Summary
The c-Myc HLH-bZIP protein has been implicated in physiological or pathological growth, proliferation, apoptosis, metabolism and differentiation at the cellular, tissue or organismal levels via regulation of numerous target genes. No principle yet unifies Myc action due partly to an incomplete inventory and functional accounting of Myc’s targets. To observe Myc target expression and function in a system where Myc is temporally and physiologically regulated, the transcriptomes and the genome-wide distributions of Myc, RNA polymerase II and chromatin modifications were compared during lymphocyte activation and in ES cells as well. A remarkably simple rule emerged from this quantitative analysis: Myc is not an on-off specifier of gene activity, but is a non-linear amplifier of expression, acting universally at active genes, except for immediate early genes that are strongly induced before Myc. This rule of Myc action explains the vast majority of Myc biology observed in literature.
Introduction
The c-Myc oncogene, identified three decades ago, is associated with many human cancers (Dang, 2010; Wasylishen and Penn, 2010). Numerous chromatin and transcription regulating factors interact with Myc (Cheng et al., 1999; Cowling and Cole, 2006; Eilers and Eisenman, 2008; Rahl et al., 2010; Wasylishen and Penn, 2010). mRNA expression and DNA-binding studies, in vitro and in vivo, have nominated an ever increasing number of genes as Myc targets including a core constituting a Myc signature (Ji et al., 2011; Margolin et al., 2009; Shaffer et al., 2006; Wasylishen and Penn, 2010). However, no single subset of Myc targets accounts for its oncogenic activity (Berns et al., 2000; Nikiforov et al., 2002); the diversity of Myc targets between systems, has further confounded the explication of discrete, linear pathway(s) for Myc-driven neoplasia.
Myc is often associated with cell activation. Typically a pulse of Myc is induced starting from a very low baseline during the G0–G1 transition or in response to numerous signals and stresses (Rabbitts et al., 1985). Thereafter, in steady-state cycling cells, c-myc output is stably maintained. In some settings, a second Myc peak ensues 12–24 hrs later (Kelly et al., 1983; Nepveu et al., 1987; Tonini et al., 1987). The relationship between Myc targets in these primary and secondary peaks has not been investigated. Although Myc pathology has been extensively studied in lymphoid neoplasms including Burkitt lymphoma, large cell lymphoma, multiple myeloma and plasmacytoma, Myc action in primary lymphocytes, has been less studied making it difficult to compare the physiological versus pathological Myc networks. Since most cancer lines or transgenic models do not recapitulate the physiologic regulation of Myc expression (Levens, 2010), we decided to investigate Myc function in primary lymphocytes using a mouse line that fuses endogenous Myc to EGFP. The Myc network was then interrogated in related, but physiologically distinct situations, and the profiles of global gene expression and of Myc-binding to its target genes were examined. The genome-wide patterns of Myc-recruitment, RNA polymerase binding and chromatin modifications were overlaid to reveal the dynamics of Myc up-regulation and its relationship to lymphocyte gene expression. These same genome-wide patterns were assessed in ES-cells to gain insight into the cell-type and differentiation specific roles of c-Myc. Putting these data together revealed that physiologically, Myc is not an on-off specifier of a particular transcriptional program(s), but is a universal amplifier of gene expression increasing output at all active promoters. This rule predicts and explains many features of Myc biology.
Results
A model to study physiological Myc function
EGFP was homologously recombined with c-myc exon 3 in mouse ES cells (Figure S1A) to provide a tag for c-Myc-immunoprecipitation and to monitor c-Myc levels in living cells. This chimera preserves all known regulatory and structural features of the endogenous c-myc gene (Liu and Levens, 2006), including the multiple transcription and translation start sites [unlike NH2-terminal fusion (Huang et al., 2008)], miRNA binding sites and 3′-UTR (Ingolia et al., 2011; Liu and Levens, 2006; Sampson et al., 2007). EGFP provided a well-characterized and efficient tag for ChIP (Poser et al., 2008) without compromising any surfaces that might interact with c-Myc’s many partners (Agrawal et al., 2010).
This c-Myc-EGFP cooperated with RAS to transform cells (Land et al., 1983) (Figure S1B) similar to the unmodified protein and had the same short half-life (Hann and Eisenman, 1984). Crosses between mice generated from ES cells heterozygous for this allele yielded unremarkable Myc-EGFP homozygotes that bred without difficulty indicating that the fusion protein functions properly from embryonic development through adulthood.
Immunoblots of mouse embryonic fibroblasts wild-type, heterozygous or homozygous for c-Myc-EGFP probed with anti-GFP or anti-c-Myc (Figures S1C-F), displayed the expected patterns. A pulse of nuclear fluorescence occurred when serum-starved Myc-EGFP MEFs were re-stimulated (Mehmet et al., 1997); fluorescence was exaggerated upon proteasome inhibitor MG132 treatment (Figures S1G&1H). The mean fluorescence intensity of heterozygotes at the population and cellular levels was between that of wild-types and homozygotes proving bi-allelic c-myc expression (Figure S1H).
c-Myc-EGFP activation in lymphocytes
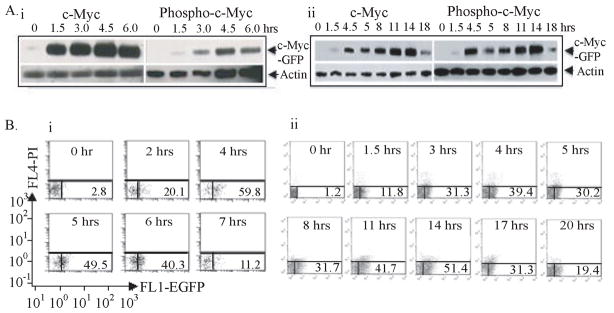
To observe the interplay of Myc with the factors and pathways activating lymphocytes, purified B- or T-cells were stimulated with lipopolysaccharide (LPS) or concanavalin A, respectively and monitored for EGFP-fluorescence, and total or phosphorylated c-Myc by immunoblotting (Figure 1). Activated B-cells displayed the stereotypical immediate-early peak of Myc (Kelly et al., 1983) accompanied by the T58/S62 phosporylation (Thomas and Tansey, 2011). In T-cells, c-Myc peaked biphasically 4 and 14 hrs post-stimulation (Kelly et al., 1983) by fluorescence and immunoblot; cell-cycle entry followed the second peak. c-Myc levels were somewhat higher in the second peak; both peaks were efficiently phosphorylated. In all respects c-Myc-EGFP lymphocytes functioned normally, so patterns of gene activation and Myc target site selection were compared in resting and stimulated B- and T-cells.
Figure 1. Myc-EGFP expression during lymphocyte activation (see also Figure S1).
(A) Activation and phosphorylation of the c-Myc-EGFP knock-in. Immunoblot analysis of extract from (i) B splenocytes at 0, 1.5, 3.0, 4.5 and 6.0 hours post-LPS activation and (ii) from ConA activated T splenocytes at 0, 1.5, 4, 5, 8, 11, 14 and 18 hours.
(B) Flow cytometric analysis of (i) LPS activated c-Myc GFP/GFP splenic B cells at 0, 2, 4, 5, 6 and 7 hours and (ii) of ConA activated c-Myc GFP/GFP splenic T cells at 0, 1.5, 3, 4, 5, 8, 11, 14, 17 and 20 hours. The scale and axes are indicated in the left bottom corner.
Quiescent B-cells (B0) were treated with LPS for 4 hrs (B4), and resting T-cells (T0) were activated with conA for 4 hrs (T4) and for 14 hrs (T14). RNA was harvested and chromatin was prepared at these times. RNA was analyzed by hybridization with Affymetrix microarrays, and Myc-EGFP-bound chromatin, immunoprecipitated with anti-EGFP was analyzed by ChIP-Seq. Chromatin immunoprecipitation with antibodies against a variety of post-translational histone modifications and RNA polymerase was performed on quiescent and activated B-cells.
