A differentiation-promoting micro-RNA regulates actin cable dynamics, intercellular adhesion, and cell migration in human and mouse epidermis.
Abstract
During keratinocyte differentiation and stratification, cells undergo extensive remodeling of their actin cytoskeleton, which is important to control cell mobility and to coordinate and stabilize adhesive structures necessary for functional epithelia. Limited knowledge exists on how the actin cytoskeleton is remodeled in epithelial stratification and whether cell shape is a key determinant to trigger terminal differentiation. In this paper, using human keratinocytes and mouse epidermis as models, we implicate miR-24 in actin adhesion dynamics and demonstrate that miR-24 directly controls actin cable formation and cell mobility. miR-24 overexpression in proliferating cells was sufficient to trigger keratinocyte differentiation both in vitro and in vivo and directly repressed cytoskeletal modulators (PAK4, Tks5, and ArhGAP19). Silencing of these targets recapitulated the effects of miR-24 overexpression. Our results uncover a new regulatory pathway involving a differentiation-promoting microribonucleic acid that regulates actin adhesion dynamics in human and mouse epidermis.
Introduction
The epidermis is a self-renewing stratified tissue, which originates as a planar polarized monolayer of basal keratinocytes. Proliferating basal keratinocytes can either divide in a vertical plane to provide a flux of differentiating cells that migrate upwards through the suprabasal layers or divide horizontally to sustain the basal cell pool, and they therefore need both high proliferation potential and high mobility (Lechler and Fuchs, 2005; Williams et al., 2011; Wu et al., 2011). These functions are crucial for organizing the cells into a three-dimensional tissue during development, for epidermal barrier function and tissue integrity, and during wound healing (Vasioukhin et al., 2000; Vaezi et al., 2002; Watt, 2002; Candi et al., 2005; Fuchs, 2007). Switching from basal to suprabasal layers, keratinocytes undergo extensive remodeling of their keratin and actin cytoskeleton as well as their ECM and cell–cell contacts. The basal keratinocytes-ECM contacts are mediated mainly by integrin receptors, whereas the cell–cell contacts are mediated by cadherins (Jamora and Fuchs, 2002). An important role in modulating changes in epithelial shape and mobility is also played by the actin cytoskeleton, which is required to generate the mechanical forces necessary to stabilize the adhesive structure. The Rho family of small GTPases is primarily involved in the reorganization of the actin cytoskeleton. In different cell types, each member of the family can induce specific types of organization of actin filaments: stress fibers (Rho), lamellae/ruffles (Rac), or filopodia (Cdc42; Schwartz, 2004; Heasman and Ridley, 2008). Rho GTPases affect many cellular processes by interacting with and stimulating various downstream targets, including actin nucleators, protein kinases, and phospholipases (Bishop and Hall 2000; Schwartz 2004). In this study, we implicate micro-RNAs (miRNAs) in the dynamics and regulation of the actin cytoskeleton during epithelial differentiation and stratification.
miRNAs are a class of small noncoding RNAs, which have been shown to regulate a variety of cellular processes, although their role in epithelial development is not fully understood. They bind to the 3′ untranslated regions (UTRs) of target mRNAs, destabilizing them and/or inhibiting their translation (Ambros, 2004; Bartel, 2004; Kim, 2005). miRNAs are also emerging as master regulators of terminal differentiation. In particular, miR-24 is one of the miRNAs up-regulated in several differentiated cell types, for example, in postmitotic differentiation of hematopoietic cell lines (Lal et al., 2009b) and during myoblast (Sun et al., 2009) and neuronal differentiation (Fukuda et al., 2005). miR-24 is an abundant miRNA, which is conserved between various species, and is clustered with miR-23 and miR-27, on chromosome 9 (cluster 1: miR-23b, miR-27b, and miR-24-1) and on chromosome 19 (cluster 2: miR-23a, miR-27a, and miR-24-2; Lal et al., 2009b). In mice, it is highly expressed in heart and skin and also plays a role in erythropoiesis by regulating ALK4 (Wang et al., 2008). Recently, miR-24 has also been shown to suppress expression of two crucial cell cycle control genes, E2F2 and Myc, via binding to seedless 3′-UTR recognition elements (Lal et al., 2009a).
Here, using keratinocytes and mouse epidermis as models, we indentify a role for miR-24 in keratinocyte differentiation and dissect the molecular mechanism. We show that miR-24 is induced in skin concomitantly with stratification and differentiation, and miR-24 overexpression in proliferating cells is sufficient to trigger keratinocyte differentiation both in vitro and in vivo. We show that miR-24 promotes actin cable dynamics and modulates intercellular junction formation and cell migration through the repression of three cytoskeletal modulators, PAK4, Tsk5, and ArhGAP19. Our results demonstrate that the actin cytoskeleton remodeling and cell shape/mobility in which miR-24 plays an important role are key determinants in switching from a proliferating to a differentiating keratinocyte.
Results
miR-24 is expressed during keratinocytes terminal differentiation
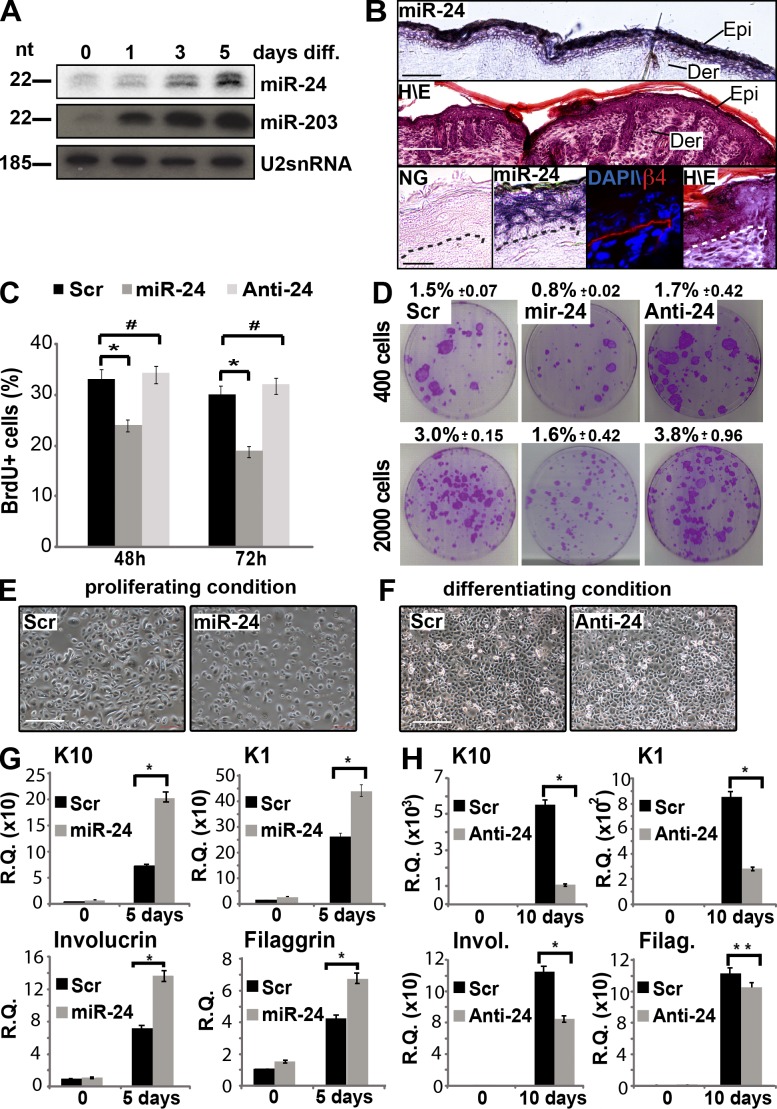
In the epidermis, miR-24 expression was rapidly increased when either mouse or human primary keratinocytes were induced by calcium to differentiate in vitro (Fig. 1 A and Fig. S1 D). Accordingly, miR-24 expression was shown to be confined to the suprabasal layers of the mouse and human epidermis by in situ hybridization (Fig. 1 B and Fig. S1 A). To investigate the function of miR-24 in keratinocyte differentiation, we performed a set of experiments in primary human keratinocytes. First of all, to address whether miR-24 restricts the proliferative potential of epidermal basal cells, we evaluated BrdU incorporation upon miR-24 overexpression, and we found a significant reduction of cell proliferation compared with controls, whereas miR-24 silencing did not show any short-term effects on cell proliferation (Fig. 1 C and Fig. S1 B). Moreover, clonogenicity assays showed a long-term effect of miR-24 in inhibiting proliferation (decreased from 3% in control to 1.6% with miR-24), whereas silencing resulted in an increased number of colonies (from 3% in control to 3.8% when the antagomir was transfected; Fig. 1 D). Although miR-24 is associated to a changed susceptibility to cell death (Qian et al., 2011; Srivastava et al., 2011; Singh and Saini, 2012), we excluded the cell death as a possible explanation for reduced proliferation and colony formation by evaluating the cell death fraction by sub-G1 peak quantification in miR-24–overexpressing keratinocytes (Fig. S1 C). We further excluded an altered plating efficiency caused by miR-24 overexpression or silencing by performing an adhesion assay upon transfection (Fig. S1 F). Next, we investigated the ability of miR-24 to regulate the switch between proliferative and differentiating keratinocytes. Overexpression of miR-24 in proliferating keratinocytes was sufficient to induce epidermal differentiation as assessed by expression of differentiation markers (K1, K10, involucrin, and filaggrin; Fig. 1, E–H). We observed a significant mRNA induction of early and late differentiation markers (K10, 2.9-fold; K1, 1.7-fold; involucrin, twofold; and filaggrin, 1.7-fold). The mean overexpression in all miR-24 transfection experiments in human keratinocytes was within the physiological range (Fig. S1, D and E). miR-24 silencing in confluent differentiated keratinocytes resulted in a significant reduction of mRNA levels of early and late differentiation markers (K10, 5.2-fold reduction; K1, threefold reduction; involucrin, 1.7-fold reduction; and filaggrin, 1.2-fold reduction). Reduction of K10 and involucrin expression in differentiating conditions was also confirmed by Western blot analysis (Fig. S1 G). These results demonstrate that increasing miR-24 expression stops proliferation and triggers keratinocyte differentiation in vitro.
Figure 1.
