). Because the potential of 5-ITu as a Haspin inhibitor in living cells has not been assessed, we set out to test its function in human cells.
Results
5-ITu is a specific Haspin inhibitor
5-ITu potently targets the ATP-binding site of Haspin (IC50 5–9 nM; Eswaran et al., 2009; Balzano et al., 2011). To evaluate its selectivity, we screened 5-ITu against a commercial panel of 180 protein kinases at two inhibitor concentrations (0.1 and 1.0 µM; Table S1). 5-ITu had significant cross-reactivity with only a small number of kinases in this panel (Table S1, highlighted in red), including the members of the CMGC kinase family CLK (Cdc2-like kinase) and DYRK (dual-specificity tyrosine (Y) regulated kinase). Modest cross-reactivity was observed for another relatively small set of kinases (highlighted in yellow), including Cdk1/Cyclin B. The selectivity screening data also showed that 5-ITu is inactive against all three human Aurora kinases. In addition, we screened 5-ITu against 117 highly diverse kinases using temperature shift (ΔTm) assays (Fedorov et al., 2012; Table S2). This assay measures the affinity of inhibitors indirectly through the increase in protein stability upon binding. ΔTm data typically correlate well with enzymatic assay data (Fedorov et al., 2011). ΔTm values larger than 6°C typically indicate inhibitors with nM potency. The ΔTm screening data confirmed the cross-reactivity of 5-ITu against CLK and DYRK and detected only weak ΔTm shifts for all protein kinases screened. Together with our previous in vitro kinase assays, in which we proved that 5-ITu at 1-µM concentration is inactive against Aurora A–TPX2, Aurora B–INCENP, Bub1, Cdk1/Cyclin B, Mps1, Plk1, and Nek2A (Balzano et al., 2011), the selectivity data generated against a large panel of kinases with distinct screening methodologies identifies 5-ITu as an attractive chemical tool for targeting Haspin.
5-ITu inhibits histone 3 Thr3 phosphorylation
To test if 5-ITu inhibits Haspin in vivo, we applied growing concentrations of 5-ITu to HeLa cells. Incubation with high concentrations of 5-ITu at early stages of the cell cycle can inhibit the entry of HeLa cells into mitosis. To avoid this problem, whose causes are unclear but likely caused by inhibition of one or more interphase targets of Haspin, we directed our analysis to cells that were already in mitosis at the time of addition of 5-ITu.
At concentrations between 0.1 and 0.5 µM, 5-ITu caused a strong decrease in the immunofluorescence levels of P-T3-H3 (Fig. S1). Depletion of P-T3-H3 was complete at 10 µM 5-ITu (Figs. 1 B and S1 A, quantitation in Fig. 1 C). A 50% reduction in the levels of P-T3-H3 was also observed when cells were treated with the Aurora B inhibitors Hesperadin and ZM447439 (Fig. 1, D and E, and Fig. S1 D), in agreement with a previous report (F. Wang et al., 2011). The relatively high concentrations of 5-ITu required for ablating P-T3-H3 may appear to contrast with the efficacy of the compound on Haspin in vitro (IC50 = 9 nM; Balzano et al., 2011). This behavior likely reflects the rather hydrophilic character of 5-ITu, whose partition coefficient, reported in PubChem (http://pubchem.ncbi.nlm.nih.gov), is −0.7, in comparison with values of 3.9, 4.2, and 4.2 for more lipophilic compounds such as Reversine, Hesperadin, and ZM447439, respectively.
To rule out down-regulation of P-T3-H3 levels by 5-ITu inhibition of adenosine kinase we tested the potent adenosine kinase inhibitor ABT-702 (IC50 = 1.7 nM; Lee et al., 2012). The mitotic levels of P-T3-H3 remained normal at ABT-702 concentrations as high as 5 µM, excluding effects mediated by adenosine kinase on P-T3-H3 phosphorylation levels (Fig. 1 F).
5-ITu displaces the CPC from centromeres
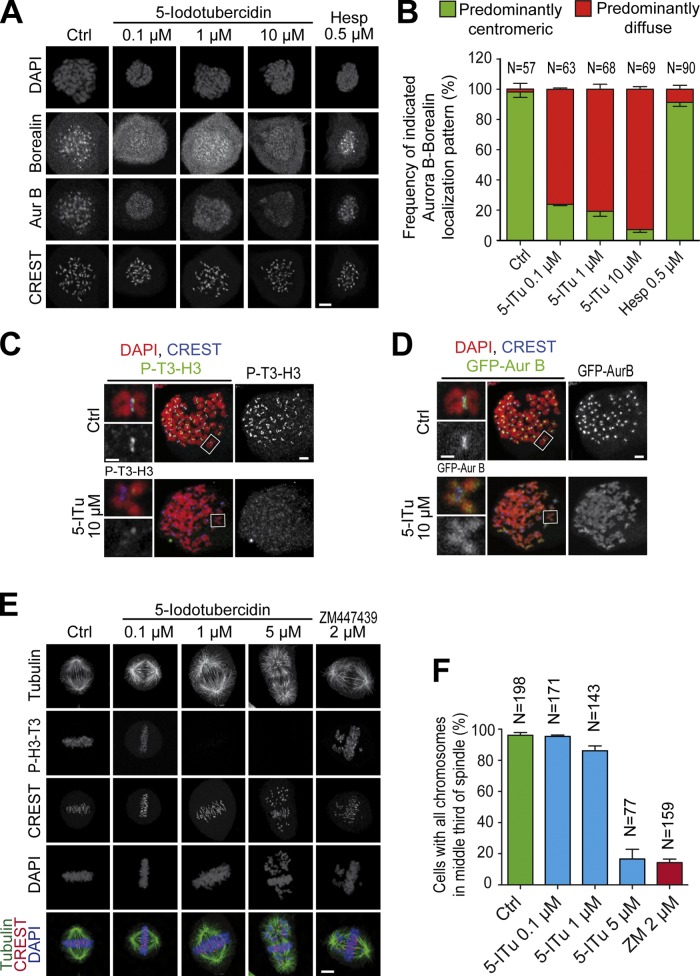
P-T3-H3 promotes localization of the CPC in human cells and in frog extracts through a direct interaction with Survivin (Kelly et al., 2010; Wang et al., 2010; Yamagishi et al., 2010; Jeyaprakash et al., 2011; Du et al., 2012; Niedzialkowska et al., 2012). We tested if treatment with 5-ITu, by preventing the accumulation of P-T3-H3, displaced the CPC from centromeres. Indeed, 5-ITu caused dose-dependent displacement of the CPC subunits Borealin and Aurora B from centromeres, and their apparent relocalization to chromosome arms (Fig. 2, A and B; and Fig. S1 B). The analysis of chromosome spreads emphasized the strong reduction of centromeric P-T3-H3 and the mislocalization of Aurora B in 5-ITu (Fig. 2, C and D).
Figure 2.
5-ITu displaces the CPC from centromeres. (A) Cells treated as in Fig. 1 B were supplemented with the indicated inhibitors and processed for immunofluorescence. Bar, 5 µm. (B) Quantitation of data shown in A. Localization of Aurora B and Borealin was analyzed for the indicated number of cells. Graphs report mean ± SEM. (C and D) Cultures of GFP-Aurora B HeLa cells were treated with 3.3 µM nocodazole for 12 h, after which 10 µM MG132 was added for 30 min. After a 90-min incubation with 10 µM 5-ITu, the mitotic population was isolated by mitotic shake-off and cells were prepared for immunofluorescence. Chromosome spreads were processed to visualize DAPI, CREST, P-T3-H3, and GFP-Aurora B. For presentation purposes, the P-T3-H3 and GFP-Aurora B signals are shown in two distinct panels with the same DAPI and CREST staining. Bar, 2 µm (insets, 1 µM). (E) HeLa cells were released from a double-thymidine arrest. After 6.5 h, the indicated inhibitors were added. After 30 min, 10 µM MG132 was added and after 90 min cells were harvested and processed for immunofluorescence. Bar, 5 µm. All images in this panel were acquired within the same experiment. (F) Quantitation of data from the experiment in E. The indicated number of cells for each condition was scrutinized for chromosome alignment defects. Graphs report mean ± SEM.
Contrarily to these extensive effects on CPC localization, inhibition of Aurora B with Hesperadin caused a relatively modest displacement of CPC subunits from centromeres and relocalization to chromosome arms (Fig. 2, A and B, and Fig. S1, B and C). Thus, Aurora B activates Haspin by phosphorylation to enhance T3-H3 phosphorylation and to promote its own recruitment to the centromere (F. Wang et al., 2011), but its inhibition does not reduce the levels of P-T3-H3 to the point of preventing centromeric localization of the CPC.
In agreement with the extensive effects of 5-ITu on Aurora B localization, HeLa cells treated with 5-ITu displayed significant defects in sister chromatid bi-orientation. At 1 µM 5-ITu most chromosomes congressed to the metaphase plate, but the plates often appeared broader and less focused (Fig. 2, E and F; and Fig. S1 D). At 5 µM 5-ITu a metaphase plate was still recognizable, but a large proportion of chromosomes failed to align at the equator and remained stuck near the poles, indicative of bi-orientation problems. These observations are in line with the previously described phenotype of RNAi-based ablation of Haspin (Dai et al., 2005, 2006).
5-ITu affects Aurora B activity at inner kinetochores
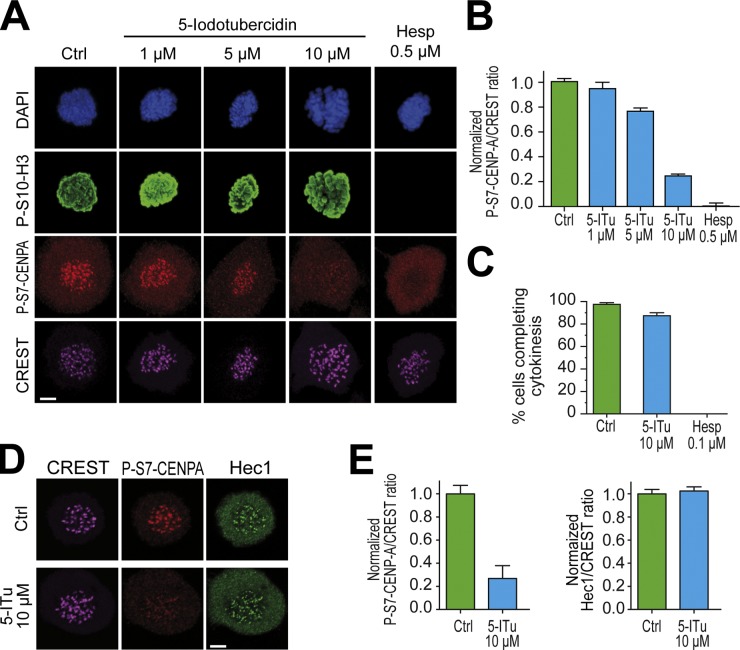
Serine 7 of CENP-A is a kinetochore substrate of Aurora B (Zeitlin et al., 2001a,b). The levels of this phosphomark at the inner kinetochore were only largely affected at 5-ITu concentrations comprised between 5 and 10 µM (Fig. 3, A and B). When considering that 5-ITu causes substantial reductions in the levels of P-T3-H3 already at 1 µM, this observation suggests the possibility that small residual levels of Aurora B are sufficient for the phosphorylation of kinetochore substrates of this kinase.
