Summary
Together with peptides, T lymphocytes respond to hydrophobic molecules, mostly lipids, presented by the non-classical CD1 family (CD1a-e). These molecules have evolved complex and diverse binding grooves in order to survey different cellular compartments for self and exogenous antigens, which are then presented for recognition to T-cell receptors (TCRs) on the surface of T cells. In particular, most CD1d-presented antigens are recognized by a population of lymphocytes denominated natural killer T (NKT) cells, characterized by a strong immunomodulatory potential. Among NKT cells, two major subsets (type I and type II NKT cells) have been described, based on their TCR repertoire and antigen specificity. Here we review recent structural and biochemical studies that have shed light on the molecular details of CD1d-mediated antigen recognition by type I and II NKT cells, which are in many aspects distinct from what has been observed for peptide MHC-reactive TCRs.
Keywords: CD1d, antigen presentation, natural killer T cell, lipids, T-cell receptor
Introduction
The αβ or γδ T-cell receptors (TCRs) expressed on T lymphocytes recognize antigens only when presented by an appropriate antigen-presenting molecule. While the recognition of peptides requires presentation by major histocompatibility complex (MHC) class I or II molecules, T lymphocytes can also recognize lipid antigens presented by the MHC I-like CD1 family (1–3). Despite the common evolutionary origin of MHC and CD1 protein families (4), the latter are mostly non-polymorphic, and their antigen-binding grooves have evolved to present hydrophobic molecules. The T lymphocytes restricted by CD1 molecules have been involved in pathogen recognition, tumor immunity, and autoimmune disease pathology (5–7). In particular, the lymphocyte population restricted by the CD1d isotype is denominated natural killer T (NKT) cells due to the initial finding that these cells express both markers typical for natural killer cells and a TCR, characteristic of conventional T cells. While the expression of NK markers does not apply to all NKT cell subsets, NKT cells represent one of the most well characterized populations of innate-like T cells, with a strong immunomodulatory potential. Here we review our current understanding of how lipid antigens are presented by CD1d molecules and recognized by NKT cell TCRs.
The CD1 antigen-presenting molecules
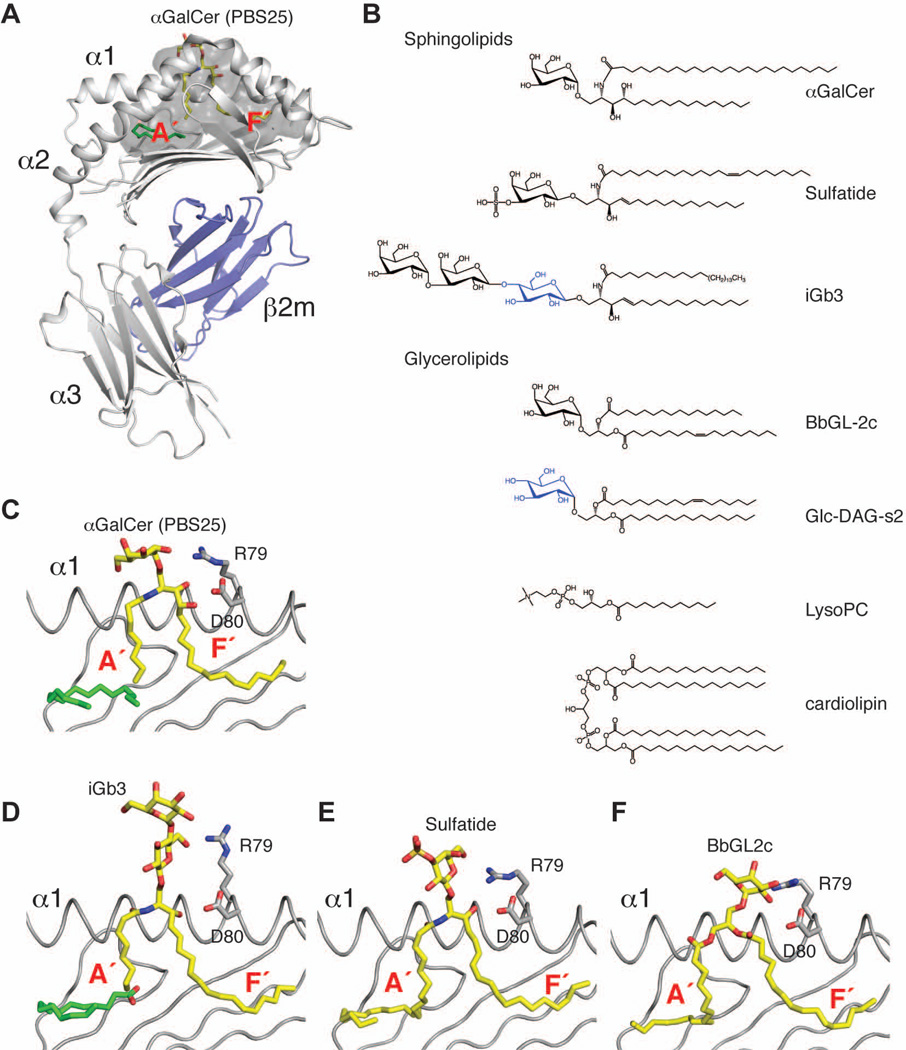
CD1 molecules are assembled in the endoplasmatic reticulum (ER) as non-covalently linked heterodimers of an isotype-specific heavy chain (CD1a-e) and β–2-microglobulin (β2m) (Fig. 1A), although CD1d can also be expressed in a β2m-independent form (8). During its assembly in the ER, CD1 incorporates endogenous lipids and traffic to the plasma membrane. While certain lipids can load onto CD1 directly at the cell surface, generally CD1 has to recycle into endosomal compartments for efficient antigen exchange and loading. Upon trafficking back to the cell surface, the antigen is then presented by CD1 to cognate T cells (9, 10). Structurally, CD1 molecules show similarity with MHC class I molecules (11), as their antigen-binding groove is defined by two α helices (denominated α1 and α2) that sit above an eight-stranded antiparallel β sheet platform (12, 13). The relatively conserved α3 domain pairs with β2m, while a single transmembrane domain connects the extracellular domain to a short cytoplasmic tail that contains an amino acid motif necessary for receptor mediated endocytosis (except for CD1a which lacks any internalization motif). Five CD1 isotypes have been identified, divided in two major groups: Group 1 (CD1a-c) and Group 2 (CD1d) (14, 15). The fifth CD1 molecule, CD1e, is not involved in antigen presentation but instead enhances processing of CD1b antigens in late endosomes (16). While studies of the CD1 molecules in birds revealed what could represent an archetypal lipid-binding groove (17, 18), all mammals studied so far express some combination of CD1 molecules (19, 20). In humans, where one gene for each isotype is expressed, differences in size and shape of the antigen-binding groove, as well as different intracellular trafficking and expression patterns of CD1 result in non-overlapping roles for these molecules in antigen presentation. The last few years have seen a dramatic advancement of our understanding of the differences and specificities within the CD1 family, and structural information is now available for each isotype. While CD1a has a relatively small binding groove and recycle through early endosomes (21, 22), CD1b has the biggest groove of the family and travels through early and late endosomes (23–25), while CD1c has an intermediate size groove and travels to early and late endosomes (26). CD1e has a relatively wide binding groove, suited for rapid lipid exchange (27). The only member of the Group 2, CD1d, represents the only CD1 molecule found in mice and rats, due to a deletion of the Group 1 CD1 members (4). Interestingly, two highly similar (95% sequence identity) copies of CD1d are found in mouse, CD1d1 and CD1d2 (28). While the expression of CD1d2 appears to be limited and dispensable in mice, it is possible that the two isoforms plays different roles in antigen presentation as several of the sequence differences could result in an altered antigen-binding groove shape and therefore specificity (28, 29). However, most studies on CD1d have insofar involved only CD1d1 (hereafter CD1d). CD1d has a groove of intermediate size and travels to late endosomes (11, 30, 31). The CD1d antigen-binding groove is characterized by two pockets, denominated A′ and F′, roughly corresponding to the position of the terminal A and F pockets in MHC I molecules (11) (Fig. 1A). The deeply buried A′ pocket is located toward the N-terminus of the α1 helix and adopts a unique donut-like shape, while the F′ pocket is rather straight, less deep, and located toward the C-terminal end of the α1 helix. Each pocket can bind one alkyl chain from the antigen and spacer molecules, such as fatty acids, are recruited when the alkyl chain of the antigen is too short to satisfactorily fill either pocket (32).