MiR-24 affects proliferation and induces differentiation of primary keratinocytes. (A) Northern blot highlights miR-24 up-regulation in differentiating (diff.) mouse keratinocytes. As a positive control, Northern blot for miR-203 is included. (B) In situ hybridization of skin from newborn mice reveals restriction of miR-24 expression to suprabasal layers. As a reference, hematoxylin–eosin (H/E) staining and anti–β4 integrin immunostaining for the basal lamina are included. epi, epidermis; der, dermis; NG, negative control of in situ hybridization. (C) Overexpression of miR-24 for the indicated time points reduces BrdU incorporation in HEKn. *, P < 0.01; #, not statistically significant compared with the control. Histograms of BrdU incorporation show the means ± SD from three independent experiments. (D) Colony formation assay performed on 400 or 2,000 cells plated after 48 h of transfection of scrambled control, miR-24, or anti–miR-24. Images are representative of three independent experiments. Numbers represent the relative percentage of clones (P < 0.01). (E) Morphology of posttransfection keratinocytes grown in proliferating conditions. (F) Morphology of posttransfection keratinocytes grown in differentiating conditions (high confluence). (G) Real-time qPCR analysis of early epidermal differentiation markers K10, K1, and involucrin and the late epidermal differentiation marker filaggrin in cells grown in proliferating conditions. qPCR results are shown as means ± SD from three independent experiments. *, P < 0.01. (H) Real-time qPCR analysis of mRNA of the early epidermal differentiation markers K10, K1, and involucrin (Invol.) and late epidermal differentiation marker filaggrin (Filag.) in cells grown in differentiating conditions. qPCR results are shown as means ± SD from three independent experiments. *, P < 0.01; **, P < 0.05. R.Q., relative quantification; Scr, scramble. Bars: (B, top and middle) 100 µm; (B, bottom) 25 µm; (E and F) 500 µm.
miR-24 controls keratinocytes proliferation in vivo
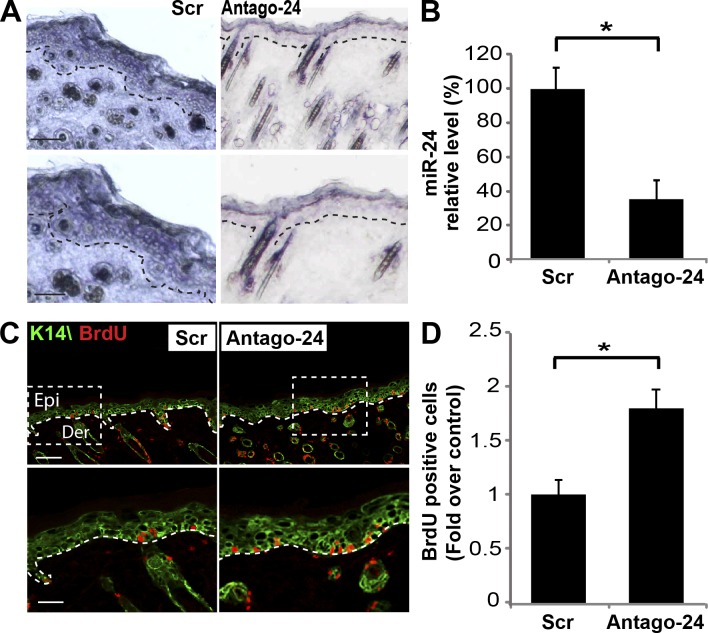
To define in vivo the functional role of miR-24 in controlling keratinocyte differentiation, we performed antagomir-24 subcutaneous injections to repress miR-24 in dorsal skin of newborn mice (Krützfeldt et al., 2005; Yi et al., 2008). 1 d after three dorsal injections (postnatal day 4 [P4]), antagomir-24, but not a scrambled control sequence, specifically repressed miR-24 expression in epidermis by 70% (Fig. 2, A and B; and Fig. S2 A). In keeping with the aforementioned in vitro experiments, in vivo silencing of miR-24 was accompanied by an expansion of proliferating cells in P4 mouse dorsal skin as shown by BrdU staining (Fig. 2, C and D) and Ki67-positive cells (Fig. S2 B). Quantification of BrdU-positive cells revealed that proliferating cells were significantly more abundant (1.8-fold increase) after antagomir-24 injection when compared with antagomir scramble–injected mice (Fig. 2 D). We also evaluated whether, during the short-term in vivo miR-24–silencing experiment, normal skin architecture and differentiation marker expression were altered. No defects were detected in the skin by hematoxylin–eosin staining (Fig. S2 A). Moreover, in immunofluorescence staining for K14 (Fig. 2 C), loricrin, and filaggrin (Fig. S2 C), no major differences were observed, possibly because of the short duration of anatagomir exposure.
Figure 2.
In vivo inhibition of miR-24 expression increases cell proliferation. (A) In situ hybridization analysis reveals reduction of endogenous miR-24 by antagomir-24 subcutaneous injections in newborn mice dorsal skin. Dotted lines delineate the basal layer of the epidermis. Bars, 100 µm. (B) Real-time qPCR performed on RNA extracts from skin of treated mice quantifies endogenous miR-24 silencing. Histogram shows means ± SD of miR-24 relative levels in treated dorsal skin (n = 4 mice for each group). (C and D) In vivo miR-24 silencing results in the induction of proliferation. Skin from antagomir-24–treated mice showed an increase in BrdU-positive cells in the basal layer (n = 4 mice for each group). Dotted boxes indicate the insets magnified on the bottom. Bars, 30 µm. Epi, epidermis; Der, dermis; Scr, scramble. *, P < 0.001.
Early expression of miR-24 in vivo restricts the proliferative potential of basal keratinocytes and induces premature differentiation
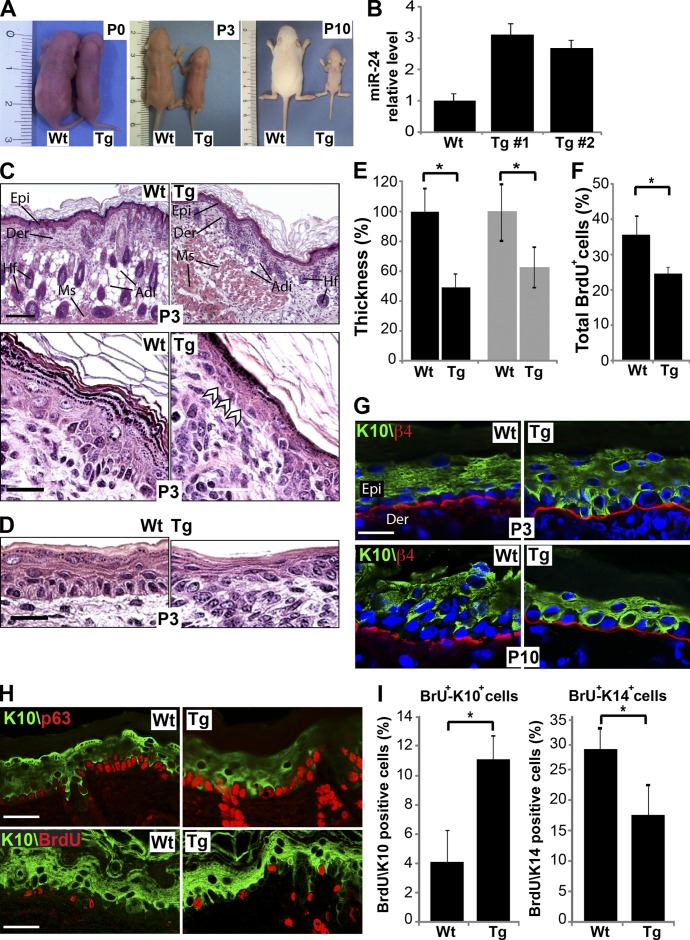
If, in our in vitro human model, miR-24 controls the switch between proliferation and differentiation in basal/suprabasal layers, we should expect that, in the in vivo mouse model, its overexpression in basal layer keratinocytes is sufficient to block proliferation and induce differentiation. To test this hypothesis, we generated transgenic mice overexpressing miR-24 under the control of the K5 (keratin 5) promoter (transgene [Tg];K5::miR-24; Candi et al., 2006, 2007) acting on basal layer cells by embryonic developmental stage 14.5 (E14.5; Fig. S2, D and E). We obtained six founders overexpressing miR-24 by 2.7–3.2-fold over wild type (wt) as evaluated by real-time PCR on skin from ear biopsies (Fig. 3, A and B). 90% of the Tg;K5::miR-24 newborn mice died shortly after birth. Transgenic mice were significantly smaller: at postnatal day 3 (P3), their weight was <40% of their wt littermates (Fig. 3 A and Fig. S2 F). This may be secondary to malnutrition because the transgenic animals showed a marked reduction in subcutaneous adipose tissue deposition (Fig. 3 C and Fig. S2 G). This failure to thrive may result from the thinner mouth and upper digestive tract epithelium (40% reduction) in the transgenics (Fig. 3, D and E).
Figure 3.
Early activation of miR-24 in epidermis restricts epithelial functions. (A) Gross morphology of K5::miR-24 transgenic mice at postnatal day 0, 3, and 10 (P0, P3, and P10). (B) qPCR quantified miR-24 overexpression in transgenic mice skin. (C and D) Hematoxylin-eosin staining of skin (C) and epithelial lining of upper digestive tract (D) sections of P3 mice. Arrowheads indicate basal cells with a flat appearance. Epi, epidermis; Der, dermis; Hf, hair follicles; Adi, adipose tissue; Ms, muscle. (E) Epidermis (black bars; *, P < 0.00001) and digestive epithelia (gray bars; *, P < 0.0001) of miR-24 transgenic mice are thinner than the wt (n = 4 mice for each group). (F) Early expression of miR-24 in stratified epithelia decreases epidermal proliferation, as evaluated by quantification of BrdU-positive cells over the basal layer cells. *, P < 0.001 (n = 4 mice for each group). (G) Basal layer keratinocytes of miR-24 transgenic mice show positive staining for the keratinocyte differentiation marker K10. K10-positive cells are in contact with the basal membrane (red). (H) Coimmunostaining for p63 and K10 (top) and BrdU and K10 (bottom) in P3 mouse epidermis. (I) Histograms show an increased number of BrdU+/K10+ cells and BrdU+/K14+ cells in transgenic mouse epidermis after 4 h of BrdU pulse chase. *, P < 0.001 (n = 4 mice for each group). Error bars show means ± SD. Bars: (C) 500 µm; (D and H) 30 µm; (G) 20 µm.
The epidermis of P3 Tg;K5::miR-24 mice was thinner (50% less) as compared with wt mice (Fig. 3, C and E), and this difference was maintained at P10 (Fig. S2 J). Otherwise, the architecture of the transgenic epidermis was similar to that of the wt except for basal layer cells, which appeared flat rather than cubical (Fig. 3 C, arrowheads). No alteration in apoptosis was revealed in basal cells of transgenic mice as detected by TUNEL assay staining, excluding any involvement of cell death in the phenotype of Tg;K5::miR-24 mice (Fig. S2, H and I). Confocal analysis of differentiation markers showed that K10-expressing keratinocytes were directly juxtaposed to the basement membrane (Fig. 3 G), and in many cells, K10 was coexpressed with K14 (Fig. S2 K). In an attempt to discriminate whether the phenotype in the transgenic mice is caused by repression of the proliferation status of the epidermal progenitor cells or to an acceleration of the differentiation rate, we further analyzed the basal layer of the mice. We found that both p63 and K14, basal layer markers, are consistently and continuously expressed in the basal layer (Fig. 3 H and Fig. S2 K), demonstrating that in the Tg;K5::miR-24 mice, this particular layer is intact, although with ∼15% reduction in proliferation rate (Fig. 3 F). To clearly demonstrate a change in the differentiation rate in the skin of Tg;K5::miR-24 mice, we performed a BrdU pulse chase labeling of 4 h and a costain with BrdU/K10 and BrdU/K14 to evaluate how the BrdU distribution, and therefore the dynamic of the cells moving from the basal to the spinous layer, was changed during the 4-h pulse. Our analysis demonstrated an accelerated differentiation in the epidermis of transgenic mice. The differentiation rate, indeed, changed from the 4.1% of BrdU/K10-positive cells in the control mice to the 11.3% of BrdU/K10-positive cells in transgenic mice, with a parallel reduction in BrdU/K14-positive cells (from 31 to 18.6%, respectively; Fig. 3, H and I). Collectively, these results indicate that expression of miR-24 in the basal layer in vivo reduced proliferation and was sufficient to induce differentiation, increasing the differentiation rate of the keratinocytes.
