Abstract
The XPB DNA helicase, a subunit of the basal transcription factor TFIIH, is also involved in nucleotide excision repair (NER). We examined recruitment of NER proteins in XP-B cells from patients with mild or severe xeroderma pigmentosum (XP) having different XPB mutations using local UV-irradiation through filters with 5 µm pores combined with fluorescent antibody labeling. XPC was rapidly recruited to UV damage sites containing DNA photoproducts (cyclobutane pyrimidine dimers, CPD) in all the XP-B and normal cells, thus reflecting its role in damage recognition prior to the function of XPB. Cells from the mild XP-B patients, with a missense mutation, showed delayed recruitment of all NER proteins except XPC to UV damage sites, demonstrating that this mutation impaired localization of these proteins. Surprisingly, in cells from severely affected patients, with a C-terminal XPB mutation, XPG and XPA proteins were normally recruited to UV damage sites demonstrating that this mutation permits recruitment of XPG and XPA. In marked contrast, in all the XP-B cells recruitment of XPF was absent immediately after UV and was delayed by 0.5 and 3h in cells from the mild and severely affected XP patients, respectively. Redistribution of NER proteins was nearly complete in normal cells by 3h but by 24h redistribution was only partially present in cells from mild patients and virtually absent in cells from the severely affected patients. Ineffectual repair of UV-induced photoproducts resulting from delayed recruitment and impaired redistribution of NER proteins may contribute to the markedly increased frequency of skin cancer in XP patients.
Keywords: Xeroderma pigmentosum, Confocal microscopy, Helicase, TFIIH
1. Introduction
Nucleotide excision repair (NER) removes a wide range of DNA lesions caused by ultraviolet (UV)-induced photoproducts and some chemical mutagens [1,2]. Two major photoproducts induced by UV-irradiation are cyclobutane pyrimidine dimers (CPD) and 6-pyrimidine–4-pyrimidone photoproducts (6–4PP). The NER system involves two distinct pathways: transcription-coupled repair (TCR), which rapidly removes lesions from the transcribed strand of active genes, and global genome repair (GGR), which processes lesions in the non-transcribed strand of active genes and from DNA of the genome overall [3]. Removal of damaged DNA by NER is a multistep process involving recognition of the DNA damage followed by opening up of the DNA helix around the lesion, dual incision, and subsequent excision of the oligonucleotide containing the DNA lesion [4–8].
Xeroderma pigmentosum (XP) and Cockayne syndrome (CS), autosomal recessive diseases, are associated with defects in NER of DNA damage [1,4]. Defects in XP have been assigned to seven complementation groups (XP-A through XP-G) corresponding to proteins involved in different stages of the NER. Another form, the XP variant has a defect in DNA polymerase eta. The human XPB DNA helicase is a gene which is essential to life [4]. XPB is a component of the basal transcription factor TFIIH and is also involved in the early steps of NER. TFIIH is composed of a 10-subunit protein complex that opens the DNA in the context of transcription initiation and NER [4].
The clinical phenotype of the patients with XPB gene mutations is varied. While three patients had severe XP/CS syndrome complex, two siblings have mild XP with only deafness without CS, and two other siblings had mild XP/CS complex [9]. The mild patients had the same missense mutation (c.296T > C, p.F99S) in one allele and different nonsense mutations in the other allele [9]. The severe patients had the same splice mutation (c.2218-6C > A, p.Q739insX42) resulting in alteration of the C-terminal 41 amino acids and deletion of the final amino acid. They had different nonsense mutations in the second allele [9]. We used these cells to examine the order of assembly of NER proteins at sites of UV damage. Our findings indicate that there is a complex relationship between the XPB protein and the recruitment and redistribution of XPG, XPA and XPF proteins.
2. Materials and methods
2.1. Cell lines, and culture conditions isolation
Normal skin fibroblasts (AG13145) and XP-B cells from XP patients with mild disease (XP33BR-GM21071 and XPCS1BA-GM13025) or with the severe XP/CS complex (XP131MA-GM21153 and XP183MA-GM21072) [9,10], or with mutations in the XPC (XP21BE-GM09943) [11] and XPG (XP96TA-GM16180) [12] genes were obtained from the Human Genetic Mutant Cell Repository, Camden, NJ. XP-A (XP315BE) cells with a deletion in exon 3 [13] were established by NCI. The cells were grown in Dulbecco’s Modified Eagle Medium (DMEM; Invitrogen Corp.) containing 40 mM glutamine, and 10% fetal bovine serum (FBS; Invitrogen Corp.) in an 8% CO2 humidified incubator at 37 °C.
2.2. ELISA
Confluent human normal and XP-B fibroblasts were irradiated with 10 J/m2 UVC from a germicidal lamp and incubated for various time periods. Repair of 6–4PP and CPD was determined using an ELISA assay with TDM-2 and 6-4M-2 monoclonal antibodies as described previously [14–17].
2.3. Cell culture and local UV-irradiation
Normal and XP patient cells were grown for 3 days to allow efficient uptake of the different size of beads (Carboxylate Microspheres, Polysciences): normal fibroblasts, 0.8 µm; XPB cells, 2.0 µm. Cells with different sized beads were grown (in a 1:1 ratio) for 1 day on coverslips in a culture dish [18]. The cells on coverslips were covered with an isopore polycarbonate filter with pores of 5 µm diameter (Millipore) during UV-irradiation [15]. Subsequently, the filter was removed, and the cells were incubated under normal culture conditions for appropriate post-irradiation incubation times.
2.4. Immunofluorescence
Cells were fixed in 1.6% formaldehyde for 20 min at room temperature and permeabilized by PBS/0.5% Triton X-100 for 10 min on ice. After washing with PBS, the cells were incubated for 1h with pairs of primary antibodies (one monoclonal antibody and one polyclonal antibody). Then cells were incubated with Alexa Flour 488 (green) goat anti-rabbit IgG conjugate for staining polyclonal antibodies or Alexa Flour 568 (red) goat anti-mouse IgG conjugate for staining monoclonal antibodies (Molecular Probes, Eugene, OR) at room temperature for 1h [19].
For double staining of NER proteins together with DNA photoproducts, the cells were sequentially labeled with an NER primary polyclonal antibody and Alexa Fluor 488 goat anti-rabbit IgG conjugate [20]. Antigen–antibody complexes were fixed with 1.6% formaldehyde for 10 min, and DNA was denatured with 2N HCL at room temperate for 20 min [21]. Cells were then labeled with TDM-2 and Alexa Flour 568 goat anti-mouse IgG. Cells were mounted in mounting medium containing 4′-6′-diamidino-2-phenylindole (DAPI) as a DNA counter stain (Vector Laboratories, Burlingame, CA). Fluorescence images were obtained using LSM 510 Confocal microscope (Carl Zeiss).
The number of nuclei containing at least one localized area of immunofluorescence was determined by examination of the confocal images. A total of at least 100 nuclei were scored for each incubation time point for localized area quantification. The ratio of NER areas to CPD areas was determined upon scoring >100 individual nuclei for merged fluorescent signals of NER areas and CPD areas.
2.5. Antibodies
Primary rabbit polyclonal antibodies used were anti-XPA (Santa Cruz, 1:50), anti-XPB (Santa Cruz, 1:100), anti-XPC (1:100, a gift from N.G.J. Jaspers, Rotterdam, The Netherlands), and anti-XPD (Santa Cruz, 1:100). Primary mouse monoclonal antibodies used were anti-XPG (Santa Cruz, 1:100), anti-XPF (NeoMarkers, 1:100), anti-CPD (TDM-2, 1:1500) and anti-6–4PP 1:200, (both gifts from T. Mori, Nara, Japan).
2.6. Cell transfection
Exponentially growing normal and XP183MA cells were plated prior to plasmid transfection. Cells were transiently transfected with lipofectamine (Invitrogen Co.) for 5h with either pSGM5 empty vector or normal pSG5M/XPBcDNA (a gift from W. Vermeulen, Rotterdam, The Netherlands). After 48h post-incubation, transfected cells were analyzed by Western blot and immunofluorescence staining [9].
