Abstract
Microfluidic encapsulation methods have been previously utilized to capture cells in picoliter-scale aqueous, monodisperse drops, providing confinement from a bulk fluid environment with applications in high throughput screening, cytometry, and mass spectrometry. We describe a method to not only encapsulate single cells, but to repeatedly capture a set number of cells (here we demonstrate one- and two-cell encapsulation) to study both isolation and the interactions between cells in groups of controlled sizes. By combining drop generation techniques with cell and particle ordering, we demonstrate controlled encapsulation of cell-sized particles for efficient, continuous encapsulation. Using an aqueous particle suspension and immiscible fluorocarbon oil, we generate aqueous drops in oil with a flow focusing nozzle. The aqueous flow rate is sufficiently high to create ordering of particles which reach the nozzle at integer multiple frequencies of the drop generation frequency, encapsulating a controlled number of cells in each drop. For representative results, 9.9 μm polystyrene particles are used as cell surrogates. This study shows a single-particle encapsulation efficiency Pk=1 of 83.7% and a double-particle encapsulation efficiency Pk=2 of 79.5% as compared to their respective Poisson efficiencies of 39.3% and 33.3%, respectively. The effect of consistent cell and particle concentration is demonstrated to be of major importance for efficient encapsulation, and dripping to jetting transitions are also addressed.
Introduction
Continuous media aqueous cell suspensions share a common fluid environment which allows cells to interact in parallel and also homogenizes the effects of specific cells in measurements from the media. High-throughput encapsulation of cells into picoliter-scale drops confines the samples to protect drops from cross-contamination, enable a measure of cellular diversity within samples, prevent dilution of reagents and expressed biomarkers, and amplify signals from bioreactor products. Drops also provide the ability to re-merge drops into larger aqueous samples or with other drops for intercellular signaling studies.1,2 The reduction in dilution implies stronger detection signals for higher accuracy measurements as well as the ability to reduce potentially costly sample and reagent volumes.3 Encapsulation of cells in drops has been utilized to improve detection of protein expression,4 antibodies,5,6 enzymes,7 and metabolic activity8 for high throughput screening, and could be used to improve high throughput cytometry.9 Additional studies present applications in bio-electrospraying of cell containing drops for mass spectrometry10 and targeted surface cell coatings.11 Some applications, however, have been limited by the lack of ability to control the number of cells encapsulated in drops. Here we present a method of ordered encapsulation12 which increases the demonstrated encapsulation efficiencies for one and two cells and may be extrapolated for encapsulation of a larger number of cells.
To achieve monodisperse drop generation, microfluidic "flow focusing" enables the creation of controllable-size drops of one fluid (an aqueous cell mixture) within another (a continuous oil phase) by using a nozzle at which the streams converge.13 For a given nozzle geometry, the drop generation frequency f and drop size can be altered by adjusting oil and aqueous flow rates Qoil and Qaq. As the flow rates increase, the flows may transition from drop generation to unstable jetting of aqueous fluid from the nozzle.14
When the aqueous solution contains suspended particles, particles become encapsulated and isolated from one another at the nozzle. For drop generation using a randomly distributed aqueous cell suspension, the average fraction of drops Dk containing k cells is dictated by Poisson statistics, where Dk = λk exp(-λ)/(k!) and λ is the average number of cells per drop. The fraction of cells which end up in the "correctly" encapsulated drops is calculated using Pk = (k x Dk)/Σ(k' x Dk'). The subtle difference between the two metrics is that Dk relates to the utilization of aqueous fluid and the amount of drop sorting that must be completed following encapsulation, and Pk relates to the utilization of the cell sample. As an example, one could use a dilute cell suspension (low λ) to encapsulate drops where most drops containing cells would contain just one cell. While the efficiency metric Pk would be high, the majority of drops would be empty (low Dk), thus requiring a sorting mechanism to remove empty drops, also reducing throughput.15
Combining drop generation with inertial ordering provides the ability to encapsulate drops with more predictable numbers of cells per drop and higher throughputs than random encapsulation. Inertial focusing was first discovered by Segre and Silberberg16 and refers to the tendency of finite-sized particles to migrate to lateral equilibrium positions in channel flow. Inertial ordering refers to the tendency of the particles and cells to passively organize into equally spaced, staggered, constant velocity trains. Both focusing and ordering require sufficiently high flow rates (high Reynolds number) and particle sizes (high Particle Reynolds number).17,18 Here, the Reynolds number Re =uDh/ν and particle Reynolds number Rep =Re(a/Dh)2, where u is a characteristic flow velocity, Dh [=2wh/(w+h)] is the hydraulic diameter, ν is the kinematic viscosity, a is the particle diameter, w is the channel width, and h is the channel height. Empirically, the length required to achieve fully ordered trains decreases as Re and Rep increase. Note that the high Re and Rep requirements (for this study on the order of 5 and 0.5, respectively) may conflict with the need to keep aqueous flow rates low to avoid jetting at the drop generation nozzle. Additionally, high flow rates lead to higher shear stresses on cells, which are not addressed in this protocol. The previous ordered encapsulation study demonstrated that over 90% of singly encapsulated HL60 cells under similar flow conditions to those in this study maintained cell membrane integrity.12 However, the effect of the magnitude and time scales of shear stresses will need to be carefully considered when extrapolating to different cell types and flow parameters. The overlapping of the cell ordering, drop generation, and cell viability aqueous flow rate constraints provides an ideal operational regime for controlled encapsulation of single and multiple cells.