Fig. 1. Antigen presentation by CD1d.
(A). Cartoon representation of the CD1d-β2m heterodimer (PDB ID 1Z5L) with CD1d in grey, β2m in blue, the αGalCer analog PBS25 in yellow and a lipid spacer in green. The antigen-binding groove is shown as a transparent dark grey surface. (B). Several lipid antigens recognized by CD1d-restricted T cells. Glucosyl moieties are shown in blue. (C–F). Detailed view of the antigen binding groove with the α2 helix removed for clarity. Ligands (C: PBS25, PDB ID 1Z5L; D: iGb3, PDB ID 2Q7Y; E: sulfatide, PDB ID 2AKR; F: BbGL2c, PDB ID 3ILQ) are shown in yellow, spacers in green.
Antigen transport and processing
The mechanism of antigen transport and lipid antigen generation have been previously reviewed extensively (33–35), and here we present only a brief summary focused on CD1d. The nature of the lipid antigens presented by CD1 molecules requires the presence of particular mechanisms to induce uptake of these molecules by antigen-presenting cells (APCs) and their loading onto CD1 molecules. Lipid transfer protein such as apolipoprotein E and fatty acid amide hydrolase (FAAH) have been shown to enhance the presentation of certain antigens by CD1d (36, 37). While antigens can be loaded on the cell surface (38), specific proteins present in the endosomal and lysosomal compartments can improve loading efficiency by promoting lipid antigen exchange. Among these, saposins (39–42) and microsomal triglyceride transfer protein (43) have been reported to increase loading of CD1 antigens with a certain degree of specificity. Similar to MHC antigens, lipid antigens can also be processed by lysosomal enzymes to yield active compounds, as demonstrated in case of CD1d for synthetic antigens (44), microbial antigens (16), and self-antigens (39).
Antigen presentation by CD1d
While CD1d traffics through different cellular compartments, it surveys a wide range of lipid molecules of the secretory pathway (45). Accordingly, the CD1d binding groove can bind a variety of different chemical moieties, as reported by mass spectrometric identification of endogenous lipids bound to mouse and human CD1d molecules (45–47). Among these, sphingolipids and glycerolipids with one, two, or four acyl chains represent the major ligand classes (Fig. 1B). Moreover, exogenous lipids such as microbial antigens from Sphingomonas spp. (48–50), Borrelia burgdorferi (51), Streptococcus pneumonia and Group B Streptococcus (52), Helicobacter pylori (53), and even hydrophobic peptides (54–56) and small nonlipidic molecules (57) have been reported to bind CD1d and stimulate NKT cells. While it should be noted that antigen binding to the CD1 groove has not been structurally characterized for nonlipidic antigens, a wealth of crystal structures of mouse and human CD1d in complex with a variety of lipids have been determined in the last few years. Taken together, these structural data allows us to define the molecular rules of CD1d to present antigens for recognition by T lymphocytes.
Sphingolipid antigens
Sphingolipids are the first class of CD1d ligands that have been identified, based on the discovery of glycosylceramides as antigens for NKT cells (58). One of the most well-characterized CD1d ligand is α–galactosyl ceramide (αGalCer), a glycosphingolipid that consists of a phytosphingosine base (C18) that is N-amide linked to a C26 fatty acid to form the phytoceramide backbone, which carries an α-anomeric galactose sugar (Fig. 1B). Structures of αGalCer in complex with human CD1d (30) and of several structurally related αGalCer analogues [PBS25 (31), OCH (59), and variants carrying a phenyl group on the sphingosine chain (60)] in complex with the mouse CD1d ortholog have been described. The orientation of the lipid alkyl chains in the binding groove appeared to be determined by conserved polar contacts between CD1d residues (Asp80, Asp153, Thr156) and the polar moieties of the phytoceramide backbone. As a result, the acyl chain always binds inside the F′ pocket, regardless of the chain length. Accordingly, the size of the F′ pocket has evolved to bind alkyl chains of 16–18 methylene units, typical of sphingosine or phytosphingosine, even though slightly longer chains can also be fitted in a tightly compact conformation (61). In the case of shorter alkyl chains, as found in the αGalCer analog OCH and PBS-25, spacer lipids can be recruited to both A′ and F′ pocket presumably to avoid collapse of the hydrophobic groove (31, 59) (Fig. 1C). The rules described for the synthetic antigen αGalCer and its analogues appear quite conserved among microbial sphingolipid antigens, such as α-galacturonosyl ceramide (GalAGSL) from Sphingomonas spp. (62) and self-antigens, such as isoglobotrihexosyl ceramide (iGb3) (58) (Fig. 1D) and sulfatide (63) (Fig. 1E), further suggesting that the ceramide backbone of the ligand dictates its binding orientation within the CD1d groove.
Glycerolipids
The second major class of antigens presented by CD1d molecules includes diacylglycerolipids formed by a glycerol that is esterified at position sn-1 and sn-2 with fatty acids, while carrying a polar group at position sn-3. Diacylglycerolipids represent the dominant lipids in the ER and are incorporated into CD1d during folding (45–47). Consistent with their increased flexibility, diacylglycerol ligands can adopt different binding orientations in the groove, with the sn-1 or sn-2 linked acyl chains being bound in either pocket. The binding mode is likely dictated by the preference of each pocket for a particular combination of chain length and/or unsaturation degree, as demonstrated in the case of the closely related Borrelia burgdorferi lipids BbGL2c and BbGL2f, which bind in opposite orientations in the mouse CD1d binding groove (64). In particular, the presence of limited (1–2) unsaturations appears to promote binding to CD1d (46, 65), as it introduces a kink in the acyl chain that helps binding in the curved A’ pocket (Fig. 1F). Cardiolipin is a tetra-acyl chain containing phosphoglycerolipid and a major lipid of mitochondrial membranes, as well as bacterial cell walls. This lipid can bind to mouse CD1d using two of the four acyl chains, thereby exposing the charged phosphate groups and the additional two acyl chains for recognition by γδ TCRs (66).
Headgroup positioning by CD1d
While the hydrophobic portion of the antigen is generally buried deep within the CD1d binding groove, the polar portion of the molecule is exposed at the entrance of the groove for recognition by the TCR. The stabilization and orientation of the polar moiety is achieved via several polar residues located at the center of the α1 and α2 helices and has been extensively characterized for glycolipid antigens. In particular, structures of CD1d in complex with sphingolipids carrying an α-linked sugar moiety showed that the sugar is oriented so that the plane of the hexose ring is parallel to the β-sheet defining the bottom of the binding groove (31, 62). Polar contacts with Asp153 and Thr156 in mouse CD1d orient the 2’, 3’ of the galactose (α-GalCer) or galacturonic acid (GalAGSL) and the oxygen of the glycosidic bond, consistent with previous mutagenesis data (67, 68), and resulting in a generally well ordered conformation. Diacylglycerol lipids carrying an α-linked sugar such as the microbial antigens from Borrelia spp. (64) and Streptococcus spp. (52) generally show a more extended conformation that lacks interaction with Asp153 while contacting residues on the α1 helix, especially Arg79 and Asp80 (Fig. 1F). Interestingly, one of the most striking differences between mouse and human CD1d molecules is found at position 155 (equivalent to position 153 in human) of the α2 helix, where a glycine residue is substituted with a tryptophan residue. The presence of this bulky sidechain in human CD1d results in a shift of the hexose residue of approximately 1Å when αGalCer ligands are compared (30, 31). In the case of glycolipids carrying a β-linked sugar, the hexose moiety is generally adopting a highly extended conformation, projecting away from the binding groove and interacting with both α1 and α2 residues, often through water-mediated hydrogen bonds (63, 69–71) (Fig. 1D,E).