3. Results
3.1. Reduced repair of photoproducts in XP-B cells
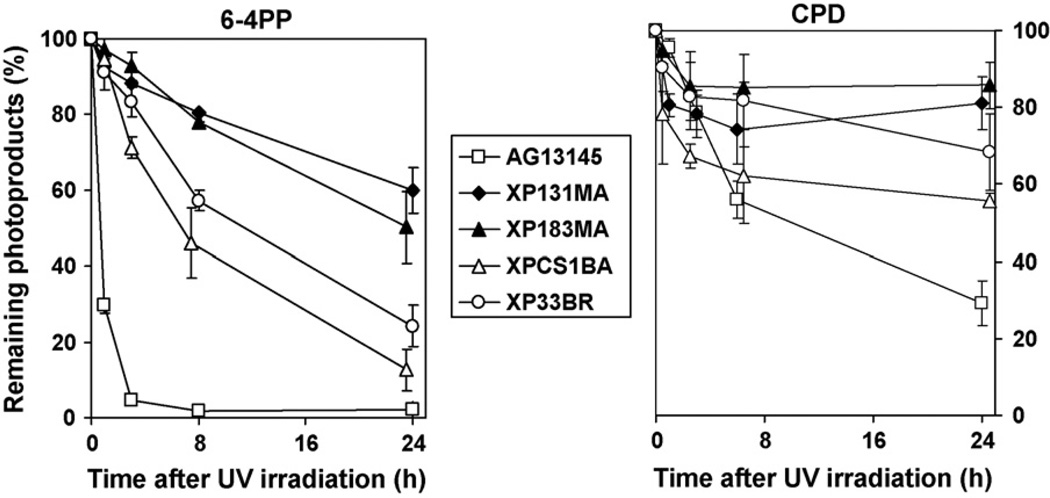
For the determination of DNA repair kinetics, we measured the removal of the 6–4PP (Fig. 1A) and CPD (Fig. 1B) in XP-B and normal cells at various times after UV-irradiation using the ELISA. As noted previously [15,22] in normal cells, almost 95% of 6–4PP were removed at 3h post-UV-irradiation. In contrast, we found that cells from the XP-B patients removed only 7–30% of 6–4PP at this time. At 24h post-UV, cells from the severely affected XP/CS complex XP131MA and XP183MA patients removed 40–50% of 6–4PP, whereas cells from mildly affected XPCS1BA and XP33BR patients removed 75–90% of 6–4PP.
Fig. 1.
Delayed repair of 6–4 photoproducts (6–4PP) and cyclobutane pyrimidine dimers (CPD) in cells from XP-B patients. XP and normal cells were irradiated with 10 J/m2 UV and incubated for the indicated times. The levels of 6–4PP (A) and CPD (B) were measured by ELISA using either anti-6–4PP or anti-CPD antibodies. Each point represents the mean from four independent experiments. The points are displaced slightly to permit visualization of the S.D. error bars.
As previously reported [15,22], in normal cells the rate of CPD repair was slower than that of 6–4PP with only 75% removed at 24h post-UV. We found that by 24h only 15–20% of the CPD were removed from cells from the severely affected XP131MA and XP183MA patients while 30–45% of the CPD were removed from cells from the milder XPCS1BA and XP33BR patients (Fig. 1B).
3.2. Delayed recruitment of NER proteins to damage sites in cells from mildly affected patients
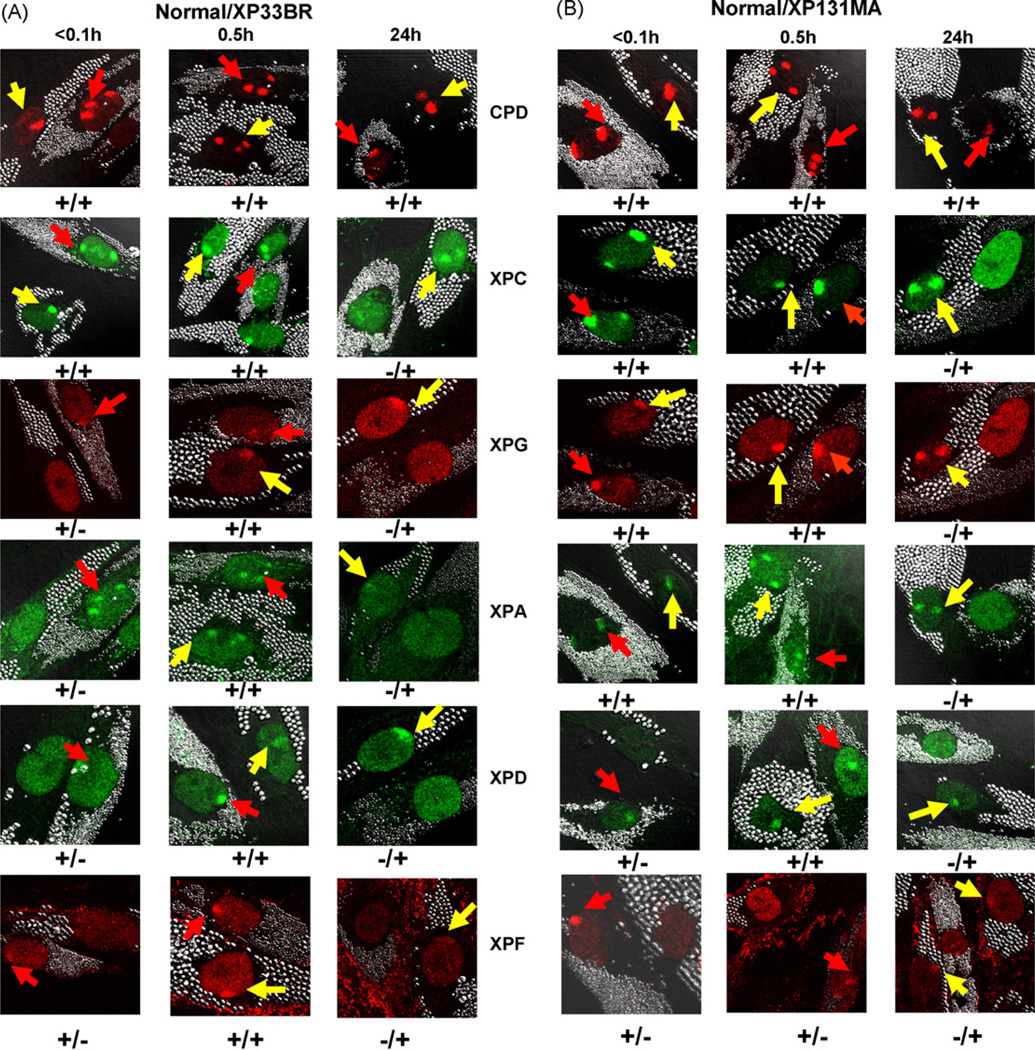
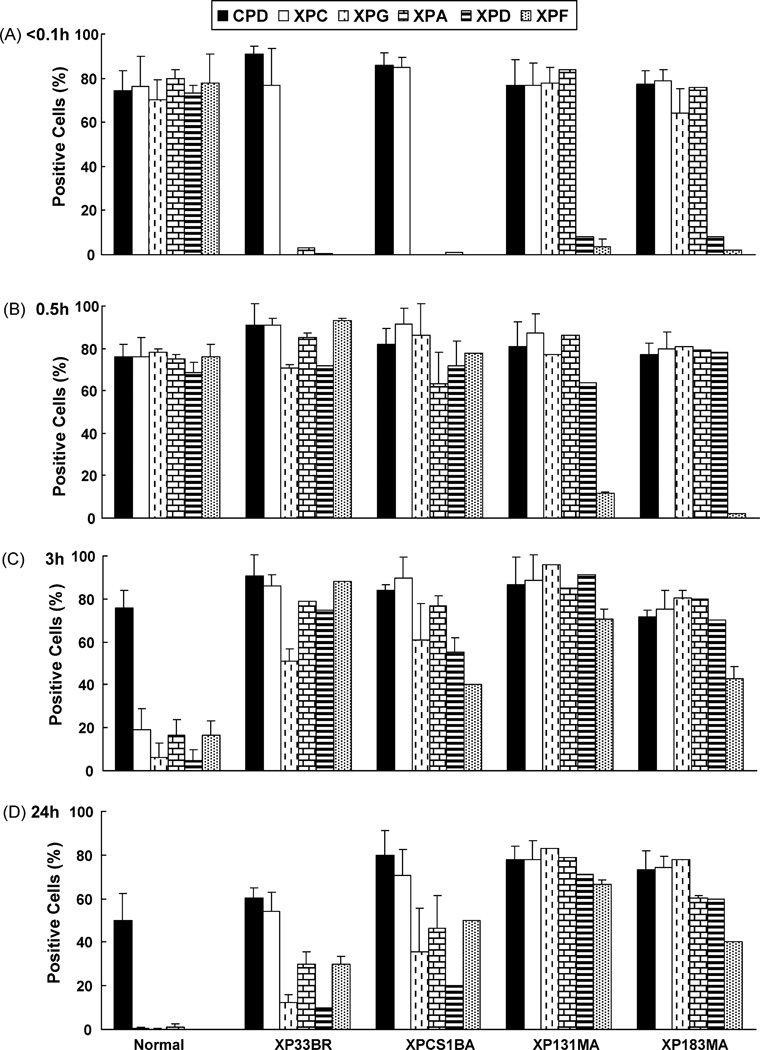
To determine whether UV-irradiation leads to intranuclear translocation of repair proteins to the sites of DNA damage, we investigated the recruitment of the NER proteins to sites of damage using a local UV-irradiation technique combined with fluorescent antibody labeling, as described previously [9,15,18,21]. Immunofluorescent double labeling was performed using an antibody against CPD and antibodies against XPC, XPD, XPG, XPA or XPF. Immediately (<0.1 h) after UV-irradiation, normal and XP-B cells showed localized areas in the nuclei, which were positive for CPD staining (Figs. 2A and B, and 3A). In normal cells the NER proteins (XPC, XPG, XPA, XPD and XPF) (Figs. 2A and B, and 3A) and XPB (data not shown) rapidly co-localized to the CPD positive regions immediately after localized UV exposure. In the mild XPB cells, XPC protein was localized immediately after UV exposure (Figs. 2A and 3A). In contrast, the NER proteins XPG, XPA, XPD and XPF were not recruited to damage sites in XP33BR (Figs. 2A and 3A) and XPCS1BA cells (Fig. 3A) at this time. The recruitment of XPG, XPA, XPD and XPF was observed in XP33BR and XPCS1BA cells 30 min after local UV damage. We counted the proportion of cells containing one or more positive areas of CPD or NER proteins immediately (<0.1 h), 0.5, 3 and 24h after local UV-irradiation in XP33BR, XPCS1BA and normal cells (Fig. 3A–D). Immediately after UV exposure about 75–80% of the normal cells showed positive areas for CPD as well as for XPC, XPG, XPA, XPD and XPF proteins. In contrast, immediately after UV exposure the cells from the mild XPB patients showed about 75–90% positive cells with CPD and XPC protein but had 0–3% positive for XPG, XPA, XPD or XPF proteins. At the 0.5h time point, the proportion of cells that were positive for NER proteins remained about 70–80% in the normal cells. At 0.5 h, 64–93% of the cells from the mild XPB patients were positive for NER proteins XPG, XPA, XPD and XPF demonstrating an approximately 30 min delay in recruitment of these proteins. These results indicate that the reduced functioning of XPB in these cells resulted in low levels of TFIIH [9,23,24] with impaired recruitment of XPG, XPA XPD and XPF but not XPC proteins.
Fig. 2.