Figure 3.
5-ITu affects Aurora B activity at inner kinetochores. (A) HeLa cells were released from a double-thymidine arrest and after 5 h, 3.3 µM nocodazole and the indicated inhibitors were added. After 2 h, 10 µM MG132 was added to prevent mitotic exit by the inhibitors. After 90 min, cells were harvested, processed for immunofluorescence, and stained with DAPI (DNA), CREST sera (kinetochores), and antibodies against P-S10-H3 and P-S7–CENP-A; respectively, chromatin- and kinetochore-based substrates of Aurora B. Bar, 5 µm. (B) Quantitation of data from the experiment in A. Graphs represent mean ± SEM. Data were normalized to the indicated control ratio. (C) HeLa cells were treated with 3.3 µM nocodazole for 12 h. Nocodazole was washed out with or without the indicated inhibitors and mitotic cells were imaged during mitotic exit to monitor cytokinesis. (D) Kinetochore levels of Hec1 were not affected by 10 µM 5-ITu. Bar, 5 µm. (E) Quantitation of data from the experiment illustrated in D. Graphs represent mean ± SEM. Data were normalized to the indicated control ratio.
By comparison, the levels of phosphorylated serine 10 of histone 3 (P-S10-H3), another substrate of Aurora B localizing all along chromosome arms (Gurley et al., 1978; Paulson and Taylor, 1982), remained apparently constant in experiments of indirect immunofluorescence (see Fig. S2 and Discussion). Thus, Aurora B activity at the inner kinetochore is strongly impaired upon addition of 10 µM 5-ITu. Furthermore, 5-ITu might cause a more global reduction in Aurora B activity with effects extending beyond the centromere and kinetochores, possibly to chromosome arms and other regions of the cell (Lampson and Cheeseman, 2011; Tan and Kapoor, 2011; E. Wang et al., 2011). To investigate this important point, we asked if the cellular levels of Aurora B activity in the presence of 10 µM 5-ITu were sufficient to complete cytokinesis, the most inhibitor-sensitive function of Aurora B (Santaguida et al., 2010; Xu et al., 2010). Cytokinesis in the presence of 5-ITu was essentially normal (Fig. 3 C), indicating that 5-ITu at 10-µM concentration does not cause strong global inhibition of Aurora B. Moreover, 10 µM 5-ITu did not cause obvious defects in kinetochore assembly because the outer kinetochore protein Hec1/Ndc80 localized normally (Fig. 3, D and E).
Effects of 5-ITu on the recruitment of checkpoint proteins
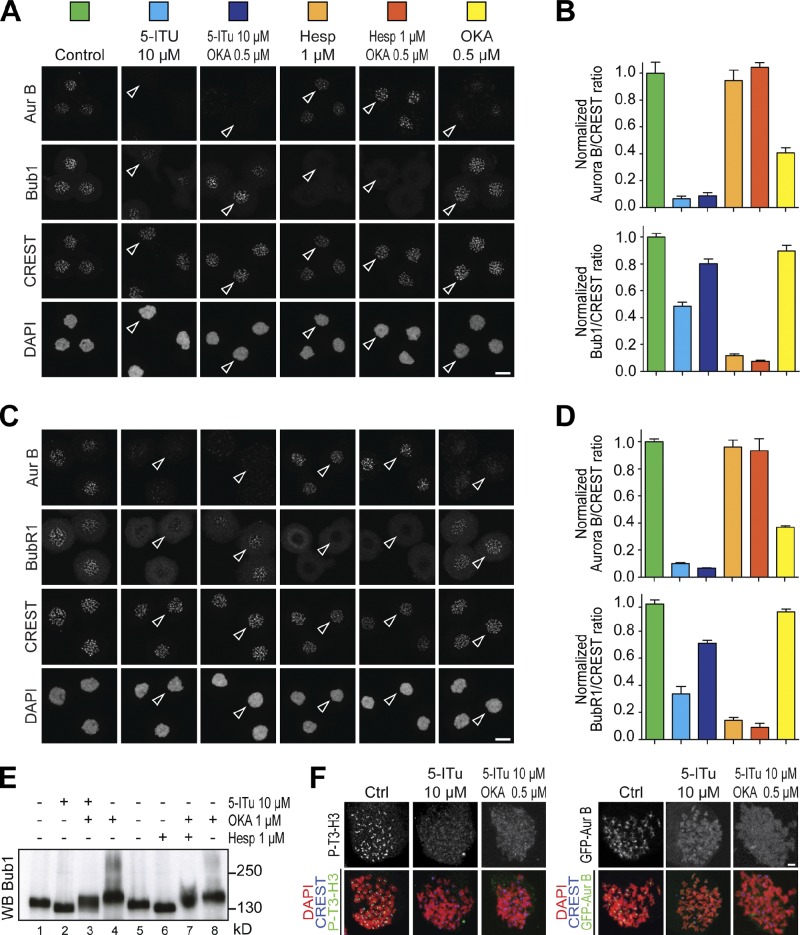
We next asked if 5-ITu perturbed kinetochore recruitment of SAC components, which depends on Aurora B (Ditchfield et al., 2003; Hauf et al., 2003; Vigneron et al., 2004). To simplify the interpretation of our results, we used a concentration of nocodazole (3.3 µM) previously shown to cause apparently complete microtubule depolymerization (Yang et al., 2009; Santaguida et al., 2011). In cells treated with 5-ITu, two mitotic checkpoint components, Bub1 and BubR1, were partially displaced from kinetochores (Fig. 4, A–D). Furthermore, 5-ITu treatment resulted in levels of Bub1 dephosphorylation comparable to those of cells treated with doses of the Aurora B inhibitor Hesperadin (1 µM) that have strong deleterious effects on the SAC (Fig. 4 E). Inhibition of Aurora B kinase with Hesperadin also prevented kinetochore localization of Bub1 and BubR1 (Fig. 4, A–D), in agreement with previous studies (Ditchfield et al., 2003; Vigneron et al., 2004). Collectively, these observations are consistent with the hypothesis that centromere- and kinetochore-localized Aurora B activity is important for kinetochore recruitment of Bub1 and BubR1, possibly through a pathway centered on Mps1 (London et al., 2012; Shepperd et al., 2012; van der Waal et al., 2012a; Yamagishi et al., 2012).
Figure 4.
Effects of 5-ITu on recruitment of checkpoint proteins. (A) Asynchronously growing HeLa cells were treated with 3.3 µM nocodazole. After 6 h mitotic cells were isolated by mitotic shake-off. Cells were replated in 10 µM MG132, and after 30 min the indicated concentrations and combinations of inhibitors were added. Cells were fixed after 90 min and processed for immunofluorescence with DAPI (DNA), CREST sera (kinetochores), and antibodies against Aurora B and Bub1. Bar, 10 µm. (B) Quantitation of data from the experiment illustrated in A. Graphs report mean ± SEM. Data were normalized to the indicated control ratio. The histograms were color coded to match the colored squares in A, with each color referring to the condition associated with the square. (C) As in A, but monitoring localization of BubR1 instead of Bub1. (D) Quantitation of data from the experiment in C. Graphs report mean ± SEM. Data were normalized to control ratio. Color coding is as explained in B. (E) Asynchronously growing HeLa cells were treated with 3.3 µM nocodazole for 6 h. Mitotic cells were collected by shake-off and treated with the indicated inhibitors complemented with 10 µM MG132 to prevent mitotic exit in the presence of the inhibitors. The mobility of Bub1 was evaluated by Western blotting after SDS-PAGE with 7.5% acrylamide gel (with an 80:1 acrylamide/bis-acrylamide ratio). (F) Chromosome spreads were generated as for Fig. 2 (C and D). On the left, the control and 5-ITu spreads are the same shown in Fig. 2 (C and D). Spreads were obtained in the same experiment. Bar, 2 µm.
Several recent reports demonstrated mutual regulation of Aurora B and protein phosphatase activity at centromeres and kinetochores (Emanuele et al., 2008; Pinsky et al., 2009; Vanoosthuyse and Hardwick, 2009a,b; Kim et al., 2010; Liu et al., 2010; Posch et al., 2010; Foley et al., 2011; Lesage et al., 2011; Meadows et al., 2011; Qian et al., 2011; Rosenberg et al., 2011; London et al., 2012; Shepperd et al., 2012). We reasoned that in the absence of centromere- and kinetochore-localized Aurora B, phosphatase activity might become deregulated, preventing the accumulation of phosphorylated substrates of Aurora B. To test this model, we asked if the PP1 and PP2 inhibitor okadaic acid (OKA, used at 0.5–1-µM concentrations) allowed Bub1 or BubR1 to accumulate at kinetochores despite the treatment with 10 µM 5-ITu. Indeed, OKA reverted the adverse effects of 5-ITu on the localization of Bub1 and BubR1 (Fig. 4, A–D). OKA also opposed the dephosphorylation of Bub1 caused by 5-ITu (Fig. 4 E). In the absence of other agents, OKA caused an ∼50% reduction in the centromeric levels of Aurora B, in agreement with a previous report (Takemoto et al., 2009).
Addition of OKA did not restore P-T3-H3, suggesting that 5-ITu achieves a very penetrant inhibition of Haspin that cannot be reverted even if PP1 (known to target P-T3-H3 for dephosphorylation; Qian et al., 2011) is inhibited (Fig. 4 F). Accordingly, Aurora B remained mislocalized when 5-ITu and OKA were used in combination (Fig. 4, A and F). Conversely, inhibition of Aurora B with 1 µM Hesperadin did not result in Aurora B mislocalization, and concomitant inhibition of phosphatase activity did not promote the recovery of Bub1 or BubR1 kinetochore localization (Fig. 4, A–D). Thus, kinetochore recruitment of Bub1 and BubR1 requires Aurora B activity, and such activity needs to be delivered near centromeres and kinetochores. The residual Aurora B activity observed in cells treated with Haspin inhibitors is sufficient to execute cytokinesis but might be insufficient for kinetochore accumulation of Bub1 and BubR1 (unless phosphatase activity is concomitantly inhibited). These results are consistent with the hypothesis of a reciprocal regulation of Aurora B and phosphatase activity at centromeres (Emanuele et al., 2008; Pinsky et al., 2009; Vanoosthuyse and Hardwick, 2009a,b; Kim et al., 2010; Liu et al., 2010; Posch et al., 2010; Foley et al., 2011; Lesage et al., 2011; Meadows et al., 2011; Qian et al., 2011; Rosenberg et al., 2011; London et al., 2012; Shepperd et al., 2012).