Cytoskeletal regulator genes are miR-24 direct targets
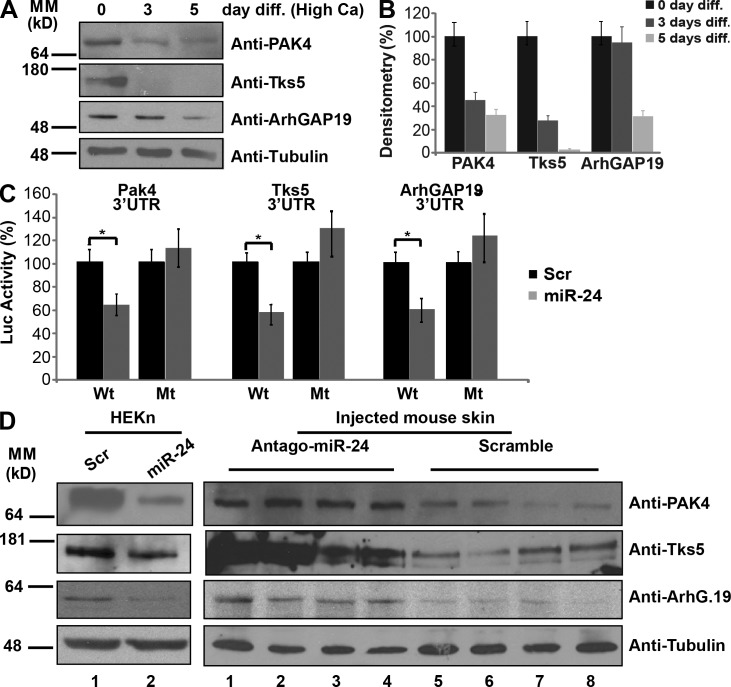
We next investigated which were the molecular targets of miR-24. First, we analyzed miR-24 target mRNAs by comparing mRNA microarrays of keratinocytes transfected with miR-24 or with a scrambled control. We found 499 mRNAs significantly down-regulated in the miR-24–expressing cells (P < 0.01). From the complete list of deregulated mRNAs, we selected 261 that contained at least one miR-24 binding site predicted by TargetScan 5.1 software (Lewis et al., 2003) in their 3′UTRs fully or partial conserved (Table S1 and Fig. S3 A). By real-time quantitative PCR (qPCR), we validated the microarray for seven of these candidate targets, expression of all of which was significantly reduced by miR-24, thus validating the microarray data (Fig. S3 B). By performing a cluster analysis on the 261 mRNAs, we identified significant enrichment for candidate targets involved in four specific categories: cellular assembly organization, cellular movement, cell morphology, and cell to cell signaling and interaction (Table S2). Many of the targets in these categories were related to actin cytoskeleton remodeling. We selected as putative targets PAK4, Tks5, and ArhGAP19 mRNAs. PAK4, Tks5, and ArhGAP19 (a) have conserved seed sequences for miR-24 in their 3′UTRs (Fig. S3 C), (b) are significantly down-regulated during calcium-induced differentiation (Fig. 4, A and B), and (c) are involved in cytoskeleton remodeling. Indeed, PAK4, a group II p21-activated kinase, is a RhoGTPase/Cdc42 effector and plays a pivotal role in organizing actin cytoskeleton and filopodia formation (Abo et al., 1998), cell migration via integrin ανβ5 (Zhang et al., 2002; Eswaran et al., 2008), and upstream inhibition of RhoA–ROCK2 via guanine nucleotide exchange factor (GEF)-H1 inhibition (Barac et al., 2004). Tks5 is a substrate of Src kinase (Lock et al., 1998) that acts as scaffold protein for many factors, including Wiskott–Aldrich syndrome protein (WASP), thereby promoting actin polymerization and podosome formation and, consequently, cell migration and invasion (Abram et al., 2003; Seals et al., 2005; Oikawa et al., 2008). ArhGAP19, a Rho GTPase-activating protein of the Arh GTPase-activating proteins family, with unknown function, is also involved in cell migration and actin regulation (Lv et al., 2007). By introducing PAK4, Tks5, and ArhGAP19 3′UTRs into a luciferase reporter vector, we confirmed that miR-24 directly targets these mRNAs (Fig. 4 C). Moreover, removal of the recognition sequence in their 3′UTRs abrogated the inhibition (Fig. 4 C). Overexpression of miR-24 in cells also induced a significant reduction in expression of these proteins (Fig. 4 D), whereas an increase in their expression was found in vivo in protein extracts from the dorsal skin of antagomir-24–injected mice (Fig. 4 D).
Figure 4.
miR-24 targets PAK4, Tks5, and ArhGAP19 3′UTRs. (A) PAK4, Tks5, and ArhGAP19 are down-regulated during calcium-induced keratinocyte differentiation (diff.). (B) Histograms show densitometry analysis of the Western blot in A. (C) Insertion of the 3′UTR from miR-24 putative targets decreases luciferase reporter activity (*, P < 0.01), whereas deletion of miR-24 seed sequences from target 3′UTRs abrogates the repression. (D) miR-24 in vitro and in vivo regulates PAK4, Tks5, and ArhGAP19 protein levels. (left) 48 h after transfection, HEKn were collected to perform Western blot analysis. Skin samples of antagomir-24–treated mice (from the experiment shown in Fig. 2) were used to perform Western blot analysis. Four antagomir-24–-injected mice (lanes 1–4) and four scramble-injected mice (lanes 5–8) were analyzed. Error bars show means ± SD. MM, molecular mass; Scr, scramble; Luc, luciferase; Mt, mutant.
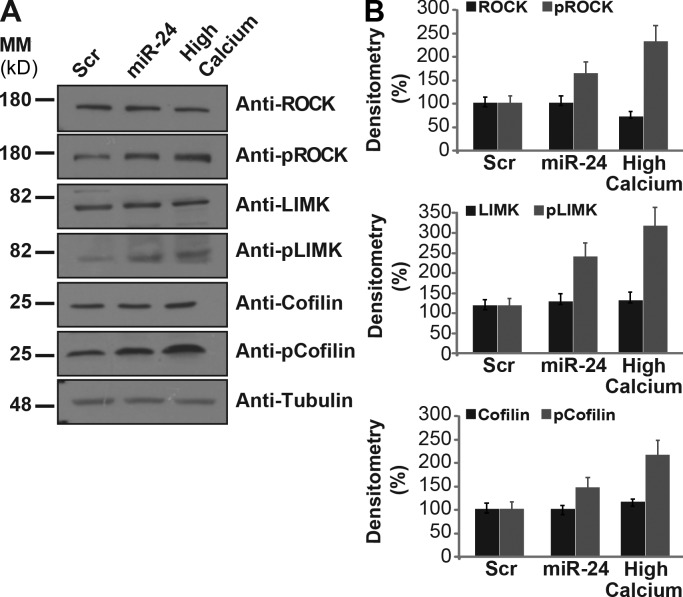
To further verify that miR-24 is involved in actin cytoskeleton remodeling, we transfected miR-24 in proliferating keratinocytes and evaluated its effect on the phosphorylation status of ROCK2, LIMK1, and cofilin, important regulators of actin filament assembly and disassembly (Abo et al., 1998). By Western blotting, we found that miR-24 affects the phosphorylation status of ROCK2, LIMK1, and cofilin as well as their cellular localization (Fig. 5, A and B), supporting a role for miR-24 in actin dynamics.
Figure 5.
miR-24 changes the phosphorylation state of actin cytoskeletal modulators. (A) Western blot analysis of ROCK2, LIMK1, and cofilin expression (basal and phosphorylated status) reveals that miR-24 affects actin dynamics modulators comparable with high calcium treatment in keratinocytes. This is one representative experiment of three. (B) Histograms show densitometry analysis of Western blot in A normalized to actin. Error bars show means ± SD. MM, molecular mass; Scr, scramble.
miR-24 induces differentiation by controlling actin cytoskeleton remodeling, cell mobility, and cell–cell adhesion
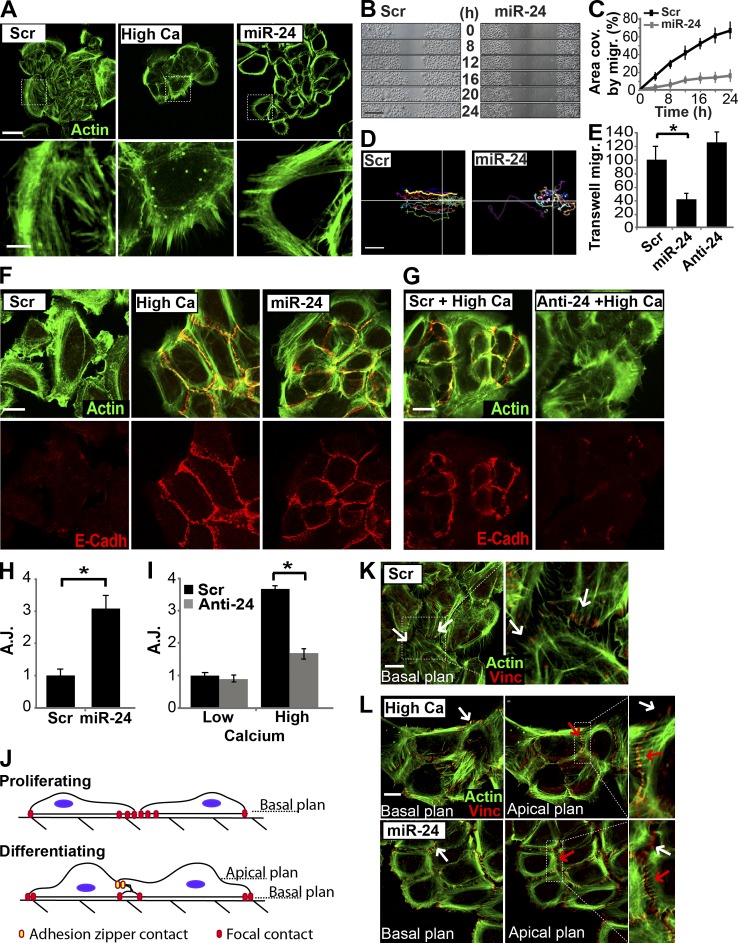
Considering that most miR-24 targets are cytoskeletal regulators and that overexpression of miR-24 induces phosphorylation of ROCK2, LIMK1, and cofilin (Fig. 5), we compared the effect of pre–miR-24 and calcium on actin remodeling, cell shape, migration, and adhesion in human primary keratinocytes. Proliferating keratinocytes showed a complex network of actin stress fibers and marked lamellipodia extensions (Fig. 6 A, scramble-transfected cells). Immunostaining for paxillin, vinculin, and actinin highlighted cell planar polarity, which is required for directional migration (Fig. S3, D–F, scramble-transfected cells). When we switched keratinocytes to differentiating medium, they became rearranged as packed groups of cells (Fig. 6 A, high calcium). Concurrently, extensive actin reorganization occurred. No actin stress fibers were detected, but actin filaments appeared organized in concentric rings around the cells, in an apical–basal (top down) polarized fashion, as described previously (Vaezi et al., 2002). Planar polarity, typical of proliferating keratinocytes, evaluated by paxillin, vinculin, and actinin staining disappeared (Fig. S3, D–F, high calcium). 48 h after transfection, miR-24–overexpressing keratinocytes showed changes in actin cytoskeleton, resembling that observed in cells in high calcium medium. Cells assumed a packaged organization; stress fibers disappeared and concentric actin ring organization became prevalent (Fig. 6 A, miR-24). Planar polarization was also lost as shown by paxillin, vinculin, and actinin staining (Fig. S3, D–F). Video time-lapse microscopy of an in vitro scratch assay showed a reduction in the ability of miR-24–overexpressing keratinocytes to migrate into the scratched area. After 24 h, miR-24–overexpressing keratinocytes had migrated into only 15% of the scratched area compared with ≤70% by the scramble-transfected keratinocytes (Fig. 6, B and C). To exclude that this difference in migration was caused by an effect on proliferation, we performed the scratch assay in the presence and in the absence of mitomycin C (Fig. S3, G and H). The reduced migratory efficiency of miR-24–transfected cells was also confirmed by transwell migration assay, in which miR-24–overexpressing keratinocytes showed a >50% reduction of migration through the porous membranes of transwell chambers (Fig. 6 E). Finally, we monitored, by video microscopy, the path of single cells during the scratch assay. As shown in Fig. 6 D, miR-24–overexpressing keratinocytes appeared to lose their directional migration, according to the loss of planar polarity shown by immunofluorescence analysis (Fig. S3, D–F). To evaluate the effect of miR-24 on stratification, we studied the formation of adherens junctions (AJs), evaluating E-cadherin localization at cell–cell contacts (Jamora and Fuchs 2002; Perez-Moreno and Fuchs 2006). We found that miR-24 overexpression induced E-cadherin localization at cell–cell contacts, corresponding to the assembly of mature AJs, similar to calcium-induced differentiated cells (Fig. 6 F). Quantification of AJs, by confocal analysis of cell–cell contacts marked by E-cadherin, showed a threefold increase in miR-24–overexpressing keratinocytes (Fig. 6 H). Conversely, we assessed AJ formation after calcium addition together with miR-24 silencing. Scramble-transfected keratinocytes treated with high calcium medium showed a 3.7-fold increase of AJs compared with low calcium medium (Fig. 6, G and I, black bars). In contrast, anti–miR-24–transfected keratinocytes showed only a very low and scattered signal of E-cadherin without AJ formation in low calcium (Fig. 6 G). AJ induced by calcium showed a significant reduction when endogenous miR-24 was knocked down (from 3.7- to 1.6-fold; Fig. 6 I).