Recruitment and redistribution of NER proteins to sites of UV damage. Normal (0.8 µm beads) and XP-B cells (2.0 µm beads) on same slide were irradiated with 100 J/m2 UV through 5 µm pore size filters and immediately fixed (<0.1 h) or subsequently cultured for various time points before fixation. Immunofluorescent double labeling was performed using antibodies against CPD, XPC, XPG, XPA, XPD or XPF. The arrows on the confocal images indicate sites of localized damage (red arrows, normal cells; yellow arrows, XP-B cells). Symbols + or − indicate localization or non-localization of NER protein in normal cells/XP-B cells. (A) Delayed recruitment of XPG, XPA, XPD and XPF proteins to damage sites in XP33BR. (B) Normal recruitment of XPC, XPG and XPA proteins to damage sites in XP131MA cells. The recruitment of XPD was delayed and no XPF recruitment was seen 0.5h after UV-irradiation. NER proteins in all XP-B cells remained visible at damage sites even 24h after local UV-irradiation. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of the article.)
Fig. 3.
Quantification of recruitment and redistribution of NER proteins to sites of UV damage at various times post-UV-irradiation. Images of slides as in Fig. 2 were analyzed. At least 100 nuclei were scored for each time point. Bars indicate mean ± S.E. of the percent positive cells for the indicated antibody stain (CPD, XPC, XPG, XPA, XPD or XPF, respectively).
3.3. Normal recruitment of XPG and XPA in cells from severely affected patients
Cells from severely affected XP183MA and XP131MA cells have mutations that would produce a 781 amino acid protein with alteration of the 42 last C-terminal amino acids from one allele and premature termination codons from the other allele [9]. As in the cells from the mild patients, XPC protein was normally recruited after localized UV in XP183MA cells (Figs. 2B and 3A). In contrast to the absence of recruitment of XPG and XPA proteins in the cells from the mild patients, XPG and XPA proteins were normally recruited to the damage sites immediately after local UV damage in cells from the severe patients (Figs. 2B and 3A). There were 64–84% of the XP131MA and XP183MA cells that showed localized areas of XPG and XPA proteins immediately after UV exposure. Thus, this alteration of XPB helicase protein did not impair recruitment of XPG and XPA to sites of UV damage.
However, XPD and XPF proteins showed very little recruitment immediately after UV with 8% of cells showing XPD positive areas and 2–4% showing XPF positive areas (Figs. 2B and 3A). By 0.5h the proportion of cells showing XPD protein increased to normal levels (64–78%). However, the proportion of cells showing XPF recruitment was only 2–12% at 0.5h in the cells from the severely affected patients. By 3h post-UV, 43–71% of the cells from the severely affected patients showed XPF positive areas. XPC and XPF strongly co-localized 3h after UV (data not shown). Our results indicate that the XPB mutations in these cells resulted in reduced levels of TFIIH [9,23,24] with partial delay of recruitment of XPD and marked delay of XPF to sites of UV-induced DNA damage.
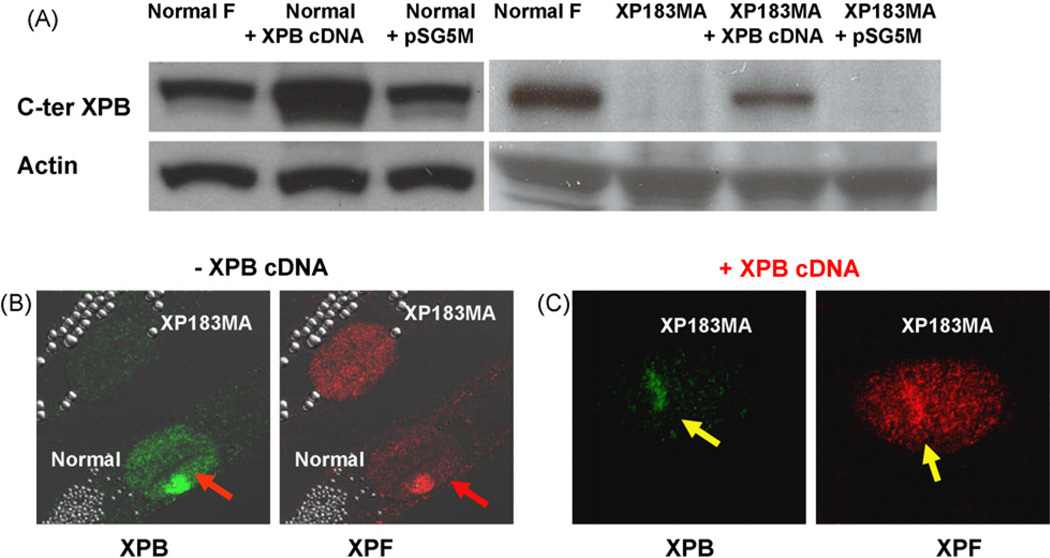
3.4. Over-expression of XPB protein in cells from severely affected patient restores XPF recruitment
Cells from the severe XP/CS patient XP183MA encode XPB protein that was not detectable by immunofluorescence or Western blotting using an antibody to the C-terminal region (Fig. 4A). Immediately (<0.1 h) after UV exposure, in contrast to normal cells, the recruitment of XPB and XPF proteins was not seen in XP183MA cells (Figs. 2B and 4B). To examine whether the rapid recruitment of XPF required XPB, a plasmid containing wild-type XPB cDNA was transfected into XP183MA and normal cells in a transient transfection assay. In transfected XP183MA cells, over-expressed XPB proteins were detected by Western blotting (Fig. 4A) and were visible through the whole nucleus using immunofluorescence (data not shown). XPF proteins were recruited immediately after local UV damage to sites in the cells expressing wild-type XPB cDNA(Fig. 4C). Thus, over-expression of the XPB protein restored the ability of the cells from the severely affected patient to recruit XPF protein to sites of UV damage.