Forcing Aurora B localization at centromeres restores P-S7–CENP-A and kinetochore localization of SAC proteins
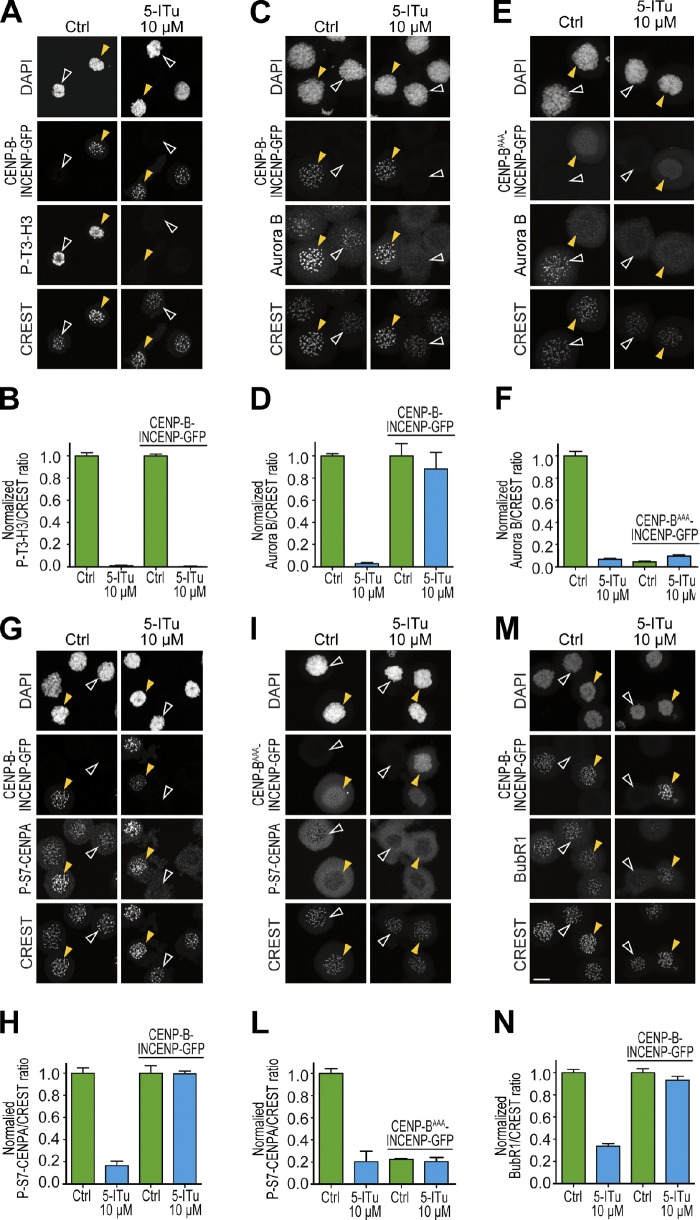
Insufficient Aurora B activity at centromeres and kinetochores in cells treated with 5-ITu likely is the cause of reduced kinetochore accumulation of P-S7–CENP-A, Bub1, and BubR1. To substantiate this idea, we forced localization of Aurora B to inner kinetochores by expressing a construct in which a fragment of INCENP containing the Aurora B binding site but devoid of the centromere-targeting domain (INCENP48–918) is fused to the C terminus of the DNA-binding domain of CENP-B (CENP-B1–158), an inner kinetochore protein (Liu et al., 2009). Transfection of this construct can be visualized via a GFP moiety fused to the C terminus of the CENP-B–INCENP fusion. We introduced three point mutations in the DNA-binding domain of CENP-B (Arg5-Ala, Lys70-Ala, Arg125-Ala, abbreviated as CENP-BAAA–INCENP) to generate a negative control.
The CENP-B–INCENP fusion correctly targeted kinetochores in the presence of 5-ITu, but as expected given the continued inhibition of Haspin, it failed to restore the P-T3-H3 signal (Fig. 5, A and B; yellow arrowheads indicate cells transfected with the CENP-B–INCENP construct, and white arrowheads indicate untransfected control cells). The chimera, however, drove Aurora B accumulation at kinetochores despite 5-ITu (Fig. 5, C and D), indicating that the CENP-B–INCENP fusion binds to Aurora B and can overcome the loss of P-T3-H3. Such accumulation was strictly dependent on the DNA-binding domain of CENP-B, because a CENP-BAAA–INCENP fusion failed to rescue kinetochore accumulation of Aurora B (Fig. 5, E and F, right). Indeed, expression of the mutant chimera had a dominant-negative effect on kinetochore accumulation of Aurora B even in the absence of 5-ITu, probably because it subtracts Aurora B from endogenous INCENP, preventing its localization to centromeres. Conversely, wild-type CENP-B–INCENP caused an increase in the levels of Aurora B in the centromere/kinetochore area in comparison to the endogenous levels measured in cells not expressing the construct (Fig. S4 A).
Figure 5.
Forcing Aurora B localization at centromeres restores P-S7–CENP-A and kinetochore localization of SAC proteins. (A, C, E, G, I, and M) HeLa cells were transfected with CENP-B–INCENP-GFP or CENP-BAAA–INCENP-GFP fusion construct for 24–48 h. Cells were subsequently treated with 3.3 µM nocodazole. After 6 h mitotic cells were isolated by mitotic shake-off, replated in 10 µM MG132, and after 30 min 5-ITu (at 10 µM) was added. Cells were harvested after 90 min and processed for immunofluorescence to visualize DAPI (DNA), CREST (kinetochores), GFP, and the indicated antigens. Bar, 10 µm. (B, D, F, H, L, and N) Quantitation of data from the experiment illustrated in A, C, E, G, I, and M, respectively. Graphs report mean ± SEM. Data were normalized to the respective indicated control ratio, except in F and L, where normalization was performed against the untransfected control because the expression of the CENP-BAAA–INCENP-GFP fusion has a dominant-negative effect on Aurora B localization even in the absence of 5-ITu. Note that CREST signal increases in cells expressing CENP-B–INCENP-GFP due to reactivity of CREST to CENP-B.
Next, we tested if the CENP-B–INCENP fusion was able to support kinetochore accumulation of the P-S7–CENP-A phosphoantigen in the presence of 10 µM 5-ITu. Indeed, CENP-B–INCENP promoted substantial reaccumulation of P-S7–CENP-A despite the continued presence of 5-ITu (Fig. 5, G and H). This effect was strictly dependent on kinetochore localization of the chimera because the triple mutant in the CENP-B DNA-binding domain abrogated it (Fig. 5, I–L). Also in this case, wild-type CENP-B–INCENP increased the levels of P-S7–CENP-A in comparison to the endogenous levels, and such increase correlated with the increased levels of Aurora B (Fig. S4 B). Finally, cells expressing CENP-B–INCENP also had apparently normal levels of kinetochore BubR1 or Bub1 (Fig. 5, M and N; and Fig. S3) despite 5-ITu.
Overall, these observations indicate that localization of Aurora B activity at centromeres promoted by Haspin via P-T3-H3 is essential for Aurora B–mediated phosphorylation of kinetochore substrates and for checkpoint function. Forced localization of Aurora B activity at kinetochores or inhibition of phosphatases that oppose Aurora B activity at kinetochores is sufficient to restore the phosphorylation status of substrates or the localization of SAC components.
5-ITu affects Aurora B activity on the outer kinetochore
An ∼80-residue N-terminal segment of Hec1/Ndc80 is important for kinetochore–microtubule binding and is a well-established prometaphase target of Aurora B (Ciferri et al., 2005; Cheeseman et al., 2006; DeLuca et al., 2006, 2011; Guimaraes et al., 2008; Miller et al., 2008; Welburn et al., 2010). We tested if 5-ITu suppressed the accumulation of the phosphorylated form of Ser44 of Hec1/Ndc80 (P-S44-Hec1), which depends on Aurora B (Cheeseman et al., 2006; DeLuca et al., 2006, 2011; Ciferri et al., 2008). Indeed, 5-ITu caused an ∼50% reduction in the kinetochore levels of P-S44-Hec1 (Fig. 6, A and B). This effect was inferior to that caused by direct inhibition of Aurora B kinase with Hesperadin. However, contrarily to the latter, it was rescued by the CENP-B–INCENP construct (Fig. 6, A and B), suggesting that the accumulation of normal levels of P-S44-Hec1 at kinetochores requires Aurora B activity locally.
Figure 6.
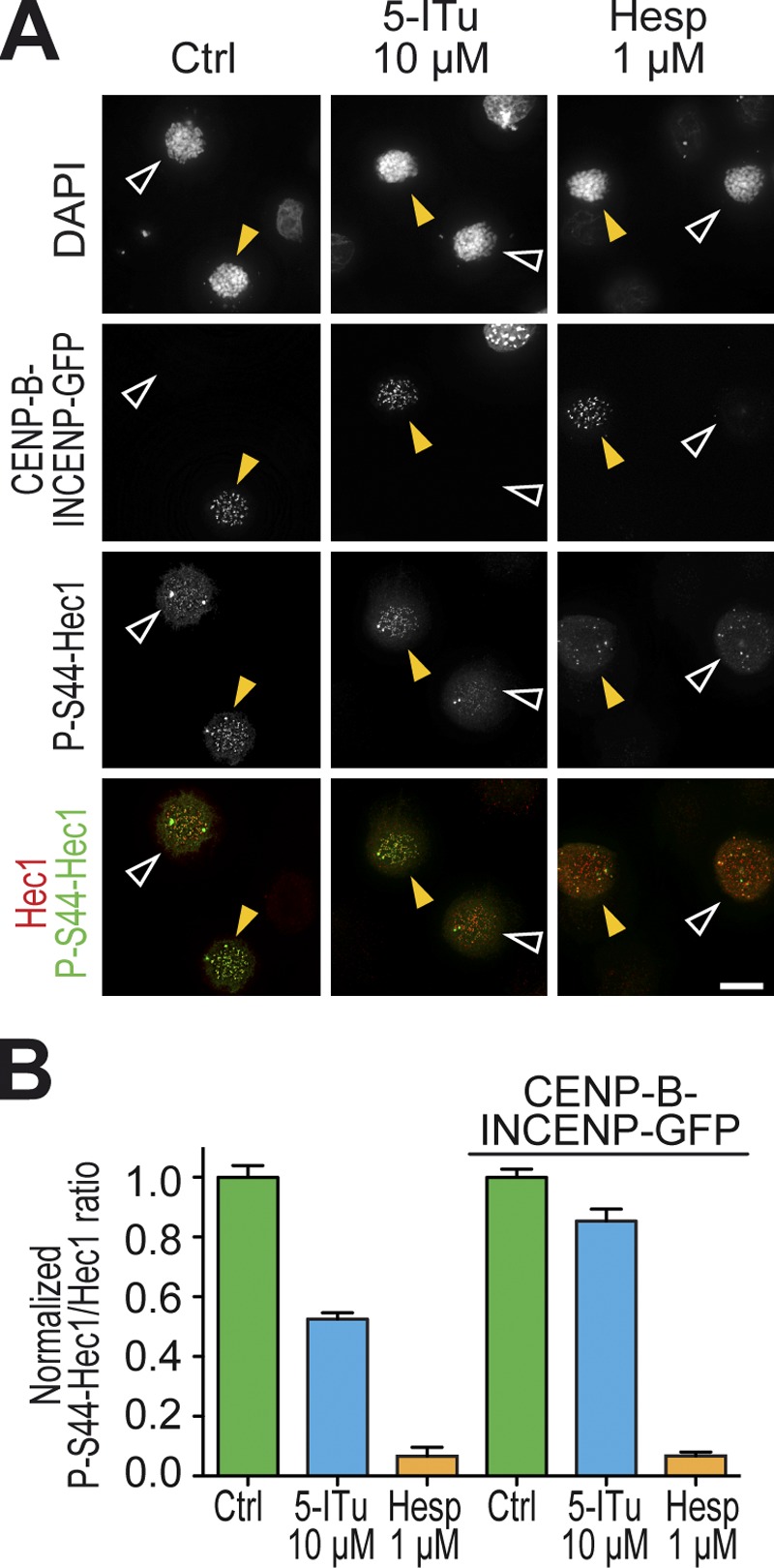
5-ITu affects Aurora B activity on the outer kinetochore. (A) HeLa cells were transfected with CENP-B–INCENP-GFP fusion construct for 24–48 h. Cells were subsequently treated with 3.3 µM nocodazole. After 6 h, 10 µM MG132 was added, and after 30 min 10 µM 5-ITu was added. Cells were fixed after 90 min and processed for immunofluorescence to visualize DAPI (DNA), Hec1 (kinetochores), GFP, and P-S44-Hec1 (DeLuca et al., 2011). Bar, 10 µm. (B) Quantitation of data from the experiment illustrated in A. Graphs report mean ± SEM. Data were normalized to the indicated control ratio.