Because very few studies address inter-particle train spacing,19,20 determining the spacing is most easily done empirically and will depend on channel geometry, flow rate, particle size, and particle concentration. Nonetheless, the equal lateral spacing between trains implies that cells arrive at predictable, consistent time intervals. When drop generation occurs at the same rate at which ordered cells arrive at the nozzle, the cells become encapsulated within the drop in a controlled manner. This technique has been utilized to encapsulate single cells with throughputs on the order of 15 kHz,12 a significant improvement over previous studies reporting encapsulation rates on the order of 60-160 Hz.4,15 In the controlled encapsulation work, over 80% of drops contained one and only one cell, a significant efficiency improvement over Poisson (random) statistics, which predicts less than 40% efficiency on average.12
In previous controlled encapsulation work,12 the average number of particles per drop λ was tuned to provide single-cell encapsulation. We hypothesize that through tuning of flow rates, we can efficiently encapsulate any number of cells per drop when λ is equal or close to the number of desired cells per drop. While single-cell encapsulation is valuable in determining individual cell responses from stimuli, multiple-cell encapsulation provides information relating to the interaction of controlled numbers and types of cells. Here we present a protocol, representative results using polystyrene microspheres, and discussion for controlled encapsulation of multiple cells using a passive inertial ordering channel and drop generation nozzle.
Keywords: Bioengineering, Issue 64, Drop-based microfluidics, inertial microfluidics, ordering, focusing, cell encapsulation, single-cell biology, cell signaling
Protocol
The protocols in this section describe the materials and equipment utilized specifically to obtain the experimental results presented. Note that alternative suppliers for chemicals and equipment may be utilized.
1. Device Fabrication and Soft Lithography
Standard soft lithography techniques,21 a number of which have been featured in previous JOVE articles,22 were used for creating polydimethylsiloxane (PDMS) microchannel networks bonded to glass substrates. Aside from master replica mold fabrication by SU-8 photolithography, the processes may be performed outside a clean room or clean hood; however, dust and particulates should still be minimized to achieve consistent results.
Design a micro-channel pattern as shown in Figure 1 in AutoCAD (AutoDesk Inc.). Employ a third party manufacturer (Fineline Imaging Inc.) to print a high resolution (50,000 dpi) transparency mask on Mylar film or quartz where channels are transparent on a dark background.
Create a silicon and SU-8 photoresist master for replica molding. Briefly, spin SU-8 2050 (MicroChem) negative photoresist with manufacturer's recommended rpm on a spin-coater to create a 52 μm thick layer on a clean 7.5 cm or 10 cm silicon wafer. After soft bake, edge bead removal, UV exposure through a contact mask, post-exposure bake, development, and flood exposure, measure the actual thickness of the SU-8 layer using a Dektak profilometer (Veeco). Tape the master mold onto the bottom of a 4" or 5" Petri dish to prepare for PDMS replica molding.
Mix PDMS elastomer base with elastomer curing agent (Dow Corning) in a 10:1 ratio w/w base to curing agent. Pour well-mixed PDMS precursor onto the silicon master to create a 2-3 mm final thickness layer. A mixture of 20 g elastomer base with 2 g curing agent is sufficient to cover a 4" diameter surface.
Place the master mold and PDMS in vacuum desiccator (Jencons) to de-gas the uncured PDMS. Using a pressure regulator (Cole Parmer), slowly decrease the chamber gage pressure from 0" Hg to -27" Hg over 20 minutes to avoid excessive foaming. Leave device in vacuum chamber at -27" Hg for 30 minutes or until air bubbles disappear.
Release vacuum and move master mold and PDMS to a 65 °C oven (Thermo Scientific) for a minimum of four hours. The device may be left in the oven overnight to improve curing.
Remove the device from oven and allow to cool. Carefully cut PDMS around circular wafer using a precision knife and peel out PDMS. Cut out device outline as shown in Figure 1 with a scalpel.
Punch fluidic ports (three per device) in the three round regions shown in Figure 1 using a biopsy punch. For this device, use a 0.75 mm outer diameter punch (Harris).