Fig. 4.
Enhanced recruitment of XPB and XPF in XP183MA cells transiently transfected with wild-type XPB-cDNA. Normal (AG13145) and XP183MA cells from a severe XPB patient were either not over-expressed or over-expressed with wild-type XPB cDNA. (A) Western blotting of XPB protein in normal and XP183MA cells transiently transfected with wild-type XPB-cDNA or empty vector. Blots were probed with antibody to the C-terminal end of XPB or to β-actin. (B) Immediately (<0.1 h) after local UV-irradiation recruitment of XPB or XPF was detected in normal cells (red arrows) but not in XP183MA cells treated with the empty vector. (C) In contrast, normal recruitment of XPB and XPF was detected in XP183MA cells transiently over-expressing wild-type XPB-cDNA (yellow arrows). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of the article.)
3.5. Effect of XPG and XPA mutations on recruitment of XPB and other NER proteins
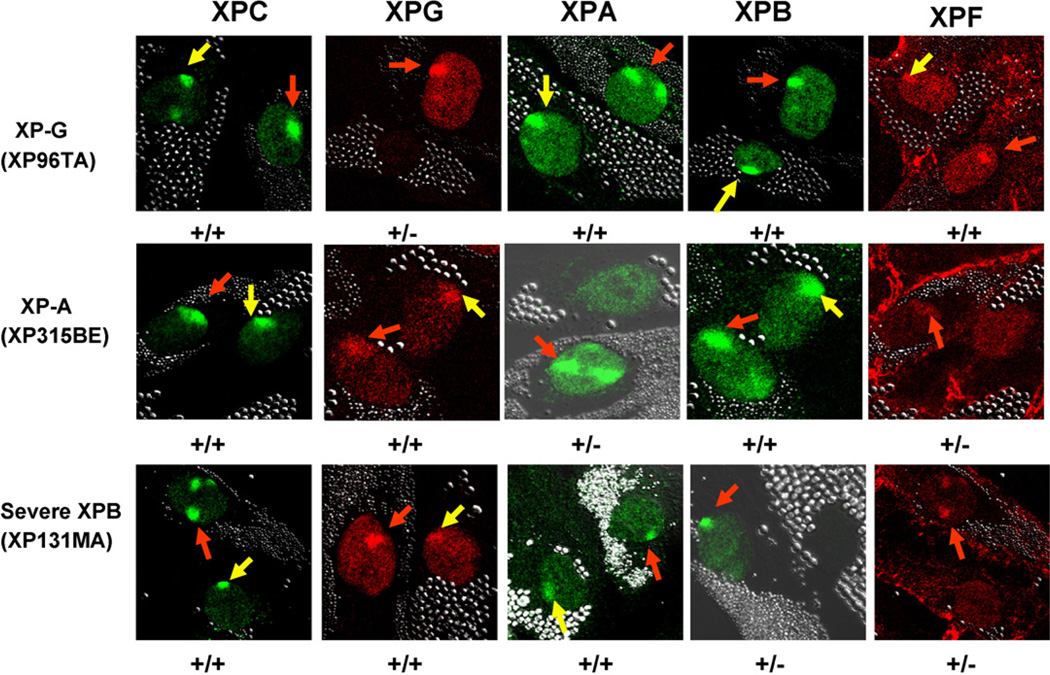
Since mutations in the XPB gene affected recruitment of XPG and XPA proteins to sites of localized UV damage, we investigated the effect of mutations in the XPG and XPA genes on immediate (<0.1 h) recruitment of XPB to sites of localized UV damage (Fig. 5). Cells with a homozygous frameshift mutation in the XPG gene (XP96TA) [12] from a severely affected XP/CS patient showed normal recruitment of XPB as well as XPC, XPA and XPF proteins to the site of localized UV damage (Fig. 5, top row). As expected, since these cells had no detectable XPG mRNA on Northern blotting [12], there was no staining with XPG antibody (Fig. 5, top row). These findings for XPB recruitment are similar to those previously reported for another XPG cell line [25]. Cells with a mutation in the XPA gene (XP315BE) [13] from a mildly affected XP patient showed normal recruitment of XPB as well as XPC and XPG proteins to the site of localized UV damage and reduced recruitment of XPF protein (Fig. 5, middle row.) These findings are similar to those previously reported for another XPA cell line [19]. These cells have reduced XPA staining without localization to the site of UV damage (Fig. 5, middle row). The normal cells showed recruitment of XPC, XPG, XPA, XPB and XPF proteins immediately after UV exposure. As in Fig. 2B, the cells from the severely affected XPB patient (XP131MA) showed normal recruitment of XPC, XPG, and XPA immediately after UV exposure and no recruitment of XPF protein. As expected from the mutations in the XPB gene [9] there was no staining with the C-terminal XPB antibody (Fig. 5, bottom row).
Fig. 5.
Effect of XPG and XPA mutations on recruitment of NER proteins. Normal (0.8 µm beads) and NER deficient XP-G, XP-A or XP-B cells (2.0 µm beads) were treated as in Fig. 2 and fixed immediately (<0.1 h) after UV exposure. Normal cells showed rapid nuclear recruitment of XPC, XPG, XPA, XPB and XPF proteins to the UV damage sites (red arrows). XP-G (XP96TA) cells showed normal recruitment of XPC, XPA, XPB and XPF (yellow arrows) but no staining for XPG protein. In XP-A (XP315BE) cells, normal recruitment of XPC, XPB and XPG was detected (yellow arrows). There was no staining for XPA protein and no recruitment of XPF to the sites of UV damage. In the XP-B cells (XP131MA) from the severe patient, normal recruitment of XPC, XPG and XPA was detected (yellow arrows) (see also Fig. 2B). There was no staining for XPB and no recruitment of XPF protein was detected. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of the article.)
3.6. Impaired redistribution of NER proteins from sites of UV damage in XP-B cells
To investigate whether the rapid redistribution of NER factors after accumulation at damage sites is due to the completion of damage repair, we measured the percentage of cells with positive immunofluorescent areas at different times after UV exposure (Fig. 3A–D). In normal cells, NER proteins were recruited to damage sites immediately after localized UV damage. By 3h after UV, 6–20% of the normal cells showed localized NER proteins and by 24h these proteins were localized in only 0.2–1% of the normal cells. However, as reported previously [15,22] CPD staining was present in about 50% of the normal cells at 24h after UV exposure (Fig. 3D). This correlated with the persistence of CPD in the normal cells as measured by the ELISA assay at 24h (Fig. 1B), a time when there were no detectable 6–4 photoproducts in these cells (Fig. 1A).
In marked contrast to the normal cells, NER proteins in the XP-B cells remained localized at damage sites even 24h after local UV-irradiation (Figs. 2A and B, and 3D). Thus, 54–78% of the XP-B cells were positive for XPC protein at 24 h. A similar proportion (60–80%) of the XP-B cells were positive for CPD staining at 24 h. Cells from the severely affected XPB patients showed a greater proportion of cells with XPG, XPA, XPD and XPF proteins (40–83%) than the cells from the milder patients (10–50%) at 24 h. The ratio of NER protein immunofluorescent areas per nucleus versus CPD positive immunofluorescent areas per nucleus also confirmed these results (data not shown). These results indicate that persistence of XPG, XPA, XPD and XPF proteins to sites of damage may be associated with clinical severity of XP-B patients.
4. Discussion
4.1. Reduced repair of photoproducts in XP-B cells: the levels of repaired CPD and 6–4PP correlate with clinical severity
Cells from the milder XP-B patients (XP/CS1BA and XP33BR) have the same heterozygous single base substitution in exon 3 (c.296 T > C, p.F99S) [9,26]. However, this missense mutation is not located in the essential core region of XPB (amino acids 243–686 [27]). A cDNA expression vector with this mutation showed partial DNA repair function [9]. The level of full length XPB protein as assessed by C-terminal XPB antibody was about half of normal in these XP-B cells [9]. The second allele in each of these patients coded for nonsense mutations [9] which likely would not be expressed due to nonsense mediated mRNA decay [28].