Effects of 5-ITu on the spindle assembly checkpoint
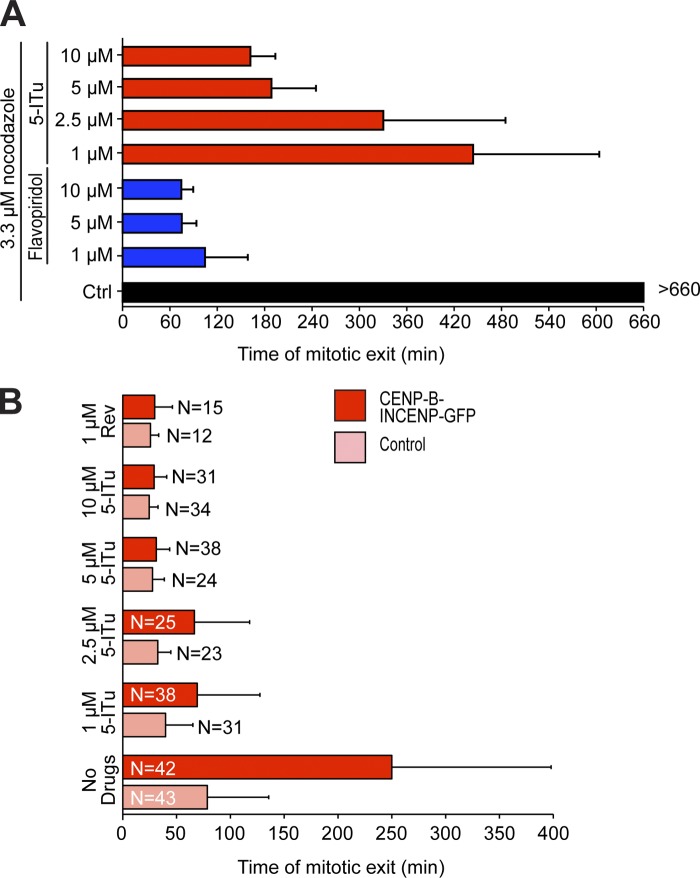
Kinetochore recruitment of SAC proteins is considered important for optimal checkpoint operation (Musacchio and Salmon, 2007). Because 5-ITu appeared to affect kinetochore recruitment of at least two SAC proteins, we asked if cells challenged with 5-ITu were able to maintain a checkpoint-dependent arrest in mitosis. We therefore tested the effects of 5-ITu on the SAC in two distinct assays. First, we generated a population of mitotic cells by mitotic shake-off of cycling cells treated with 3.3 µM nocodazole for 6 h. Control cells maintained the arrest for at least 11 h (Fig. 7 A). Conversely, cells treated with 5-ITu underwent checkpoint override at times that depended on drug concentration, with effects that became evident between 0.5 and 1.0 µM (Fig. 7 A and Fig. S5). At 10 µM, the adverse effect of 5-ITu on checkpoint duration was as pervasive as that caused by 1 µM Hesperadin (Fig. S5). As a positive control for these experiments, we also monitored the effects of the Cdk1 inhibitor flavopiridol (Losiewicz et al., 1994), which caused mitotic exit in less than 2 h at 1-µM or higher concentrations (Fig. 7 A).
Figure 7.
Effects of 5-ITu on spindle checkpoint signaling. (A) Asynchronously growing HeLa cells were treated with 3.3 µM nocodazole. After 6 h, mitotic cells were isolated by shake-off, replated, and inhibitors were added at the indicated concentrations under continued presence of nocodazole. Low-resolution live-cell imaging was then started. Cell flattening indicated mitotic exit. At least 50 cells per condition were analyzed. (B) HeLa cells expressing H2B-Cherry were transfected with the CENP-B–INCENP-GFP fusion construct, and 38 h after transfection they were exposed to 330 nM nocodazole for 4.5 h to cause mitotic arrest. Mitotic cells were harvested and nocodazole washed out. After 50 min, cells were either left untreated, or were treated with Reversine or 5-ITu. The timing of anaphase onset was monitored by high-resolution time-lapse video microscopy. The number of analyzed cells is indicated.
Second, we asked if 5-ITu was able to override the SAC arrest caused by expression of the CENP-B–INCENP chimera in the absence of spindle poisons. This effect, which has been discussed before, is probably caused by alignment errors after relocalization of Aurora B close to the outer kinetochores (Liu et al., 2009). Cells expressing the CENP-B–INCENP chimera and control cells were first arrested in nocodazole. Nocodazole was then washed out, and the time of anaphase onset was monitored by time-lapse video microscopy. Control cells underwent anaphase at 79 ± 57 min, whereas cells expressing CENP-B–INCENP underwent anaphase at 250 ± 148 min (Fig. 7 B), confirming that expression of CENP-B–INCENP causes a mitotic arrest in otherwise normally dividing cells. We therefore asked if 5-ITu was able to override such arrest. As a control, we used Reversine, a small-molecule ATP-competitive inhibitor of Mps1 (Santaguida et al., 2010). Both Reversine and 5-ITu abrogated the mitotic arrest caused by expression of CENP-B–INCENP, with mean times of mitotic exit of 30 ± 16 min and 29 ± 12 min, respectively (Fig. 6 D). Thus, 5-ITu overrides the mitotic arrest caused by the expression of the CENP-B–INCENP fusion protein. Collectively, these results indicate that 5-ITu is a rather potent SAC inhibitor.
Forcing centromeric localization of Aurora B does not restore a functional SAC
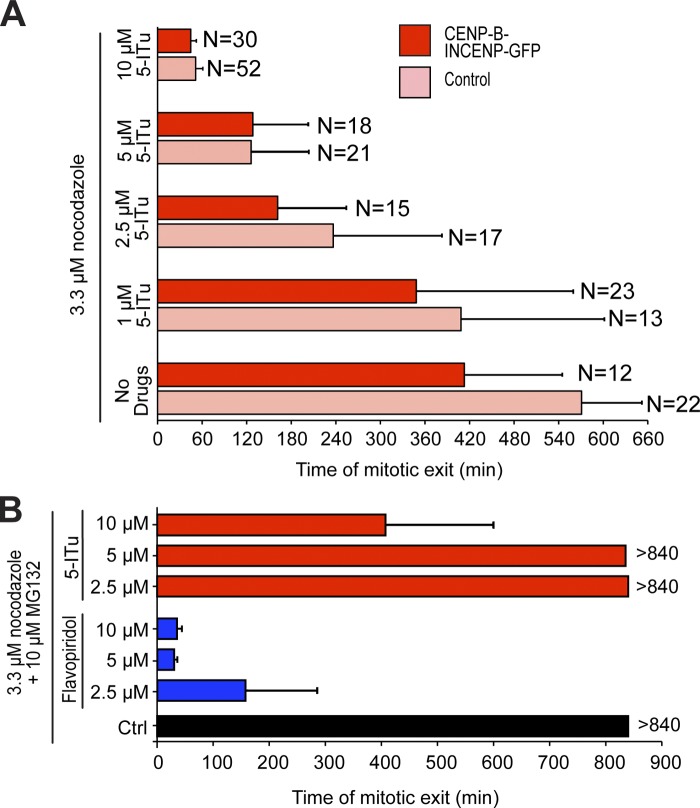
Aurora B contributes to the SAC independently of its well-established role in error correction (Kallio et al., 2002; Ditchfield et al., 2003; Hauf et al., 2003; Petersen and Hagan, 2003; King et al., 2007; Vader et al., 2007; Vanoosthuyse and Hardwick, 2009a; Maldonado and Kapoor, 2011; Santaguida et al., 2011; Saurin et al., 2011; Matson et al., 2012). Thus, its mislocalization from centromeres might be sufficient for SAC impairment. On the other hand, the ability of 5-ITu of overriding the mitotic delay caused by expression of the CENP-B–INCENP suggested that 5-ITu might inhibit certain aspects of kinetochore signaling even when Aurora B localizes at the inner kinetochore region. To test this possibility formally, we asked if restoration of Aurora B localization at inner kinetochores with the CENP-B–INCENP construct rescued the SAC defect caused by growing concentrations of 5-ITu in the presence of high doses of nocodazole (Fig. 8 A). Expression of the CENP-B–INCENP construct did not rescue the SAC defect caused by 5-ITu at any concentration tested. Indeed, expression of CENP-B–INCENP induced a slightly faster exit compared with control HeLa cells treated with the same concentration of 5-ITu.
Figure 8.
Forcing centromeric localization of Aurora B does not restore a functional SAC. (A) HeLa cells expressing H2B-Cherry were transfected with CENP-B–INCENP-GFP fusion construct for 36 h. 7 h after addition of 3.3 µM nocodazole, mitotic cells were isolated by shake-off and replated, and then the indicated concentrations of 5-ITu were added (or not added in the control). The time of mitotic exit was recorded by high-resolution live-cell video microscopy in the indicated number of cells. (B) Cycling HeLa cells expressing H2B-Cherry were treated with 3.3 µM nocodazole, and after 7 h mitotic cells were collected by shake-off. After addition of 10 µM MG132 the indicated drugs were added and cells were filmed by low-resolution time-lapse video microscopy. At least 50 cells per condition were analyzed.
In Table S1, we show that 5-ITu can target cyclin-dependent kinases in vitro, albeit at much higher concentrations compared with Haspin (see also Discussion). To test the hypothesis that inhibition of Cdk1/Cyclin B may cause mitotic exit in cells treated with nocodazole and 5-ITu, we asked if MG132, a proteasome inhibitor, antagonized the mitotic exit observed in these cells. By opposing the destruction of Cyclin B, MG132 prevents mitotic exit when cells with an active SAC are challenged with a SAC inhibitor. On the other hand, MG132 is unable to prevent mitotic exit when cells are treated with a Cdk1 inhibitor, a consequence of the decline of Cdk1/Cyclin B activity required to maintain the mitotic state (Potapova et al., 2006). In agreement with this notion, mitotic HeLa cells treated with nocodazole (3.3 µM), MG132, and the Cdk1 inhibitor flavopiridol exited mitosis in ∼30 min (Fig. 8 B).