Figure 6.
miR-24 modifies the actin cytoskeleton similarly to calcium. (A) HEKn overexpressing miR-24 or grown in high calcium medium (1.2 mM) showed similar modifications in actin cytoskeletal organization, as shown by Alexa Fluor 488–-phallotoxin staining (green). Dotted boxes indicate the insets magnified on the bottom. (B and C) In vitro scratch wound assay performed by video microscopy. miR-24–overexpressing keratinocytes showed a significant reduction in migration ability, reported in the graph in C as area covered (area cov.) by migration (migr.) during 24 h. (D) Tracking analysis of a single cell path revealed loss of directional migration in miR-24–overexpressing keratinocytes. Lines indicate the x-y axis in the cell migration plan. (E) Transwell migration assay confirmed the reduced migration ability in miR-24–transfected keratinocytes. *, P < 0.001. (F) HEKn overexpressing miR-24 or differentiated in high calcium showed similar localization of E-cadherin at cell–cell contacts (AJs). (G) Silencing of endogenous miR-24 after high calcium treatment reduces formation of cell–cell contacts (AJs) as indicated by E-cadherin (E-Cadh) staining. (H and I) Quantification of AJs, marked by E-cadherin staining in the experiment shown in Fig. 5 (F [miR-24 overexpression] and G [miR-24 silencing]), confirmed the ability of miR-24 to directly modulate AJ formation during differentiation. *, P > 0.01. (J) Model of keratinocyte cell–cell contact formation. Proliferating keratinocytes form stable focal contacts in the basal plane. Basal–apical architecture of differentiating keratinocytes allows formation of top-down intercellular junctions (adhesion zippers). Adhesion zipper assembly occurs in an apical plane. (K and L) Keratinocyte cell–cell contact formation. Transfected or high calcium–treated HEKn were stained with Alexa Fluor 488–phallotoxin (green) and vinculin (Vinc) antibody. In basal plane views (0 µm), vinculin staining marks focal contact in focus (white arrows). In apical plane views (0.6 µm), vinculin staining marks adhesion zippers in focus (red arrows), and out of focus focal contacts are displayed as white arrows. Dotted boxes indicate the insets magnified on the right. Scr, scramble. Error bars show means ± SD. Bars, 20 µm.
We also examined the formation of adhesion zippers (Vasioukhin et al., 2000; Vaezi et al., 2002). In the early stage of differentiation, keratinocytes form adhesion zippers in their apical planes, whereas in the basal planes, focal contacts are maintained as shown by positive staining in the lamellipodia (Fig. 6 J). Proliferating keratinocytes, transfected with scrambled control, showed only focal contacts (see vinculin staining) as shown in a view corresponding to the basal plane of the cells (Fig. 6 K, white arrows); no basal–apical polarization was observed. miR-24–overexpressing keratinocytes and calcium-treated keratinocytes showed an apical–basal polarization. Indeed, in both cases, focal contacts were also present in the basal plane (indicated by vinculin staining in lamellipodia), whereas in apical plane views, strong adhesion zipper structures were detected (Fig. 6 L, white arrows indicate focal contacts, red arrows indicate adhesion zipper structures, and white arrows in apical plane views indicate focal contact out of focus). These results indicate that miR-24 has a role in coordinating the changes occurring in the cytoskeleton necessary to achieve stratification and differentiation.
Down-regulation of actin cytoskeleton regulators triggers epidermal differentiation
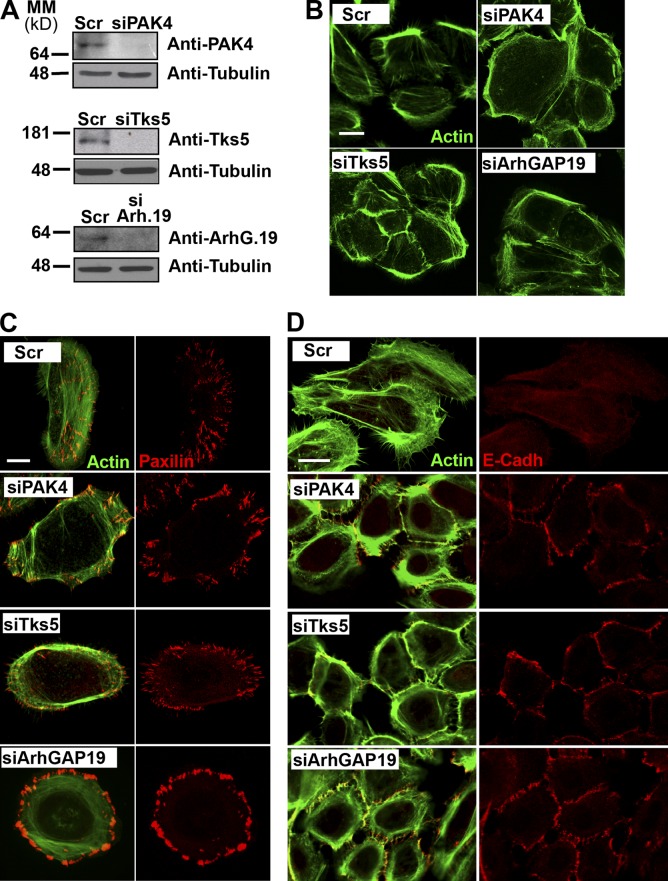
To determinate whether miR-24 effects on keratinocyte differentiation/proliferation were mainly mediated by PAK4, Tks5, or ArhGAP19 down-regulation, we selectively silenced miR-24 targets in human epidermal keratinocyte, neonatal (HEKn) and achieved efficient knockdown of all three targets by siRNA (Fig. 7 A). Immunostaining of actin filaments showed that PAK4, Tsk5, or ArhGAP19 silencing induced actin filament reorganization: no actin stress fibers were seen, as in the control, but actin filaments appeared organized in concentric rings around the periphery of the cells (Fig. 7 B). Furthermore, silencing of these miR-24 targets resulted in the loss of planar polarization as shown by paxillin (Fig. 7 C), actinin, and vinculin (Fig. S4, A and B) staining. In addition, silencing of miR-24 targets also resulted in the formation of mature AJs as highlighted by the localization of E-cadherin at the cell–cell interface (Fig. 7 D). Next, we analyzed the effect of silencing the same miR-24 targets on keratinocyte migration. In a scratch assay, performed after siRNA transfections, there was a reduction in migration of ≤70% in the space covered by scramble-transfected control cells (Fig. 8, A and B). Furthermore, a transwell assay showed a consistent reduction in migration (≤65%) for all targets silenced (Fig. 8 C). These results clearly show that silencing of PAK4, Tsk5, or ArhGAP19 recapitulates the results shown in Fig. 6.
Figure 7.
Silencing of miR-24 targets, PAK4, Tks5, and ArhGAP19 phenocopies miR-24 overexpression in cells. (A) Western blot analysis confirmed silencing of PAK4, Tsk5, and ArhGAP19. Tubulin was used as a loading control. (B) Silencing of PAK4, Tsk5, and ArhGAP19 causes remodeling of actin filaments. (C) Specific silencing of PAK4, Tsk5, or ArhGAP19 induces loss of planar polarization in HEKn as shown by Alexa Fluor 488–phallotoxin (green) and paxillin staining. (D) HEKn were fixed and stained with Alexa Fluor 488–phallotoxin (green) and E-cadherin (E-Cadh) antibody to highlight the formation of AJs. MM, molecular mass; Scr, scramble. Bars: (B) 20 µm; (C) 10 µm; (D) 30 µm.
Figure 8.