CD1d-restricted T cells
Several different T-lymphocyte populations respond to antigens presented by CD1d. Despite their relatively small number compared to MHC class I-, class II-, and even Group I CD1-reactive T cells, they have been found to play important roles in several different aspects of the immune response. Among CD1d-restricted T cells, subsets expressing either αβ or γδ TCRs have been reported. In particular, subsets of γδ T cells responding to phospholipids such as phosphoatidylethanolamine, phosphatidylcholine, and cardiolipin have been described (66, 72–74), but, as our current understanding of how γδ TCRs recognize CD1d-presented antigens is minimal, they will be not discussed further here.
NKT cells
The best characterized population of CD1d-restricted T cells is formed by NKT cells. While initially defined by the expression of both natural killer receptors such as NK1.1/CD161 and αβ TCRs, a substantial proportion of NKT cells do not express NK1.1 (75). Here we define NKT cells as T cell lymphocytes expressing an αβ TCR restricted to the antigen-presenting molecule CD1d. Among NKT cells, two major subsets have been described, denominated type I and type II NKT cells (75). Despite sharing many features typical of innate-like immune cells, the two subsets differ in the repertoire employed by their TCR receptors and therefore in their antigen specificity (7). Type I NKT cells are characterized by expressing a conserved TCR α chain with an invariant germline-encoded rearrangement (Vα14Jα18 in mice, Vα24Jα18 in humans) that pairs with a limited repertoire of β chains (Vβ8.2, Vβ7, Vβ2 in mice, Vβ11 in humans) and by their reactivity to αGalCer. Because of the invariant α chain, type I NKT cells are also known as invariant (iNKT) or Vα14i NKT cells. Interestingly, the limited repertoire of Vβ genes used by this population does not appear to be derived from a preferential pairing with the invariant chain, but it is likely the result of the positive selection by a growing list of self-antigens (76, 77). Type II NKT cells, instead, express a more variable TCR repertoire, with enrichment of certain V genes such as Vα3 and Vβ8 (78–80) and do not respond to stimulation with αGalCer. Both populations are conserved in mice and humans, although the relative frequency appears to be different, with type I NKT cells more abundant in mice and type II in humans. In mouse, type I NKT cells represent ~0.5% of the T lymphocytes in blood and peripheral lymph nodes, ~2.5% of the T cells in the spleen, mesenteric, and pancreatic lymph nodes, and comprise ~30% of the T cells in the liver (7). In humans, type I NKT cells appear to be approximately 10 times less abundant than in mice, although there is marked variability among individuals (7). The distribution of type II NKT cells is generally less understood, although comparison of MHC II−/− and CD1d1−/− mice suggests they represent a fraction of the number of type I NKT cells (~1/10 in spleen) (78, 79). In humans, type II NKT cells appear to constitute a significant proportion of the T cells in bone marrow, liver, and gut (81–84). Both subsets can express either CD4 or CD8 or be double negative (DN), with the exception of murine type I NKT cells that are never CD8+ (85). This is consistent with the hypothesis that CD1d cannot bind CD4 or CD8 (85), likely due to a single residue deletion on its α3 domain (11). Moreover, both populations exhibit features that suggest an innate-like nature, such as constitutive expression of activation markers like CD69 and rapid secretion of both Th1 and Th2 cytokine upon activation (75). In particular, the availability of CD1d tetramers loaded with the potent type I NKT antigen αGalCer (86) allows to readily identify this population in vivo, while Jα18−/−, Cd1d1−/−, Cd1d1−/−Cd1d2−/− and conditional Cd1d1−/− mice allowed to dissect the role of pathological and normal conditions. Despite their relatively low numbers, type I NKT cells appear to be implicated in an astonishing range of physiological processes, including pathogen recognition, tumor immunity, allergy, atherosclerosis, and autoimmune diseases (reviewed in 5–7). Because of the lack of a prototypical antigen equivalent to the type I NKT cell antigen αGalCer, our understanding of the physiological role of type II NKT cells is currently limited. Studies performed using Jα18−/− mice (lacking type I NKT cells) and CD1d1−/− mice (lacking both type I and type II NKT cells) showed that this population has a protective role in EAE (a mouse model of multiple sclerosis) (87), Con-A induced hepatitis (88), ischemic riperfusion (89, 90), diabetes (91, 92), and during hepatitis B infections (93). Interestingly, type II NKT cells appear to have an immunosuppressive role in tumor immunity, in contrast to type I NKT cells (94). Following this observation, it has been suggested that these two populations constitute an immunoregulatory axis influencing tumor immunity development and outcome (95, 96).
Antigen recognition by Type I NKT cells
A conserved binding footprint
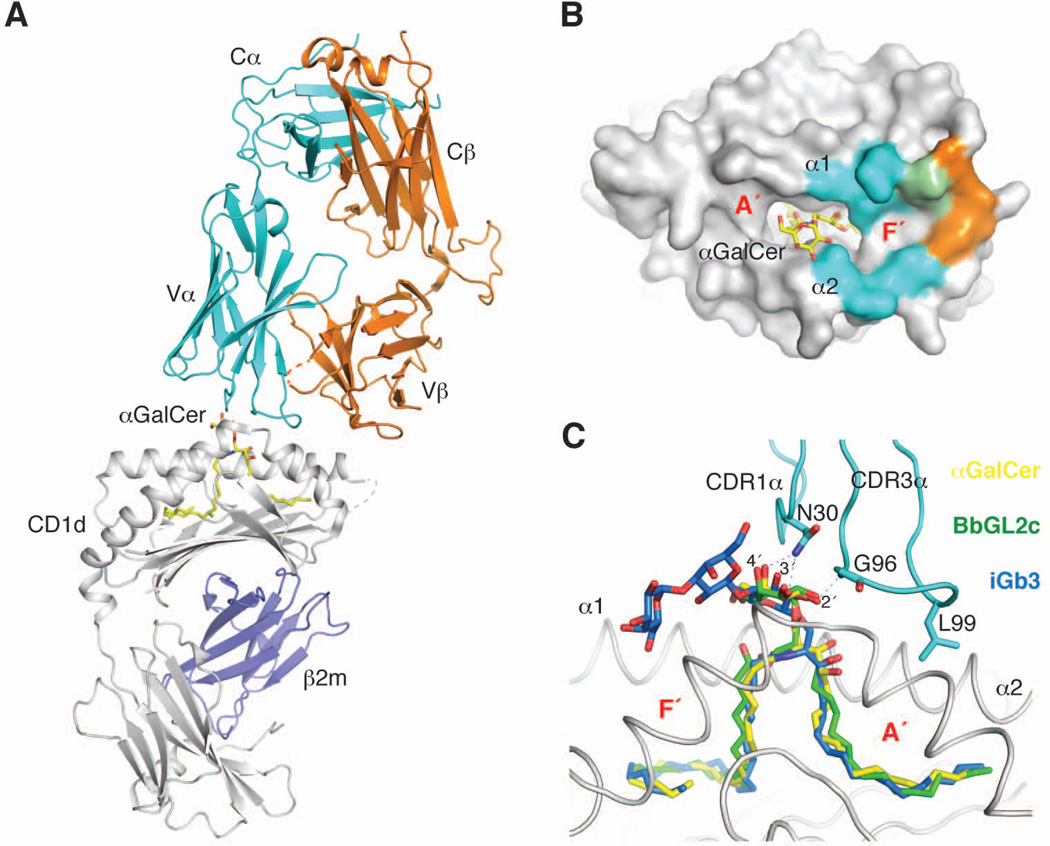
Structural analysis of uncomplexed human and mouse type I NKT TCRs showed that they tend to be relatively rigid structures, with well-ordered complementarity determining region (CDR) loops (70, 97, 98). This in turn raised the question of how such a rigid sequence and structure could bind the wide range of different lipid moieties recognized by type I NKT cells. Mutational studies provided the first insights, suggesting that the TCR uses the same conserved, germline contacts to recognize a variety of antigens, with the CDR1α and CDR3α loops playing critical roles (99, 100). These findings were confirmed by the crystal structures of several ternary complexes between CD1d, lipid antigens, and the TCR (71, 101–111) (Fig. 2A). The type I NKT TCR binds above the CD1d with a conserved footprint dominated by the invariant α chain, while the β chain docks toward the C-terminal part of the α1 helix (Fig. 2B). Because of this docking mode, the TCR adopts a unique parallel docking mode above the antigen-binding groove, radically different from what has been observed for MHC-reactive TCRs, which generally bind diagonally above the antigen presenting molecule (112). Human and mouse type I NKT TCRs docks with the same footprint (101, 102), consistent with their observed cross-reactivity (113). Moreover, the binding mode is generally conserved between mouse TCRs containing Vβ8.2, Vβ7, or Vβ2 chains, suggesting that the overall docking orientation is dictated by the invariant α chain (102, 103). In particular, residues on the CDR1α (Asn30α in mouse, Phe29α in human) and CDR3α (Gly96α) loops contact the 2′, 3′, and 4′ hydroxyl groups of the glycolipids via H bonds, therefore explaining