Adhere scotch tape to the patterned side of the PDMS and peel to remove any dust. As a cost-saving but viable alternative to conventional oxygen plasma apparatuses,21,22 plasma treat the patterned side of the PDMS and a clean 3" x 1" glass microscope slide using a hand-held laboratory corona treater (Electro-Technic Products Inc.).23 Note that this device should be used in a fume hood or well-ventilated area due to ozone discharge, and all watches and cell phones should be kept at least ten feet away. Adjust the corona discharge to attain a stable corona with minimal sparking. Slowly wave the electrode about 1/4" above each surface for around 20 seconds and then immediately bring the treated surfaces into contact to form a strong permanent bond before the PDMS surfaces return to their native state.
Place the device on a metal plate, place in a cool oven, set the oven to 120 °C, and bake overnight to complete bonding and to return the PDMS to its original hydrophobic state.24 During this high temperature baking, the glass surface of the channel will also be rendered hydrophobic due to deposition of a thin hydrophobic layer onto the glass. Alternatively, hydrophobic coatings such as Aquapel (PPG Industries) may be injected into the fluidic ports using a 1 mL syringe and a syringe needle.12 Carefully but firmly inject the Aquapel followed by purging air into the fluidic ports without breaking the PDMS to glass bond. Aggressively repeat the air purge on all inlet and outlet ports while wiping off any excess Aquapel in order to avoid any deposits that may clog the channels upon drying.
2. Sample Preparation
Prepare a cell culture according to established procedures for your chosen cell type. For the particular device used in this study, 8-15 μm particles or cells should adequately order for encapsulation. Smaller or larger cell types may require changing the dimensions of the focusing channel to achieve adequate Rep. For the method demonstration results shown in this paper, 9.9 μm polystyrene microspheres (G1000, Thermo Scientific) are utilized as cell surrogates.
Prepare the aqueous particle or cell suspension through gentle mixing. When using cells or polystyrene particles, concentration control is essential (see Figure 4) to achieve ideal ordered encapsulation. Using previous data12 as a guide, calculate the desired cell or particle concentration based on the ordered train spacing and micro-channel size as: one cell or particle per expected longitudinal train spacing times the focusing channel cross-sectional area. If the stock concentration (1% w/w) is inadequate, increase the concentration (here to 1.5% w/w) by gently centrifuging the stock sample, removing supernatant liquid, and re-suspending the particles by vortex mixing, or gentler mixing when using cells. Prepare an adequate volume to account for desired collection volume and for run time associated with flow tuning.
Both cells and polystyrene particles have a specific gravity greater than one. Although not demonstrated in this protocol, for long-term experiments lasting on the order of many minutes to hours, buoyancy match the solution by adding a solute such as CaCl2 for particles or OptiPrep (Sigma-Aldrich) for cells.
Prepare a 10 mL sample of the continuous fluorocarbon oil phase by mixing the fluorocarbon oil FC-40 (3M) and PFPE-PEG block copolymer surfactant25 (2.5% w/w) (RainDance Technologies) in a 15 mL centrifuge tube. Alternatively, light mineral oil (PTI Process Chemicals) can be utilized with ABIL-EM 90 surfactant (2.5% w/w) (Evonik Goldschmidt Corporation).
3. Experimental Setup
Power on the inverted optical microscope (Axio Observer, Zeiss) and high speed camera (Phantom V310, Vision Research). Focus and inspect the channels for clogs and debris by either manually moving the device or by using a motorized microscope stage. Some small debris may be pushed out when liquid flows through. For large debris or obvious clogs, select another channel on the device as debris in the focusing channel can degrade ordering quality significantly. Note that clogs can often be removed under flow by pressing firmly on the PDMS surface above the affected region with blunt tweezers.
Cut three lengths of PVC tubing (0.01" ID/0.03" OD, Tygon) for the aqueous inlet, oil inlet, and emulsion outlet. To minimize dead volume, cut just enough tubing to reach from the syringe pumps to the microscope stage. Cut tubing ends at a 45° angle to facilitate insertion into fluidic ports.
Use tweezers to press fit the tube ends into the fluidic ports punched in Step 1 and then press fit two 30 gauge blunt-tip stainless steel syringe needles (SmallParts) into the free ends of the respective aqueous and oil inlet tubes (no adhesive necessary). Place the outlet tubing into a waste reservoir. This tube will later be moved into a collection reservoir.
Move the device and attached tubing to the microscope stage, align, and focus on the device nozzle using an available objective (20x was used for this experiment). Adjust for K hler illumination and other microscope settings as required for optimal recording.
Fill a 1 mL syringe (BD) with the well-mixed aqueous phase and a 3 mL syringe (BD) with the oil phase solution prepared in Step 2. Note that any syringes of any volume may be used and should be carefully selected depending on the desired run times and minimization of any pulsatility. Tilt one syringe vertically and flick to move air bubbles to the syringe outlet. Slowly depress plunger enough to push the air to the syringe tip. Holding the syringe vertically, connect the syringes to the respective syringe needle already attached to the device in Step 3.3. Depress plunger to force the air through the syringe needle dead volume until fluid is pushed through the tubing almost to the device. Securely mount the syringe to a syringe pump (Nexus 3000, Chemyx) and engage the plunger block. Repeat connections for the second syringe and mount to a second syringe pump.