Overexpression of PAK4, Tks5, and ArhGAP19 cDNAs abolishes the effects of miR-24. (A) Silencing of miR-24 targets reduces in vitro keratinocyte migration. 48 h after transfection, confluent HEKn were subjected to the in vitro scratch wound assay. Cell migration was observed into the scratch at the time indicated. Micrographs are taken at 0 and 24 h after scratch. Red bars denote the distance between the edges of the scratch wounds. Bar, 500 µm. (B) Histogram shows the relative percentage of distance (Dist.) covered by migration (migr.) 24 h after scratch. *, P < 0.001. (C) Transwell migration assay confirmed the reduced migration ability of siRNA-transfected keratinocytes. *, P < 0.001. (D) Specific silencing of PAK4, Tsk5, or ArhGAP19 reduces human primary keratinocyte BrdU incorporation. Histogram shows the relative percent reductions of BrdU-incorporating cells. (E) Specific silencing of PAK4, Tks5, or ArhGAP19 increases keratinocyte differentiation as evaluated by real-time qPCR of specific markers (K10, K1, and involucrin). (F) cDNA of miR-24 targets were introduced into miR-24–transfected keratinocytes by retrovirus infection. Expression of miR-24 targets abrogates miR-24 induction of differentiation as evaluated by real-time qPCR. (G) Expression of miR-24 targets abrogates miR-24–mediated AJ formation as evaluated by confocal analysis of E-cadherin staining. Bar, 50 µm. (H) Histogram shows quantification of AJs of the experiment shown in G. *, P < 0.01. (I) Proposed model for the role of miR-24 in keratinocyte differentiation showing that miR-24 acts on pro-proliferation and actin cable dynamic factors. miR-24 inhibits proliferation repressing Myc, E2F2, etc. expression and induces differentiation by inhibiting the actin cytoskeletal modulators PAK4, Tks5, and ArhGAP19. The overall effect of miR-24 on actin dynamics results in reduced cell motility and directional migration, in changes in cell shape, and in increased formation of AJs, promoting keratinocyte differentiation. Error bars show means ± SD. EV, empty vector; Scr, scramble.
To further investigate the contribution of miR-24 targets on keratinocyte differentiation, we evaluated proliferation and expression of differentiation markers after PAK4, Tsk5, or ArhGAP19 silencing. Silencing of the three targets causes 20–25% inhibition in BrdU incorporation (Fig. 8 D) and significant induction (3.5–10-fold) of differentiation markers (Fig. 8 E). To prove that these are indeed the crucial targets, we overexpressed the PAK4, Tks5, or ArhGAp19 cDNAs lacking their 3′UTRs concurrently with miR-24. Retrovirus infection of the target cDNAs inhibited the effects of miR-24 on expression of differentiation markers (Fig. 8 F) and on AJ formation (Fig. 8, G and H). Finally, silencing of miR-24 targets (PAK4, Tks5, or ArhGAp19) in anti–miR-24–transfected keratinocytes reverted the decrease in AJ formation produced by miR-24 knockdown (Fig. S4, C and D). Together, these results demonstrated that PAK4, Tks5, and ArhGAP19 are important miR-24 targets involved in the control of differentiation, proliferation, and cell shape in keratinocytes.
miR-24 also targets Myc and E2F2, which are important regulators of the mammalian epidermis (Pelengaris et al., 1999; Flores et al., 2004; Watt et al., 2008; Berta et al., 2010). To test their involvement in our system, we evaluated the effects of Myc and E2F2 on actin filaments. miR-24 overexpression in proliferating keratinocytes induced a significant reduction (60 and 20%, respectively) of Myc and E2F2 protein levels (Fig. S5 A). siMyc- and siE2F2-transfected keratinocytes showed no alteration in the actin cytoskeleton (Fig. S5 B), and cellular planar polarity was still present as shown by paxillin staining (Fig. S5 C). In addition, down-regulation of Myc and E2F2 expression did not contribute to AJ formation (Fig. S5 D). In contrast, Myc and E2F2 silencing had a strong effect on proliferation as evaluated by a 60–80% inhibition of BrdU incorporation (Fig. S5 E) without significant effects on the induction of differentiation (K10, K1, and involucrin; Fig. S5 F).
In summary, we have shown that overexpression of miR-24 (Fig. 6) or silencing of its targets PAK4, Tsk5, and ArhGAP19 (Fig. 7 and Fig. 8) produces very similar effects: (a) alteration of actin cables (Fig. 6 A and Fig. 7 B), (b) loss of cell polarity and directional migration (Fig. 6, B–E; Fig. 7 C; Fig. 8, A–C; Fig. S3, D–H; and Fig. S4, A and B), (c) formation of mature AJ (Fig. 6, F–I; and Fig. 7 D), (d) inhibition of proliferation (Fig. 1, C and D; and Fig. 8 D), and (e) induction of keratinocyte differentiation markers (Fig. 1, E and G; and Fig. 8 E). Furthermore, restoration of PAK4, Tsk5, and ArhGAP19 expression blocked all the effects caused by miR-24 overexpression (Fig. 8, F–H).
Discussion
Despite extensive analysis on the mechanism controlling epithelial morphogenesis, the involvement of actin cytoskeleton during keratinocyte differentiation and stratification has not been completely clarified. Several studies have shown that skin morphogenesis is governed by differentially expressed sets of miRNAs (Andl et al., 2006; Aberdam et al., 2008; Lena et al., 2008; Yi et al., 2008), and our results clearly indicate that during the keratinocyte migration from the basal to the suprabasal layer, a specific miRNA, miR-24, induces differentiation in vivo by controlling actin adhesion dynamics (Fig. 8 I). These results parallel the effects obtained by calcium treatment, supporting the role of miR-24 and cytoskeletal modification during keratinocyte differentiation. The most striking result is that early expression of miR-24 in the basal layer of the epidermis of Tg;K5::miR-24 mice strongly triggers basal cells to differentiate as shown by the expression of K10, a marker of the spinous layer, parallel to a reduction of proliferation rate (Fig. 3). Although the antiproliferative effect of miR-24 has been already described in in vitro conditions (Lal et al., 2009a), its effects in vivo and on the actin cytoskeleton, which in turn controls cell shape, cellular polarization, cell adhesion, and migration, have not been previously described. We identify PAK4, Tsk5, and ArhGAP19 as important miR-24 targets able to control proliferation, differentiation, and migration of human keratinocytes. For the first time, these targets have been associated with epithelial morphogenesis. One of the key targets of miR-24 is PAK4, which is essential for embryonic development. PAK4-null mice show defects in heart, neural tube formation, and in the differentiation and migration of neurons (Abo et al., 1998). PAK4, a group II p21-activated kinase, is a Rac1 and RhoGTPase/Cdc42 effector and plays a pivotal role in organizing the actin cytoskeleton and filopodia formation (Abo et al., 1998) and inhibits Rho–ROCK2 via GEF-H1 phosphorylation (Barac et al., 2004). In particular, PAK4 is the only member of the family that regulates the dynamic process of actin depolymerization through the actin-depolymerizing factor/cofilin complex. Moreover, in agreement with our finding, recently, PAK4 has been described as a miR-24 target in endothelial cells, during the neoangiogenesis after myocardial infarction (Fiedler et al., 2011). The Tks5 adaptor protein is a substrate of Src kinase (Lock et al., 1998) involved in podosome formation and in cell migration and invasion (Lock et al., 1998; Abram et al., 2003). Tks5 acts as a scaffold for the recruitment of WASP, thereby promoting actin polymerization (Seals et al., 2005). Very little is known about the functions of the other target ArhGAP19. ArhGAP19 is a Rho GTPase-activating protein of the Arh GTPase-activating protein family that is involved in cell migration and actin regulation (Lv et al., 2007). The crucial effect of miR-24 action appears to be the inhibition of actin filament polymerization/depolymerization. miR-24 acts downstream of Rho, Rac1, and Cdc42 by repressing PAK4 and Tks5. PAK4 repression results in accumulation of inactive phosphocofilin involved in actin filament depolymerization (Abo et al., 1998). In parallel, miR-24, by repressing PAK4, inhibits GEF-H1 and activates ROCK2 and LIMK1, also resulting in phosphocofilin accumulation. On the other hand, miR-24–dependent Tks5 repression may deprive WASP/Arp2/3, although active, of its scaffold protein required for actin filament polymerization. miR-24 overexpression also produces accumulation of phospho-ROCK2 through the reduction of the upstream inhibition exerted by PAK4 on GEF-H1/RhoA (Zanet et al., 2005). This finding is in agreement with a previous study, which demonstrates a crucial role for the Rho–ROCK pathway in formation of cell–cell junctions during keratinocyte stratification (Vaezi et al., 2002). Therefore, phospho-ROCK2 accumulation, upon miR-24 overexpression, may possibly be implicated in the formation of AJs.
miR-24 also controls Myc and E2F2 expression (Lal et al., 2009a). The transcriptional factor Myc has a crucial role in the mammalian epidermis (Pelengaris et al., 1999; Flores et al., 2004; Watt et al., 2008; Yi et al., 2008; Berta et al., 2010). We found that the antiproliferative effect of miR-24 is also mediated by Myc down-regulation (80% inhibition; Fig. S5), although PAK4, Tsk5, and ArhGAP19 silencing also reduces proliferation (20%; Fig. 8 D). However, the actin cytoskeletal remodeling and differentiation effects mediated by miR-24 are Myc independent (Fig. S5, A–D). Other studies confirm that Myc is not involved in cytoskeletal remodeling. For instance, Myc deletion in the epidermis (using a K5-cre promoter), possibly resembling our Tg-K5::miR-24 mouse model, results in a thinner epidermis caused by impairment of stem cell self-renewal as measured by a reduction in the number of DNA label–retaining cells (Watt et al., 2008), but no abnormalities have been described in Myc-null mice in the overall architecture of the epidermis and in basal layer cell morphology. Despite the debatable role of Myc in also promoting keratinocyte differentiation depending on dose, time, and context as proposed by Watt et al. (2008), we did not detect a significant effect on induction of keratinocyte differentiation upon Myc silencing (Fig. S5 F). In addition, Lal et al. (2009a) showed that the increase in cell proliferation induced by anti–miR-24 was not abrogated by knocking down endogenous Myc (Zanet et al., 2005), suggesting that other miR-24 targets, possibly including those identified in this paper, must have an antiproliferative role. Thus, although we cannot rule out the possibility that other miR-24 targets affect differentiation and actin adhesion dynamics, it would seem likely that the correct expression of miR-24 and PAK4, Tsk5, and ArhGAP19 in keratinocytes has a critical role for skin morphogenesis.
Although the mechanisms underlying the balance between proliferating and differentiating cells are complex and likely to be regulated by transcriptional mechanisms, the mutually exclusive expression patterns and the evolutionarily conserved regulation strongly suggest a close relationship between miR-24 and its targets in regulating the boundary between proliferation and differentiation in the epidermis. Our results demonstrate that actin cytoskeletal dynamics and cell shape are strong intracellular signals at can influence keratinocytes toward different cellular fates (proliferation vs. differentiation); these signals are, at least in part, controlled by miR-24. Notably, the actin cytoskeleton also transduces physical signals from the microenvironment to regulate epidermal stem cell fate decisions (Connelly et al., 2010). This represents a new mechanism for regulating epidermal differentiation and may have implications for hyperproliferative disorders, including psoriasis and cancers, in which modulation of miR-24 expression might be an attractive tool for future therapeutic approaches. This pathway may also be important in wound healing, in which delay of terminal differentiation and increased cell spreading and migration may allow more rapid reepithelialization.
Materials and methods
Generation of transgenic mice and animal maintenance
A 434-bp DNA fragment flanking pre–miR-24 hairpin was cloned from mouse genomic DNA with 5′ primer 5′-ACACGTGATGGGTGGGGGTGTG-3′ and 3′ primer 5′-AATGCTCAGCGCGTGCCCTACC-3′ and subsequently inserted in pBluScript-K5 between the bovine K5 promoter and poly(A)+ cassette (vector was provided by M. Blessing, Joannes Gutemberg University, Mainz, Germany). K5–miR-24 transgenic mice were generated in the Friend leukemia virus B mice strain by standard transgenic protocols. Founder mice were determined by PCR genotyping with 5′ primer 5′-CCCCGCTGTAAAGAAAGATTTG-3′ and 3′ primer 5′-CCCTTCATCTTCTTCTCCGTG-3′. Two lines exhibited a similar phenotype. 50 mg/kg BrdU was administrated in sterile PBS by intraperitoneal injection 4 h before sacrifice. Quantification of BrdU-positive cells was performed by counting three sections of three injected mice over the nuclei of the basal layer.