Conventional analysis fails to define a general principle for Myc action
Because Myc regulates various synthetic and metabolic processes, its binding was expected at genes that must be differentially induced during the G0 to G1/S transition, for example genes involved in RNA and protein biosynthesis, cell-cycle regulation and metabolism. Because the spectrum of Myc-action is known to be complex and idiosyncratic in different systems, strict criteria were applied in an attempt to highlight core Myc-targets while minimizing false positives. Myc-binding sites were assessed using SICER and MACs (Zang et al., 2009; Zhang et al., 2008); the former algorithm identifies broad regions of factor binding, whereas the latter finds sharp peaks; with stringent thresholds their union predicts 8020, 3053 and 6623 peaks in the B4, T4 and T14 datasets, respectively (examples shown in Figure S2A). c-Myc peaks were enriched for E-boxes as expected (Figure S2B). Forty-50% of the Myc B-cell targets overlapped with those from an earlier study of Burkitt’s lymphoma (Li et al., 2003) (Figure S2C). Gene ontology of c-Myc targets revealed an amalgam of various cellular processes (Figure S2D). A Venn diagram showed that 12–29% of targets were unique to a single sample, while the 1045 universally shared targets constituted 13 to 34% of each sample (Figure S2E). A global inspection of promoters revealed that many sample-specific targets displayed sub-threshold Myc peaks in the other samples underestimating the number of shared targets (Figure S2F). Although largely promoter-associated (Figure S2G), intra- and intergenic Myc-binding sites were not rare. Microarray expression analysis of RNA normalized between datasets, showed responsive genes to be roughly 2/3 up- versus 1/3 down-regulated (Figure S2H); the magnitudes of the relative expression changes of c-Myc-target were modest (Figure S2I). Only 10–20% of total binding targets were up- and 4–10% were down-regulated greater than 2-fold (p<10−5), leaving the importance of c-Myc binding at most genes undefined (Figure S2H). These complex patterns of binding, expression, and function, typical for studies of this oncogene (Chen et al., 2008b; Dang, 2010; Eilers and Eisenman, 2008; Ji et al., 2011), failed to illuminate a principle for Myc action. The discordance of targets between datasets was difficult to rationalize in terms of cell type specificity, stage of activation or function.
In a further attempt to distill the essentials of Myc action, the binding and expression changes amongst a set of genes accepted as bona fide Myc targets were highlighted (Shaffer et al., 2006). This Myc-signature set (defined in human cells) is comprised of Myc-binding genes that tend to be highly expressed (Figure S3A) and whose abundance changes with perturbation of Myc levels in a variety of systems (Ji et al., 2011; Shaffer et al., 2006). Signature genes were highly enriched in the common sectors between the datasets. To test whether Myc-expression is unconditionally sufficient to enforce coherent two-fold up-regulation of its signature genes, binding of Myc at their promoters was compared between B-cells activated for 4 hours versus T-cells activated for 4 or 14 hours (Figure S3B). In B-cells, the responsive signature genes (37/50) were induced as a cohort at 4 hours. In contrast, the signature genes appeared to be induced biphasically in T-cells (26/50 at 4 hrs increasing to 44/50 at 14 hrs. No functional or ontological rationale for the differential early versus late co-regulation of Myc targets in T-cells was evident. Because late-activated targets were collectively less expressed within each of the T0, T4 and T14 datasets, the apparent late specificity might simply reflect the kinetics at which these genes surmount arbitrary experimental thresholds for scoring during a global ramp-up of expression rather than precise temporal switching. Conventional peak calling algorithms might have been biasing the identification of Myc-target genes.
Promoter output is related to c-Myc binding at all active genes
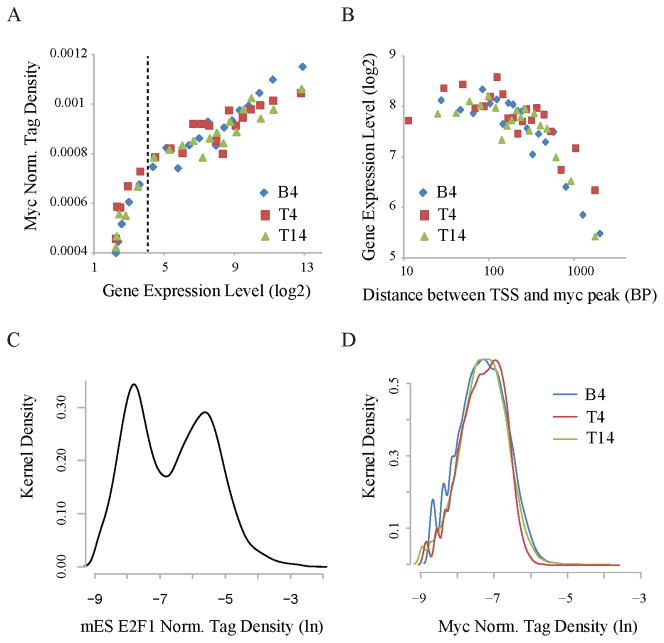
To see if c-Myc-binding related to target output without arbitrary thresholds, all genes were binned according to expression level, and the means of the bins were plotted against promoter-bound c-Myc density. Remarkably, the B4, T4 and T14 datasets each revealed a monotonic linear relationship between the logarithm of the expression and the density of c-Myc binding from the highest expression levels down to background (Figure 2A); no landmark in this plot demarcated boundaries between weak and strong c-Myc-targets. If Myc-binding is functionally proportional to the logarithm of a gene’s expression, then perturbation of Myc levels would preferentially disturb high output promoters. However, the dispersion among the data indicated that other factors modify this relationship at the level of individual promoters (Figure S4A). To ascertain whether the location of the bound c-Myc was one such factor, overall expression levels were plotted against the distance of the bound c-Myc from the TSS revealing that Myc is most closely associated with high output if bound within ~250 bp of the TSS (Figure 2B; Figure S4B). Highly expressed Myc target genes also tended to be associated with the Myc cognate sequence CACGTG, the E-box, although this association was so loose as to preclude a rigid E-box requirement (Figure S4C). Indeed, canonical E-boxes themselves were relatively more restricted to the vicinity of TSSs than were non-canonical E-boxes, although both occurred frequently, as expected for a hexanucleotide (Figure S4D). In principle, a 4/6 match to the canonical E-box would occur every 30 nucleotides in random 50% AT/GC DNA. Empirically, even after restricting wobble to the E-box’s central two C-G base pairs, ~77% and 97% of promoters still have E-boxes within 400 bp and 1000 bp, respectively of the TSS. Degenerate E-boxes are so common as to be essentially ubiquitous.
Figure 2. Myc binds to all promoters according to their outputs (see Figure S2 also).
(A) Myc-binding at promoters strongly correlates with expression. Genes were sorted into 20 equal size bins based on expression levels. The averages of Myc ChIP-Seq tag densities at promoters and expression levels are shown for each bin. Dashed vertical lines separate expressed genes from silent or minimally expressed genes.
(B) Myc-binding is associated with high expression within ~250 bp of the transcription start site (TSS). Myc targets were sorted into 20 equal-size bins based on the distance of TSS to the nearest peak of c-Myc binding. The averages of gene expression levels (y-axis) and distances (x-axis) are shown for each bin.
(C) The distribution of normalized E2F1 ChIP-Seq tag density at mouse ES cell promoters. The y-axis shows the Gaussian kernel density for each tag density point shown in the x-axis.
(D) The distribution of normalized Myc tag density at B4, T4 and T14 promoters.