Thus, if 5-ITu at the concentrations used in our studies inhibited Cdk1, then MG132 should not be able to counteract mitotic exit, as illustrated by flavopiridol. Treatment of HeLa cells with nocodazole (3.3 µM), MG132, and concentrations of 5-ITu up to 5 µM did not cause visible effects on the timing of mitotic exit (monitored by measuring DNA decondensation of live HeLa cells expressing H2B-mCherry) compared with control cells treated only with nocodazole and MG132 (Fig. 8 B). At 10 µM 5-ITu, an effect on the timing of mitotic exit (mean time of mitotic exit of 417 ± 197 min, Fig. 8 B), consistent with the possibility of modest Cdk1 inhibition, was observed.
Discussion
In this study, we extended previous analyses on the role of the Haspin substrate P-T3-H3 in the mitotic localization and activation of the CPC (Kelly et al., 2010; Wang et al., 2010; Yamagishi et al., 2010; F. Wang et al., 2011; Qian et al., 2011). In an accompanying paper, Wang et al. (2012) also report a detailed characterization of the effects of 5-ITu, reaching conclusions that are almost invariably in line with ours (see Wang et al. in this issue).
The effects of 5-ITu on HeLa cells are consistent with the observed specificity of this inhibitor for Haspin in vitro (Tables S1 and S2; Balzano et al., 2011). Acute inhibition of Haspin in mitotic cells caused CPC eviction from centromeres and its redistribution to chromosome arms. Consistently, P-S7-CENP-A underwent dose-dependent removal from kinetochores. The kinetochore levels of the SAC proteins Bub1 and BubR1, as well as the levels of the P-S44-Hec1 phosphoantigen were also strongly reduced, and the SAC response was severely weakened.
Concomitant impairment of protein phosphatase activity rescued kinetochore localization of Bub1 and BubR1. This suggests that centromere localization of Aurora B is required to oppose phosphatase function at centromeres and kinetochores, in agreement with recent studies (Emanuele et al., 2008; Pinsky et al., 2009; Vanoosthuyse and Hardwick, 2009a,b; Kim et al., 2010; Liu et al., 2010; Posch et al., 2010; Foley et al., 2011; Lesage et al., 2011; Meadows et al., 2011; Qian et al., 2011; Rosenberg et al., 2011; London et al., 2012; Shepperd et al., 2012). Consistently, kinetochore localization of Bub1 and BubR1 in 5-ITu was also rescued by a kinetochore-targeting CENP-B–INCENP construct, but not by an equivalent construct impaired in DNA binding. PP1-targeting motifs conforming to the Arg-Val-Ser/Thr-Phe (RVS/TF) consensus (Hendrickx et al., 2009) can be phosphorylated by Aurora B to counteract PP1 binding (Kim et al., 2010; Liu et al., 2010; Lesage et al., 2011; Meadows et al., 2011; Rosenberg et al., 2011), and it is plausible that PP1 activity prevails at kinetochores when Aurora B localization is perturbed.
We documented the effects of 5-ITu over a range of concentrations. For the localization experiments, we opted for the 10-µM 5-ITu concentration because the effects on Aurora B substrates at kinetochores (P-S7–CENP-A and P-S44–Hec1) become especially evident at this concentration. Such effects were invariably rescued by ectopic Aurora B localization mediated by CENP-B–INCENP. We therefore assume that the effects of 5-ITu, at least for what concerns the localization of phosphoantigens and SAC proteins that depend on Aurora B for their appearance at kinetochores, are caused exclusively by Aurora B mislocalization. It may seem puzzling that the decrease in the centromeric levels of Aurora B becomes apparent at lower concentrations of 5-ITu (Fig. 2 A) than those required for the eviction of kinetochore phosphoantigens attributed to Aurora B (e.g., P-S7–CENP-A in Fig. 3 A). This observation might reflect a nonlinear relationship between the levels of Aurora B and its kinase output, so that even very small residual amounts of Aurora B might be sufficient to phosphorylate kinetochore substrates.
When using 5-ITu at a concentration of 10 µM we failed to observe an obvious decrease in the levels of P-S10-H3 in immunofluorescence experiments (Fig. 3). Western blot analysis of cell lysates, however, demonstrated a noticeable decrease of the P-S10-H3 signal in cells treated with 10 µM 5-ITu (Fig. S2). Although we are at present unable to provide a conclusive explanation for such a discrepancy, we infer that it might reflect the presence of a nonmitotic fraction of cells in the population subjected to Western blot analysis. At 10-µM concentration, 5-ITu forced mitotic exit in the presence of nocodazole and MG132, possibly indicative of partial Cdk1 inhibition (Fig. 8 B). However, this effect typically manifested itself on average 400 min after addition of 5-ITu, whereas cells were harvested for Western blot analysis only 90 min after addition of 5-ITu. Future studies will have to address this issue in greater depth.
Given the importance of kinetochore localization of Bub1 and BubR1 for their SAC function, our observations are consistent with the idea that centromere localization of Aurora B is necessary for SAC function (E. Wang et al., 2011). However, we failed to observe significant rescue of the checkpoint defect caused by 5-ITu simply by targeting Aurora B to the kinetochore with a CENP-B–INCENP construct. Failure to rescue the adverse effects of 5-ITu on the SAC might indicate that the CENP-B–INCENP construct does not fully recapitulate Aurora B localization. For instance, recent studies identified the active form of Aurora B specifically at kinetochores (Posch et al., 2010; DeLuca et al., 2011; Petsalaki et al., 2011). Targeting Aurora B to other kinetochore domains might provide more realistic localization conditions that mimic more effectively the localization of the endogenous protein.
Although we previously excluded known checkpoint kinases as targets of 5-ITu in vitro (Balzano et al., 2011), it remains possible that the adverse effects of 5-ITu on the SAC may result from off-target inhibition of a different SAC component. Besides setting the stage for the execution of mitosis, Cdk1 is also considered important for the SAC (D’Angiolella et al., 2003; Wong and Fang, 2007; Morin et al., 2012). In our previous studies, where 5-ITu was tested at 1 µM, we failed to observe inhibition of Cdk1/Cyclin B activity with 5-ITu (Balzano et al., 2011). Discrepancy with analyses described here (Tables S1 and S2) might be due to differences in the preparation of kinase samples for the in vitro assay. A comparison of the experiments displayed in Figs. 7 A and 8 B, however, suggest that inhibition of Cdk1 is not a major concern in the interpretation of the effects of 5-ITu at the concentrations used in this study. Specifically, the much longer mitotic delay in the presence of MG132 in these experiments strongly suggests that Cdk1 is not a crucial target in the SAC override effects of 5-ITu.
The effects of 5-ITu on localization of Aurora B substrates were invariably rescued by the CENP-B–INCENP construct, but those on the SAC were not, even at intermediate concentrations of 5-ITu. A possible interpretation of this is that Haspin itself is required for an aspect of SAC signaling distinct from centromere and kinetochore localization of Aurora B and other proteins whose localization is controlled by Aurora B. Alternatively, 5-ITu might be inhibiting a distinct SAC pathway, in addition to that represented by the Haspin–Aurora B axis. Future studies will have to address this issue in detail.
Due to its function as a regulator of Aurora B, Haspin is recognized as a potential target for pharmacological intervention in anti-tumor therapy (Patnaik et al., 2008; Cuny et al., 2012; Huertas et al., 2012). Characterization of 5-ITu as a Haspin inhibitor discussed here will provide the research community with a readily available, public domain inhibitor for chemical biology investigations of this important kinase.
Materials and methods
Mammalian plasmids and cell lines
To express the CENP-B(1–158)-INCENP(47–920)-GFP construct in HeLa cells we used the SL417 vector, which was obtained from the pEGFP-N1 backbone [SL417 was provided by S.M.A. Lens, University Medical Center, Utrecht, Netherlands, who has previously provided a detailed description of this vector (Liu et al., 2009)]. The CENP-BAAA mutant (Arg5-Ala, Lys70-Ala, Arg125-Ala) was obtained by site-directed mutagenesis using the following primers: R5A_F (5′-TTCGGCCCCAAGGCGCGACAGCTGACGTTC-3′), R5A_R (5′-GAACGTCAGCTGTCGCGCCTTGGGGCCGAA-3′), K70A_F (5′-TGCCGCAAGACCAACGCGCTGTCTCCCTACGAC-3′), K70A_R (5′-GTCGTAGGGAGACAGCGCGTTGGTCTTGCGGCA-3′), R125A_F (5′-GGCTGGCTGGACGCCTTCCGCCGGCGCCAC-3′), and R125A_R (5′-GTGGCGCCGGCGGAAGGCGTCCAGCCAGCC-3′). HeLa cells stably expressing H2B-Cherry used in high-resolution time-lapse experiments were a gift of Sara Barozzi (Imaging Facility, IFOM-IEO Campus, Milan, Italy).
Cell culture and transfection
HeLa cells were grown in DME (Euroclone) supplemented with 10% fetal bovine serum (FBS, Hyclone) and 2 mM l-glutamine. Nocodazole (Sigma-Aldrich) was used at a concentration of 3.3 µM unless differently specified. Thymidine (2.5 mM) was purchased from Sigma-Aldrich. For transfection, FuGENE 6 Transfection Agent (Roche) was used at a 3:1 ratio with plasmid DNA. Cells were analyzed 24–48 h after transfection.
Immunoblotting
Cells were resuspended in lysis buffer (75 mM Hepes, pH 7.5, 150 mM KCl, 1.5 mM EGTA, 1.5 mM MgCl2, 10% glycerol, 0.1% NP-40, protease inhibitors [EMD Millipore], and phosphatase inhibitors [PhosSTOP tablets; Roche]). Cells were lysed by sonication and the lysates were cleared by centrifugation at 45,000 g for 30–60 min. The total protein amount in the cell lysates was quantified using the Bradford protein assay (Bio-Rad Laboratories). The following antibodies were used for immunoblotting: mouse anti-Actin (AC-40, working dilution 1:1,000; Sigma-Aldrich), rabbit anti-Bub1 (Ab9000, working dilution 1:3,000; Abcam), rabbit anti-phospho-H3-Ser10 (06–570, working dilution 1:1500; EMD Millipore), and mouse anti-CyclinB1 (sc-205, working dilution 1:1,000; Santa Cruz Biotechnology, Inc.).