why mannose- and glucose-containing glycolipids are generally weaker antigens (58, 114) (Fig. 2C) and why deoxy αGalCer analogs have weaker affinities for the iNKT TCR (115). Comparison of the structure of the type I NKT TCR before and after complex formation did not highlight major conformational changes, confirming the rigid nature of this TCR (101). Therefore, to achieve its conserved footprint on CD1d, the TCR must induce conformational changes in both the antigen and/or CD1d. While α-linked sphingolipids such αGalCer and the antigens isolated from Sphingomonas spp. are already positioned in an orientation ideal for TCR binding, diacylglycerol ligands such as BbGL2c and GlcDAG-s2 require a rearrangement of their polar group to allow for complex formation (104, 107) (Fig. 1C). This re-orientating ability has been previously described for MHC-reactive TCRs (116); however, the extent of antigen repositioning exerted by the type I NKT TCR is unprecedented, as exemplified in the case of the self-antigen iGb3 (Fig 1D, Fig 2C). This antigen is characterized by the presence of an extended trisaccharide headgoup β-linked to the ceramide backbone (Galα1–3–Galβ1–4–Glcβ1–1Cer), with the terminal sugar being important for antigenicity (117). Previous studies suggested that iGb3 is recognized with a conserved TCR footprint (99), and this was confirmed by crystal structures of the ternary complex (106, 108). Comparison of the mCD1d-iGb3 structures in the presence or absence of the iNKT TCR (70, 106, 108) showed how the TCR squashes the β-linked trisaccharide so that the proximal sugar adopts the conformation previously observed for α-linked antigens (Fig. 2C). Interestingly, the terminal α-linked sugar is not directly contacted by the TCR but instead is locked in position by a small pocket on the α2 helix of CD1d, next to Met162, therefore explaining the requirement of this particular terminal linkage for this lipid to be antigenic. The conserved footprint used by the type I NKT TCR extends to phospholipids antigens such as phosphatidylinositol (PI) (71) and lysophosphatidylcholine (LPC) (111), the latter showing slight differences in the conformation of CDR loops that do not contact the antigen. Intriguingly, in addition to the canonical contacts formed with CDR1α and CDR3α, both PI and iGb3 are also in contact with CDR2α (Val 50α, Lys68α) of the iNKT TCR, possibly constituting a second layer of antigen specificity involving complex self-antigens.
Fig. 2. Antigen recognition by the type I NKT TCR.
(A). Ternary complex (PDB ID 3HE6) between CD1d/b2m (grey and blue), αGalCer (yellow), and the type I NKT TCR (α chain in cyan, β chain in orange). (B). Footprint of the type I NKT TCR on CD1d. Residues on the CD1d surface contacting the α chain are shown in cyan, residues contacting the β chain are shown in orange, shared residues in green. (C). Details of the antigen-binding groove showing the superposition of the bound conformations of αGalCer (yellow), BbGL2c (green, PDB ID 3O9W), and iGb3 (blue, PDB ID 3RZC), after being flattened by the TCR. The CDR loops (cyan) contact the antigens exclusively through Asn30α and Gly96α. Polar contacts are shown as dashed lines.
Note how all the bound ligands adopt similar conformation for their first sugar upon TCR binding.
Flexibility of CD1d upon TCR binding has also been reported. The antigen-binding groove of CD1d can readjust to accommodate particular lipid moieties, as in the case of α-linked glycolipids carrying bulky modifications at the 6′ position of the hexose sugar (105). In the case of the ligand naphtyl urea (NU)-αGalCer, a urea linker connects the 6′ hydroxyl group of galactose to a naphtyl group. In the crystal structure of the ternary complex, the NU moiety serves as a third anchor, in addition to the two alkyl chains and binds in a small pocket within the A′ roof that is formed by repositioning of the Met69 side chain, as well as a widening of the groove through an increase of the distance between both α-helices by over 1 Å. Moreover, rearrangement of the side chains in the area above the F′ pocket have been observed upon TCR binding (104). In particular the sidechains of Leu84, Val149 and Leu150 undergo a conformational change to bind the TCR CDR3α loop, especially the side chain of Leu99. These movements results in the formation of an hydrophobic roof above the F′ pocket that appears to play a critical role for the stability of the ternary complex (108, 116).
Binding affinity/kinetics of type I NKT TCRs and implication for binding mechanism and signaling potency
Type I NKT TCR affinity and binding kinetics for CD1d-antigen complexes have been measured by several different techniques, including surface plasmon resonance (SPR) (reviewed in 32), single molecule force spectroscopy (119) and equilibrium tetramer binding (120, 121), the latter giving information of the avidity of the binding. In mouse, the type I NKT TCR binds CD1d-αGalCer complexes with high affinity characterized by a slow dissociation rate (32). Antigens requiring a rearrangement of CD1d or their polar head generally exhibit slower association rates and faster dissociation kinetics, resulting in equilibrium affinities in the micromolar range (64, 104). These results are consistent with a model of complex formation where the rigid TCR CDR loops contact the antigen first, rearranging the position of its polar head, followed by contacts with the CD1d α1 and α2 helices that rearrange the area above the F′ pocket (104). This binding mode is radically different from what is thought to happen with MHC-reactive TCRs (122), although exceptions have been reported (116). Human type I NKT TCRs appear to have a weaker affinity (10–50 times) for antigens presented by human CD1d (32). HuCD1d-αGalCer complexes bind the type I NKT TCR with micromolar affinity (32, 97, 98) and a fast dissociation rate. Interestingly, the TCR affinity generally correlates well with the potency of the NKT cell antigens, as measured in an cell-free antigen presentation assay using type I NKT cell hybridoma lines (52, 64, 108). An important role in modulating the affinity and the potency of type I NKT cell antigens appear to be played by the CDR3β loop of the TCR. This loop has been found to present high diversity in sequence and length in both mouse (123–125) and human (126, 127) type I NKT TCRs, consistent with the lack of conserved contacts required for the formation of the ternary complex. However, differences in the sequence of this loop have been shown to affect the affinity of the TCR for CD1d (128). Finally, binding of the type I NKT TCR to CD1d-αGalCer multimers has been reported to show cooperativity, allowing this TCR to recognize small amount of antigenic complexes on the cell surface (121, 129).