Myc target selection is more “analog” than “digital”
In principle, sequence-specific transcription factor binding occurs against a background of non-specific binding to the rest of the genome; so transcription factor binding sites should resolve into two populations; a high-affinity, high-occupancy, lower abundance population of specific binding sites versus low affinity, low occupancy, but highly abundant non-specific binding sites comprising the bulk of the genome. In this scenario, a histogram of the amount of transcription factor in promoter regions would be bi-modal due to non-specific versus specific binding as seen with E2F1 (Figure 2C). Other factors, for example GABP-α and CTCF may populate more complex distributions (Figure S4E). In contrast, histograms of Myc-density were unimodal (Figure 2D); the absence of any local minima demarcating low- versus high-density c-Myc binding sites renders arbitrary any threshold selected to discriminate between these two populations. Myc-binding more resembles a continuous, analog process, rather than the binary (digital) switch often observed with other factors (Zhang et al., 2008). Supportive of this notion, relaxing the stringency for peak-calling using SICER returned a much larger number of potentially significant peaks, ranging up to 30–40 thousands at E-value =1000 (Figure S2J), a threshold where the same number of sequence tags, if randomly distributed across unique sequences in the genome would yield ~1000 peaks. Genes bearing canonical E-boxes near promoters populated the leading edge of this unimodal distribution, but failed to separate from non-E box Myc targets (Figure S4F). By expression and by binding, authentic Myc-target genes fail to resolve from the rest of the transcriptome/genome.
If Myc mainly partitions unimodally between target sites, then at reduced levels, Myc should populate this same distribution of peaks, but with reduced amplitude. Alternatively, if there were several classes of Myc sites, only the highest affinity sites would fill when Myc levels are severely restricted. In fact, ChIP-Seq for Myc-EGFP of B0 cells that express very low levels of Myc, revealed a proportional scaling down of all Myc peaks down consistent with a unimodal population of binding sites paralleling expression levels (Figure S4G).
Myc binds to open chromatin
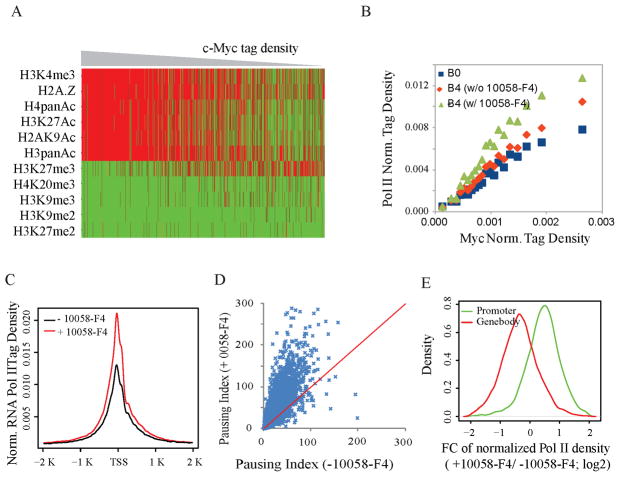
As c-Myc levels rise from baseline to high levels during lymphocyte activation, what features anticipate and dictate its recruitment to promoters? Chromatin immunoprecipation using antibodies against a variety of histone modifications revealed that Myc-binding sites in naïve cells were prefigured with active chromatin marks including H3K4Me3 and H3K27Ac (Figure 3A); conversely Myc was excluded from regions with repressive histone modifications (Figure 3A). At weak binding sites, no set of histone modifications seemed to compel Myc-binding.
Figure 3. Myc is recruited to promoters according to the amount of RNA polymerase II loaded.
(A) Presence (red) and absence (green) of histone markers for heterochromatin (H3K27me2/3, H3K9me2/3 and H4K20me3) and euchromatin (histone acetylation, H2A.Z and H3K4me3) at naive B cell promoters. Genes are sorted by promoter c-Myc tag density. Each line represents a gene. Columns are hierarchically clustered. Only chromosome 1genes are shown.
(B) Correlation between normalized c-Myc ChIP-Seq tag density at promoters of B4 cells and RNA Pol II ChIP-Seq tag density at promoters of B resting cells (B0) or B4 cells, with or without 10058-F4 treatment during LPS activation. The promoters are sorted into 20 equal-size groups based on the c-Myc ChIP-Seq tag densities, and the averages of the two sorts of tag densities are shown for each bin.
(C) Normalized RNA Pol II ChIP-Seq tag density around TSS in B4 cells treated with or without 10059-F4 during B cell LPS activation.
(D) Scatter plot for RNA Pol II pausing index (Muse et al., 2007) in B4 cells treated with and without 10058-F4.
(E) The distribution of fold-change of normalized RNA Pol II ChIP-Seq tag density (with 10058-F4/without 10058-F4) in promoter regions (+/− 2K bps around TSS) and in gene body regions (excluding the first 2K bps).
RNA polymerase loading in resting B-cells anticipates Myc-recruitment after activation
Because c-Myc-binding correlated expression for all genes, we explored the relationship between Myc recruitment and RNA polymerase II (RNAP II) loading. Ranking promoters in naïve B-cells according to the amount of RNAP II loaded, and comparing with the amount of c-Myc subsequently recruited 4 hours post-activation, revealed a surprising relationship: c-Myc was recruited according to the amount of RNA polymerase pre-loaded at these promoters (Figure 3B, blue). Because the gene expression profiles of resting and activated cells are highly correlated with each other (Figure S5) and with RNA polymerase loading, these results indicate that c-Myc is drawn to genes already expressed in resting cells; if so, then Myc would be a global amplifier of all expressed genes and not a switch turning targets on or off. Thus the association noted between c-MYC and polymerase in lymphoma (Li et al., 2003) is found physiologically during B-cell activation and is not due to MYC over-expression in cancer. This same relationship between TSS-bound Myc and promoter-loaded RNA polymerase is maintained at the peak of Myc expression during B-cell activation (Figure 3B; red) indicating that high Myc-levels do not re-specify or redistribute RNA polymerase loading at promoters.
Myc potentiates pause release at all promoters
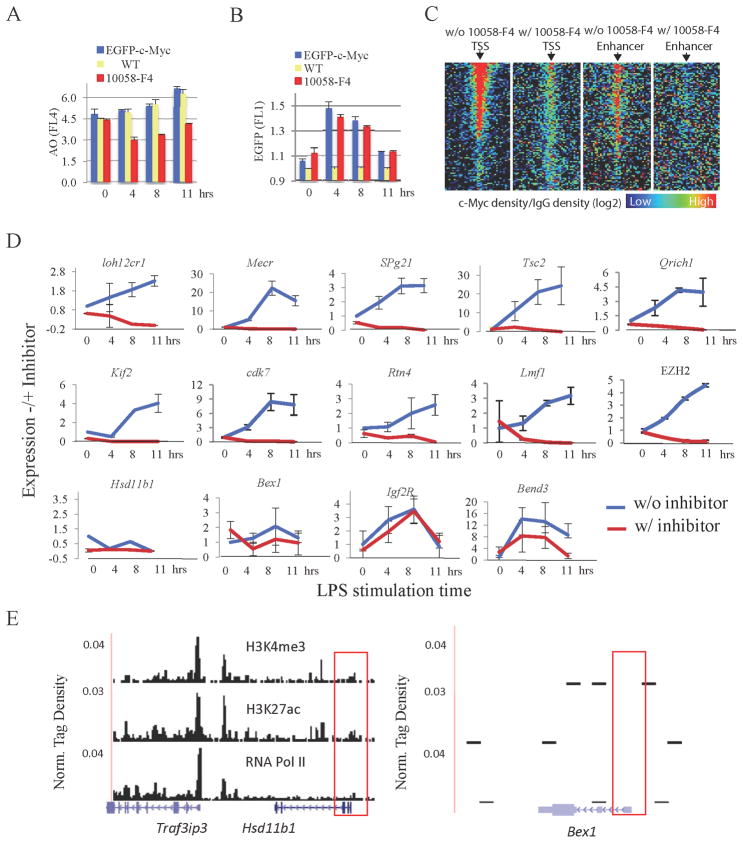
Myc-binding at active, RNA polymerase-loaded promoters might merely reflect access to open, actively transcribed chromatin, but lack biological significance. Or Myc recruited to TSSs might modify the RNA polymerase profile at active promoters. Myc has been reported to facilitate the release of RNA polymerases paused at the promoters of its targets (Eberhardy and Farnham, 2002; Rahl et al., 2010). To test if Myc modifies the RNA polymerase distribution at all active genes, the Myc-Max dimerization inhibitor 10058-F4 (Wang et al., 2007) was applied to cells and the loading of polymerase at promoters and within gene bodies was examined genome-wide. The inhibitor increased RNA polymerase at all active promoters, commensurate with the amount of promoter-associated Myc in uninhibited cells (Figure 3B red vs. green & Figure 3C). There was a general increase in the pausing index (Figure 3D) reflecting both increased RNA polymerase at TSSs and decreased polymerase in gene bodies (Figure 3E) indicating that Myc is a universal potentiator of pause-release at all actively transcribing promoters.