Power on each syringe pump and program using the pump manufacturer's protocols. Set the initial flow rates to Qoil = 50 μL/min and Qaq = 5 μL/min for the oil phase and aqueous phase, respectively. Start the pumps.
Wait for each fluid to enter the device and fill the channels, pushing out remaining dead air. This may take several minutes. If there is a large amount of air in the inlet tubing, temporarily increase each flow rate until the air is expelled. Do not increase the flow rates so high that large pressures occur in the channel, potentially leading to PDMS-to-glass bond failure.
Using the initial flow rates, observe the formation of drops at the nozzle (results shown here: 20x magnification, frame rate 21005 fps, exposure 3 μs). Reduce the camera field of view to only the nozzle to maximize the frame rate and reduce memory requirements if possible. Capture sample videos and confirm that the sampling rate is adequate to avoid aliasing.
To avoid jetting (see Figure 2), start with low aqueous flow rates. Slowly increase the aqueous flow rate to observe ordering of particles in the long aqueous solution channel as the flow rate increases.
If the particle concentration is too low to provide trains with relatively few "missing" particles and the sample was not buoyancy matched, physically tilt the syringe pump toward the syringe outlet to provide gradual settling of particles toward the syringe outlet. This method is demonstrated in the video protocol. Periodically rotating the syringe along its axis may also reduce undesired settling.
Once adequate ordering occurs, adjust the oil flow rate to tune the generation frequency and size of drops. The mean drop volume may be calculated using the aqueous flow rate divided by the drop generation frequency as measured by video capture. Iteratively adjust both flow rates to achieve desired encapsulation rates and drop volumes.
Once stable ordered encapsulation is confirmed, move the outlet tubing from the waste reservoir into a collection reservoir or feed it into another device for subsequent testing.
Determine the collection time based on desired number of droplets and the calculated generation frequency.
Record the fraction of drops containing 0, 1, 2,…, N particles to quantify efficiency using either drop generation video results or by pipetting a sample of collected emulsion for inspection.
4. Representative Results
Results are presented which achieve both controlled single-particle and controlled double-particle encapsulation (Figure 3). By cutting the FC-40 oil flow rate in half, single-particle encapsulation becomes two-particle encapsulation. Conversely, we could have increased the aqueous flow rate to deliver particles to the nozzle more quickly, but we also would have increased the risk of jetting of the aqueous stream. Histograms in Figure 3 present the fractional number of particles per drop for the two cases, along with comparisons to Poisson statistics. The occasional drops with zero particles are primarily due to "missing" particles in the ordered trains, while the cases where there are more encapsulated particles than desired result from local high particle concentrations and particles which sometimes migrate toward one of the two vertical focusing positions. Note that buoyancy matching as described in Section 2 was not utilized. Instead, the syringe pump was physically tilted to allow settling of particles toward the syringe outlet, leading to a high concentration of particles during the run.
An experimental run illustrating the need for proper particle and cell concentrations is shown in Figure 4. Without full ordering, localized groups of particles order and are encapsulated, but many drops are without particles. A histogram shows the decreased encapsulation efficiency for the desired two particle encapsulation.
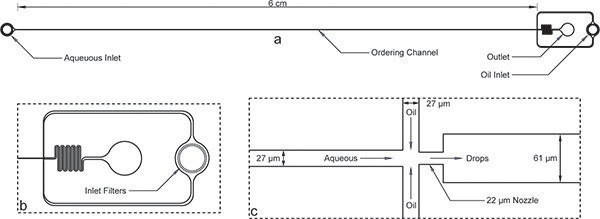 Figure 1. Encapsulation device. a) Overall device with inlets, outlet, and long ordering channel. The device height is 52 μm and the ordering channel width is 27 μm. b) Both aqueous and oil inlets have large debris filters with gaps on the order of the ordering channel width for the enlarged view of the oil inlet. c)The enlarged nozzle view shows equal channel widths of 27 μm for the aqueous and oil channels, followed by the nozzle contraction of 22 μm and sudden expansion to a wider 61 μm channel. Note that the dimensions of the device shown here have been verified using a profilometer after microfabrication and differ slightly from the nominal dimensions on the mask. A true image of the ordering channel and nozzle are available online as Supplemental Figure 1. The AutoCAD mask file has also been included online as a supplement to this manuscript.
Figure 1. Encapsulation device. a) Overall device with inlets, outlet, and long ordering channel. The device height is 52 μm and the ordering channel width is 27 μm. b) Both aqueous and oil inlets have large debris filters with gaps on the order of the ordering channel width for the enlarged view of the oil inlet. c)The enlarged nozzle view shows equal channel widths of 27 μm for the aqueous and oil channels, followed by the nozzle contraction of 22 μm and sudden expansion to a wider 61 μm channel. Note that the dimensions of the device shown here have been verified using a profilometer after microfabrication and differ slightly from the nominal dimensions on the mask. A true image of the ordering channel and nozzle are available online as Supplemental Figure 1. The AutoCAD mask file has also been included online as a supplement to this manuscript.