Immunofluorescence
HeLa cells were plated onto coverslips coated with 15 µg/ml poly-d-lysine (Sigma-Aldrich). Cells were fixed using 4% PFA in PBS, permeabilized using 0.1% Triton X-100 in PBS, then treated with 4% BSA in PBS as a blocking agent and incubated with the appropriate antibodies diluted in 4% BSA in PBS. Alternatively cells were fixed through incubation with 4% paraformaldehyde in PHEM buffer, pH 7.0 (100 mM Pipes, 50 mM Hepes, 20 mM EGTA, and 16 mM MgSO4) for 20 s, followed by incubation in PHEM containing 0.5% Triton X-100 for 5 min, and by further 10 min fixation with 4% paraformaldehyde in PHEM. For Fig. 6, cells were fixed as above but for 20 min, then treated with PHEM containing 1% Triton X-100 for 5 min and prepared for antibody staining. The following antibodies were used for immunofluorescence: anti-centromeric antibody (working dilution 1:50–1:100; Antibodies Inc.), mouse anti Hec1 (clone 9G3.23, working dilution 1:1,000; Genetex), mouse anti–α-tubulin (clone B512, working dilution 1:2,000; Sigma-Aldrich), mouse anti-Borealin (M147-3, working dilution 1:200; MBL International), rabbit anti-Aurora B (AB2254, working dilution 1:1,000; Abcam), mouse anti-phospho-H3 Ser10 (working dilution 1:1,000; Abcam), rabbit anti-phospho-CENP-A-Ser7 (04–792, working dilution 1:200–1:300; EMD Millipore); rabbit anti-phospho-H3-Thr3 (9714S, working dilution 1:50–1:100; Cell Signaling Technology). Sheep anti-Bub1 (residues 336–489) and sheep anti-BubR1 (residues 2–211; a gift from S. Taylor, University of Manchester, Manchester, UK; working dilution 1:1,000 for both) and rabbit P-S44-Hec1 (a gift from J. DeLuca, Colorado State University, Fort Collins, CO; working dilution 1:3,000) have been described previously (Taylor et al., 2001; Tighe et al., 2008; DeLuca et al., 2011). Cy3-, RhodamineRed-X–, Cy5-, and DyLigth649-conjugated secondary antibodies were purchased from Jackson ImmunoResearch Laboratories. Alexa 488–labeled secondary antibodies were from Invitrogen. DNA was stained with DAPI. The coverslips were mounted using Mowiol mounting media or ProLong Gold Antifade reagent (Life Technologies). Cells were imaged at 25°C using a confocal microscope (model TCS SP2; Leica) equipped with a 63× NA 1.4 objective lens and using LCS 3D software (Leica) or, alternatively, using a DeltaVision Elite imaging system (Applied Precision) and microscope (model IX-71; Olympus) controlled by SoftWoRx software (Applied Precision) and a 100× 1.49 NA objective lens with a CoolSNAP HQ2 camera (Photometrics). Images were acquired as z-sections at 0.2442 µm (SP2; Leica) or 0.3 µm (DeltaVision), respectively, and converted into maximal intensity projections using ImageJ (National Institutes of Health) or SoftWoRx (Applied Precision) software. Deconvolution was performed using a constrained-iterative algorithm in SoftWoRx.
Quantitation of immunofluorescence data
Quantitation of immunofluorescence was performed using SoftWoRx. Pixel intensity was measured for each channel within a mask encompassing individual kinetochores; the average background pixel intensity was measured from three different cytosolic regions. After background subtraction, kinetochore signals were normalized to CREST (with the exception of P-S44-Hec1, where the signal was normalized to Hec1). Data analysis was performed using Prism 5.0 (GraphPad Software); final figures were assembled in Illustrator CS5.1 (Adobe). Graphs show mean ± SEM from at least two independent experiments. 18–20 kinetochores from 5–6 cells were used in each case.
Chromosome spreads
HeLa cells stably expressing mouse-Aurora B-LAP (MCB 73, a gift of I. Poser and A.A. Hyman, Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany) were treated for 12 h with 3.3 µM nocodazole, incubated with 10 µM MG132 for 30 min and then treated with the indicated drugs for 90 min. Mitotic cells were harvested by shake-off, incubated for 20 min in 75 mM KCl, 3.3 µM nocodazole, MG132 10 µM and the indicated drugs, and then dropped on a coverslip to be processed for immunofluorescence. Primary antibodies used were: rabbit anti phospho-H3-Thr3 (1:75); rabbit anti-phospho-CENP-A-Ser7 (1:300); and human anti-centromeric antibodies (1:50). Images were acquired on a confocal microscope (model TCS SP2; Leica) equipped with a 100× NA 1.4 objective lens and using LCS 3D software (Leica).
Live-cell imaging
For high-resolution live-cell imaging, cells were plated after mitotic shake-off onto Lab-Tek II chambered coverglasses (Thermo Fisher Scientific) coated with 15 µg/ml poly-d-lysine (Sigma-Aldrich), in DME without phenol red (Gibco). Live-cell imaging was performed using a DeltaVision Elite imaging system (Applied Precision) with an inverted microscope (model IX71; Olympus), a PlanApo 60× (1.42 NA) oil immersion objective (Olympus), and a CoolSNAP HQ2 camera (Photometrics). The system was equipped with an environmental chamber maintained at 37°C in an atmosphere of 5% CO2. Images were acquired as z-stacks (1.5-µm step) and converted into maximal intensity projections using ImageJ. For low-resolution live-cell imaging cells were plated after mitotic shake-off onto 12-well plastic dishes coated with 15 µg/ml poly-d-lysine. Imaging was performed using a ScanR system (Olympus) equipped with an inverted microscope (model IX70; Olympus) using 20× (0.45 NA) or 40× (0.6 NA) dry objectives and an ORCA-ER camera (Hamamatsu Photonics). Cells were maintained at 37°C and 5% CO2 in a humidified incubation chamber (Solent Scientific).
Online supplemental material
Fig. S1 shows additional points of concentration series for experiments displayed in Fig. 1 and Fig. 2. Fig. S2 reports Western blots that complement observations displayed in Fig. 3. Fig. S3 reports additional localization experiments that complete the series shown in Fig. 5. Fig. S4 reports additional quantitation experiments that complete the series shown in Fig. 5. Fig. S5 reports additional experiments related to Fig. 7 B. Tables S1 and S2 report 5-ITu inhibition data for two sets of kinases in two distinct assays. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201205119/DC1.
Acknowledgments
We thank Fabrizio Villa, Gerben Vader, Holger Bastians, Mathieu Bollen, Alessio Maiolica, and the members of A. Musacchio’s laboratory for discussions; Jonathan Higgins for sharing unpublished results; Silvia Monzani, Tiziana Melis, and Nicoletta Caridi for technical help; Sara Barozzi and Amanda Oldani in the Imaging Core Facility at the IFOM-IEO campus for help with time-lapse experiments; Henning Artl and Christian Ungermann for sharing equipment; and Susanne Lens, Jennifer DeLuca, Stephen S. Taylor, Ina Poser, and Anthony A. Hyman for reagents.
Work in A. Musacchio’s laboratory is funded by the European Union’s seventh Framework Program ERC agreement KINCON and the Integrated Project MitoSys, the Italian Association for Cancer Research (AIRC), and the Human Frontier Science Program. S. Santaguida was supported by a fellowship of the Italian Foundation for Cancer Research (FIRC). S. Knapp receives support from the SGC, a registered charity (number 1097737) that receives funds from the Canadian Institutes for Health Research, the Canada Foundation for Innovation, Genome Canada, GlaxoSmithKline, Abbott, Takeda, Pfizer, Eli Lilly, the Novartis Research Foundation, the Ontario Ministry of Research and Innovation, and the Wellcome Trust.
Footnotes
Abbreviations used in this paper:
- 5-ITu
- 5-iodotubercidin
- CPC
- chromosome passenger complex
- OKA
- okadaic acid
- SAC
- spindle assembly checkpoint
References
- Balzano D., Santaguida S., Musacchio A., Villa F. 2011. A general framework for inhibitor resistance in protein kinases. Chem. Biol. 18:966–975 10.1016/j.chembiol.2011.04.013 [DOI] [PubMed] [Google Scholar]
- Becker M., Stolz A., Ertych N., Bastians H. 2010. Centromere localization of INCENP-Aurora B is sufficient to support spindle checkpoint function. Cell Cycle. 9:1360–1372 10.4161/cc.9.7.11177 [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Chappie J.S., Wilson-Kubalek E.M., Desai A. 2006. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 127:983–997 10.1016/j.cell.2006.09.039 [DOI] [PubMed] [Google Scholar]
- Ciferri C., De Luca J., Monzani S., Ferrari K.J., Ristic D., Wyman C., Stark H., Kilmartin J., Salmon E.D., Musacchio A. 2005. Architecture of the human ndc80-hec1 complex, a critical constituent of the outer kinetochore. J. Biol. Chem. 280:29088–29095 10.1074/jbc.M504070200 [DOI] [PubMed] [Google Scholar]
- Ciferri C., Pasqualato S., Screpanti E., Varetti G., Santaguida S., Dos Reis G., Maiolica A., Polka J., De Luca J.G., De Wulf P., et al. 2008. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 133:427–439 10.1016/j.cell.2008.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuny G.D., Ulyanova N.P., Patnaik D., Liu J.-F., Lin X., Auerbach K., Ray S.S., Xian J., Glicksman M.A., Stein R.L., Higgins J.M.G. 2012. Structure-activity relationship study of beta-carboline derivatives as haspin kinase inhibitors. Bioorg. Med. Chem. Lett. 22:2015–2019 10.1016/j.bmcl.2012.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angiolella V., Mari C., Nocera D., Rametti L., Grieco D. 2003. The spindle checkpoint requires cyclin-dependent kinase activity. Genes Dev. 17:2520–2525 10.1101/gad.267603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J., Sultan S., Taylor S.S., Higgins J.M. 2005. The kinase haspin is required for mitotic histone H3 Thr 3 phosphorylation and normal metaphase chromosome alignment. Genes Dev. 19:472–488 10.1101/gad.1267105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J., Sullivan B.A., Higgins J.M.G. 2006. Regulation of mitotic chromosome cohesion by Haspin and Aurora B. Dev. Cell. 11:741–750 10.1016/j.devcel.2006.09.018 [DOI] [PubMed] [Google Scholar]
- DeLuca J.G., Gall W.E., Ciferri C., Cimini D., Musacchio A., Salmon E.D. 2006. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 127:969–982 10.1016/j.cell.2006.09.047 [DOI] [PubMed] [Google Scholar]