Increased Myc binding at promoters in ES cells is associated with increased RNA polymerase loading and higher promoter output
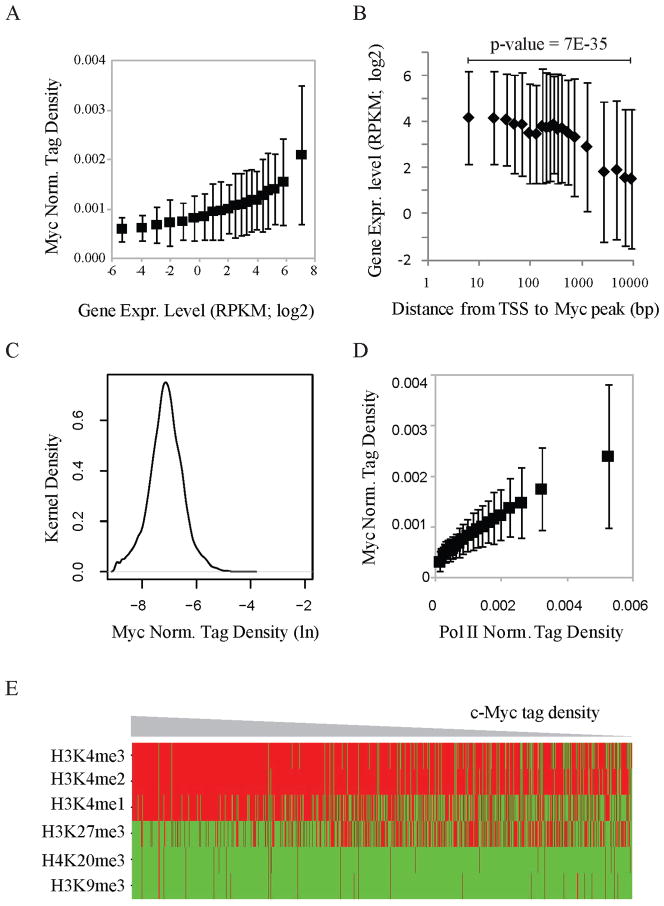
Is the universal association of c-Myc with levels of expression and RNA polymerase II promoter loading specific to the G0–G1 transition and/or lymphocytes or is it general, occurring in other cells and different physiological situations? To address this issue, c-Myc-binding, RNA-expression and RNA polymerase II binding at all transcription start sites were examined in mouse embryonic stem cells (mESCs). Again, c-Myc binding correlated with expression (Figure 4A), and expression inversely correlated with the distance of the Myc-binding site from the TSS (Figure 4B). The upward bow in the graph of expression (logarithm) versus c-Myc-binding (linear) for the most highly expressed genes might suggest that these targets are beginning to saturate (Figure 4A). As with lymphocytes (Figure 2D), the distribution of c-Myc-density at all c-Myc peaks was unimodal (Figure 4C) and c-Myc binding correlated with RNA polymerase II loading at TSSs (Figure 4D). c-Myc was recruited to promoters with histone marks for open chromatin (Figure 4E). That the same associations occurred in lymphocytes and mESCs suggests that c-Myc is preferentially associated with the transcription levels of whatever genes are on in any cell.
Figure 4. The association of Myc with gene expression, Pol II loading, and open chromatin is conserved in mouse ES cells.
(A) Myc binding levels at promoters strongly correlate with gene expression levels (RNA-Seq). Genes were sorted into 20 equal size bins based on gene expression level. Shown for each bin are the averages of Myc ChIP-Seq tag densities (Y-axis) at promoters and of gene expression levels (X-axis).
(B) Myc is associated with high gene expression if bound within ~250 bp of TSS. Data analysis as in Figure 2(B).
(C) The distribution of normalized Myc tag density at promoters in mouse ES cells.
(D) Correlation of Pol II tag densities at promoters and c-Myc ChIP-Seq tag densities at promoters in mouse ES cells. Data analysis as in Figure 3(B).
(E) Presence (red) and absence (green) of histone markers for heterochromatin (H3K27me3, H3K9me3 and H4K20me3) and euchromatin (histone acetylation H3K4me) at promoters from mouse ES cells. Genes are sorted by the c-Myc tag density at promoters. Heat-map as in Figure 3(A).
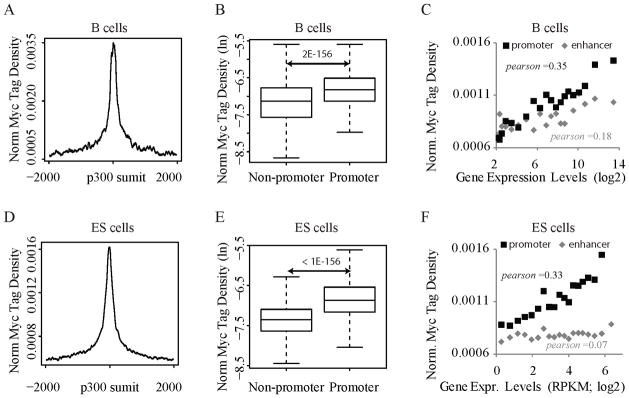
Myc acts more strongly at promoters than enhancers
Because Myc binding at TSSs so closely paralleled expression, we wondered whether this relationship extended to enhancers. c-Myc binding was compared between p300-loaded intergenic enhancers and their associated promoters in B4 activated lymphocytes and in ES cells. In each case c-Myc was preferentially bound at promoters (Figures 5AB, 5D–E). Moreover, the RNA output of those promoters better correlated with TSS- than enhancer-bound c-Myc in both systems (Figures 5C,F). RNA output was especially insensitive to enhancer-bound Myc in mESCs (Figure 5F). The data up to this point suggest that Myc is a transcription amplifier operating at promoters.
Figure 5. Myc prefers promoters over enhancers.
(A) Distribution of Myc ChIP-Seq tag densities (B4 cells) around putative enhancers in resting B cells. Enhancers are defined as p300 binding sites in non-promoter regions.
(B) Myc occupancy (B4 cells) at p300 binding sites (resting B cells) is lower in non-promoter vs. promoter regions.
(C) Enhancer binding correlates weakly with target expression. The gene nearest an enhancer site is defined as its target. p300 binding sites were sorted by target expression levels. For each group, the average Myc tag density near enhancers (+/− 2K bps) is plotted vs. the average target expression level. The correlation between the levels of Myc enhancer binding and target gene expression was measured by Pearson Coefficient r, in which +1 means a perfect correlation, −1 perfect negative anti-correlation, and 0 no correlation. As a positive control, Myc binding levels at promoters of target genes are plotted against their expression levels for each group.
(D), (E), and (F) are similar to (A), (B), (C), respectively, except that the data analysis was done for mouse ES cells.
Myc increases cellular RNA content
Is Myc an amplifier of all expressed genes? If so, then cells with more Myc should make more of the RNAs present before Myc up-regulation. Importantly, global amplification of RNA expression would be missed in studies comparing equal amounts of RNA versus equal cell-equivalents of RNA. Several approaches were used to explore the relationship of Myc with total cellular RNA and mRNA content.