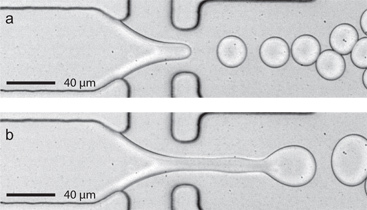 Figure 2. Hysteresis of a dripping to jetting transition using a wider device (80 μm wide x 22 μm high). a) At constant FC-40 flow rate (Qoil = 45 μL/min), steady drop formation occurs at 10 kHz using an aqueous flow rate Qaq = 8 μL/min. As the aqueous flow rate is slowly increased to 10 μL/min, jetting of the aqueous fluid stream is triggered. b) When the flow rate is returned to 8 μL/min jetting continues. Note that steady drop formation can be re-established by briefly pausing the aqueous flow pump (a 1 second pause is typical).
Figure 2. Hysteresis of a dripping to jetting transition using a wider device (80 μm wide x 22 μm high). a) At constant FC-40 flow rate (Qoil = 45 μL/min), steady drop formation occurs at 10 kHz using an aqueous flow rate Qaq = 8 μL/min. As the aqueous flow rate is slowly increased to 10 μL/min, jetting of the aqueous fluid stream is triggered. b) When the flow rate is returned to 8 μL/min jetting continues. Note that steady drop formation can be re-established by briefly pausing the aqueous flow pump (a 1 second pause is typical).
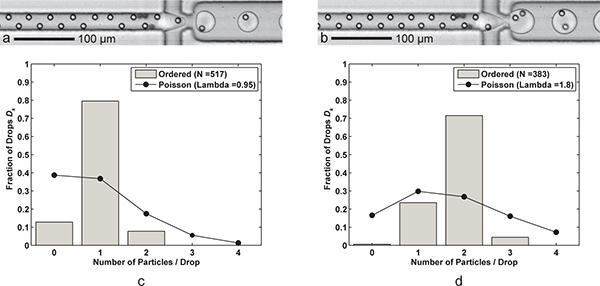 Figure 3. Single- and double- particle encapsulation. a) Drop formation with one cell per drop (Qoil = 60 μL/min, Qaq = 9 μL/min) with a drop generation rate of 6.1 kHz, average drop size of 24.4 pL, and a single-cell capture efficiencies Dk = 79.5% and Pk = 83.7% (λ = 0.95) for a sample size of nd = 517 drops and np = 491 particles. b) Drop formation with two cells per drop is achieved simply by reducing the FC-40 flow rate Qoil to 30 μL/min. The larger (39.8 pL) drops are formed at a rate of 3.8 kHz with a two-cell capture efficiency Dk = 71.5% and Pk = 79.5% (λ = 1.80) for a sample size of nd = 383 drops and np = 689 particles. c-d) Two histograms compare the drop encapsulation particle efficiencies Dk of ordered single- and double- particle encapsulation with Poisson statistics (random encapsulation). Note that for both cases, particle spacing in the direction of flow is about 17-18 μm for fully ordered, alternating particles. Supplemental videos showing both single- and double- particle encapsulation are available online. Click here to view Supplemental Movie 3a. Click here to view Supplemental Movie 3b.
Figure 3. Single- and double- particle encapsulation. a) Drop formation with one cell per drop (Qoil = 60 μL/min, Qaq = 9 μL/min) with a drop generation rate of 6.1 kHz, average drop size of 24.4 pL, and a single-cell capture efficiencies Dk = 79.5% and Pk = 83.7% (λ = 0.95) for a sample size of nd = 517 drops and np = 491 particles. b) Drop formation with two cells per drop is achieved simply by reducing the FC-40 flow rate Qoil to 30 μL/min. The larger (39.8 pL) drops are formed at a rate of 3.8 kHz with a two-cell capture efficiency Dk = 71.5% and Pk = 79.5% (λ = 1.80) for a sample size of nd = 383 drops and np = 689 particles. c-d) Two histograms compare the drop encapsulation particle efficiencies Dk of ordered single- and double- particle encapsulation with Poisson statistics (random encapsulation). Note that for both cases, particle spacing in the direction of flow is about 17-18 μm for fully ordered, alternating particles. Supplemental videos showing both single- and double- particle encapsulation are available online. Click here to view Supplemental Movie 3a. Click here to view Supplemental Movie 3b.