Antagomir synthesis and injection
Antagomir-24 and scrambled control were designed and synthesized (Thermo Fisher Scientific). The antagomir-24 sequence was 5′-CpsUpsGUUCCUGCUGAACUGAGpsCpsCpsAps-Chol-3′ and scrambled control was 5′-UpsCpsACAACCUCCUAGAAAGAGUpsApsGpsAps-Chol-3′ (ps represents a phosphorothioate linkage, and Chol represents cholesterol linked through a hydroxyprolinol linkage). Subcutaneous injection was performed in newborn CD-1 mice at a dosage of 80 mg/kg every day for 3 d as described previously (Krützfeldt et al., 2005).
Cell culture and transfection
Human primary epidermal keratinocytes from neonatal foreskin of four to five donors, HEKn (Cascade Biologics), were grown in EpiLife medium supplemented with human keratinocyte growth supplement (HKGS; Cascade Biologics; Invitrogen). Cells were transfected with pre–miR-24, anti–miR-24, and scrambled negative control (Ambion) using the transfection agent (siPORT NeoFX; Ambion). Keratinocytes were transfected with siRNA (FlexiTube; QIAGEN) of PAK4 (product code SI02660315), Tsk5 (product code SI04309403), or ArhGAP19 (product code SI04147234), Myc (product code SI00300902), E2F2 (product code SI00375389), and negative control (product code 1027280) using RNAiMAX according to the manufacturer’s protocol (Invitrogen). Mouse primary keratinocytes were isolated according to Yuspa et al. (1989) from CD-1 strain newborn skin. Primary keratinocytes were induced to differentiate by adding 1.2 mM CaCl2 to the culture medium. The SAOS-2 cell line was grown in DME-F12 (1:1) medium, 10% FBS, 100 U penicillin, and 100 µg streptomycin (Gibco; Invitrogen).
Retroviral vector generation and infection
To express human Pak4, Tks5, and ArhGAP19 cDNA in primary keratinocytes by retroviruses infection, their mRNA had been transcribed and amplified from keratinocyte RNA exacts by RT-PCR using the following primers: PAK4-HA-HindF, 5′-GGCCAAGCTTATGGCTTACCCATACGATGTTCCAGATTACGCTTTTGGGAAGAGGAAGAAGCGGGTGG-3′; PAK4-NotR, 5′-GCGCGCGGCCGCTCATCTGGTGCGGTTCTGGCGCATGAGG-3′; Tks5-HA-HindF, 5′-GGCCAAGCTTATGGCTTACCCATACGATGTTCCAGATTACGCCTCTGCGCCTACTGCGTGCAGGATGC-3′; Tks5-NotR, 5′-GCGCGCGGCCGCCTAGTTCTTTTTCTCAAGGTAGTTGG-3′; ArhGAP19-HA-HindF, 5′-GGCCAAGCTTATGGCTTACCCATACGATGTTCCAGATTACGCTGCGACTGAGGCACAGAGTGAAGG-5′; and ArhGAP19-NotR 5′-GCGCGCGGCCGCTCAGAGAAATCCTTTTTTCCCCTCTTTCTTCCC-3′. Forward primers include the HA tag sequence. The resulting fragments were restricted and ligated to pLNCX2 retrovirus vector (Takara Bio Inc.) HindIII–NotI linearized. Packaging GP-293 cells (Takara Bio Inc.) were transfected by Effectene (QIAGEN) with retroviral plasmids. After 24 h, DME medium, 10% FBS, 100 U penicillin, and 100 µg streptomycin (Gibco; Invitrogen) were replaced with EpiLife medium supplemented with HKGS for retroviral collection. After 48 h, virus-containing medium was collected with 8 µg/ml polybrene (Sigma-Aldrich). Human primary keratinocytes were infected by replacing the culture medium with the viral supernatant for 4 h (Peschiaroli et al., 2006).
FACS analysis and clonogenicity assay
Human epidermal keratinocytes were pulse labeled for 4 h with 10 µM of the BrdU analogue EdU and then processed with EdU Alexa Fluor 488 flow cytometry assay kit (Click-iT; Invitrogen). Proliferating cells were analyzed and quantified with a flow cytometer (FACSCalibur; BD). Sub-G1 cells have been evaluated by performing propidium iodide staining and FACS analysis after 48 and 72 h of transfection. For the clonogenicity assay, human epidermal keratinocytes were transfected in 6-well plates. 48 h after transfection, keratinocytes were trypsinized and plated in 24-well plates in complete keratinocyte medium. At day 12, colonies were visualized by rhodamine staining followed by extensive washing. Plating efficiency was evaluated by adhesion assay: after plating, the cells were washed with PBS and fixed with 4% formaldehyde for 15 min before being stained with 0.25% crystal violet in 20% methanol for 10 min. After removing the crystal violet solution, the plates were washed extensively and dried, and the stain was released using 2% SDS in PBS. Stain intensity was quantified by spectrophotometry (570 nm) using a plate reader.
miRNA extraction and Northern blotting
The miRNA fraction was extracted with the miRNA isolation kit using the miRNA enrichment protocol (mirVana; Ambion) and quantified by spectrophotometric analysis. For Northern blotting, RNA was transferred by electroblotting onto a membrane (GeneScreen Plus; PerkinElmer). Prehybridization was performed for 1 h at 42°C in hybridization buffer (ULTRAhyb-Oligo; Ambion), and the membrane was hybridized overnight at 42°C in the same buffer with the labeled probe. Antisense RNA probes were 3′-end labeled with γ-[32P]ATP by T4 polynucleotide kinase, miR-24, antisense probe sequence 5′-CUGUUCCUGCUGAACUGAGCCA-3′, miR-203 antisense probe sequence 5′-CTAGTGGTCCTAAACATTTCAC-3′, or U2 small nuclear RNA antisense probe sequence 5′-GGGTGCACCGTTCCTGGAGGTAC-3′. The membranes were washed three times for 30 min in 0.5% SDS and 2× SSC and exposed on an Imaging Screen-K (Bio-Rad Laboratories) at RT. Image scanning and analysis were performed on a Personal Molecular Imager using the QuantityOne software (Bio-Rad Laboratories).
RNA extraction and quantitative real-time RT-PCR
Total RNA was extracted with the miRNA isolation kit (mirVana) and quantified by spectrophotometric analysis. The relative expression levels of miR-24 were measured by a two-step TaqMan assay according to the manufacturer’s instructions (Applied Biosystems). The relative quantification of differentiation marker mRNAs was obtained by real-time PCR using Platinum SYBR green qPCR SuperMix uracil-DNA glycosylase (Invitrogen). All real-time primers used are reported in Table S3. Relative quantification of gene expression was calculated according to the method of 2−DDCt described in the User Bulletin no. 2 and the Relative Quantification software version 1.3 of Applied Biosystems.
Microarray gene expression analysis
RNA was extracted from human primary keratinocytes after 24 h of pre–miR-24 or scrambled transfection using TRIZOL (Invitrogen). cRNAs were hybridized to the GeneChip U133 Plus 2.0 (Affymetrix), which contains probes for ∼47,000 mRNA species. Scanned output files were inspected for hybridization artifacts and further analyzed using GeneChip 3.3 software. Ratios were obtained by dividing the mean difference of scrambled and pre–miR-24 samples.
Western blotting
Total cell extracts were resolved on SDS-polyacrylamide gels and blotted onto polyvinylidene difluoride membranes (Hybond; GE Healthcare). Membranes were blocked and incubated with primary and secondary antibodies in PBST (PBS with Tween 20) and 5% nonfat dry milk. The antibodies used were anti–β-tubulin, anti-Tks5, anti-ArhGAP19 (Santa Cruz Biotechnology, Inc.), anti-PAK4, anti–c-Myc (Cell Signaling Technology), anti-E2F2 (Sigma-Aldrich), anti-ROCK2, anti-pROCK2, anti-Arp2/3, anticofilin, anti–p-cofilin, anti-WASP, and anti-pWASP (ECM Bioscience).
Cell migration assays and time-lapse video microscopy
For time-lapse experiments of the scratch wound-healing assay, human primary keratinocytes were transfected and grown to confluency in 12-well multiwell plates in standard culture medium and conditions. 48 h after transfection, the cells were washed with PBS and pretreated with 10 mg/ml mitomycin C (Sigma-Aldrich) 2 h before wounding to prevent cellular proliferation. After 2 h, the mitomycin C was removed, and cells were washed three times with PBS. Cells were then scratched. Cells were then incubated in a heated frame incubator (DGTCO2BX; Okolab; Nikon), and images were acquired for video microscopy analysis by a camera (DS-Fi1; Nikon) on a microscope (TE Eclipse; Nikon) and then elaborated by NIS-Element 3.0 software (Nikon). Tracking analysis was performed manually by using NIS-Element 3.0 software. Images of the areas were collected with a microscope (Eclipse TE200; Nikon) equipped with a camera (DS-Fi1) using Plan Fluor 4×/0.13 NA or Plan Fluor 10×/0.25 NA objective lenses at RT. The transwell migration assay was performed according to the guidelines of the transwell chamber (BD). In brief, 48 h after transfection, cells were seeded on an 8-µm porous membrane (upper chamber) at 105 cells/ml in serum-free medium and were allowed to migrate through the membrane under the effect of a chemoattractant (HKGS) added into the medium of lower chamber. After 16 h at 37°C, nonmigrating cells were removed from the upper chamber by scrubbing with a cotton swab, whereas invading cells were fixed to the bottom surface with 4% paraformaldehyde and then stained with crystal violet and counted by microscope. Images of the areas were collected with a microscope (Eclipse TE200) equipped with a camera (DS-Fi1) using Plan Fluor 4×/0.13 NA or Plan Fluor 10×/0.25 NA objective lenses at RT.
Immunofluorescence and confocal analysis
Mouse tissues were embedded in frozen specimen medium (Criomatix; Shandon) or fixed in 4% paraformaldehyde (48 h) and embedded in paraffin. Nonspecific antigens were blocked by incubation in 5% goat serum in PBS for 1 h in a humidified atmosphere at RT. Subsequently, sections were incubated for 1 h with primary antibodies: anti-K14, anti-K10, antiloricrin, antifilaggrin (1:1,000 dilution; Covance), anti-p63 (Ab4; 1:500 dilution; NeoMarkers), anti-BrdU (1:50 dilution; Dako), and then washed three times with PBS and incubated for 1 h with Alexa Fluor 488 or 568 secondary antibodies (dilution of 1:1,000; Molecular Probes; Invitrogen) and anti–β4 integrin (dilution of 1:100; BD). Hematoxylin-eosin staining was performed as previously described (Candi et al., 2007) on paraffin-embedded sections to allow morphological analysis of tissue samples. In brief, after deparaffinization and rehydration, sections were stained for 5 min with hematoxylin and then washed in distilled water for 3 min and incubated 1 min in 1% HCl/70% alcohol. They were, then, washed in distilled water and stained 1 min in 1% eosin. Slides were mounted by using an antifade kit (ProLong; Invitrogen). Immunofluorescence on cells was performed by fixing in 4% paraformaldehyde (10 min). After 0.5% Triton X-100 permeabilization (10 min) and 5% goat serum PBS blocking (1 h), cells were incubated 1 h with Alexa Fluor 488 phallotoxin (1:40 dilution; Molecular Probes; Invitrogen) or with primary antibodies: antivinculin (dilution of 1:4,000; Sigma-Aldrich), antipaxillin (dilution of 1:200; BD), anti–E-cadherin (dilution of 1:200; Takara Bio Inc.), and antiactinin (dilution of 1:200; Abcam). Then, cells were washed three times with PBS and incubated for 1 h with Alexa Fluor 488 or 568 secondary antibodies (dilution of 1:1,000). Then, cells were washed three times with PBS and incubated for 1 h with Alexa Fluor 488 or 568 secondary antibodies (dilution of 1:1,000). After two washes in PBS, the tissue sections or cells were counterstained with DAPI to highlight nuclei. Slides were then mounted by using ProLong antifade kit. TUNEL assay was performed using the In Situ Cell Death Detection kit, TMR red (Roche) on paraffin-embedded sections according the manufacturer’s instructions. Fluorescence was evaluated by confocal microscopy (C1 on Eclipse Ti; EZ-C1 software; Nikon) fitted with an argon laser (488-nm excitation), He/Ne laser (542-nm excitation), and UV excitation at 405 nm using Plan Fluor 4×/0.13 NA, Plan Fluor 10×/0.25 NA, or Plan Apochromat 20×/0.75 NA or oil immersion Plan Fluor 40×/1.30 NA or Plan Apochromat 60×/1.4 objective lenses at RT. EZ-C1 images were exported as full resolution TIF files and processed in Photoshop CS5 (Adobe) to adjust brightness and contrast. Acquisition was performed at RT using Plan Fluor 4×/0.13 NA, Plan Fluor 10×/0.25 NA, or Plan Apochromat 20×/0.75 NA or oil immersion Plan Fluor 40×/1.30 NA or Plan Apochromat 60×/1.4 NA objective lenses.
miRNA in situ hybridization analysis
Mouse and human tissues were embedded in frozen specimen medium Criomatix after an overnight incubation in 4% paraformaldehyde followed by an overnight incubation in 0.5 M sucrose. 14-µm-thin sections were cut and mounted on Superfrost glass slides. Slides were then fixed for 10 min in 4% paraformaldehyde and acetylated for 10 min in triethanolamine/acetic anhydride. Slides were then hybridized in incubation chambers overnight at 50°C using 30 nM detection digoxigenin probes (miRCURY LNA; Exiqon). After hybridization, slides were washed (20 min in 5× SSC, two times for 30 min in Tween 20/SSC at 50°C, and twice for 15 min in 0.2× SSC and 15 min in PBS at RT). After 1-h incubation in blocking solution at RT, slides were hybridized for 2 h in alkaline phosphatase–conjugated antidigoxigenin Fab fragment (1:1,500 dilution; Roche) at RT. After two washes of 20 min, detection was performed by incubating 250 µl nitroblue tetrazolium/BCIP (1-STEP; Thermo Fisher Scientific) together with 2 mM levamisole on the slides for 16 h in the dark at RT. Images of the areas were collected with an microscope (Eclipse TE200) equipped with a camera (DS Fi1) using Plan Fluor 4×/0.13 NA or Plan Fluor 10×/0.25 NA objective lenses or Plan Apochromat 20×/0.75 NA at RT.
Luciferase assay and constructs
A total of 2 × 105 SAOS-2 cells were seeded in 12-well dishes 24 h before transfection. 100 ng pGL3 vectors, 50 nM pre–miR-24 or scrambled control, and 10 ng pRL-cytomegalovirus vector were cotransfected using Lipofectamine 2000 (Invitrogen). Luciferase activities of cellular extracts were measured 24 h after transfection by using a Dual-Luciferase Reporter Assay System (Promega). Light emission was measured over 10 s using a luminometer (OPTOCOMP I; MGM Instruments, Inc.). Efficiency of transfection was normalized using Renilla luciferase activity. Details for cloning target 3′UTRs are as follows: A 650-bp fragment from human PAK4 3′UTR was amplified with 5′ primer 5′-GGCCCAGCGCCCTTCCCCTC-3′ and 3′ primer 5′-TAGACTTCTAGAAAAGAGGAG-3′. A 620-bp fragment from human Tks5 3′UTR was amplified with 5′ primer 5′-CAGGATGTTGAGTTAATCTG-3′ and 3′ primer 5′-AGGGGATGGGATAGAGACCTC-3′. A 523-bp fragment from human ArhGAP19 3′UTR was amplified with 5′ primer 5′-TATAGCTGGTCTCCTTGGGTGG-3′ and 3′ primer 5′-CACACAGTACAGTCTGGACC-3′. The 3′UTR fragments were subjected to restriction and then ligated to pGL3 control vector (Promega). The miR-24 predicted target site was deleted by PCR using two overlapping primers: PAK4 3′UTR mutant 5′ primer, 5′-ACACCCAGCCCCCTCTCCCCATTGTGGGGGTCGATCATGA-3′; and 3′ primer, 5′-TCATGATCGACCCCCACAATGGGGAGAGGGGGCTGGGTGT-3′; Tks5 3′UTR mutant 5′ primer, 5′-TCGCTCCTCAGATGTCTGCAGTTCAGTCACTAGTGCGCCA-3′; and 3′ primer, 5′-TGGCGCACTAGTGACTGAACTGCAGACATCTGAGGAGCGA-3′; and ArhGAP19 3′UTR mutant, 5′-TCGCTCCTCAGATGTCTGGCAGTTCAGTCACTAGTGCGCCA-3′; and 3′ primer, 5′-TGGCGCACTAGTGACTGAACTGCCAGACATCTGAGGAGCGA-3′.
Statistics
All experiments are presented as means ± SDs. Statistical analyses were performed using unpaired, two-tailed Student’s t tests.
Bioinformatics
miR-24 target sites on PAK4, Tks5, and ArhGAP19 3′UTR were predicted by TargetScan 5.1 software. Functional analysis, on the array dataset result, was performed by using Ingenuity Pathway Analysis software (Ingenuity Systems).
Online supplemental material
Fig. S1 shows the expression profile of miR-24 in human and mouse epidermis and its effects on keratinocytes proliferation and differentiation. Fig. S2 shows histological and immunofluorescence analysis of the epidermis from antagomir-24–treated mice and K5::miR-24 transgenic mice. Fig. S3 shows results of microarray analysis after miR-24 overexpression. Fig. S4 shows effects of knockdown of miR-24 targets on keratinocyte actin cytoskeleton. Fig. S5 shows effects of Myc and E2F2 knockdown on keratinocyte actin cytoskeleton. Table S1 includes the list of genes down-regulated in miR-24–overexpressing keratinocytes that present miR-24 binding sites. Table S2 includes the results of Ingenuity Pathway Analysis analysis performed on genes down-regulated in miR-24–overexpressing keratinocytes to identify the most significant categories of biological functions. Table S3 shows the list of primers used for real-time qPCR. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201203134/DC1.
Acknowledgments
We thank Dr. Marco Ranalli and Rossella Loria for technical support and Dr. Angelo Peschiaroli for scientific discussion.
The work reported in this manuscript has been supported by grants from Telethon (GGP09133), Associazione Italiana per la Ricerca sul Cancro (2743), Ministero dell’Università e della Ricerca, Ministero della Sanità, and Alleanza contro il Cancro (ACC12) to G. Melino and partially supported by grants (RF73, RF57, and ACC12) to E. Candi.
Author contributions: I. Amelio performed and analyzed the majority of the experiments and contributed to manuscript preparation. A.M. Lena performed mouse injections, generated transgenic mice, and contributed to figure preparation. G. Viticchiè helped with the migration assay and Western blots. R. Shalom-Feuerstein performed the clonogenic assay. D. Dinsdale, C. Fortunato, E. Bonanno, and L.G. Spagnoli performed tissue staining and contributed to the phenotype analysis of the transgenic mice. D. Aberdam contributed to the editing of the paper. A. Terrinoni and G. Russo performed gene array analysis. G. Melino contributed to the development of project and pointed out the in vivo strategy. E. Candi designed and analyzed the majority of the experiments, wrote the paper, and supervised the project.