Interplay between lipid and sugars in determining the potency of type I NKT antigens
While initial studies focused on the effect of modifications of the polar portion of αGalCer and its analogs on the potency of these lipids, recent studies showed how the antigenicity is influenced directly by the composition of its hydrophobic moieties. This is exemplified by the previously mentioned Borrelia burgorferi antigens BbGL2c and BbGL2f (51). The first antigen carries a C18:1 oleic acid on position sn-1 and a palmitic acid (C16:0) in the sn-2 position while the latter has the oleic acid in position sn-2 and a linoleic acid (C18:2) chain on sn-1. Importantly, only BbGL2c is able to activate mouse type I NKT cells, while BbGL2f exclusively activate human type I NKT cells. Structural analysis of the binding of these two compounds to mouse CD1d showed how the lipids are bound in opposite orientations in the binding groove, resulting in a different presentation of the galactosyl moiety to the TCR and therefore explaining the potency difference observed between these two highly related lipids (64). However, how both borrelial lipids are presented by human CD1d and also recognized differently by human NKT cells has not been established yet.
Perhaps even more striking, antigenicity can be the result of a very specific combination of a generally weak sugar moiety and a uncommon lipid chain, as in the case of the Streptococcus antigen α-glucosyl diacylglycerol (Glc-DAG-s2) (52, 107). This diacylglycerol antigen carries a palmitic acid in position sn-1, a cis-vaccenic acid (C18:1, n-7) in position sn-2, and an α-anomeric glucose sugar in sn-3 (Fig. 1B). Positional isomers of this antigen with the vaccenic acid in position sn-1, or versions carrying a galactose sugar (which differ from glucose for the orientation of its 4′-OH group) were not active (52, 107). As glycosphingolipids carrying a glucose sugar are generally weaker than their galactose-containing counterparts, it is surprising that GlcDAG-s2 is at least as antigenic as BbGL2c, both in terms of NKT cell activation and affinity for the TCR (64, 107). The structure of the corresponding ternary complex showed how, in presence of the sn-2 vaccenic acid, the equatorial 4′-OH group of the glucose allows the ligand to make a new contact with Gly155 on CD1d, therefore stabilizing the bound conformation of the antigen (107). This novel contact depends on the correct orientation of the glucose sugar, which in turn is influenced by the lipid moieties of the antigen, therefore providing a clear example of the interplay between the different portions of the molecule in determining antigenicity.
Type I NKT cell agonists with immunomodulatory properties
The ability of type I NKT cells to rapidly release both Th1 and Th2 cytokines make them particularly interesting targets for the development of immunomodulatory therapies. The prototypical antigen αGalCer was identified during a screen for compounds with anti-tumoral activity (130, 131). αGalCer induces NKT cell-dependent suppression of tumor growth, mainly through IFN-γ-mediated mechanisms (132–134), and these effects are enhanced by using αGalCer-pulsed DCs (135). NKT cells can also respond to cytokines such as IL-12 and IL-18 and mediate anti-tumor effects in a TCR-independent way (136, 137). However, due to the contrasting effects of the Th1 and Th2 cytokines released by αGalCer-activated type I NKT cells and the induction of post-activation cell anergy (138), phase I clinical trials of αGalCer showed only weak antitumor effects (139–141). Therefore, several attempts have been made to generate αGalCer analogs able to induce preferential expression of T-helper 1 (Th1) or Th2 cytokines. Two of the earliest candidates identified were OCH (142), a Th2 inducer, and C-glycoside (143), a Th1 cytokine inducer. OCH differs from αGalCer by having a truncated sphingosine chain (C8), which results in the recruitment of spacer lipids that fill the F’ pocket of CD1d (59). C-glycoside has its glycosidic O replaced by a methylene group, and as a consequence is resistant to degradation by lysosomal α-galactosidases. Interestingly, the binding mode of the two lipids to CD1d in the type I NKT ternary complex is essentially identical to that of αGalCer (59, 105, 109, 115), suggesting that factors other than their binding mode by the iNKT TCR are responsible for the cytokine profile they induce. Among these, the stability of the ligand-CD1d complex (105), the site of lipid loading (144, 145), their localization in the membrane, and the particular APC presenting the lipid (146) have all been implicated in determining the cytokine profile induced by the ligand. Following the discovery of OCH and C-glycoside, a number of ligands with promising properties have been developed (reviewed in 147). Modifications of either the sphingosine chain, the acyl chain and the sugar moiety [as in the case of NU-αGalCer (105)] have been reported. Interestingly, the moiety contacting the TCR does not need to be an hexose sugar, as threitol (148) and aminocyclitol (110) groups can also activate NKT cells. Moreover, αGalCer and its analogs have also shown potential as a new vaccine adjuvants (149). In particular, a variant of αGalCer carrying a short acid chain terminating in with a p-phenyl group is currently undergoing clinical studies and showed promising results (150, 151).
Antigen recognition by type II NKT cells
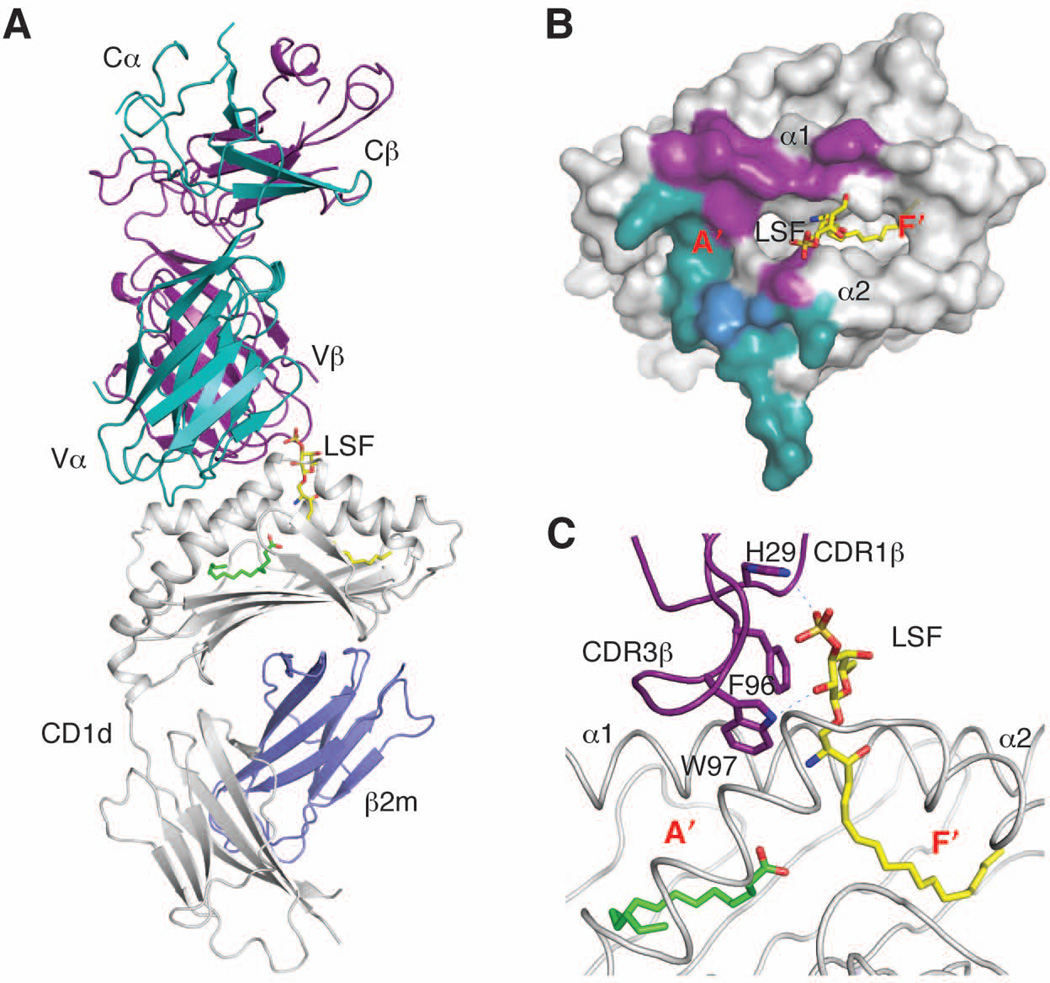
In comparison to type I NKT cells, our understanding of how type II NKT cells recognize antigens is rather limited. However, the recently solved crystal structures of the TCR from the type II NKT hybridoma line XV19/Hy19.3 (38, 78, 87, 152) and of the ternary complexes between mouse CD1d, sulfatide antigens and the same TCR (153, Patel et al., manuscript in preparation) offer the first insight into how the variable repertoire of this population recognizes CD1d-bound antigens (Fig. 3A). Comparison of the unbound and complexed TCR structures shows that the CDR loops undergo conformational changes for the complex to be formed, in contrast to what has been observed for the type I NKT TCR (101). The type II NKT TCR binds diagonally at one end of the CD1d molecule, right above the A′ pocket (Fig. 3B), similar to the diagonal footprint observed for MHC-reactive TCRs (112) but drastically different from the F′ pocket-centered and parallel binding mode of the type I NKT TCR (101) (Fig. 2B). TCR α and β chains contribute equally to the CD1d-TCR interface, with all CDR loops contacting