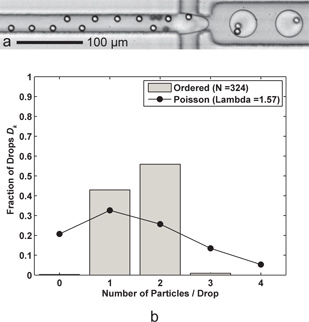 Figure 4. Concentration greatly affects encapsulation efficiency. a) As the concentration decreases, full ordering does not occur, and thus "holes" in the trains emerge, leaving some drops with fewer than anticipated particles. b) The histogram shows the decreased efficiency (Dk = 55.9%, Pk = 70.9%) for two-particle encapsulation due to a lower value of λ = 1.57 where there are nearly as many single-particle drops as there are double-particle drops. This figure results from Qoil = 30 μL/min and Qaq = 9 μL/min, the same flow conditions as for Figure 3b. A representative supplemental video is available online. Click here to view Supplemental Movie 4.
Figure 4. Concentration greatly affects encapsulation efficiency. a) As the concentration decreases, full ordering does not occur, and thus "holes" in the trains emerge, leaving some drops with fewer than anticipated particles. b) The histogram shows the decreased efficiency (Dk = 55.9%, Pk = 70.9%) for two-particle encapsulation due to a lower value of λ = 1.57 where there are nearly as many single-particle drops as there are double-particle drops. This figure results from Qoil = 30 μL/min and Qaq = 9 μL/min, the same flow conditions as for Figure 3b. A representative supplemental video is available online. Click here to view Supplemental Movie 4.
Discussion
Despite relatively high degrees of ordering, not all drops will contain the proper number of particles or cells. Encapsulation efficiency may be calculated as the number of cells or particles that become encapsulated in drops with the desired occupancy divided by their total number. These raw data can be obtained either from an automated high speed video algorithm or from imaging a sample of collected emulsion. This can be compared to the fraction of particles Pk encapsulated in a drop containing k particles and the fraction of drops Dk that would contain k particles. From Figure 3, both the single and double particle encapsulation efficiencies outperform random encapsulation efficiencies by over a factor of two and greatly reduce the number of drops with more than the desired number of particles. Figure 4 demonstrates the need for proper concentrations for high efficiency; that is, λ, a function of both particle concentration and drop volume, should be equal or close to the number of desired cells per drop to maximize correctly-encapsulated particles or cells. Note that a higher concentration of particles or cells is usually a good thing for full ordering as dense trains tend to spread out over time and fill emptier regions between trains. On the other hand, if concentration is too high, the high number of particles may cause interfacial instabilities that induce jetting at the nozzle. In specific studies (such as single-cell encapsulation, for example), it may be more advantageous to avoid multiple-cell droplets at the expense of introducing a few more empty droplets, so a slightly lower λ will be desired. This would also apply for studies aimed at interactions between two cells or between a cell and a particle, where single-particle or single-cell droplets are more tolerable than droplets with two or more of one kind of cell or particle.
Maintaining a constant λ over time is critical for consistent encapsulation. Buoyancy matching assists in long-term concentration control by reducing settling of cells and particles in the syringe and tubing. However, buoyancy matching also results in a higher aqueous viscosity that may delay ordering (resulting in longer focusing channel requirements), increase the channel pressure drop, and change the flow rates required for drop generation. An alternative to buoyancy matching used in this experiment is to physically tilt the syringe pump so that the syringe outlet is pointed almost vertically downward (to minimize adhesion of cells or particles to the syringe interior). Here, we used 9.9 μm diameter microspheres with a particle volume fraction of 1.3% (approximately 25 million particles per mL), but we utilized tilting to increase volume fractions to 2% for the data shown in Figure 3. A second alternative is to mix the aqueous fluid intermittently with an enclosed stainless steel ball bearing (Teflon coated for working with cells) using a small external magnet. Care is required however to avoid letting the ball bearing settle to the syringe tip where it may occlude the entrance to the inlet tubing. However, these alternatives are more labor intensive and less repeatable than buoyancy matching, so buoyancy matching is most suitable for larger scale experiments occurring over long time frames. While inertial ordering requires high Re and Rep to operate, when the aqueous and oil flows are pushed higher and higher, steady dripping of drops turns to jetting14 (see Figure 2) and uncontrolled encapsulation results. For cells smaller than the 10 μm particles used here, smaller channel dimensions may be required to achieve sufficient Rep if the flow rates cannot be increased without jetting. One peculiarity of jetting in microfluidic systems is that hysteresis effects can occur which make it difficult to stop jetting by simply lowering the aqueous flow rate once it occurs back to a point where it was not observed. Based on experimental results, one could develop a dimensional or non-dimensional dripping to jetting flow map like those previously developed for axial co-flowing nozzles14 and T-junctions26-28 with additional contours for drop generation rate, cells per drop, and encapsulation efficiency. This map would provide a robust roadmap from which the drop generation rate can be predicted to calculate λ and thus provide an estimated flow rate for water and oil streams a priori.
While not directly demonstrated here, additional reductions in oil flow rate Qoil from those presented in Figure 3b would further increase the number of particles per drop to three, four, and so on. To achieve more particles per drop, either Qoil must decrease or the aqueous flow rate Qaq must increase. As an aside, we have included an online supplemental MATLAB script which models the encapsulation efficiency of capturing any number of particles in drops. The user inputs the average particle spacing and an particle spacing standard deviation, which models the degree of ordering. For ordered trains, the standard deviation will be small. Additionally, the user inputs the average drop size and drop size standard deviation, which accounts for the polydispersity of drop sizes. Refer to the script documentation for additional information.
When increasing the aqueous flow rate or decreasing the oil flow rate to increase the number of particles or cells per drop, the risk of unstable jetting increases as the respective flow rates near extreme values. Thus, the maximum number of achievable particles/cells per drop will depend on device geometry and fluid properties. Given the particle/cell concentration and oil flow rate, the number of particles/cells per drop is constrained by upper limits on aqueous flow rates, which must be large enough to induce ordering but must be small enough to avoid unstable jetting (and limit shear stresses on cells to ensure viability). Alternatively, given an aqueous flow rate at which ordering occurs, the oil flow rate must remain sufficiently large enough to remain in the dripping regime.
Note that drop generation and the dripping to jetting transition are very sensitive to surfactant concentration. High surfactant concentrations increase the viscosity of the oil, changing the drop generation parameters. As an aside, the scarcity of widely available biocompatible surfactants for fluorocarbon oils presents a major challenge. Currently, one commercial supplier (RainDance Technologies) exists for PFPE-PEG block copolymer surfactants,25 but studies demonstrate small-scale synthesis techniques of a number of surfactant groups such as PFPE-HEG.29,30 Alternatives such as light mineral oil have been utilized in biological drop generation applications to access a wider range of available surfactants,24,31 but note that the accompanying increase in viscosity as compared to fluorocarbon oil alters the drop generation parameters. A recent review32 describes a large number of published continuous phase oils and surfactants.
Disclosures
JE is an inventor on a pending patent based on the technology utilized in this manuscript.
Acknowledgments
We thank RainDance Technologies for the sample of PFPE-PEG surfactant utilized in this study, and we thank the BioMEMS Resource Center (Mehmet Toner, director) for the silicon wafer mold used to create PDMS channel replicas.
References
- Zagnoni M, Lain GLe, Cooper JM. Electrocoalescence mechanisms of microdroplets using localized electric fields in microfluidic channels. Langmuir : the ACS journal of surfaces and colloids. 2010;26:14443–14449. doi: 10.1021/la101517t. [DOI] [PubMed] [Google Scholar]
- Niu XZ, Gielen F, Edel JB, deMello AJ. A microdroplet dilutor for high-throughput screening. Nat. Chem. 2011;3:437–442. doi: 10.1038/nchem.1046. [DOI] [PubMed] [Google Scholar]
- Vincent ME, Liu W, Haney EB, Ismagilov RF. Microfluidic stochastic confinement enhances analysis of rare cells by isolating cells and creating high density environments for control of diffusible signals. Chemical Society reviews. 2010;39:974–984. doi: 10.1039/b917851a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebner A. Quantitative detection of protein expression in single cells using droplet microfluidics. Chemical communications. 2007. pp. 1218–1220. [DOI] [PubMed]
- Love JC, Ronan JL, Grotenbreg GM, van der Veen AG, Ploegh HL. A microengraving method for rapid selection of single cells producing antigen-specific antibodies. Nature biotechnology. 2006;24:703–707. doi: 10.1038/nbt1210. [DOI] [PubMed] [Google Scholar]
- Bradshaw EM. Concurrent detection of secreted products from human lymphocytes by microengraving: Cytokines and antigen-reactive antibodies. Clin. Immunol. 2008;129:10–18. doi: 10.1016/j.clim.2008.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WS, Kim HJ, Lucchetta EM, Du WB, Ismagilov RF. Isolation, incubation, and parallel functional testing and identification by FISH of rare microbial single-copy cells from multi-species mixtures using the combination of chemistrode and stochastic confinement. Lab on a chip. 2009;9:2153–2162. doi: 10.1039/b904958d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boedicker JQ, Li L, Kline TR, Ismagilov RF. Detecting bacteria and determining their susceptibility to antibiotics by stochastic confinement in nanoliter droplets using plug-based microfluidics. Lab on a chip. 2008;8:1265–1272. doi: 10.1039/b804911d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster S. Drop-based microfluidic devices for encapsulation of single cells. Lab on a chip. 2008;8:1110–1115. doi: 10.1039/b802941e. [DOI] [PubMed] [Google Scholar]
- Kelly RT, Page JS, Marginean I, Tang K, Smith RD. Dilution-free analysis from picoliter droplets by nano-electrospray ionization mass spectrometry. Angew Chem. Int. Ed. Engl. 2009;48:6832–6835. doi: 10.1002/anie.200902501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J, deMello AJ, Jayasinghe SN. Bio-electrospraying and droplet-based microfluidics: control of cell numbers within living residues. Biomedical materials. 2010;5:21001. doi: 10.1088/1748-6041/5/2/021001. [DOI] [PubMed] [Google Scholar]
- Edd JF. Controlled encapsulation of single-cells into monodisperse picolitre drops. Lab on a chip. 2008;8:1262–1264. doi: 10.1039/b805456h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anna SL, Bontoux N, Stone HA. Formation of dispersions using "flow focusing" in microchannels. Applied Physics Letters. 2003;82:364. [Google Scholar]
- Utada A, Fernandez-Nieves A, Stone H, Weitz D. Dripping to Jetting Transitions in Coflowing Liquid Streams. Physical Review Letters. 2007;99 doi: 10.1103/PhysRevLett.99.094502. [DOI] [PubMed] [Google Scholar]
- Chabert M, Viovy JL. Microfluidic high-throughput encapsulation and hydrodynamic self-sorting of single cells. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:3191–3196. doi: 10.1073/pnas.0708321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segrí G, Silberberg A. Radial Particle Displacements in Poiseuille Flow of Suspensions. Nature. 1961;189:209–210. [Google Scholar]
- Carlo DDi. Inertial microfluidics. Lab on a chip. 2009;9:3038–3046. doi: 10.1039/b912547g. [DOI] [PubMed] [Google Scholar]
- Carlo DDi, Edd J, Humphry K, Stone H, Toner M. Particle Segregation and Dynamics in Confined Flows. Physical Review Letters. 2009;102 doi: 10.1103/PhysRevLett.102.094503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphry KJ, Kulkarni PM, Weitz DA, Morris JF, Stone HA. Axial and lateral particle ordering in finite Reynolds number channel flows. Physics of Fluids. 2010;22:081703. [Google Scholar]
- Lee W, Amini H, Stone HA, Carlo DDi. Dynamic self-assembly and control of microfluidic particle crystals. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:22413, 22418. doi: 10.1073/pnas.1010297107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy DC, McDonald JC, Schueller OJA, Whitesides GM. Rapid prototyping of microfluidic systems in poly(dimethylsiloxane. Anal. Chem. 1998;70:4974–4984. doi: 10.1021/ac980656z. [DOI] [PubMed] [Google Scholar]
- Kotz K, Cheng X, Toner M. PDMS Device Fabrication and Surface Modification. J. Vis. Exp. 2007. p. e319. [DOI] [PMC free article] [PubMed]
- Haubert K, Drier T, Beebe D. PDMS bonding by means of a portable, low-cost corona system. Lab on a chip. 2006;6:1548–1549. doi: 10.1039/b610567j. [DOI] [PubMed] [Google Scholar]
- Hatch AC. 1-Million droplet array with wide-field fluorescence imaging for digital PCR. Lab on a chip. 2011. pp. 3838–3845. [DOI] [PubMed]
- Holtze C. Biocompatible surfactants for water-in-fluorocarbon emulsions. Lab on a chip. 2008;8:1632–1639. doi: 10.1039/b806706f. [DOI] [PubMed] [Google Scholar]
- Garstecki P, Stone H, Whitesides G. Mechanism for Flow-Rate Controlled Breakup in Confined Geometries: A Route to Monodisperse Emulsions. Physical Review Letters. 2005;94 doi: 10.1103/PhysRevLett.94.164501. [DOI] [PubMed] [Google Scholar]
- Garstecki P, Fuerstman MJ, Stone HA, Whitesides GM. Formation of droplets and bubbles in a microfluidic T-junction-scaling and mechanism of break-up. Lab on a chip. 2006;6:437–446. doi: 10.1039/b510841a. [DOI] [PubMed] [Google Scholar]
- Nie Z. Emulsification in a microfluidic flow-focusing device: effect of the viscosities of the liquids. Microfluidics and Nanofluidics. 2008.
- Holt DJ, Payne RJ, Chow WY, Abell C. Fluorosurfactants for microdroplets: interfacial tension analysis. Journal of colloid and interface science. 2010;350:205–211. doi: 10.1016/j.jcis.2010.06.036. [DOI] [PubMed] [Google Scholar]
- Holt DJ, Payne RJ, Abell C. Synthesis of novel fluorous surfactants for microdroplet stabilisation in fluorous oil streams. Journal of Fluorine Chemistry. 2010;131:398–407. [Google Scholar]
- Hatch AC, Fisher JS, Pentoney SL, Yang DL, Lee AP. Tunable 3D droplet self-assembly for ultra-high-density digital micro-reactor arrays. Lab on a chip. 2011;11:2509–2517. doi: 10.1039/c0lc00553c. [DOI] [PubMed] [Google Scholar]
- Baret JC. Surfactants in droplet-based microfluidics. Lab on a chip. 2012;12:422–433. doi: 10.1039/c1lc20582j. [DOI] [PubMed] [Google Scholar]