First, transcriptomes were compared across a time course of B-cell activation. Despite dramatic changes in cell size (Figure S6A) and increases in mRNA and total RNA of 350% and 200% per cell, respectively, by 11 hrs (and up to 8.5-fold each at 48 hrs, data not shown) (Figures S6B–D), transcriptomes were similarly composed at all time points (Figure S5; left most panel). Total RNA levels in splenic B-cells activated with LPS from c-Myc-EGFP homozygous or wild-type mice were monitored by flow cytometry using acridine orange fluorescence (James and Eisenman, 2002). Pretreatment of LPS-activated B-cells with 10058-F4 blocked this increase in RNA (Figure 6A), but did not affect the induction of c-Myc, an immediate early gene (Figure 6B). Because Myc is a direct activator of the genes transcribed by RNA polymerases I and III (Felton-Edkins et al., 2003; Grandori et al., 2005), as well as by RNA polymerase II, increased rRNA was expected to account for most Myc-elevated RNA; this was confirmed by using a BioAnalyzer (Agilent). Estimation of non-rRNA amounts and direct measurement of poly-A mRNA dramatized the rapid increase of RNA polymerase II transcription products. It should be noted that Myc may influence RNA levels by modulating rates of synthesis or degradation directly or indirectly aside from controlling pause release.
Figure 6. c-Myc amplifies all expressed genes in B-splenocytes (see also Figure S7).
(A) and (B) Flow cytometric analysis of total acridine orange (AO) stained RNA (A) and c-Myc-EGFP (B) in LPS-activated B-splenocytes at 0, 4, 8 and 11 hrs. The cells from wild type (yellow) or c-Myc -EGFP mice were treated with (red), or without (blue) Myc-Max inhibitor 10058-F4.
(C) Heatmap of c-Myc tag density (against IgG) near TSS (+/− 2Kbps; 40 windows) for LPS-activated B4 cells treated with and without 10058-F4. Genes are sorted into 100 equal size bins based on expression levels. Shown are the averaged c-Myc tag densities for each bin. The analysis was repeated for p300 binding sites pre-established in resting B cells in non-promoter regions, serving as a proxy for enhancers. The p300 binding sites are sorted into 100 bins based on the H3k27ac level, an estimate of enhancer activity (Creyghton et al., 2010; Rada-Iglesias et al., 2011)).
(D) Q-RT-PCR analysis of genes selected randomly from expression array data. Cells were cultured with or without 10058-F4. Two immediate-early genes, Bend3 and Igf2R, expressed with or before c-Myc were 10058-F4 insensitive. Genes Hsd11b1, Bex1 reside in heterochromatin and are inactive.
(E) The heterochromatin vs. euchromatin features of 10058-F4 (Traf3ip3) and insensitive genes (Hsd11b1 & Bex1).
c-Myc-dependent amplification of mRNA from cells treated or not with 10058-F4 was evaluated by qPCR of randomly chosen mRNAs (Figure S6E) expressed at different levels and selected without consideration of c-Myc-binding,. ChIP-Seq (and ChIP-qPCR at several TSSs, not shown) using anti-EGFP confirmed that the inhibitor globally compromised c-Myc-EGFP binding at TSSs and enhancers (Figure 6C). Evaluation after normalizing mRNA yield to cell number revealed that almost every mRNA increased after B-cell activation and 10058-F4 prevented these increases (Figure 6D).
To confirm that Myc regulates total cellular RNA and mRNA levels, c-Myc null naïve B-cells were recovered from mice carrying a conditional Myc allele (Mycflox/flox) and a tamoxifen-inducible Cre recombinase CreERTam) (Wang et al., 2011) that were treated with tamoxifen for three days. Naïve splenic B-cells essentially devoid of c-Myc were activated with LPS, and analyzed for total RNA and mRNA levels as above (Wang et al., 2011). Naïve B-cells from the spleens of Myc knockout mice were smaller (Figure S7A) and contained less total RNA (Figure S7B) than their wild-type littermates. So, the small amount of Myc in resting cells still augments RNA levels. Upon activation, c-Myc knockout B-cells not only failed to increase the panel of randomly selected mRNAs, total RNA or mRNA (Figure S7C), but the levels of these molecules actually declined, as with 10058-F4 treatment. Evidently Myc helps to maintain a cell’s full kit. Conditional knockout of this flox/flox c-myc allele with C19-Cre whose expression commences in pro-B-cells also yields c-Myc-less cells stuck in the early stages of activation (de Alboran et al., 2001).
Silent and immediate-early genes are not Myc-dependent
Silent genes lacking active histone marks and RNA Pol II (e.g., Hsd11b1 & Bex1) were unaffected by Myc inhibition (Figures 6D, E). The expression of several immediate early genes (that in naïve cells often reside in heterochromatin), such as c-fos, IGF2R, BEND3 (Figure 6D), and c-Myc itself (Figure 6B), was unperturbed by the Myc inhibitor. c-fos expression peaks 15–30 minutes post-activation (Figure S6F) but is already shutoff by 4 hours accompanied by H3K27-trimethylation, a marker for repressive chromatin. A pulse of promoter expression that precedes the Myc peak cannot be effectively amplified.
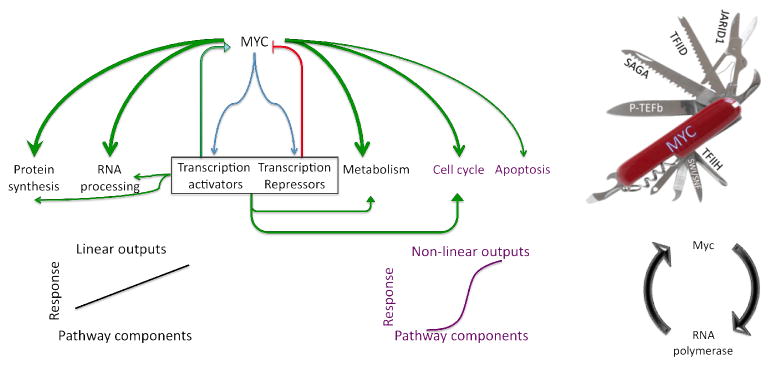
Several genes displayed more complex temporal profiles, initially rising and then falling. Acting universally during transcription, Myc would amplify gene specific activators and repressors that secondarily modify target expression via emergent feedforward or feedback circuitry (Figure 7) and control RNA half-life.
Figure 7. Myc controls cellular subnetworks by transcriptionally varying their component concentrations.
Left--Global up-regulation of MYC provokes linear and non-linear changes in outputs of cellular subsystems according to network architecture. By inducing activators and repressors, a panoply of feedback and coherent and incoherent feedforward loops are employed according to programs pre-existing in the cells. Right—Myc’s multiple partners have the potential to expedite passage through multiple stages of the transcription cycle and to create kinetic synergy (Chung and Levens, 2005; Herschlag and Johnson, 1993). Myc stimulated pause release and promoter reloading would amplify expression according to output levels.
Discussion
This study reports that c-Myc binding is positively correlated with the expression levels of the vast majority of active genes and also with Pol II binding in two primary cell types as well as mouse ES cells. In activated B-cells, Myc binding provoked a redistribution of RNA polymerase from promoters into gene bodies. The simplest interpretation of all these results is that c-Myc is a universal amplifier of transcription that drives the transcription machinery through pause release. The discordance of Myc targets between cell types, and the concordance of expressed genes irrespective of Myc levels within a single cell type, dictates that Myc is neither a specifier nor re-programmer of cell fate. Most simply, c-Myc is a universal amplifier of expression. Total RNA levels, not just differential expression, must be compared between samples to appreciate this effect. Some genes may be exempted from Myc amplification via c-Myc-interacting repressors such Miz-1(Herkert and Eilers, 2010) or via negative feedback through Myc induced repressors (Liu and Levens, 2006). Whether Myc primarily up-regulates targets via PTEF-b stimulated release of promoter paused RNAPII (Rahl et al., 2010), or exploits additional mechanisms (Cheng et al., 1999; Cowling and Cole, 2006; Eilers and Eisenman, 2008; Rahl et al., 2010; Wasylishen and Penn, 2010) may depend on whether all promoters follow a universal reaction scheme. If different promoters are limited at several distinct or multiple kinetically equivalent steps, then a universal activator such as Myc, must be a molecular Swiss Army knife functioning at different steps (Figure 7, right) to generate kinetic synergy (Chung and Levens, 2005; Herschlag and Johnson, 1993), rationalizing the plethora of activities marshaled by c-Myc (Cowling and Cole, 2006). If recruited Myc stimulates pause-release and facilitates promoter reloading, then it would operate preferentially at highly expressed genes, perhaps enhancing their nonlinear preferential amplification. c-Myc also activates RNA polymerases I and III, rRNA transcribed by the former accounting for the bulk of the RNA (James and Eisenman, 2002). c-Myc also directly augments DNA replication (Dominguez-Sola et al., 2007). Considering Myc to be a universal amplifier may help to explain and predict its role in diverse biological systems.
Metabolism and Cell size
Up-regulating all active genes, c-Myc increases the flux through cellular networks (Figure 7). In an anabolic cell, such increased throughput would drive cell growth yielding bigger cells. To tune the expression of specific genes, specific regulation must be superimposed over this global up-regulation as described for densely overlapping regulons (Alon, 2007). While the net output of many pathways would scale linearly and monotonically as the synthesis of their components increases, other processes are inherently non-linear and would respond according to thresholds wired into their pathways (Figure 7).
Proliferation and Apoptosis
The differential responses of different pathways to changes in Myc abundance do not demand differential up-regulation of their components. For example, cell division is all-or-none. Until cell-cycle components exceed a mitogenic threshold, amplification by Myc is irrelevant. Myc amplification is also irrelevant if proliferation genes are off when Myc is expressed explaining why enforced Myc expression is tumorigenic in growing or regenerating, but not adult mouse livers (Beer et al., 2004). Similarly, up-regulating the apoptosis apparatus across critical thresholds eventually triggers cell death. Adjustments in the settings for such critical thresholds depend on the inventory of regulatory factors in any given cell; the composition of this inventory is largely not under Myc’s purview.
Sustained Myc expression and implications for cancer
Physiologically, c-Myc is usually expressed as an immediate early pulse before returning to baseline. Such a pulse would drive the accumulation of the macromolecules needed for proliferation or other pre-programmed pathways; without Myc these pathways would labor as cells gradually ramped up their synthetic capacity. Thus Myc would provide a bolus of material supporting all pathways until control is assumed by dedicated regulatory factors. Sustained Myc over-expression in cancer would leave cells in a state of chronic overdrive through all cellular networks. At pathological levels, when Myc invades enhancers, many cellular subsystems may be driven across critical thresholds (Lin, 2012*). Under these circumstances, even a modest reduction in Myc may be sufficient to deprive cells of the net anabolic, metabolic and mitogenic impulse necessary to sustain unchecked proliferation.
Myc maybe pathologically up-regulated by a host of mechanisms such as chromosomal rearrangements or unchecked stimulation directing transcription factors to the c-myc regulatory sequences that serve as an antenna for signals from many cellular subsystems. Because amplification of the factors driving Myc would create a dangerous positive feedback loop, Myc must also induce repressors to limit its own synthesis (Levens, 2010; Liu and Levens, 2006; Wierstra and Alves, 2008); abrogation of this negative feedback would also enforce Myc over-expression. Mutations that increase Myc levels and overdrive the apoptotic machinery must be balanced by the over-production of survival factors. Weaning cells from high Myc levels could potentially create an imbalance between longer-lived pro- and shorter-lived anti-apoptotic factors (Sharma et al., 2006) and contribute to oncogene addiction by Myc.
Differentiation
c-Myc expression almost always declines, at least transiently during differentiation when large batteries of genes must be turned on and off to enable reprogramming. As an amplifier, Myc would reinforce whatever state a cell is in. Suspending Myc-driven amplification would enable more efficient and rapid reprogramming. Thereafter increased Myc levels would reinforce the new cell state. Exactly such differentiation-linked biphasic Myc expression has been described in several models of erythroleukemia cells (Dmitrovsky et al., 1986; Nepveu et al., 1987; Tonini et al., 1987). In ES cells, high Myc levels reinforce the undifferentiated state to prevent stochastic differentiation. During the generation of iPS cells, Myc may help to trap genes in the on-state as they are transiently activated by reprogramming factors.
What about Myc repressed genes?
If Myc is a universal amplifier of gene activation, why do previous studies estimate ~1/3 of Myc targets to be downregulated? Two reasons may explain repression by Myc. First, when comparing RNA expression between samples normalized for equal amounts of RNA (versus equal numbers of cells), “repressed” genes may actually be up-regulated by Myc at the cellular level, just less so than the average gene. Second, repression may be indirect as transcriptional or chromatin repressors activated by Myc are recruited to Myc target genes. For example during B-cell activation, c-Myc bound and up-regulated the EZH2 promoter (Figure 6D); EZH2 mediates transcriptional repression across the genome by catalyzing methylation of histone H3 lysine 27. Myc also induces a number of miRNAs that limit the amplification of their targets (Bui and Mendell, 2010).
In summary, Myc is a universal of amplifier of gene activation; to predict precisely the response of cells and tissues to physiological, pathological, or therapeutic manipulation of Myc, it will be necessary to elucidate how Myc-amplification changes the flux through cellular compartments and sub-networks to determine cell fate in health and disease.
Experimental Procedures
Isolation and Activation of Mouse Splenocytes
Naïve mouse B or T splenocytes from 8–11 week mice were negatively selected with CD-43-(Ly48) MicroBeads (MACS, Miltenyi Biotech, Cat.no. 130-049-801 ) or Pan T cell isolation kit (MACS, Cat.no. 130-095-130), respectively. Isolated splenocytes were cultured at 0.5 × 106 cells /ml in RPMI 1640 (Gibco-Invitrogen) with Hepes, L-glutamine, sodium pyruvate and 50 mM β-mercaptoethanol. B-cells were activated with lipopolysaccharide (LPS, 25mg/ml) (SIGMA) for different times. For Myc inhibition, cells were treated with 66.5 mM 10058-F4 (SIGMA) for 2hrs before LPS stimulation. T-splenocytes were cultured at 1 × 106 cells/ml in the same medium as B cells and activated with Concanavalin A (ConA, 7.5μg/ml) (SIGMA) for different times.
Antibodies
Full length A.V. polyclonal anti-GFP (Clontech, 632460) was used for mouse B-cell ChIP analysis, MEF immunoprecipitation and blot analysis. Anti-Myc (Santa Cruz, SC-41, C-8) was also used for the MEF immunoprecipitation and blot analysis. Normal mouse IgG (Santa Cruz SC-2025) and normal rabbit IgG (Santa Cruz SC-2027) were used for the ChIP control. Anti-RNA polymerase II CTD repeat YSPTSPS [4H8] - ChIP Grade (ab5408) was used for the Pol II ChIP analysis.
c-Myc (N-term) Antibody (Epitomics, 1472-1) and c-Myc Phospho (pT58/pS62) Antibody (Epitomics, 1203-1) were used for immunoblot analysis of c-Myc expression and phosphorylation in B- and T-cells, respectively. The antibodies for histone modifications were described previously (Kuchen et al., 2010; Yamane et al., 2011).
Flow cytometry
Steady state, serum starved and re-stimulated MEFs; propidium iodide stained naïve, LPS or ConA stimulated splenocytes; or AO stained B-cells were analyzed by flow cytometry to detect c-Myc-EGFP and/or RNA intensity (Acridine Orange) on Cyflow ML Instrument (PARTEC) using FloMAX and/or FlowJo (Treestar version 7.6.1) software. At least 25,000 events were acquired for each sample.