Cells from the severely affected XP/CS complex patients (XP131MA and XP183MA) both have the same splice acceptor site mutation in intron 14 in one allele leading to a 781 amino acid protein with alteration of the 42 last C-terminal amino acids. Thus, an antibody to the N-terminal portion of XPB showed about 63–65% of normal levels of protein in cells from the severe patients while an antibody to the C-terminal portion revealed no detectable normal sized XPB protein [9]. This mutation leads to a severe defect in NER and subtle transcription defects in XP11BE [9,29–31]. The C-terminal end of XPB interacts with ERCC1-XPF which makes a incision 5′ to the DNA lesion [4,32,33]. The XP131MA cells have an impaired helix opening in NER [32]. These severe XPB/CS cells also had nonsense mutations in the second allele [9] which likely would not be expressed due to nonsense-mediated mRNA decay [28].
As in earlier studies [24], all the XP-B cells we studied were deficient in the repair of UV-induced CPD and 6–4PP photoproducts (Fig. 1). However, the levels of repair of 6–4PP and CPD in cells from the milder patients were substantially greater than in cells from the severe XP/CS complex patients (Fig. 1). Thus, these results indicate that repair of CPD and 6–4PP are related to XPB function, and the different levels of repair of CPD and 6–4PP correlate with different mutations in the XPB gene. Further, the levels of repair for CPD and 6–4PP in XP-B cells are correlated with clinical severity between relatively mild and severe syndromes [9].
4.2. Early recruitment of the XPC protein complex
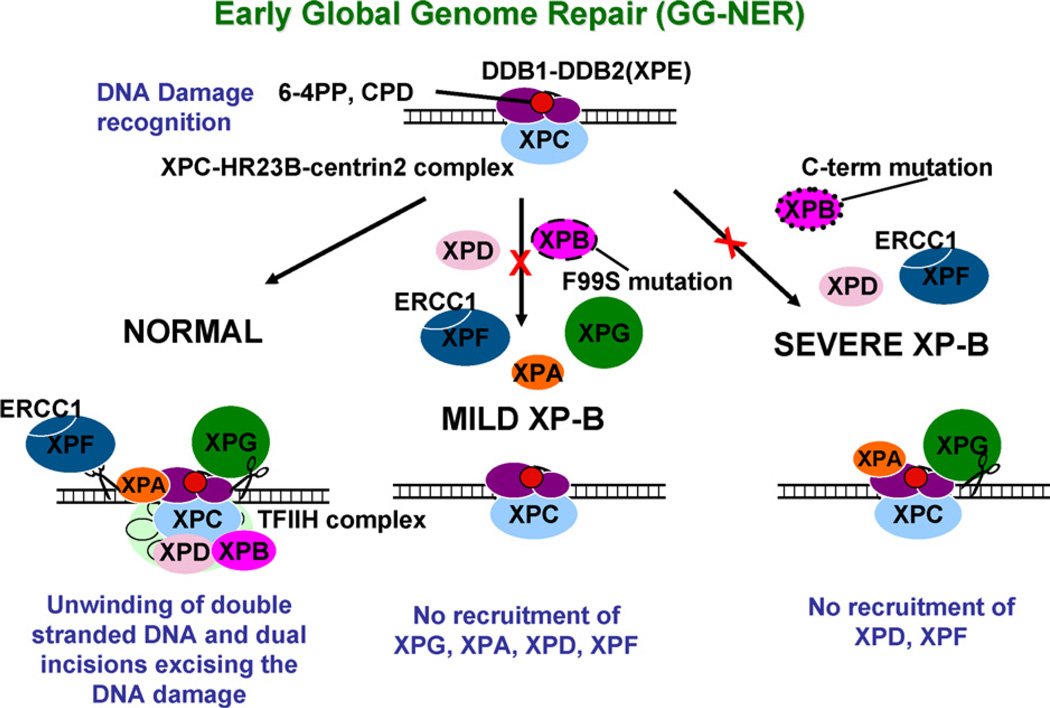
NER proteins which form the NER complex move to sites of DNA damage in a specific sequence [4,6,19,34,35]. The XPC protein is tightly complexed with HR23B, one of two mammalian homologs of Saccharomyces cerevisiae NER factor RAD23, and centrin 2 [36]. This XPC-HR23B-centrin 2 complex is known to play a distinct role in lesion recognition and initiation of assembly of repair machinery in global genome repair. Functional XPC is essential for the formation of the pre-incision complex [37] in vitro and recruitment of TFIIH to DNA damage sites in vivo [19]. Our results show that in normal fibroblasts XPC and the other NER proteins rapidly move to DNA damage sites within 0.1h and are redistributed within 3–4h after UV-irradiation (Table 1 and Fig. 6). In the cells from the XP-B patients, as well as in cells deficient in XPA [19] or XPG [25] protein (Fig. 5 and Table 1), XPC was recruited normally to sites of DNA damage within 0.1h of local irradiation. Conversely, in the absence of XPC protein there was no binding of XPB, XPD, XPG, XPA or XPF proteins (Table 1, data not shown and [19]). This indicates that binding of the XPC-HR23B-centrin 2 complex is independent of XPB, XPG, and XPA binding. These observations support the theory that XPC-HR23B-centrin 2 complex is the first to bind to UV DNA damage sites and initiates assembly of pre-incision complexes (Fig. 6 and discussion in ref. [4]).
Table 1.
Sequential assembly of NER proteins in NER deficient XP-C, XP-B, XP-G or XP-A cells
| Normal | XP-C | XP-B - Mild | XP-B - Severe | XP-G | XP-A | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.1 hr | 0.5 hr | 3 hr | 24 hr | 0.1 hr | 0.5 hr | 24 hr | 0.1 hr | 0.5 hr | 3 hr | 24 hr | 0.1 hr | 0.5 hr | 3 hr | 24 hr | 0.1 hr | 0.5 hr | 24 hr | 0.1 hr | 0.5 hr | 24 hr | |
| XPC | +++ | +++ | + | − | − | − | − | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | + | +++ | +++ | ++ |
| XPB | +++ | +++ | + | − | − | − | − | − | +++ | +++ | + | − | − | − | − | +++ | +++ | ++ | +++ | +++ | ++ |
| XPD | +++ | +++ | + | − | − | − | − | − | +++ | +++ | + | − | +++ | +++ | +++ | +++ | +++ | ++ | +++ | +++ | ++ |
| XPG | +++ | +++ | + | − | − | − | − | − | +++ | +++ | + | +++ | +++ | +++ | +++ | − | − | − | +++ | +++ | ++ |
| XPA | +++ | +++ | + | − | − | − | − | − | +++ | +++ | + | +++ | +++ | +++ | +++ | +++ | +++ | ++ | − | − | − |
| XPF | +++ | +++ | + | − | − | − | − | − | +++ | +++ | + | − | − | +++ | +++ | +++ | +++ | + | − | − | − |
Normal (AG13145), XP-C (XP21BE), XP-B mild (XP33BR, XPCS1BA), XP-B severe (XP131MA, XP183MA), XP-G (XP96TA), XP-A (XP315BE).
Fig. 6.
Schematic diagram of early stages (<0.1 h) of global genome nucleotide excision repair in cells from normal, mild XP-B and severe XP-B patients. DNA damage recognition is accomplished by binding of DDB2 (XPE) – DDB1 proteins (purple ovals) to the 6–4 PP or the CPD (red circle) along with the XPC-HR23B-centrin2 complex (blue oval). In the normal cells (left column) this is followed by unwinding of double stranded DNA which is associated with binding of XPG (green circle), XPA (orange oval), and the TFIIH complex (light green) including XPB (dark pink oval) and XPD (pink oval). Dual incisions excising the DNA damage in an approximately 30 nucleotide segment are carried out by the ERCC1-XPF 5′ endonuclease (gray ovals with scissors) and the XPG 3′ endonuclease (green circle with scissors). In the cells from the mild XP-B patients with a F99S mutation (middle column) there is normal recruitment of XPC to the site of the DNA damage but there is no recruitment of XPG, XPA, XPD or XPF. In contrast, the cells from the severe XP-B patients (right column) with a C-terminal mutation show normal recruitment of XPC, XPG and XPA but no recruitment of XPD or XPF to the site of the DNA damage. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of the article.)
4.3. Impaired recruitment of XPG, XPA and XPF in cells from the mild patients
In normal cells, XPC, XPG, XPA, XPD, XPB and XPF proteins are rapidly recruited to the DNA damage sites (Figs. 2, 3, 5, 6 and Table 1 and discussion in ref. [4]). In cells from mild XPB patients the recruitment of XPG, XPA and XPF was delayed until 0.5h (Figs. 2 and 3 and Table 1). Our results indicate that functional XPB protein was required for rapid recruitment of these NER proteins and that the F99S mutation in the cells from mild XP-B patients impaired this activity. The presence of reduced levels of this defective XPB protein resulting in low levels of TFIIH [9,23,24] delayed the normal recruitment of other NER proteins. This is in agreement with the recent report of delayed recruitment of XPG to local UV damage in XPCS1BA (XP-B) cells [38].
4.4. Delayed recruitment of XPD and XPF but normal recruitment of XPG and XPA proteins in cells from severely affected patients
In the XP-B cells from the severely affected patients, the recruitment of XPD was delayed to about 0.5h and recruitment of XPF did not reach normal levels during the 24h of observation in the XP183MA cells (Fig. 3) indicating that functional XPB was required for rapid recruitment of XPD and XPF in these cells. The XP131MA cells did not show any accumulation of ERCC1 in the DNA damage sites within 3min [39] suggesting that the XPB helicase activity is required for the recruitment of the ERCC1-XPF complex. Coin et al. [40] showed that phosphorylation of S751 of XPB controls the 5′ incision by ERCC1/XPF whereas the 3′ incision by XPG is unaffected. This C-terminal region of XPB is altered in the cells from the severely affected patients [9]. We found that over-expression of XPB restored the ability of the XP-B cells to recruit XPB and XPF proteins to UV damage sites (Fig. 4C) clearly linking the level of the XPB protein to the rapid recruitment of NER proteins. Thus, XPB is essential for XPF recruitment.
XPG is the human endonuclease that cuts 3′ to DNA lesions during NER [4]. In contrast to the results in the cells from the mild XP-B patients, the recruitment of XPG and XPA proteins was strongly induced immediately after local-UV damage in the cells from the severely affected XP/CS complex patients (Figs. 2, 3 and 5 and Table 1). The XPB protein in these cells is mutated with alterations of the final 42 C-terminal amino acids [9]. This indicates that the recruitment of XPG and XPA to DNA damage sites was unaffected by this alteration of the C-terminus of the XPB helicase but that the presence of XPB protein with the F99S missense mutation delayed NER protein recruitment.
XPB is a component of TFIIH. It is unclear if XPG function requires functional TFIIH for NER assembly [41,42]. As in our experiments, Zotter et al. [38] found that the XPCS1BA cells from the mild XPB patient show delayed recruitment of XPG protein to sites of localized DNA damage. They concluded that XPG binding required TFIIH helicase activity. In contrast to this conclusion we found that the cells from the severe XPB patients have normal XPG localization (Figs. 2, 3, 5 and 6 and Table 1). In cells lacking detectable XPG, XPB protein was normally recruited to damage sites (Table 1 and Fig. 5 and [25]). This indicates that strong XPG–XPB interaction is not required for the recruitment of XPB or XPG to sites of UV damage in DNA. Similarly, the recruitment of XPA and XPF was normal in cells lacking detectable XPG protein (Table 1 and Fig. 5). XPB, XPA and XPF recruitment to sites of UV damage does not require the presence of XPG suggesting that XPG and XPF are recruited to DNA damage independent of each other and that XPB and XPA can associate with the complex without the functional XPG endonuclease.
4.5. Recruitment of XPB and XPG does not depend on functional XPA
In cells lacking detectable XPA protein, XPB and XPG were recruited after local UV treatment (Table 1; Fig. 5 and [19]) demonstrating that the recruitment of XPB and XPG to UV damage sites does not require XPA. The assembly of a functional incision complex requires the binding of structure-specific endonucleases XPG and ERCC1-XPF responsible for 3′ and 5′ incision in NER, respectively [4]. While XPG was recruited normally, XPF was not recruited to damaged sites in cells lacking XPA (Table 1; Fig. 5 and [19]). Thus, XPA is needed for the assembly of the 5′ endonuclease XPF-ERCC1 in the incision complex but the 3′ endonuclease XPG can associate with the complex independently of functional XPA. In locally irradiated XP-F (XP24KY) cells, XPG and XPA recruitment was unaffected by the absence of functional XPF [19] supporting the theory that binding of XPF occurs later than binding of XPG and XPA at sites of DNA damage [4].
4.6. Assembly of the incision complex
Some features of the early phase of sequential assembly of the NER proteins in global genome repair (reviewed in ref. [4]) in normal and NER deficient XP cells are described in Fig. 6 and Table 1. Repair of photolesions in damage sites requires the assembly of the NER incision complex and hence the recruitment of NER proteins [19]. To examine whether the rapid recruitment of NER proteins to damage sites requires other NER proteins, we examined the co-localization patterns of NER proteins in various repair deficient XP-C, XP-A, XP-B and XP-G cells immediately after local UV damage (Table 1 and Fig. 5). Normal cells showed rapid nuclear recruitment of NER proteins at the damage sites. In NER deficient XP-G, XP-A and XP-B cells, rapid recruitment of XPC was seen immediately after UV-irradiation (Table 1; Figs. 2, 3 and 5). Thus, XPC (a part of the XPC-HR23B-centrin 2 complex)was first recruited to site of localized DNA damage as indicated by the co-localization of CPD and XPC immunofluorescence and is independent of XPB, XPG and XPA binding (Fig. 6, left column). In cells from the severe XPB patients, with a C-terminal alteration in the XPB protein, the recruitment of XPG and XPA to DNA damage sites was normal at this time (Figs. 2B, 3C, 5 and 6, right column). The region of the XPB protein from amino acids 198–387 was reported to be involved in binding of XPG [43]. This may not be affected by the C-terminal mutation in the cells from the severe XP-B patients. However, XPF was not recruited to damaged sites in these cells. Thus, XPB protein was not essential for recruitment of XPG and XPA, but XPB was required for the subsequent recruitment of XPF. However, cells from the mild XP-B patients with the F99S mutation showed delayed binding of XPG and XPA indicating a possible modulating role for XPB in binding of XPG and XPA to TFIIH (Fig. 5, middle). There was rapid recruitment of XPG in cells lacking XPA and conversely there was rapid binding of XPA in cells lacking XPG, thus these proteins were not essential for the recruitment of each other (Table 1 and Fig. 5 and [19,25]). Similarly, there was normal localization of XPB, XPD and XPF in cells lacking XPG (Table 1 and Fig. 6). It has been reported [25] that XPB recruitment to damage sites does not require the presence of XPG. Thus, recruitment of XPC, XPB, XPA and XPF to DNA damage sites does not require of XPG. Cells lacking XPA showed normal recruitment of XPB, XPD and XPG, but the recruitment of XPF was affected in these cells (Table 1 and Fig. 6 and [19]). Thus, XPF recruitment occurs after that of XPB and XPA and requires both these proteins.
4.7. The NER proteins are unable to redistribute in the absence of damage repair
NER proteins were greatly reduced by 3 and 24h they were no longer found in nucleus of normal fibroblast cells suggesting that the redistribution of NER proteins from local UV damage sites coincides with completion of repair synthesis activity [41]. In this study, antibodies to 6–4PP and CPD were used to identify the recruitment of NER proteins. In normal cells using the ELISA assay, removal of 6–4PP was complete by 6h while about 30% of the CPD were detectable at 24h (Fig. 1 and [15,22]). CPD were visualized by immunofluorescence in about 75% of the normal cell nuclei immediately after UV and fell to about 50% by 24h (Fig. 3). However, the redistribution of NER proteins from sites of DNA damage seems to correlate with repair of 6–4PP rather than CPD suggesting that 6–4PP are main stimulus for recruitment of NER protein and XPC-HR23B has a higher affinity for 6–4PP [44,45]. Our findings were also corroborated by the observation that 24h after exposure to 100 J/m2 UV, localized areas of 6–4PP were not detected in normal cells (data not shown), indicating virtually complete repair of these 6–4PP (Fig. 1A).
In contrast to normal cells, all of NER proteins persisted up to 24h post-UV-irradiation in XP-B cells (Figs. 2 and 3 and Table 1). XP131MA cells from a severely affected XPB patient, showed higher persistent accumulation of NER proteins than XP patients with milder disease suggesting that persistent accumulation correlated with clinical severity of XP-B patients. Thus, the overall data indicate that unless DNA damage is excised, initial factors of NER assembly remain physically associated with the DNA damage. Ineffectual repair of UV-induced photoproducts resulting from delayed recruitment and impaired redistribution of NER proteins may contribute to the markedly increased frequency of skin cancer in XP patients.
Acknowledgements
This research was supported by the Intramural Research Program of the NIH and the Center for Cancer Research of the National Cancer Institute. We thank Susan Garfield and Stephen Wincovitch of NCI for assistance with the confocal microscopy and Dr. John J. DiGiovanna for helpful discussions.
REFERENCES
- 1.Bootsma D, Kraemer KH, Cleaver JE, Hoeijmakers JHJ. Nucleotide excision repair syndromes: xeroderma pigmentosum, Cockayne syndrome, and trichothiodystrophy. In: Vogelstein B, Kinzler KW, editors. The Genetic Basis of Human Cancer. 2nd ed. New York: McGraw-Hill; 2002. pp. 211–237. [Google Scholar]
- 2.Lindahl T, Wood RD. Quality control by DNA repair. Science. 1999;286:1897–1905. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- 3.Moriwaki S, Kraemer KH. Xeroderma pigmentosum—bridging a gap between clinic and laboratory. Photodermatol. Photoimmunol. Photomed. 2001;17:47–54. doi: 10.1034/j.1600-0781.2001.017002047.x. [DOI] [PubMed] [Google Scholar]
- 4.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. Washington, DC: ASM Press; 2006. [Google Scholar]
- 5.Petit C, Sancar A. Nucleotide excision repair: from E. coli to man. Biochimie. 1999;81:15–25. doi: 10.1016/s0300-9084(99)80034-0. [DOI] [PubMed] [Google Scholar]
- 6.Tapias A, Auriol J, Forget D, Enzlin JH, Scharer OD, Coin F, Coulombe B, Egly JM. Ordered conformational changes in damaged DNA induced by nucleotide excision repair factors. J. Biol. Chem. 2004;279:19074–19083. doi: 10.1074/jbc.M312611200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Laat WL, Jaspers NGJ, Hoeijmakers JHJ. Molecular mechanism of nucleotide excision repair. Genes Dev. 1999;13:768–785. doi: 10.1101/gad.13.7.768. [DOI] [PubMed] [Google Scholar]
- 8.Sarker AH, Tsutakawa SE, Kostek S, Ng C, Shin DS, Peris M, Campeau E, Tainer JA, Nogales E, Cooper PK. Recognition of RNA polymerase II and transcription bubbles by XPG, CSB, and TFIIH: insights for transcription-coupled repair and Cockayne Syndrome. Mol. Cell. 2005;20:187–198. doi: 10.1016/j.molcel.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 9.Oh KS, Khan SG, Jaspers NG, Raams A, Ueda T, Lehmann A, Friedmann PS, Emmert S, Gratchev A, Lachlan K, Lucassan A, Baker CC, Kraemer KH. Phenotypic heterogeneity in the XPB DNA helicase gene (ERCC3): xeroderma pigmentosum without and with Cockayne syndrome. Hum. Mutat. 2006;27:1092–1103. doi: 10.1002/humu.20392. [DOI] [PubMed] [Google Scholar]
- 10.Bartenjev I, Butina MR, Potocnik M. Rare case of Cockayne syndrome with xeroderma pigmentosum. Acta Derm. Venereol. 2000;80:213–214. doi: 10.1080/000155500750043032. [DOI] [PubMed] [Google Scholar]
- 11.Khan SG, Oh KS, Emmert S, Shahlavi T, Baker CC, Schmidt D, DiGiovanna JJ, Kraemer KH. An idenitical mutation in the human XPC DNA repair gene in two patients with and without neurological symptoms. J. Investig. Dermatol. 2004;122:A85. (ref. type: abstract). [Google Scholar]
- 12.Emmert S, Slor H, Busch DB, Batko S, Albert RB, Coleman D, Khan SG, Abu-Libdeh B, DiGiovanna JJ, Cunningham BB, Lee MM, Crollick J, Inui H, Ueda T, Hedayati M, Grossman L, Shahlavi T, Cleaver JE, Kraemer KH. Relationship of neurologic degeneration to genotype in three xeroderma pigmentosum group G patients. J. Investig. Dermatol. 2002;118:972–982. doi: 10.1046/j.1523-1747.2002.01782.x. [DOI] [PubMed] [Google Scholar]
- 13.Imoto K, Oh KS, Boyle J, Nadem C, Ueda T, Busch DB, Gozukara E, Khan SG, Tamura D, DiGiovanna JJ, Kraemer KH. Twelve novel mutations in 14 xeroderma pigmentosum group A families: phenotype–genotype correlation and haplotype analysis. J. Investig. Dermatol. 2006;126:A78. [Google Scholar]
- 14.Imoto K, Kobayashi N, Katsumi S, Nishiwaki Y, Iwamoto TA, Yamamoto A, Yamashina Y, Shirai T, Miyagawa S, Dohi Y, Sugiura S, Mori T. The total amount of DNA damage determines ultraviolet-radiation-induced cytotoxicity after uniformor localized irradiation of human cells. J. Investig. Dermatol. 2002;119:1177–1182. doi: 10.1046/j.1523-1747.2002.19514.x. [DOI] [PubMed] [Google Scholar]
- 15.Kobayashi N, Katsumi S, Imoto K, Nakagawa A, Miyagawa S, Furumura M, Mori T. Quantitation and visualization of ultraviolet-induced DNA damage using specific antibodies: application to pigment cell biology. Pigment Cell Res. 2001;14:94–102. doi: 10.1034/j.1600-0749.2001.140204.x. [DOI] [PubMed] [Google Scholar]
- 16.Mori T, Nakane M, Hattori T, Matsunaga T, Ihara M, Nikaido O. Simultaneous establishment of monoclonal antibodies specific for either cyclobutane pyrimidine dimer or (6–4)photoproduct from the same mouse immunized with ultraviolet-irradiated DNA. Photochem. Photobiol. 1991;54:225–232. doi: 10.1111/j.1751-1097.1991.tb02010.x. [DOI] [PubMed] [Google Scholar]
- 17.Nakagawa A, Kobayashi N, Muramatsu T, Yamashina Y, Shirai T, Hashimoto MW, Ikenaga M, Mori T. Three-dimensional visualization of ultraviolet-induced DNA damage and its repair in human cell nuclei. J. Investig. Dermatol. 1998;110:143–148. doi: 10.1046/j.1523-1747.1998.00100.x. [DOI] [PubMed] [Google Scholar]
- 18.Jaspers NG, Bootsma D. Genetic heterogeneity in ataxia-telangiectasia studied by cell fusion. Proc. Natl. Acad. Sci. U.S.A. 1982;79:2641–2644. doi: 10.1073/pnas.79.8.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Volker M, Mone MJ, Karmakar P, van Hoffen A, Schul W, Vermeulen W, Hoeijmakers JH, van Driel R, van Zeeland AA, Mullenders LH. Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell. 2001;8:213–224. doi: 10.1016/s1097-2765(01)00281-7. [DOI] [PubMed] [Google Scholar]
- 20.Katsumi S, Kobayashi N, Imoto K, Nakagawa A, Yamashina Y, Muramatsu T, Shirai T, Miyagawa S, Sugiura S, Hanaoka F, Matsunaga T, Nikaido O, Mori T. In situ visualization of ultraviolet-light-induced DNA damage repair in locally irradiated human fibroblasts. J. Investig. Dermatol. 2001;117:1156–1161. doi: 10.1046/j.0022-202x.2001.01540.x. [DOI] [PubMed] [Google Scholar]
- 21.Mone MJ, Volker M, Nikaido O, Mullenders LH, van Zeeland AA, Verschure PJ, Manders EM, van Driel R. Local UV-induced DNA damage in cell nuclei results in local transcription inhibition. EMBO Rep. 2001;2:1013–1017. doi: 10.1093/embo-reports/kve224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riou L, Eveno E, van Hoffen A, van Zeeland AA, Sarasin A, Mullenders LH. Differential repair of the two major UV-induced photolesions in trichothiodystrophy fibroblasts. Cancer Res. 2004;64:889–894. doi: 10.1158/0008-5472.can-03-2070. [DOI] [PubMed] [Google Scholar]
- 23.Botta E, Nardo T, Lehmann AR, Egly JM, Pedrini AM, Stefanini M. Reduced level of the repair/transcription factor TFIIH in trichothiodystrophy. Hum. Mol. Genet. 2002;11:2919–2928. doi: 10.1093/hmg/11.23.2919. [DOI] [PubMed] [Google Scholar]
- 24.Riou L, Zeng L, Chevallier-Lagente O, Stary A, Nikaido O, Taieb A, Weeda G, Mezzina M, Sarasin A. The relative expression of mutated XPB genes results in xeroderma pigmentosum/Cockayne’s syndrome or trichothiodystrophy cellular phenotypes. Hum. Mol. Genet. 1999;8:1125–1133. doi: 10.1093/hmg/8.6.1125. [DOI] [PubMed] [Google Scholar]
- 25.Thorel F, Constantinou A, Dunand-Sauthier I, Nouspikel T, Lalle P, Raams A, Jaspers NG, Vermeulen W, Shivji MK, Wood RD, Clarkson SG. Definition of a short region of XPG necessary for TFIIH interaction and stable recruitment to sites of UV damage. Mol. Cell. Biol. 2004;24:10670–10680. doi: 10.1128/MCB.24.24.10670-10680.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vermeulen W, Scott RJ, Rodgers S, Muller HJ, Cole J, Arlett CF, Kleijer WJ, Bootsma D, Hoeijmakers JH, Weeda G. Clinical heterogeneity within xeroderma pigmentosum associated with mutations in the DNA repair and transcription gene ERCC3. Am. J. Hum. Genet. 1994;54:191–200. [PMC free article] [PubMed] [Google Scholar]
- 27.Fan L, Arvai AS, Cooper PK, Iwai S, Hanaoka F, Tainer JA. Conserved XPB core structure and motifs for DNA unwinding: implications for pathway selection of transcription or excision repair. Mol. Cell. 2006;22:27–37. doi: 10.1016/j.molcel.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 28.Maquat LE. Nonsense-mediated mRNA decay in mammals. J. Cell. Sci. 2005;118:1773–1776. doi: 10.1242/jcs.01701. [DOI] [PubMed] [Google Scholar]
- 29.Weeda G, van Ham RC, Vermeulen W, Bootsma D, van der Eb AJ, Hoeijmakers JH. A presumed DNA helicase encoded by ERCC-3 is involved in the human repair disorders xeroderma pigmentosum and Cockayne’s syndrome. Cell. 1990;62:777–791. doi: 10.1016/0092-8674(90)90122-u. [DOI] [PubMed] [Google Scholar]
- 30.Weeda G, van Ham RC, Masurel R, Westerveld A, Odijk H, de Wit J, Bootsma D, van der Eb AJ, Hoeijmakers JH. Molecular cloning and biological characterization of the human excision repair gene ERCC-3. Mol. Cell. Biol. 1990;10:2570–2581. doi: 10.1128/mcb.10.6.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weeda G, Ma LB, van Ham RC, van der Eb AJ, Hoeijmakers JH. Structure and expression of the human XPBC/ERCC-3 gene involved in DNA repair disorders xeroderma pigmentosum and Cockayne’s syndrome. Nucl. Acids Res. 1991 doi: 10.1093/nar/19.22.6301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Evans E, Moggs JG, Hwang JR, Egly JM, Wood RD. Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. EMBO J. 1997;16:6559–6573. doi: 10.1093/emboj/16.21.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hwang JR, Moncollin V, Vermeulen W, Seroz T, Van Vuuren H, Hoeijmakers JHJ, Egly JM. A 3′→5′ XPB helicase defect in repair/transcription factor TFIIH of xeroderma pigmentosum group B affects both DNA repair and transcription. J. Biol. Chem. 1996;271:15898–15904. doi: 10.1074/jbc.271.27.15898. [DOI] [PubMed] [Google Scholar]
- 34.Rademakers S, Volker M, Hoogstraten D, Nigg AL, Mone MJ, van Zeeland AA, Hoeijmakers JH, Houtsmuller AB, Vermeulen W. Xeroderma pigmentosum group A protein loads as a separate factor onto DNA lesions. Mol. Cell. Biol. 2003;23:5755–5767. doi: 10.1128/MCB.23.16.5755-5767.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang QE, Zhu Q, Wani MA, Wani G, Chen J, Wani AA. Tumor suppressor p53 dependent recruitment of nucleotide excision repair factors XPC and TFIIH to DNA damage. DNA Rep. (Amst.) 2003;2:483–499. doi: 10.1016/s1568-7864(03)00002-8. [DOI] [PubMed] [Google Scholar]
- 36.Nishi R, Okuda Y, Watanabe E, Mori T, Iwai S, Masutani C, Sugasawa K, Hanaoka F. Centrin 2 stimulates nucleotide excision repair by interacting with xeroderma pigmentosum group C protein. Mol. Cell. Biol. 2005;25:5664–5674. doi: 10.1128/MCB.25.13.5664-5674.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sugasawa K, Ng JM, Masutani C, Iwai S, Van der Spek PJ, Eker AP, Hanaoka F, Bootsma D, Hoeijmakers JH. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol. Cell. 1998;2:223–232. doi: 10.1016/s1097-2765(00)80132-x. [DOI] [PubMed] [Google Scholar]
- 38.Zotter A, Luijsterburg MS, Warmerdam DO, Ibrahim S, Nigg A, van Cappellen WA, Hoeijmakers JH, van Driel R, Vermeulen W, Houtsmuller AB. Recruitment of the nucleotide excision repair endonuclease XPG to sites of UV-induced DNA damage depends on functional TFIIH. Mol. Cell. Biol. 2006;26:8868–8879. doi: 10.1128/MCB.00695-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mone MJ, Bernas T, Dinant C, Goedvree FA, Manders EM, Volker M, Houtsmuller AB, Hoeijmakers JH, Vermeulen W, van Driel R. In vivo dynamics of chromatin-associated complex formation in mammalian nucleotide excision repair. Proc. Natl. Acad. Sci. U.S.A. 2004;101:15933–15937. doi: 10.1073/pnas.0403664101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coin F, Auriol J, Tapias A, Clivio P, Vermeulen W, Egly JM. Phosphorylation of XPB helicase regulates TFIIH nucleotide excision repair activity. EMBO J. 2004;23:4835–4846. doi: 10.1038/sj.emboj.7600480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dunand-Sauthier I, Hohl M, Thorel F, Jaquier-Gubler P, Clarkson SG, Scharer OD. The spacer region of XPG mediates recruitment to nucleotide excision repair complexes and determines substrate specificity. J. Biol. Chem. 2005;280:7030–7037. doi: 10.1074/jbc.M412228200. [DOI] [PubMed] [Google Scholar]
- 42.Riedl T, Hanaoka F, Egly JM. The comings and goings of nucleotide excision repair factors on damaged DNA. EMBO J. 2003;22:5293–5303. doi: 10.1093/emboj/cdg489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iyer N, Reagan MS, Wu KJ, Canagarajah B, Friedberg EC. Interactions involving the human RNA polymerase II transcription/nucleotide excision repair complex TFIIH, the nucleotide excision repair protein XPG, and Cockayne syndrome group B (CSB) protein. Biochemistry. 1996;35:2157–2167. doi: 10.1021/bi9524124. [DOI] [PubMed] [Google Scholar]
- 44.Batty D, Rapic’-Otrin V, Levine AS, Wood RD. Stable binding of human XPC complex to irradiated DNA confers strong discrimination for damaged sites. J. Mol. Biol. 2000;300:275–290. doi: 10.1006/jmbi.2000.3857. [DOI] [PubMed] [Google Scholar]
- 45.Wakasugi M, Sancar A. Order of assembly of human DNA repair excision nuclease. J. Biol. Chem. 1999;274:18759–18768. doi: 10.1074/jbc.274.26.18759. [DOI] [PubMed] [Google Scholar]