CD1d. In particular, while both CDR3 loops are critical for the complex formation, as demonstrated by mutagenesis studies (153), the two loops appear to have different roles in binding to the CD1d-lysosulfatide complex. The CDR3β loop contacts both CD1d and the antigen through a combination of hydrophobic and polar residues. Interestingly, the lysosulfatide/sulfatide antigen is recognized in a rather unspecific way, pinned between Phe96 and Trp97 on the CDR3β loop, His29 on CDR1β, and Asp153 on the α2 helix of CD1d (Fig. 3C). Importantly, this recognition mode explains why the type II NKT TCR is not compatible with the α-anomeric conformation of type I NKT cell antigens, as the flat and relatively wide α-anomeric carbohydrate would incur in steric clashes with the TCR that would not be tolerated. Interestingly, and in contrast to type I NKT cells, the CDR3α loop does not contact the antigen but instead forms several polar contacts with CD1d residues that form the roof above the A′ pocket (Fig. 3B), involving in particular two Asn residues on CDR3α. It is likely, therefore, that CDR3α of type II NKT cells is positively selected to bind exclusively CD1d and not the antigen. While the available structural information is still limited at this time, the observation that several of the critical residues important for the formation of the mCD1d-sulfatide-Hy19.3 TCR complex are conserved in the oligoclonal repertoire of sulfatide-reactive type II NKT cells, suggests that the recognition mode observed in the complex could extend to a significant proportion of type II NKT TCRs (153). Moreover, the restricted length of CDR3α loops reported for type II NKT cells (79, 80), together with the conservation of the Asn-Asn motif, suggests that most of TCRs from sulfatide-reactive type II NKT cells use CDR3α to bind to CD1d in a conserved orientation, rather than the antigen. This contrasts with MHC-reactive TCRs, where both CDR3α and CDR3β are crucial for peptide antigen discrimination, and the type I NKT cell TCR, which recognizes the antigen exclusively with CDR1α and CDR3α. Instead, unlike any of the previous binding modes, the type II NKT TCR employs CDR1β and CDR3β for antigen recognition and discrimination (Figs 3C, 4).
Fig. 3. Antigen recognition by the type II NKT TCR.
(A). Ternary complex (PDB ID 3ELM) between CD1d/b2m (grey and blue), lysosulfatide (yellow), and the type II NKT TCR (α chain in dark cyan, β chain in purple). (B). Footprint of the type II NKT TCR on CD1d. The shared residue Met162 is shown in blue. (C). Detail of the antigen-binding groove showing the bound lysosulfatide in yellow and a spacer in green. The CDR loops and residues contacting the antigen are shown in purple. Polar contacts are shown as dashed lines.
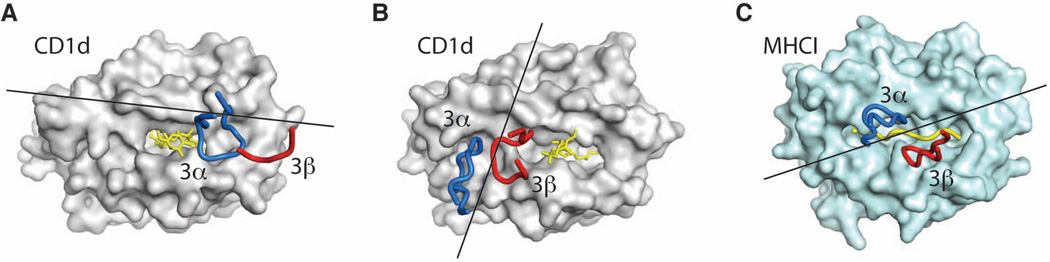
Fig. 4. Docking modes of NKT and MHC-reactive TCRs.
Role of CDR3α (blue) and CDR3β (red) in binding to CD1 (grey), MHC I (light blue), and antigen (yellow). (A). Type I NKT TCR docking to CD1d presenting αGalCer (PDB ID 3HE6). (B). Type II NKT TCR docking to CD1d presenting lysosulfatide (PDB ID 3ELM). (C). MHC-reactive TCR docking to H-2Kb –peptide (PDB ID 2CKB). The lines show the direction of the vector connecting the centroids of the conserved V domain disulfide bonds.
Conclusions
The extensive structural and biochemical data on the recognition of CD1d-presented antigens by NKT TCRs that has recently been accumulated offers an unprecedented, detailed view on how the immune system responds to lipid antigens presented by a non-classical MHC I-like molecule. Based on these studies, we now understand the general rules that control how lipid antigens bind to the CD1d-binding groove, determining how the polar portion of the antigen is exposed to the solvent for recognition. Moreover, the work of our and other groups on the type I NKT TCR provides the basis to understand how a semi-invariant TCR can recognize a range of chemically diverse ligands. This is the result of the ability of the rigid type I NKT TCR to ‘mold’ the exposed headgroup of the ligand and the antigen-presenting molecule in a conserved conformation. Interestingly, this binding mode still imposes stringent requirements on the structure of the antigens, as demonstrated by the interplay between different parts of the antigen molecule in determining potency.
Finally, the recent structures of a type II NKT TCR ternary complex helps us to complete the picture of how NKT cells recognize CD1d-presented antigens and provides us with an opportunity to compare their binding modes with MHC-reactive TCRs, raising novel interrogatives. Surprisingly, the NKT TCRs bind on opposite sides of the antigen-presenting molecule and not in the center as the vast majority of MHC-reactive TCRs (112) (Figs 2B, 3B, 4). As a consequence, only one NKT TCR chain contacts the antigen (α chain for the type I, β chain for the type II), while the second chain contacts CD1d with limited (type I) or extensive (type II) surfaces. Moreover, the two NKT TCRs adopt docking angles that mark the extremes of the range (20–70°) observed for MHC-reactive TCRs, with the type I TCR adopting a parallel docking mode (Fig. 4A), while the type II TCR binds almost perpendicularly (Fig. 4B) on the antigen-binding groove. It is tempting to speculate that this could be the result of the lack of constraints imposed by CD4 and CD8 coreceptors (which do not appear to ligate CD1d), as opposed to MHC-reactive TCRs for which only a limited range of angles result in productive signaling (154). Further studies are therefore required to determine the nature of correlation between the TCR docking mode and the signaling processes occurring in NKT cells. The study of the recognition mechanisms used by NKT cells provided unexpected insight into the variety of strategies that the immune system, and T cells in particular can adopt to respond to internal and external triggers. However, several additional questions remain unanswered, including whether the recognition mode observed for mouse type II NKT TCRs also applies in humans, how the recognition mode observed for NKT cells extends to non-lipidic molecules and how CD1-antigen complexes are recognized by γδ TCRs and Group 1 CD1-reactive T cells. Future work should therefore aim to shed light on these topics, to further expand our understanding of this relatively new field and exploit the therapeutic potential of NKT and lipid-reactive T cells.
Acknowledgments
This work was supported by NIH grant AI074952.
Footnotes
The authors have no conflicts of interest to disclose.