ChIP-Seq and Expression Arrays
Chromatin immunoprecipitation and sequencing (ChIP-Seq) were performed as described (Barski et al., 2007). ChIP-Seq datasets for histone modifications H3K4me1, H3K27me3, H4K20me3 and H3K9me3 and ChIP-Seq datasets for TFs c-Myc and E2F1 in mouse ES cells were from (Mikkelsen et al., 2007), and (Chen et al., 2008a), respectively. RNA-Seq library preparation and sequencing for mouse ES cells followed the procedure described in (Chepelev et al., 2009). Sequence reads of 25-bp for ChIP-Seq and 36-bp for RNA-Seq were generated from an Illumina Genome Analyzer, mapped to mouse genome (mm8) using Bowtie (Langmead et al., 2009). ChIP-Seq tag enriched regions were predicted by SICER (Zang et al., 2009) and MACS(Zhang et al., 2008). Heat-maps relating histone modification to Myc ChIP-Seq tag density were performed by MeV (Chu et al., 2008). The mRNA expression level of UCSC known genes was quantified by the RPKM measure from RNA-Seq dataset (read per kilobase of exon model per million reads) based on UCSC known genes annotations.
Total RNAs were isolated from B- or T- splenocytes with TRIzol Reagent (Invitrogen) and analyzed using Affymetrix expression arrays. RNA quality was checked on Agilent Bioanalyzer. All microarray samples had a high quality score (RIN >9). RNA (100 ng) was reverse-transcribed and labeled with biotin using Affymetrix 3′ IVTexpress Labeling according to the manufacturer’s protocol. Four biological replicates of each group were labeled, and hybridized to Affymetrix mouse 430 2.0 GeneChip and scanned on Affymetrix GeneChip scanner 3000. Data were collected using Affymetrix AGCC software. Quantification of mRNA expression levels, GCRMA normalization, and call of differentially expressed genes used affylmGUI software (Wettenhall et al., 2006).
Q-RT-PCR Analysis of the expression of Random Selected Genes
Total RNAs were purified from resting LPS-stimulated and 10058-F4 treated B-splenocytes at various time points. 0.25 μg of total RNA from each sample programmed first-strand c-DNA synthesis using Enhanced Avian HS RT-PCR-100 kit (SIGMA, Cat. No. HSRT100-1kt). The primers and probes for each gene were designed using Roche Universal Probelibrary Assay Design Center Web (Figure S7F). q-PCRs were performed with Roche LightCycler 480 system (LightCycler 480 Probe Master, Ref. No. 04 707 494 001; Universal Probelibrary set, Human, Ref. No. 04 683 633 001). The gene expression levels were adjusted according to the cell number used for input RNA, normalized to the RNA level of an untreated resting cell.
Supplementary Material
Acknowledgments
We thank our colleagues, especially Eric Batchelor and Dan Larson for insight and valuable discussions. This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, of NHLBI, and of NIAMS. We thank the DNA Sequencing Core Facility of NHLBI for sequencing some of the ChIP-Seq libraries.
Footnotes
Accession numbers
Microarray and ChIP-Seq data deposited with GEO (GSE37230).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agrawal P, Yu K, Salomon AR, Sedivy JM. Proteomic profiling of Myc-associated proteins. Cell Cycle. 2010;9:4908–4921. doi: 10.4161/cc.9.24.14199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alon U. An introduction to systems biology : design principles of biological circuits. Boca Raton, FL: Chapman & Hall/CRC; 2007. [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Beer S, Zetterberg A, Ihrie RA, McTaggart RA, Yang Q, Bradon N, Arvanitis C, Attardi LD, Feng S, Ruebner B, et al. Developmental context determines latency of MYC-induced tumorigenesis. PLoS Biol. 2004;2:e332. doi: 10.1371/journal.pbio.0020332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berns K, Hijmans EM, Koh E, Daley GQ, Bernards R. A genetic screen to identify genes that rescue the slow growth phenotype of c-myc null fibroblasts. Oncogene. 2000;19:3330–3334. doi: 10.1038/sj.onc.1203639. [DOI] [PubMed] [Google Scholar]
- Bui TV, Mendell JT. Myc: Maestro of MicroRNAs. Genes Cancer. 2010;1:568–575. doi: 10.1177/1947601910377491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of External Signaling Pathways with the Core Transcriptional Network in Embryonic Stem Cells. Cell. 2008a;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008b;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Cheng SW, Davies KP, Yung E, Beltran RJ, Yu J, Kalpana GV. c-MYC interacts with INI1/hSNF5 and requires the SWI/SNF complex for transactivation function. Nat Genet. 1999;22:102–105. doi: 10.1038/8811. [DOI] [PubMed] [Google Scholar]
- Chepelev I, Wei G, Tang Q, Zhao K. Detection of single nucleotide variations in expressed exons of the human genome using RNASeq. Nucleic Acids Res. 2009;37:e106. doi: 10.1093/nar/gkp507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu VT, Gottardo R, Raftery AE, Bumgarner RE, Yeung KY. MeV+R: using MeV as a graphical user interface for Bioconductor applications in microarray analysis. Genome Biol. 2008;9:R118. doi: 10.1186/gb-2008-9-7-r118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HJ, Levens D. c-myc expression: keep the noise down! Mol Cells. 2005;20:157–166. [PubMed] [Google Scholar]
- Cowling VH, Cole MD. Mechanism of transcriptional activation by the Myc oncoproteins. Semin Cancer Biol. 2006;16:242–252. doi: 10.1016/j.semcancer.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV. Enigmatic MYC Conducts an Unfolding Systems Biology Symphony. Genes Cancer. 2010;1:526–531. doi: 10.1177/1947601910378742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Alboran IM, O’Hagan RC, Gartner F, Malynn B, Davidson L, Rickert R, Rajewsky K, DePinho RA, Alt FW. Analysis of C-MYC function in normal cells via conditional gene-targeted mutation. Immunity. 2001;14:45–55. doi: 10.1016/s1074-7613(01)00088-7. [DOI] [PubMed] [Google Scholar]
- Dmitrovsky E, Kuehl WM, Hollis GF, Kirsch IR, Bender TP, Segal S. Expression of a transfected human c-myc oncogene inhibits differentiation of a mouse erythroleukaemia cell line. Nature. 1986;322:748–750. doi: 10.1038/322748a0. [DOI] [PubMed] [Google Scholar]
- Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M, Galloway DA, Gu W, Gautier J, Dalla-Favera R. Nontranscriptional control of DNA replication by c-Myc. Nature. 2007;448:445–451. doi: 10.1038/nature05953. [DOI] [PubMed] [Google Scholar]
- Eberhardy SR, Farnham PJ. Myc recruits P-TEFb to mediate the final step in the transcriptional activation of the cad promoter. J Biol Chem. 2002;277:40156–40162. doi: 10.1074/jbc.M207441200. [DOI] [PubMed] [Google Scholar]
- Eilers M, Eisenman RN. Myc’s broad reach. Genes Dev. 2008;22:2755–2766. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felton-Edkins ZA, Kenneth NS, Brown TR, Daly NL, Gomez-Roman N, Grandori C, Eisenman RN, White RJ. Direct regulation of RNA polymerase III transcription by RB, p53 and c-Myc. Cell Cycle. 2003;2:181–184. [PubMed] [Google Scholar]
- Grandori C, Gomez-Roman N, Felton-Edkins ZA, Ngouenet C, Galloway DA, Eisenman RN, White RJ. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat Cell Biol. 2005;7:311–318. doi: 10.1038/ncb1224. [DOI] [PubMed] [Google Scholar]