Footnotes
Abbreviations used in this paper:
- AJ
- adherens junction
- GEF
- guanine nucleotide exchange factor
- HEKn
- human epidermal keratinocyte, neonatal
- HKGS
- human keratinocyte growth supplement
- miRNA
- micro-RNA
- qPCR
- quantitative PCR
- Tg
- transgene
- UTR
- untranslated region
- WASP
- Wiskott–Aldrich syndrome protein
- wt
- wild type
References
- Aberdam D., Candi E., Knight R.A., Melino G. 2008. miRNAs, ‘stemness’ and skin. Trends Biochem. Sci. 33:583–591 10.1016/j.tibs.2008.09.002 [DOI] [PubMed] [Google Scholar]
- Abo A., Qu J., Cammarano M.S., Dan C., Fritsch A., Baud V., Belisle B., Minden A. 1998. PAK4, a novel effector for Cdc42Hs, is implicated in the reorganization of the actin cytoskeleton and in the formation of filopodia. EMBO J. 17:6527–6540 10.1093/emboj/17.22.6527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abram C.L., Seals D.F., Pass I., Salinsky D., Maurer L., Roth T.M., Courtneidge S.A. 2003. The adaptor protein fish associates with members of the ADAMs family and localizes to podosomes of Src-transformed cells. J. Biol. Chem. 278:16844–16851 10.1074/jbc.M300267200 [DOI] [PubMed] [Google Scholar]
- Ambros V. 2004. The functions of animal microRNAs. Nature. 431:350–355 10.1038/nature02871 [DOI] [PubMed] [Google Scholar]
- Andl T., Murchison E.P., Liu F., Zhang Y., Yunta-Gonzalez M., Tobias J.W., Andl C.D., Seykora J.T., Hannon G.J., Millar S.E. 2006. The miRNA-processing enzyme dicer is essential for the morphogenesis and maintenance of hair follicles. Curr. Biol. 16:1041–1049 10.1016/j.cub.2006.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barac A., Basile J., Vázquez-Prado J., Gao Y., Zheng Y., Gutkind J.S. 2004. Direct interaction of p21-activated kinase 4 with PDZ-RhoGEF, a G protein-linked Rho guanine exchange factor. J. Biol. Chem. 279:6182–6189 10.1074/jbc.M309579200 [DOI] [PubMed] [Google Scholar]
- Bartel D.P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 116:281–297 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- Berta M.A., Baker C.M., Cottle D.L., Watt F.M. 2010. Dose and context dependent effects of Myc on epidermal stem cell proliferation and differentiation. EMBO Mol. Med. 2:16–25 10.1002/emmm.200900047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop A.L., Hall A. 2000. Rho GTPases and their effector proteins. Biochem. J. 348:241–255 10.1042/0264-6021:3480241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candi E., Schmidt R., Melino G. 2005. The cornified envelope: a model of cell death in the skin. Nat. Rev. Mol. Cell Biol. 6:328–340 10.1038/nrm1619 [DOI] [PubMed] [Google Scholar]
- Candi E., Rufini A., Terrinoni A., Dinsdale D., Ranalli M., Paradisi A., De Laurenzi V., Spagnoli L.G., Catani M.V., Ramadan S., et al. 2006. Differential roles of p63 isoforms in epidermal development: selective genetic complementation in p63 null mice. Cell Death Differ. 13:1037–1047 10.1038/sj.cdd.4401926 [DOI] [PubMed] [Google Scholar]
- Candi E., Rufini A., Terrinoni A., Giamboi-Miraglia A., Lena A.M., Mantovani R., Knight R., Melino G. 2007. DeltaNp63 regulates thymic development through enhanced expression of FgfR2 and Jag2. Proc. Natl. Acad. Sci. USA. 104:11999–12004 10.1073/pnas.0703458104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly J.T., Gautrot J.E., Trappmann B., Tan D.W., Donati G., Huck W.T., Watt F.M. 2010. Actin and serum response factor transduce physical cues from the microenvironment to regulate epidermal stem cell fate decisions. Nat. Cell Biol. 12:711–718 10.1038/ncb2074 [DOI] [PubMed] [Google Scholar]
- Eswaran J., Soundararajan M., Kumar R., Knapp S. 2008. UnPAKing the class differences among p21-activated kinases. Trends Biochem. Sci. 33:394–403 10.1016/j.tibs.2008.06.002 [DOI] [PubMed] [Google Scholar]
- Fiedler J., Jazbutyte V., Kirchmaier B.C., Gupta S.K., Lorenzen J., Hartmann D., Galuppo P., Kneitz S., Pena J.T., Sohn-Lee C., et al. 2011. MicroRNA-24 regulates vascularity after myocardial infarction. Circulation. 124:720–730 10.1161/CIRCULATIONAHA.111.039008 [DOI] [PubMed] [Google Scholar]
- Flores I., Murphy D.J., Swigart L.B., Knies U., Evan G.I. 2004. Defining the temporal requirements for Myc in the progression and maintenance of skin neoplasia. Oncogene. 23:5923–5930 10.1038/sj.onc.1207796 [DOI] [PubMed] [Google Scholar]
- Fuchs E. 2007. Scratching the surface of skin development. Nature. 445:834–842 10.1038/nature05659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda Y., Kawasaki H., Taira K. 2005. Exploration of human miRNA target genes in neuronal differentiation. Nucleic Acids Symp. Ser. (Oxf). 49:341–342 10.1093/nass/49.1.341 [DOI] [PubMed] [Google Scholar]
- Heasman S.J., Ridley A.J. 2008. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat. Rev. Mol. Cell Biol. 9:690–701 10.1038/nrm2476 [DOI] [PubMed] [Google Scholar]
- Jamora C., Fuchs E. 2002. Intercellular adhesion, signalling and the cytoskeleton. Nat. Cell Biol. 4:E101–E108 10.1038/ncb0402-e101 [DOI] [PubMed] [Google Scholar]
- Kim V.N. 2005. MicroRNA biogenesis: coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 6:376–385 10.1038/nrm1644 [DOI] [PubMed] [Google Scholar]
- Krützfeldt J., Rajewsky N., Braich R., Rajeev K.G., Tuschl T., Manoharan M., Stoffel M. 2005. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 438:685–689 10.1038/nature04303 [DOI] [PubMed] [Google Scholar]
- Lal A., Navarro F., Maher C.A., Maliszewski L.E., Yan N., O’Day E., Chowdhury D., Dykxhoorn D.M., Tsai P., Hofmann O., et al. 2009a. miR-24 inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3’UTR microRNA recognition elements. Mol. Cell. 35:610–625 10.1016/j.molcel.2009.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal A., Pan Y., Navarro F., Dykxhoorn D.M., Moreau L., Meire E., Bentwich Z., Lieberman J., Chowdhury D. 2009b. miR-24-mediated downregulation of H2AX suppresses DNA repair in terminally differentiated blood cells. Nat. Struct. Mol. Biol. 16:492–498 10.1038/nsmb.1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechler T., Fuchs E. 2005. Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature. 437:275–280 10.1038/nature03922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lena A.M., Shalom-Feuerstein R., Rivetti di Val Cervo P., Aberdam D., Knight R.A., Melino G., Candi E. 2008. miR-203 represses ‘stemness’ by repressing DeltaNp63. Cell Death Differ. 15:1187–1195 10.1038/cdd.2008.69 [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B. 2003. Prediction of mammalian microRNA targets. Cell. 115:787–798 10.1016/S0092-8674(03)01018-3 [DOI] [PubMed] [Google Scholar]
- Lock P., Abram C.L., Gibson T., Courtneidge S.A. 1998. A new method for isolating tyrosine kinase substrates used to identify fish, an SH3 and PX domain-containing protein, and Src substrate. EMBO J. 17:4346–4357 10.1093/emboj/17.15.4346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv L., Xu J., Zhao S., Chen C., Zhao X., Gu S., Ji C., Xie Y., Mao Y. 2007. Sequence analysis of a human RhoGAP domain-containing gene and characterization of its expression in human multiple tissues. DNA Seq. 18:184–189 [DOI] [PubMed] [Google Scholar]
- Oikawa T., Itoh T., Takenawa T. 2008. Sequential signals toward podosome formation in NIH-src cells. J. Cell Biol. 182:157–169 10.1083/jcb.200801042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelengaris S., Littlewood T., Khan M., Elia G., Evan G. 1999. Reversible activation of c-Myc in skin: induction of a complex neoplastic phenotype by a single oncogenic lesion. Mol. Cell. 3:565–577 10.1016/S1097-2765(00)80350-0 [DOI] [PubMed] [Google Scholar]
- Perez-Moreno M., Fuchs E. 2006. Catenins: keeping cells from getting their signals crossed. Dev. Cell. 11:601–612 10.1016/j.devcel.2006.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschiaroli A., Dorrello N.V., Guardavaccaro D., Venere M., Halazonetis T., Sherman N.E., Pagano M. 2006. SCFbetaTrCP-mediated degradation of Claspin regulates recovery from the DNA replication checkpoint response. Mol. Cell. 23:319–329 10.1016/j.molcel.2006.06.013 [DOI] [PubMed] [Google Scholar]
- Qian L., Van Laake L.W., Huang Y., Liu S., Wendland M.F., Srivastava D. 2011. miR-24 inhibits apoptosis and represses Bim in mouse cardiomyocytes. J. Exp. Med. 208:549–560 10.1084/jem.20101547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M. 2004. Rho signalling at a glance. J. Cell Sci. 117:5457–5458 10.1242/jcs.01582 [DOI] [PubMed] [Google Scholar]
- Seals D.F., Azucena E.F., Jr, Pass I., Tesfay L., Gordon R., Woodrow M., Resau J.H., Courtneidge S.A. 2005. The adaptor protein Tks5/Fish is required for podosome formation and function, and for the protease-driven invasion of cancer cells. Cancer Cell. 7:155–165 10.1016/j.ccr.2005.01.006 [DOI] [PubMed] [Google Scholar]
- Singh R., Saini N. 2012. Downregulation of BCL2 by miRNAs augments drug-induced apoptosis - a combined computational and experimental approach. J. Cell Sci. 125:1568–1578 10.1242/jcs.095976 [DOI] [PubMed] [Google Scholar]
- Srivastava N., Manvati S., Srivastava A., Pal R., Kalaiarasan P., Chattopadhyay S., Gochhait S., Dua R., Bamezai R.N. 2011. miR-24-2 controls H2AFX expression regardless of gene copy number alteration and induces apoptosis by targeting antiapoptotic gene BCL-2: a potential for therapeutic intervention. Breast Cancer Res. 13:R39 10.1186/bcr2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F., Wang J., Pan Q., Yu Y., Zhang Y., Wan Y., Wang J., Li X., Hong A. 2009. Characterization of function and regulation of miR-24-1 and miR-31. Biochem. Biophys. Res. Commun. 380:660–665 10.1016/j.bbrc.2009.01.161 [DOI] [PubMed] [Google Scholar]
- Vaezi A., Bauer C., Vasioukhin V., Fuchs E. 2002. Actin cable dynamics and Rho/Rock orchestrate a polarized cytoskeletal architecture in the early steps of assembling a stratified epithelium. Dev. Cell. 3:367–381 10.1016/S1534-5807(02)00259-9 [DOI] [PubMed] [Google Scholar]
- Vasioukhin V., Bauer C., Yin M., Fuchs E. 2000. Directed actin polymerization is the driving force for epithelial cell-cell adhesion. Cell. 100:209–219 10.1016/S0092-8674(00)81559-7 [DOI] [PubMed] [Google Scholar]
- Wang Q., Huang Z., Xue H., Jin C., Ju X.L., Han J.D., Chen Y.G. 2008. MicroRNA miR-24 inhibits erythropoiesis by targeting activin type I receptor ALK4. Blood. 111:588–595 10.1182/blood-2007-05-092718 [DOI] [PubMed] [Google Scholar]
- Watt F.M. 2002. Role of integrins in regulating epidermal adhesion, growth and differentiation. EMBO J. 21:3919–3926 10.1093/emboj/cdf399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt F.M., Frye M., Benitah S.A. 2008. MYC in mammalian epidermis: how can an oncogene stimulate differentiation? Nat. Rev. Cancer. 8:234–242 10.1038/nrc2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S.E., Beronja S., Pasolli H.A., Fuchs E. 2011. Asymmetric cell divisions promote Notch-dependent epidermal differentiation. Nature. 470:353–358 10.1038/nature09793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Shen Q.T., Oristian D.S., Lu C.P., Zheng Q., Wang H.W., Fuchs E. 2011. Skin stem cells orchestrate directional migration by regulating microtubule-ACF7 connections through GSK3β. Cell. 144:341–352 10.1016/j.cell.2010.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R., Poy M.N., Stoffel M., Fuchs E. 2008. A skin microRNA promotes differentiation by repressing ‘stemness’. Nature. 452:225–229 10.1038/nature06642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuspa S.H., Kilkenny A.E., Steinert P.M., Roop D.R. 1989. Expression of murine epidermal differentiation markers is tightly regulated by restricted extracellular calcium concentrations in vitro. J. Cell Biol. 109:1207–1217 10.1083/jcb.109.3.1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanet J., Pibre S., Jacquet C., Ramirez A., de Alborán I.M., Gandarillas A. 2005. Endogenous Myc controls mammalian epidermal cell size, hyperproliferation, endoreplication and stem cell amplification. J. Cell Sci. 118:1693–1704 10.1242/jcs.02298 [DOI] [PubMed] [Google Scholar]
- Zhang H., Li Z., Viklund E.K., Stromblad S. 2002. p21-activated kinase 4 interacts with integrin αvβ5 and regulates αvβ5-mediated cell migration. J. Cell Biol. 158:1287–1297 10.1083/jcb.200207008 [DOI] [PMC free article] [PubMed] [Google Scholar]