References
- 1.Porcelli S, Brenner MB, Greenstein JL, Terhorst C, Balk SP, Bleicher PA. Recognition of cluster of differentiation 1 antigens by human CD4-CD8- cytolytic T lymphocytes. Nature. 1989;184:3306–3309. doi: 10.1038/341447a0. [DOI] [PubMed] [Google Scholar]
- 2.Bendelac A, Lantz O, Quimby M, Yewdell J, Bennink J, Brutkiewicz RR. CD1 recognition by mouse NK1+ T lymphocytes. Science. 1995;268:863–865. doi: 10.1126/science.7538697. [DOI] [PubMed] [Google Scholar]
- 3.Porcelli S, Morita CT, Brenner MB. CD1b restricts the response of human CD4-8- T lymphocytes to a microbial antigen. Nature. 1992;360:593–597. doi: 10.1038/360593a0. [DOI] [PubMed] [Google Scholar]
- 4.Dascher CC. Evolutionary biology of CD1. Curr Top Microbiol Immunol. 2007;314:3–26. doi: 10.1007/978-3-540-69511-0_1. [DOI] [PubMed] [Google Scholar]
- 5.Brigl M, Brenner MB. CD1: antigen presentation and T cell function. Annu Rev Immunol. 2004;22:817–890. doi: 10.1146/annurev.immunol.22.012703.104608. [DOI] [PubMed] [Google Scholar]
- 6.Kronenberg M. Toward an understanding of NKT cell biology: progress and paradoxes. Annu Rev Immunol. 2005;23:877–900. doi: 10.1146/annurev.immunol.23.021704.115742. [DOI] [PubMed] [Google Scholar]
- 7.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 8.Balk SP, et al. Beta 2-microglobulin-independent MHC class Ib molecule expressed by human intestinal epithelium. Science. 1994;265:259. doi: 10.1126/science.7517575. [DOI] [PubMed] [Google Scholar]
- 9.Sugita M, et al. Separate pathways for antigen presentation by CD1 molecules. Immunity. 1999;11:743–752. doi: 10.1016/s1074-7613(00)80148-x. [DOI] [PubMed] [Google Scholar]
- 10.Hava DL, et al. CD1 assembly and the formation of CD1-antigen complexes. Curr Opin Immunol. 2005;17:88–94. doi: 10.1016/j.coi.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 11.Zeng Z, Castaño AR, Segelke BW, Stura EA, Peterson PA, Wilson IA. Crystal structure of mouse CD1: An MHC-like fold with a large hydrophobic binding groove. Science. 1997;277:339–345. doi: 10.1126/science.277.5324.339. [DOI] [PubMed] [Google Scholar]
- 12.Moody DB, Zajonc DM, Wilson IA. Anatomy of CD1-lipid antigen complexes. Nat Rev Immunol. 2005;5:387–399. doi: 10.1038/nri1605. [DOI] [PubMed] [Google Scholar]
- 13.Zajonc DM, Wilson IA. Architecture of CD1 proteins. Curr Top Microbiol Immunol. 2007;314:27–50. doi: 10.1007/978-3-540-69511-0_2. [DOI] [PubMed] [Google Scholar]
- 14.Calabi F, Milstein C. A novel family of human major histocompatibility complex-related genes not mapping to chromosome 6. Nature. 1986;323:540–543. doi: 10.1038/323540a0. [DOI] [PubMed] [Google Scholar]
- 15.Calabi F, Jarvis J, Martin L. Two classes of CD1 genes. Eur J Immunol. 1989;19:285–292. doi: 10.1002/eji.1830190211. [DOI] [PubMed] [Google Scholar]
- 16.de la Salle H, et al. Assistance of microbial glycolipid antigen processing by CD1e. Science. 2005;310:1321–1324. doi: 10.1126/science.1115301. [DOI] [PubMed] [Google Scholar]
- 17.Zajonc DM, Striegl H, Dascher CC, Wilson IA. The crystal structure of avian CD1 reveals a smaller, more primordial antigen-binding pocket compared to mammalian CD1. Proc Natl Acad Sci USA. 2008;105:17925–17930. doi: 10.1073/pnas.0809814105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dvir H, Wang J, Ly N, Dascher CC, Zajonc DM. Structural Basis for Lipid-Antigen Recognition in Avian Immunity. J Immunol. 2010;184:2504–2511. doi: 10.4049/jimmunol.0903509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Looringh van Beeck FA, et al. Two canine CD1a proteins are differentially expressed in skin. Immunogenetics. 2008;60:315–324. doi: 10.1007/s00251-008-0297-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Rhijn I, et al. The bovine CD1 family contains group 1 CD1 proteins, but no functional CD1d. J Immunol. 2006;176:4888–4893. doi: 10.4049/jimmunol.176.8.4888. [DOI] [PubMed] [Google Scholar]
- 21.Zajonc DM, Elsliger MA, Teyton L, Wilson IA. Crystal structure of CD1a in complex with a sulfatide self antigen at a resolution of 2.15 A. Nat Immunol. 2003;4:808–815. doi: 10.1038/ni948. [DOI] [PubMed] [Google Scholar]
- 22.Zajonc DM, et al. Molecular mechanism of lipopeptide presentation by CD1a. Immunity. 2005;22:209–219. doi: 10.1016/j.immuni.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 23.Gadola SD, et al. Structure of human CD1b with bound ligands at 2.3 Å a maze for alkyl chains. Nat Immunol. 2002;3:721–726. doi: 10.1038/ni821. [DOI] [PubMed] [Google Scholar]
- 24.Batuwangala T, et al. The crystal structure of human CD1b with a bound bacterial glycolipid. J Immunol. 2004;172:2382–2388. doi: 10.4049/jimmunol.172.4.2382. [DOI] [PubMed] [Google Scholar]
- 25.Garcia-Alles LF, et al. Endogenous phosphatidylcholine and a long spacer ligand stabilize the lipid-binding groove of CD1b. EMBO J. 2006;25:3684–3692. doi: 10.1038/sj.emboj.7601244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scharf L, et al. The 2.5 Å structure of CD1c in complex with a mycobacterial lipid reveals an open groove ideally suited for diverse antigen presentation. Immunity. 2010;33:853–862. doi: 10.1016/j.immuni.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garcia-Alles LF, et al. Crystal structure of human CD1e reveals a groove suited for lipid-exchange processes. Proc Natl Acad Sci USA. 2011;108:13230–13235. doi: 10.1073/pnas.1105627108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bradbury A, Belt K, Neri T, Milstein C. Mouse CD1 is distinct from and co-exists with TL in the same thymus. EMBO J. 1988;7:3081–3086. doi: 10.1002/j.1460-2075.1988.tb03173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Y, Wang B, Chun T, Zhao L. Expression of CD1d2 on thymocytes is not sufficient for the development of NK T cells in CD1d1-deficient mice. J Immunol. 1999;162:4560–4566. [PubMed] [Google Scholar]
- 30.Koch M, et al. The crystal structure of human CD1d with and without alpha-galactosylceramide. Nat Immunol. 2005;6:819–826. doi: 10.1038/ni1225. [DOI] [PubMed] [Google Scholar]
- 31.Zajonc DM, et al. Structure and function of a potent agonist for the semi-invariant natural killer T cell receptor. Nat Immunol. 2005;6:810–818. doi: 10.1038/ni1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Joyce S, Girardi E, Zajonc DM. NKT cell ligand recognition logic: molecular basis for a synaptic duet and transmission of inflammatory effectors. J Immunology. 2011;187:1081–1089. doi: 10.4049/jimmunol.1001910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Libero G De, Mori L. Mechanisms of lipid-antigen generation and presentation to T cells. Trends Immunol. 2006;27:485–492. doi: 10.1016/j.it.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 34.De Libero G, Mori L. Novel insights into lipid antigen presentation. Trends Immunol. 2012;33:103–111. doi: 10.1016/j.it.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 35.Barral DC, Brenner MB. CD1 antigen presentation: how it works. Nat Rev Immunol. 2007;7:929–941. doi: 10.1038/nri2191. [DOI] [PubMed] [Google Scholar]
- 36.van den Elzen P, et al. Apolipoprotein-mediated pathways of lipid antigen presentation. Nature. 2005;437:906–910. doi: 10.1038/nature04001. [DOI] [PubMed] [Google Scholar]