- DeLuca K.F., Lens S.M.A., DeLuca J.G. 2011. Temporal changes in Hec1 phosphorylation control kinetochore-microtubule attachment stability during mitosis. J. Cell Sci. 124:622–634 10.1242/jcs.072629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditchfield C., Johnson V.L., Tighe A., Ellston R., Haworth C., Johnson T., Mortlock A., Keen N., Taylor S.S. 2003. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 161:267–280 10.1083/jcb.200208091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J., Kelly A.E., Funabiki H., Patel D.J. 2012. Structural basis for recognition of H3T3ph and Smac/DIABLO N-terminal peptides by human Survivin. Structure. 20:185–195 10.1016/j.str.2011.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuele M.J., Lan W., Jwa M., Miller S.A., Chan C.S.M., Stukenberg P.T. 2008. Aurora B kinase and protein phosphatase 1 have opposing roles in modulating kinetochore assembly. J. Cell Biol. 181:241–254 10.1083/jcb.200710019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswaran J., Patnaik D., Filippakopoulos P., Wang F., Stein R.L., Murray J.W., Higgins J.M.G., Knapp S. 2009. Structure and functional characterization of the atypical human kinase haspin. Proc. Natl. Acad. Sci. USA. 106:20198–20203 10.1073/pnas.0901989106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov O., Marsden B., Pogacic V., Rellos P., Müller S., Bullock A.N., Schwaller J., Sundström M., Knapp S. 2007. A systematic interaction map of validated kinase inhibitors with Ser/Thr kinases. Proc. Natl. Acad. Sci. USA. 104:20523–20528 10.1073/pnas.0708800104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov O., Huber K., Eisenreich A., Filippakopoulos P., King O., Bullock A.N., Szklarczyk D., Jensen L.J., Fabbro D., Trappe J., et al. 2011. Specific CLK inhibitors from a novel chemotype for regulation of alternative splicing. Chem. Biol. 18:67–76 10.1016/j.chembiol.2010.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov O., Niesen F.H., Knapp S. 2012. Kinase inhibitor selectivity profiling using differential scanning fluorimetry. Methods Mol. Biol. 795:109–118 10.1007/978-1-61779-337-0_7 [DOI] [PubMed] [Google Scholar]
- Foley E.A., Maldonado M., Kapoor T.M. 2011. Formation of stable attachments between kinetochores and microtubules depends on the B56-PP2A phosphatase. Nat. Cell Biol. 13:1265–1271 10.1038/ncb2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giet R., Glover D.M. 2001. Drosophila aurora B kinase is required for histone H3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. J. Cell Biol. 152:669–682 10.1083/jcb.152.4.669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimaraes G.J., Dong Y., McEwen B.F., Deluca J.G. 2008. Kinetochore-microtubule attachment relies on the disordered N-terminal tail domain of Hec1. Curr. Biol. 18:1778–1784 10.1016/j.cub.2008.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurley L.R., D’Anna J.A., Barham S.S., Deaven L.L., Tobey R.A. 1978. Histone phosphorylation and chromatin structure during mitosis in Chinese hamster cells. Eur. J. Biochem. 84:1–15 10.1111/j.1432-1033.1978.tb12135.x [DOI] [PubMed] [Google Scholar]
- Hauf S., Cole R.W., LaTerra S., Zimmer C., Schnapp G., Walter R., Heckel A., van Meel J., Rieder C.L., Peters J.-M. 2003. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell Biol. 161:281–294 10.1083/jcb.200208092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickx A., Beullens M., Ceulemans H., Den Abt T., Van Eynde A., Nicolaescu E., Lesage B., Bollen M. 2009. Docking motif-guided mapping of the interactome of protein phosphatase-1. Chem. Biol. 16:365–371 10.1016/j.chembiol.2009.02.012 [DOI] [PubMed] [Google Scholar]
- Higgins J.M. 2001. Haspin-like proteins: a new family of evolutionarily conserved putative eukaryotic protein kinases. Protein Sci. 10:1677–1684 10.1110/ps.49901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins J.M.G. 2010. Haspin: a newly discovered regulator of mitotic chromosome behavior. Chromosoma. 119:137–147 10.1007/s00412-009-0250-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu J.Y., Sun Z.W., Li X., Reuben M., Tatchell K., Bishop D.K., Grushcow J.M., Brame C.J., Caldwell J.A., Hunt D.F., et al. 2000. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell. 102:279–291 10.1016/S0092-8674(00)00034-9 [DOI] [PubMed] [Google Scholar]
- Huertas D., Soler M., Moreto J., Villanueva A., Martinez A., Vidal A., Charlton M., Moffat D., Patel S., McDermott J., et al. 2012. Antitumor activity of a small-molecule inhibitor of the histone kinase Haspin. Oncogene. 31:1408–1418 10.1038/onc.2011.335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyaprakash A.A., Basquin C., Jayachandran U., Conti E. 2011. Structural basis for the recognition of phosphorylated histone h3 by the survivin subunit of the chromosomal passenger complex. Structure. 19:1625–1634 10.1016/j.str.2011.09.002 [DOI] [PubMed] [Google Scholar]
- Kallio M.J., McCleland M.L., Stukenberg P.T., Gorbsky G.J. 2002. Inhibition of aurora B kinase blocks chromosome segregation, overrides the spindle checkpoint, and perturbs microtubule dynamics in mitosis. Curr. Biol. 12:900–905 10.1016/S0960-9822(02)00887-4 [DOI] [PubMed] [Google Scholar]
- Kawashima S.A., Yamagishi Y., Honda T., Ishiguro K.-I., Watanabe Y. 2010. Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing shugoshin. Science. 327:172–177 10.1126/science.1180189 [DOI] [PubMed] [Google Scholar]
- Kelly A.E., Sampath S.C., Maniar T.A., Woo E.M., Chait B.T., Funabiki H. 2007. Chromosomal enrichment and activation of the aurora B pathway are coupled to spatially regulate spindle assembly. Dev. Cell. 12:31–43 10.1016/j.devcel.2006.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly A.E., Ghenoiu C., Xue J.Z., Zierhut C., Kimura H., Funabiki H. 2010. Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science. 330:235–239 10.1126/science.1189505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Holland A.J., Lan W., Cleveland D.W. 2010. Aurora kinases and protein phosphatase 1 mediate chromosome congression through regulation of CENP-E. Cell. 142:444–455 10.1016/j.cell.2010.06.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King E.M.J., Rachidi N., Morrice N., Hardwick K.G., Stark M.J.R. 2007. Ipl1p-dependent phosphorylation of Mad3p is required for the spindle checkpoint response to lack of tension at kinetochores. Genes Dev. 21:1163–1168 10.1101/gad.431507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson M.A., Cheeseman I.M. 2011. Sensing centromere tension: Aurora B and the regulation of kinetochore function. Trends Cell Biol. 21:133–140 10.1016/j.tcb.2010.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.-H., Jiang M., Cowart M., Gfesser G., Perner R., Kim K.H., Gu Y.G., Williams M., Jarvis M.F., Kowaluk E.A., et al. 2012. Discovery of 4-Amino-5-(3-bromophenyl)-7-(6-morpholino-pyridin- 3-yl)pyrido[2,3-d]pyrimidine, an Orally Active, Non-Nucleoside Adenosine Kinase Inhibitor. J. Med. Chem. 44:2133–2138 10.1021/jm000314x [DOI] [PubMed] [Google Scholar]
- Lesage B., Qian J., Bollen M. 2011. Spindle checkpoint silencing: PP1 tips the balance. Curr. Biol. 21:R898–R903 10.1016/j.cub.2011.08.063 [DOI] [PubMed] [Google Scholar]
- Liu D., Vader G., Vromans M.J.M., Lampson M.A., Lens S.M.A. 2009. Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science. 323:1350–1353 10.1126/science.1167000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Vleugel M., Backer C.B., Hori T., Fukagawa T., Cheeseman I.M., Lampson M.A. 2010. Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase. J. Cell Biol. 188:809–820 10.1083/jcb.201001006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- London N., Ceto S., Ranish J.A., Biggins S. 2012. Phosphoregulation of Spc105 by Mps1 and PP1 regulates Bub1 localization to kinetochores. Curr. Biol. 22:900–906 10.1016/j.cub.2012.03.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losiewicz M.D., Carlson B.A., Kaur G., Sausville E.A., Worland P.J. 1994. Potent inhibition of CDC2 kinase activity by the flavonoid L86-8275. Biochem. Biophys. Res. Commun. 201:589–595 10.1006/bbrc.1994.1742 [DOI] [PubMed] [Google Scholar]
- Maldonado M., Kapoor T.M. 2011. Constitutive Mad1 targeting to kinetochores uncouples checkpoint signalling from chromosome biorientation. Nat. Cell Biol. 13:475–482 10.1038/ncb2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markaki Y., Christogianni A., Politou A.S., Georgatos S.D. 2009. Phosphorylation of histone H3 at Thr3 is part of a combinatorial pattern that marks and configures mitotic chromatin. J. Cell Sci. 122:2809–2819 10.1242/jcs.043810 [DOI] [PubMed] [Google Scholar]
- Massillon D., Stalmans W., van de Werve G., Bollen M. 1994. Identification of the glycogenic compound 5-iodotubercidin as a general protein kinase inhibitor. Biochem. J. 299:123–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson D.R., Demirel P.B., Stukenberg P.T., Burke D.J. 2012. A conserved role for COMA/CENP-H/I/N kinetochore proteins in the spindle checkpoint. Genes Dev. 26:542–547 10.1101/gad.184184.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadows J.C., Shepperd L.A., Vanoosthuyse V., Lancaster T.C., Sochaj A.M., Buttrick G.J., Hardwick K.G., Millar J.B.A. 2011. Spindle checkpoint silencing requires association of PP1 to both Spc7 and kinesin-8 motors. Dev. Cell. 20:739–750 10.1016/j.devcel.2011.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S.A., Johnson M.L., Stukenberg P.T. 2008. Kinetochore attachments require an interaction between unstructured tails on microtubules and Ndc80(Hec1). Curr. Biol. 18:1785–1791 10.1016/j.cub.2008.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin V., Prieto S., Melines S., Hem S., Rossignol M., Lorca T., Espeut J., Morin N., Abrieu A. 2012. CDK-dependent potentiation of MPS1 kinase activity is essential to the mitotic checkpoint. Curr. Biol. 22:289–295 10.1016/j.cub.2011.12.048 [DOI] [PubMed] [Google Scholar]
- Musacchio A., Salmon E.D. 2007. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8:379–393 10.1038/nrm2163 [DOI] [PubMed] [Google Scholar]
- Newby A.C. 1981. The interaction of inhibitors with adenosine metabolising enzymes in intact isolated cells. Biochem. Pharmacol. 30:2611–2615 10.1016/0006-2952(81)90589-X [DOI] [PubMed] [Google Scholar]
- Niedzialkowska E., Wang F., Porebski P.J., Minor W., Higgins J.M.G., Stukenberg P.T. 2012. Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres. Mol. Biol. Cell. 23:1457–1466 10.1091/mbc.E11-11-0904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patnaik D., Jun Xian M.A., Glicksman M.A., Cuny G.D., Stein R.L., Higgins J.M. 2008. Identification of small molecule inhibitors of the mitotic kinase haspin by high-throughput screening using a homogeneous time-resolved fluorescence resonance energy transfer assay. J. Biomol. Screen. 13:1025–1034 10.1177/1087057108326081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson J.R., Taylor S.S. 1982. Phosphorylation of histones 1 and 3 and nonhistone high mobility group 14 by an endogenous kinase in HeLa metaphase chromosomes. J. Biol. Chem. 257:6064–6072 [PubMed] [Google Scholar]
- Petersen J., Hagan I.M. 2003. S. pombe aurora kinase/survivin is required for chromosome condensation and the spindle checkpoint attachment response. Curr. Biol. 13:590–597 10.1016/S0960-9822(03)00205-7 [DOI] [PubMed] [Google Scholar]
- Petsalaki E., Akoumianaki T., Black E.J., Gillespie D.A.F., Zachos G. 2011. Phosphorylation at serine 331 is required for Aurora B activation. J. Cell Biol. 195:449–466 10.1083/jcb.201104023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinsky B.A., Nelson C.R., Biggins S. 2009. Protein phosphatase 1 regulates exit from the spindle checkpoint in budding yeast. Curr. Biol. 19:1182–1187 10.1016/j.cub.2009.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polioudaki H., Markaki Y., Kourmouli N., Dialynas G., Theodoropoulos P.A., Singh P.B., Georgatos S.D. 2004. Mitotic phosphorylation of histone H3 at threonine 3. FEBS Lett. 560:39–44 10.1016/S0014-5793(04)00060-2 [DOI] [PubMed] [Google Scholar]
- Posch M., Khoudoli G.A., Swift S., King E.M., Deluca J.G., Swedlow J.R. 2010. Sds22 regulates aurora B activity and microtubule-kinetochore interactions at mitosis. J. Cell Biol. 191:61–74 10.1083/jcb.200912046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potapova T.A., Daum J.R., Pittman B.D., Hudson J.R., Jones T.N., Satinover D.L., Stukenberg P.T., Gorbsky G.J. 2006. The reversibility of mitotic exit in vertebrate cells. Nature. 440:954–958 10.1038/nature04652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian J., Lesage B., Beullens M., Van Eynde A., Bollen M. 2011. PP1/Repo-man dephosphorylates mitotic histone H3 at T3 and regulates chromosomal aurora B targeting. Curr. Biol. 21:766–773 10.1016/j.cub.2011.03.047 [DOI] [PubMed] [Google Scholar]
- Rosasco-Nitcher S.E., Lan W., Khorasanizadeh S., Stukenberg P.T. 2008. Centromeric Aurora-B activation requires TD-60, microtubules, and substrate priming phosphorylation. Science. 319:469–472 10.1126/science.1148980 [DOI] [PubMed] [Google Scholar]
- Rosenberg J.S., Cross F.R., Funabiki H. 2011. KNL1/Spc105 recruits PP1 to silence the spindle assembly checkpoint. Curr. Biol. 21:942–947 10.1016/j.cub.2011.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruchaud S., Carmena M., Earnshaw W.C. 2007. Chromosomal passengers: conducting cell division. Nat. Rev. Mol. Cell Biol. 8:798–812 10.1038/nrm2257 [DOI] [PubMed] [Google Scholar]
- Santaguida S., Musacchio A. 2009. The life and miracles of kinetochores. EMBO J. 28:2511–2531 10.1038/emboj.2009.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S., Tighe A., D’Alise A.M., Taylor S.S., Musacchio A. 2010. Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. J. Cell Biol. 190:73–87 10.1083/jcb.201001036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S., Vernieri C., Villa F., Ciliberto A., Musacchio A. 2011. Evidence that Aurora B is implicated in spindle checkpoint signalling independently of error correction. EMBO J. 30:1508–1519 10.1038/emboj.2011.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurin A.T., van der Waal M.S., Medema R.H., Lens S.M.A., Kops G.J.P.L. 2011. Aurora B potentiates Mps1 activation to ensure rapid checkpoint establishment at the onset of mitosis. Nat Commun. 2:316–319 10.1038/ncomms1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepperd L.A., Meadows J.C., Sochaj A.M., Lancaster T.C., Zou J., Buttrick G.J., Rappsilber J., Hardwick K.G., Millar J.B.A. 2012. Phosphodependent recruitment of Bub1 and Bub3 to Spc7/KNL1 by Mph1 kinase maintains the spindle checkpoint. Curr. Biol. 22:891–899 10.1016/j.cub.2012.03.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storchová Z., Becker J.S., Talarek N., Kögelsberger S., Pellman D. 2011. Bub1, Sgo1, and Mps1 mediate a distinct pathway for chromosome biorientation in budding yeast. Mol. Biol. Cell. 22:1473–1485 10.1091/mbc.E10-08-0673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto A., Maeshima K., Ikehara T., Yamaguchi K., Murayama A., Imamura S., Imamoto N., Yokoyama S., Hirano T., Watanabe Y., et al. 2009. The chromosomal association of condensin II is regulated by a noncatalytic function of PP2A. Nat. Struct. Mol. Biol. 16:1302–1308 10.1038/nsmb.1708 [DOI] [PubMed] [Google Scholar]
- Tan L., Kapoor T.M. 2011. Examining the dynamics of chromosomal passenger complex (CPC)-dependent phosphorylation during cell division. Proc. Natl. Acad. Sci. USA. 108:16675–16680 10.1073/pnas.1106748108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H., Yoshimura Y., Nozaki M., Yomogida K., Tsuchida J., Tosaka Y., Habu T., Nakanishi T., Okada M., Nojima H., Nishimune Y. 1999. Identification and characterization of a haploid germ cell-specific nuclear protein kinase (Haspin) in spermatid nuclei and its effects on somatic cells. J. Biol. Chem. 274:17049–17057 10.1074/jbc.274.24.17049 [DOI] [PubMed] [Google Scholar]
- Taylor S.S., Hussein D., Wang Y., Elderkin S., Morrow C.J. 2001. Kinetochore localisation and phosphorylation of the mitotic checkpoint components Bub1 and BubR1 are differentially regulated by spindle events in human cells. J. Cell Sci. 114:4385–4395 [DOI] [PubMed] [Google Scholar]
- Tighe A., Staples O., Taylor S. 2008. Mps1 kinase activity restrains anaphase during an unperturbed mitosis and targets Mad2 to kinetochores. J. Cell Biol. 181:893–901 10.1083/jcb.200712028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukahara T., Tanno Y., Watanabe Y. 2010. Phosphorylation of the CPC by Cdk1 promotes chromosome bi-orientation. Nature. 467:719–723 10.1038/nature09390 [DOI] [PubMed] [Google Scholar]
- Vader G., Cruijsen C.W.A., van Harn T., Vromans M.J.M., Medema R.H., Lens S.M.A. 2007. The chromosomal passenger complex controls spindle checkpoint function independent from its role in correcting microtubule kinetochore interactions. Mol. Biol. Cell. 18:4553–4564 10.1091/mbc.E07-04-0328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Waal M.S., Saurin A.T., Vromans M.J.M., Vleugel M., Wurzenberger C., Gerlich D.W., Medema R.H., Kops G.J.P.L., Lens S.M.A. 2012a. Mps1 promotes rapid centromere accumulation of Aurora B. EMBO Rep. 13:847–854 10.1038/embor.2012.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Waal M.S., Hengeveld R.C.C., van der Horst A., Lens S.M.A. 2012b. Cell division control by the Chromosomal Passenger Complex. Exp. Cell Res. 318:1407–1420 10.1016/j.yexcr.2012.03.015 [DOI] [PubMed] [Google Scholar]
- Vanoosthuyse V., Hardwick K.G. 2009a. A novel protein phosphatase 1-dependent spindle checkpoint silencing mechanism. Curr. Biol. 19:1176–1181 10.1016/j.cub.2009.05.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanoosthuyse V., Hardwick K.G. 2009b. Overcoming inhibition in the spindle checkpoint. Genes Dev. 23:2799–2805 10.1101/gad.1882109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigneron S., Prieto S., Bernis C., Labbé J.-C., Castro A., Lorca T. 2004. Kinetochore localization of spindle checkpoint proteins: who controls whom? Mol. Biol. Cell. 15:4584–4596 10.1091/mbc.E04-01-0051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa F., Capasso P., Tortorici M., Forneris F., de Marco A., Mattevi A., Musacchio A. 2009. Crystal structure of the catalytic domain of Haspin, an atypical kinase implicated in chromatin organization. Proc. Natl. Acad. Sci. USA. 106:20204–20209 10.1073/pnas.0908485106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Dai J., Daum J.R., Niedzialkowska E., Banerjee B., Stukenberg P.T., Gorbsky G.J., Higgins J.M.G. 2010. Histone H3 Thr-3 phosphorylation by Haspin positions Aurora B at centromeres in mitosis. Science. 330:231–235 10.1126/science.1189435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E., Ballister E.R., Lampson M.A. 2011. Aurora B dynamics at centromeres create a diffusion-based phosphorylation gradient. J. Cell Biol. 194:539–549 10.1083/jcb.201103044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Ulyanova N.P., van der Waal M.S., Patnaik D., Lens S.M.A., Higgins J.M.G. 2011. A positive feedback loop involving Haspin and Aurora B promotes CPC accumulation at centromeres in mitosis. Curr. Biol. 21:1061–1069 10.1016/j.cub.2011.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Ulyanova N.P., Daum J.R., Patnaik D., Kateneva A.V., Gorbsky G.J., Higgins J.M.G. 2012. Haspin inhibitors reveal centromeric functions of Aurora B in chromosome segregation. J. Cell Biol. 199:251–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welburn J.P.I., Vleugel M., Liu D., Yates J.R., III, Lampson M.A., Fukagawa T., Cheeseman I.M. 2010. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol. Cell. 38:383–392 10.1016/j.molcel.2010.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong O.K., Fang G. 2007. Cdk1 phosphorylation of BubR1 controls spindle checkpoint arrest and Plk1-mediated formation of the 3F3/2 epitope. J. Cell Biol. 179:611–617 10.1083/jcb.200708044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wotring L.L., Townsend L.B. 1979. Study of the cytotoxicity and metabolism of 4-amino-3-carboxamido-1-(beta-D-ribofuranosyl)pyrazolo[3,4-d]pyrimidine using inhibitors of adenosine kinase and adenosine deaminase. Cancer Res. 39:3018–3023 [PubMed] [Google Scholar]
- Xu Z., Vagnarelli P., Ogawa H., Samejima K., Earnshaw W.C. 2010. Gradient of increasing Aurora B kinase activity is required for cells to execute mitosis. J. Biol. Chem. 285:40163–40170 10.1074/jbc.M110.181545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi Y., Honda T., Tanno Y., Watanabe Y. 2010. Two histone marks establish the inner centromere and chromosome bi-orientation. Science. 330:239–243 10.1126/science.1194498 [DOI] [PubMed] [Google Scholar]
- Yamagishi Y., Yang C.-H., Tanno Y., Watanabe Y. 2012. MPS1/Mph1 phosphorylates the kinetochore protein KNL1/Spc7 to recruit SAC components. Nat. Cell Biol. 14:746–752 10.1038/ncb2515 [DOI] [PubMed] [Google Scholar]
- Yang Z., Kenny A.E., Brito D.A., Rieder C.L. 2009. Cells satisfy the mitotic checkpoint in Taxol, and do so faster in concentrations that stabilize syntelic attachments. J. Cell Biol. 186:675–684 10.1083/jcb.200906150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlin S.G., Barber C.M., Allis C.D., Sullivan K.F., Sullivan K. 2001a. Differential regulation of CENP-A and histone H3 phosphorylation in G2/M. J. Cell Sci. 114:653–661 [DOI] [PubMed] [Google Scholar]
- Zeitlin S.G., Shelby R.D., Sullivan K.F. 2001b. CENP-A is phosphorylated by Aurora B kinase and plays an unexpected role in completion of cytokinesis. J. Cell Biol. 155:1147–1157 10.1083/jcb.200108125 [DOI] [PMC free article] [PubMed] [Google Scholar]