- Hann SR, Eisenman RN. Proteins encoded by the human c-myc oncogene: differential expression in neoplastic cells. Mol Cell Biol. 1984;4:2486–2497. doi: 10.1128/mcb.4.11.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herkert B, Eilers M. Transcriptional repression: the dark side of myc. Genes Cancer. 2010;1:580–586. doi: 10.1177/1947601910379012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschlag D, Johnson FB. Synergism in transcriptional activation: a kinetic view. Genes Dev. 1993;7:173–179. doi: 10.1101/gad.7.2.173. [DOI] [PubMed] [Google Scholar]
- Huang CY, Bredemeyer AL, Walker LM, Bassing CH, Sleckman BP. Dynamic regulation of c-Myc proto-oncogene expression during lymphocyte development revealed by a GFP-c-Myc knock-in mouse. Eur J Immunol. 2008;38:342–349. doi: 10.1002/eji.200737972. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James L, Eisenman RN. Myc and Mad bHLHZ domains possess identical DNA-binding specificities but only partially overlapping functions in vivo. Proc Natl Acad Sci U S A. 2002;99:10429–10434. doi: 10.1073/pnas.162369299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Wu G, Zhan X, Nolan A, Koh C, De Marzo A, Doan HM, Fan J, Cheadle C, Fallahi M, et al. Cell-Type Independent MYC Target Genes Reveal a Primordial Signature Involved in Biomass Accumulation. PLoS ONE. 2011;6:e26057. doi: 10.1371/journal.pone.0026057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly K, Cochran BH, Stiles CD, Leder P. Cell-specific regulation of the c-myc gene by lymphocyte mitogens and platelet-derived growth factor. Cell. 1983;35:603–610. doi: 10.1016/0092-8674(83)90092-2. [DOI] [PubMed] [Google Scholar]
- Kuchen S, Resch W, Yamane A, Kuo N, Li Z, Chakraborty T, Wei L, Laurence A, Yasuda T, Peng S, et al. Regulation of microRNA expression and abundance during lymphopoiesis. Immunity. 2010;32:828–839. doi: 10.1016/j.immuni.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land H, Parada LF, Weinberg RA. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature. 1983;304:596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- Langmead B, Schatz MC, Lin J, Pop M, Salzberg SL. Searching for SNPs with cloud computing. Genome Biol. 2009;10:R134. doi: 10.1186/gb-2009-10-11-r134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levens D. You Don’t Muck with MYC. Genes Cancer. 2010;1:547–554. doi: 10.1177/1947601910377492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Van Calcar S, Qu C, Cavenee WK, Zhang MQ, Ren B. A global transcriptional regulatory role for c-Myc in Burkitt’s lymphoma cells. Proc Natl Acad Sci U S A. 2003;100:8164–8169. doi: 10.1073/pnas.1332764100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA. Transcriptional Amplification in Tumor Cells with Elevated c-Myc. Cell. 2012* doi: 10.1016/j.cell.2012.08.026. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Levens D. Making myc. Curr Top Microbiol Immunol. 2006;302:1–32. doi: 10.1007/3-540-32952-8_1. [DOI] [PubMed] [Google Scholar]
- Margolin AA, Palomero T, Sumazin P, Califano A, Ferrando AA, Stolovitzky G. ChIP-on-chip significance analysis reveals large-scale binding and regulation by human transcription factor oncogenes. Proc Natl Acad Sci U S A. 2009;106:244–249. doi: 10.1073/pnas.0806445106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehmet H, Littlewood TD, Sinnett-Smith J, Moore JP, Evan GI, Rozengurt E. Large induction of c-Myc is not essential for the mitogenic response of Swiss 3T3 fibroblasts. Cell Growth Differ. 1997;8:187–193. [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nepveu A, Marcu KB, Skoultchi AI, Lachman HM. Contributions of transcriptional and post-transcriptional mechanisms to the regulation of c-myc expression in mouse erythroleukemia cells. Genes Dev. 1987;1:938–945. doi: 10.1101/gad.1.9.938. [DOI] [PubMed] [Google Scholar]
- Nikiforov MA, Chandriani S, O’Connell B, Petrenko O, Kotenko I, Beavis A, Sedivy JM, Cole MD. A functional screen for Myc-responsive genes reveals serine hydroxymethyltransferase, a major source of the onecarbon unit for cell metabolism. Mol Cell Biol. 2002;22:5793–5800. doi: 10.1128/MCB.22.16.5793-5800.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poser I, Sarov M, Hutchins JR, Heriche JK, Toyoda Y, Pozniakovsky A, Weigl D, Nitzsche A, Hegemann B, Bird AW, et al. BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals. Nat Methods. 2008;5:409–415. doi: 10.1038/nmeth.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbitts PH, Watson JV, Lamond A, Forster A, Stinson MA, Evan G, Fischer W, Atherton E, Sheppard R, Rabbitts TH. Metabolism of c-myc gene products: c-myc mRNA and protein expression in the cell cycle. EMBO J. 1985;4:2009–2015. doi: 10.1002/j.1460-2075.1985.tb03885.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P, Petrelli NJ, Dunn SP, Krueger LJ. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 2007;67:9762–9770. doi: 10.1158/0008-5472.CAN-07-2462. [DOI] [PubMed] [Google Scholar]
- Shaffer AL, Wright G, Yang L, Powell J, Ngo V, Lamy L, Lam LT, Davis RE, Staudt LM. A library of gene expression signatures to illuminate normal and pathological lymphoid biology. Immunol Rev. 2006;210:67–85. doi: 10.1111/j.0105-2896.2006.00373.x. [DOI] [PubMed] [Google Scholar]
- Sharma SV, Gajowniczek P, Way IP, Lee DY, Jiang J, Yuza Y, Classon M, Haber DA, Settleman J. A common signaling cascade may underlie “addiction” to the Src, BCR-ABL, and EGF receptor oncogenes. Cancer Cell. 2006;10:425–435. doi: 10.1016/j.ccr.2006.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas LR, Tansey WP. Proteolytic control of the oncoprotein transcription factor Myc. Adv Cancer Res. 2011;110:77–106. doi: 10.1016/B978-0-12-386469-7.00004-9. [DOI] [PubMed] [Google Scholar]
- Tonini GP, Radzioch D, Gronberg A, Clayton M, Blasi E, Benetton G, Varesio L. Erythroid differentiation and modulation of c-myc expression induced by antineoplastic drugs in the human leukemic cell line K562. Cancer Res. 1987;47:4544–4547. [PubMed] [Google Scholar]
- Wang H, Hammoudeh DI, Follis AV, Reese BE, Lazo JS, Metallo SJ, Prochownik EV. Improved low molecular weight Myc-Max inhibitors. Mol Cancer Ther. 2007;6:2399–2408. doi: 10.1158/1535-7163.MCT-07-0005. [DOI] [PubMed] [Google Scholar]
- Wang R, Dillon CP, Shi LZ, Milasta S, Carter R, Finkelstein D, McCormick LL, Fitzgerald P, Chi H, Munger J, et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35:871–882. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasylishen AR, Penn LZ. Myc: the beauty and the beast. Genes Cancer. 2010;1:532–541. doi: 10.1177/1947601910378024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wettenhall JM, Simpson KM, Satterley K, Smyth GK. affylmGUI: a graphical user interface for linear modeling of single channel microarray data. Bioinformatics. 2006;22:897–899. doi: 10.1093/bioinformatics/btl025. [DOI] [PubMed] [Google Scholar]
- Wierstra I, Alves J. The c-myc promoter: still MysterY and challenge. Adv Cancer Res. 2008;99:113–333. doi: 10.1016/S0065-230X(07)99004-1. [DOI] [PubMed] [Google Scholar]
- Yamane A, Resch W, Kuo N, Kuchen S, Li Z, Sun HW, Robbiani DF, McBride K, Nussenzweig MC, Casellas R. Deep-sequencing identification of the genomic targets of the cytidine deaminase AID and its cofactor RPA in B lymphocytes. Nat Immunol. 2011;12:62–69. doi: 10.1038/ni.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang C, Schones DE, Zeng C, Cui K, Zhao K, Peng W. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.