- 37.Freigang S, et al. Fatty acid amide hydrolase shapes NKT cell responses by influencing the serum transport of lipid antigen in mice. J Clin Invest. 2010;120:1873–1884. doi: 10.1172/JCI40451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roy KC, Maricic I, Khurana A, Smith TRF, Halder RC, Kumar V. Involvement of secretory and endosomal compartments in presentation of an exogenous self-glycolipid to type II NKT cells. J Immunol. 2008;180:2942–2950. doi: 10.4049/jimmunol.180.5.2942. [DOI] [PubMed] [Google Scholar]
- 39.Zhou D, et al. Editing of CD1d-Bound Lipid Antigens by Endosomal Lipid Transfer Proteins. Science. 2004;303:523–527. doi: 10.1126/science.1092009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuan W, et al. Saposin B is the dominant saposin that facilitates lipid binding to human CD1d molecules. Proc Natl Acad Sci USA. 2007;104:5551–5556. doi: 10.1073/pnas.0700617104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Winau F, et al. Saposin C is required for lipid presentation by human CD1b. Nat Immunol. 2004;5:169–174. doi: 10.1038/ni1035. [DOI] [PubMed] [Google Scholar]
- 42.León L, et al. Saposins utilize two strategies for lipid transfer and CD1 antigen presentation. Proc Natl Acad Sci USA. 2012;109:4357–4364. doi: 10.1073/pnas.1200764109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dougan SK, et al. Microsomal triglyceride transfer protein lipidation and control of CD1d on antigen-presenting cells. J Exp Med. 2005;202:529–539. doi: 10.1084/jem.20050183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prigozy TI, et al. Glycolipid antigen processing for presentation by CD1d molecules. Science. 2001;291:664–667. doi: 10.1126/science.291.5504.664. [DOI] [PubMed] [Google Scholar]
- 45.Yuan W, Kang S-J, Evans JE, Cresswell P. Natural lipid ligands associated with human CD1d targeted to different subcellular compartments. J Immunol. 2009;182:4784–4791. doi: 10.4049/jimmunol.0803981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cox D, et al. Determination of cellular lipids bound to human CD1d molecules. PLoS One. 2009;4:e5325. doi: 10.1371/journal.pone.0005325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haig NA, Guan Z, Li D, McMichael A, Raetz CRH, Xu X-N. Identification of self-lipids presented by CD1c and CD1d. J Biol Chem. 2011;286:37692–37701. doi: 10.1074/jbc.M111.267948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kinjo Y, et al. Recognition of bacterial glycosphingolipids by natural killer T cells. Nature. 2005;434:520–525. doi: 10.1038/nature03407. [DOI] [PubMed] [Google Scholar]
- 49.Mattner J, et al. Exogenous and endogenous glycolipid antigens activate NKT cells during microbial infections. Nature. 2005;434:525–529. doi: 10.1038/nature03408. [DOI] [PubMed] [Google Scholar]
- 50.Kinjo Y, et al. Natural Sphingomonas glycolipids vary greatly in their ability to activate natural killer T cells. Chem Biol. 2008;15:654–664. doi: 10.1016/j.chembiol.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kinjo Y, et al. Natural killer T cells recognize diacylglycerol antigens from pathogenic bacteria. Nat Immunol. 2006;7:978–986. doi: 10.1038/ni1380. [DOI] [PubMed] [Google Scholar]
- 52.Kinjo Y, et al. Invariant natural killer T cells recognize glycolipids from pathogenic Gram-positive bacteria. Nat Immunol. 2011;12:966–974. doi: 10.1038/ni.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chang Y-J, et al. Influenza infection in suckling mice expands an NKT cell subset that protects against airway hyperreactivity. J Clin Invest. 2011;121:57–69. doi: 10.1172/JCI44845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Castaño AR, et al. Peptide binding and presentation by mouse CD1. Science. 1995;269:223–226. doi: 10.1126/science.7542403. [DOI] [PubMed] [Google Scholar]
- 55.Tangri S, Brossay L, Burdin N, Lee DJ, Corr M, Kronenberg M. Presentation of peptide antigens by mouse CD1 requires endosomal localization and protein antigen processing. Proc Natl Acad Sci USA. 1998;95:14314–14319. doi: 10.1073/pnas.95.24.14314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu Y, Teige A, Mondoc E, Ibrahim S, Holmdahl R, Issazadeh-Navikas S. Endogenous collagen peptide activation of CD1d-restricted NKT cells ameliorates tissue-specific inflammation in mice. J Clin Invest. 2011;121:249–264. doi: 10.1172/JCI43964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Van Rhijn I, et al. CD1d-restricted T cell activation by nonlipidic small molecules. Proc Natl Acad Sci USA. 2004;101:13578–13583. doi: 10.1073/pnas.0402838101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kawano T, et al. CD1d-restricted and TCR-mediated activation of valpha14 NKT cells by glycosylceramides. Science. 1997;278:1626–1629. doi: 10.1126/science.278.5343.1626. [DOI] [PubMed] [Google Scholar]
- 59.Sullivan BA, et al. Mechanisms for glycolipid antigen-driven cytokine polarization by Valpha14i NKT cells. J Immunol. 2010;184:141–153. doi: 10.4049/jimmunol.0902880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schiefner A, Fujio M, Wu D, Wong C-H, Wilson IA. Structural evaluation of potent NKT cell agonists: implications for design of novel stimulatory ligands. J Mol Biol. 2009;394:71–82. doi: 10.1016/j.jmb.2009.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tyznik AJ, et al. Glycolipids that Elicit IFN-γ-Biased Responses from Natural Killer T Cells. Chem Biol. 2011;18:1620–1630. doi: 10.1016/j.chembiol.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu D, et al. Design of natural killer T cell activators: structure and function of a microbial glycosphingolipid bound to mouse CD1d. Proc Natl Acad Sci USA. 2006;103:3972–3977. doi: 10.1073/pnas.0600285103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zajonc DM, et al. Structural basis for CD1d presentation of a sulfatide derived from myelin and its implications for autoimmunity. J Exp Med. 2005;202:1517–1526. doi: 10.1084/jem.20051625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang J, et al. Lipid binding orientation within CD1d affects recognition of Borrelia burgorferi antigens by NKT cells. Proc Natl Acad Sci USA. 2010;107:1535–1540. doi: 10.1073/pnas.0909479107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rauch J, et al. Structural features of the acyl chain determine self-phospholipid antigen recognition by a CD1d-restricted invariant NKT (iNKT) cell. J Biol Chem. 2003;278:47508–47515. doi: 10.1074/jbc.M308089200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dieudé M, et al. Cardiolipin binds to CD1d and stimulates CD1d-restricted γδ T cells in the normal murine repertoire. J Immunol. 2011;186:4771–4781. doi: 10.4049/jimmunol.1000921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brossay L, Naidenko O, Burdin N, Matsuda J, Sakai T, Kronenberg M. Structural requirements for galactosylceramide recognition by CD1-restricted NK T cells. J Immunol. 1998;161:5124–5128. [PubMed] [Google Scholar]
- 68.Kamada N, et al. Crucial amino acid residues of mouse CD1d for glycolipid ligand presentation to V(alpha)14 NKT cells. Int Immunol. 2001;13:853–861. doi: 10.1093/intimm/13.7.853. [DOI] [PubMed] [Google Scholar]
- 69.Zajonc DM, Ainge GD, Painter GF, Severn WB, Wilson IA. Structural characterization of mycobacterial phosphatidylinositol mannoside binding to mouse CD1d. J Immunol. 2006;177:4577–4583. doi: 10.4049/jimmunol.177.7.4577. [DOI] [PubMed] [Google Scholar]
- 70.Zajonc DM, Savage PB, Bendelac A, Wilson IA, