Abstract
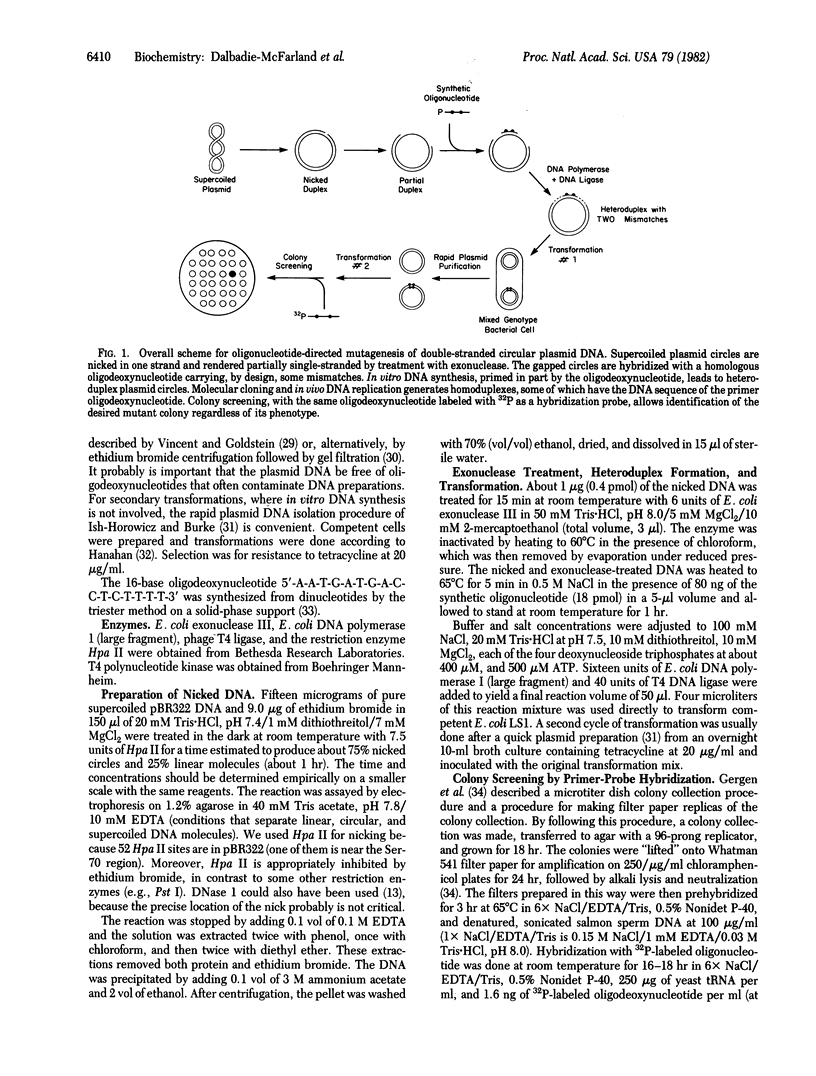
We have used oligonucleotide-directed mutagenesis to make a specific change in the beta-lactamase (EC 3.5.2.6) (ampicillin resistance) gene of the plasmid pBR322. Evidence suggests that the active site for this enzyme may include a serine-threonine dyad (residues 70 and 71). By priming in vitro DNA synthesis with a chemically synthesized 16-base oligodeoxyribonucleotide, we have inverted the Ser-Thr dyad to Thr-Ser and thereby generated a mutant with an ampicillin-sensitive phenotype. This "double-mismatch" method is relatively simple and also very general because detection of mutants is at the level of DNA and involves only colony hybridization. Accordingly, the procedure can be applied to any DNA sequence and does not depend on the phenotype of the mutant.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ambler R. P. The structure of beta-lactamases. Philos Trans R Soc Lond B Biol Sci. 1980 May 16;289(1036):321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Cohen S. A., Pratt R. F. Inactivation of Bacillus cereus beta-lactamase I by 6 beta-bromopencillanic acid: mechanism. Biochemistry. 1980 Aug 19;19(17):3996–4003. doi: 10.1021/bi00558a017. [DOI] [PubMed] [Google Scholar]
- Fisher J., Belasco J. G., Khosla S., Knowles J. R. beta-Lactamase proceeds via an acyl-enzyme intermediate. Interaction of the Escherichia coli RTEM enzyme with cefoxitin. Biochemistry. 1980 Jun 24;19(13):2895–2901. doi: 10.1021/bi00554a012. [DOI] [PubMed] [Google Scholar]
- Gergen J. P., Stern R. H., Wensink P. C. Filter replicas and permanent collections of recombinant DNA plasmids. Nucleic Acids Res. 1979 Dec 20;7(8):2115–2136. doi: 10.1093/nar/7.8.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillam S., Astell C. R., Smith M. Site-specific mutagenesis using oligodeoxyribonucleotides: isolation of a phenotypically silent phi X174 mutant, with a specific nucleotide deletion, at very high efficiency. Gene. 1980 Dec;12(1-2):129–137. doi: 10.1016/0378-1119(80)90023-2. [DOI] [PubMed] [Google Scholar]
- Gillam S., Jahnke P., Astell C., Phillips S., Hutchison C. A., 3rd, Smith M. Defined transversion mutations at a specific position in DNA using synthetic oligodeoxyribonucleotides as mutagens. Nucleic Acids Res. 1979 Jul 11;6(9):2973–2985. doi: 10.1093/nar/6.9.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillam S., Smith M. Site-specific mutagenesis using synthetic oligodeoxyribonucleotide primers: II. In vitro selection of mutant DNA. Gene. 1979 Dec;8(1):99–106. doi: 10.1016/0378-1119(79)90010-6. [DOI] [PubMed] [Google Scholar]
- Hall A., Knowles J. R. Directed selective pressure on a beta-lactamase to analyse molecular changes involved in development of enzyme function. Nature. 1976 Dec 23;264(5588):803–804. doi: 10.1038/264803a0. [DOI] [PubMed] [Google Scholar]
- Hutchison C. A., 3rd, Phillips S., Edgell M. H., Gillam S., Jahnke P., Smith M. Mutagenesis at a specific position in a DNA sequence. J Biol Chem. 1978 Sep 25;253(18):6551–6560. [PubMed] [Google Scholar]
- Ish-Horowicz D., Burke J. F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981 Jul 10;9(13):2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Ike Y., Ikuta S., Itakura K. Solid phase synthesis of polynucleotides. VI. Further studies on polystyrene copolymers for the solid support. Nucleic Acids Res. 1982 Mar 11;10(5):1755–1769. doi: 10.1093/nar/10.5.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott-Hunziker V., Orlek B. S., Sammes P. G., Waley S. G. 6 beta-Bromopenicillanic acid inactivates beta-lactamase I. Biochem J. 1979 Jan 1;177(1):365–367. doi: 10.1042/bj1770365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott-Hunziker V., Waley S. G., Orlek B. S., Sammes P. G. Penicillinase active sites: labelling of serine-44 in beta-lactamase I by 6beta-bromopenicillanic acid. FEBS Lett. 1979 Mar 1;99(1):59–61. doi: 10.1016/0014-5793(79)80248-3. [DOI] [PubMed] [Google Scholar]
- Koshland D., Botstein D. Secretion of beta-lactamase requires the carboxy end of the protein. Cell. 1980 Jul;20(3):749–760. doi: 10.1016/0092-8674(80)90321-9. [DOI] [PubMed] [Google Scholar]
- Kudo I., Leineweber M., RajBhandary U. L. Site-specific mutagenesis on cloned DNAs: generation of a mutant of Escherichia coli tyrosine suppressor tRNA in which the sequence G-T-T-C corresponding to the universal G-T-pseudouracil-C sequence of tRNAs is changed to G-A-T-C. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4753–4757. doi: 10.1073/pnas.78.8.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loosemore M. J., Cohen S. A., Pratt R. F. Inactivation of Bacillus cereus beta-lactamase I by 6 beta-bromopenicillanic acid: kinetics. Biochemistry. 1980 Aug 19;19(17):3990–3995. doi: 10.1021/bi00558a016. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Montell C., Fisher E. F., Caruthers M. H., Berk A. J. Resolving the functions of overlapping viral genes by site-specific mutagenesis at a mRNA splice site. Nature. 1982 Feb 4;295(5848):380–384. doi: 10.1038/295380a0. [DOI] [PubMed] [Google Scholar]
- Müller W., Weber H., Meyer F., Weissmann C. Site-directed mutagenesis in DNA: generation of point mutations in cloned beta globin complementary dna at the positions corresponding to amino acids 121 to 123. J Mol Biol. 1978 Sep 15;124(2):343–358. doi: 10.1016/0022-2836(78)90303-0. [DOI] [PubMed] [Google Scholar]
- Norgard M. V. Rapid and simple removal of contaminating RNA from plasmid DNA without the use of RNase. Anal Biochem. 1981 May 1;113(1):34–42. doi: 10.1016/0003-2697(81)90040-3. [DOI] [PubMed] [Google Scholar]
- Pratt R. F., Loosemore M. J. 6-beta-bromopenicillanic acid, a potent beta-lactamase inhibitor. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4145–4149. doi: 10.1073/pnas.75.9.4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A., Hirose T., Itakura K., Riggs A. D. Efficient correction of a mutation by use of chemically synthesized DNA. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4268–4270. doi: 10.1073/pnas.75.9.4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortle D., Grisafi P., Benkovic S. J., Botstein D. Gap misrepair mutagenesis: efficient site-directed induction of transition, transversion, and frameshift mutations in vitro. Proc Natl Acad Sci U S A. 1982 Mar;79(5):1588–1592. doi: 10.1073/pnas.79.5.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortle D., Koshland D., Weinstock G. M., Botstein D. Segment-directed mutagenesis: construction in vitro of point mutations limited to a small predetermined region of a circular DNA molecule. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5375–5379. doi: 10.1073/pnas.77.9.5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortle D., Nathans D. Local mutagenesis: a method for generating viral mutants with base substitutions in preselected regions of the viral genome. Proc Natl Acad Sci U S A. 1978 May;75(5):2170–2174. doi: 10.1073/pnas.75.5.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons G. F., Veeneman G. H., Konings R. N., van Boom J. H., Schoemakers J. G. Oligonucleotide-directed mutagenesis of gene IX of bacteriophage M13. Nucleic Acids Res. 1982 Feb 11;10(3):821–832. doi: 10.1093/nar/10.3.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3737–3741. doi: 10.1073/pnas.75.8.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent W. S., 3rd, Goldstein E. S. Rapid preparation of covalently closed circular DNA by acridine yellow affinity chromatography. Anal Biochem. 1981 Jan 1;110(1):123–127. doi: 10.1016/0003-2697(81)90121-4. [DOI] [PubMed] [Google Scholar]
- Wallace R. B., Johnson P. F., Tanaka S., Schöld M., Itakura K., Abelson J. Directed deletion of a yeast transfer RNA intervening sequence. Science. 1980 Sep 19;209(4463):1396–1400. doi: 10.1126/science.6997991. [DOI] [PubMed] [Google Scholar]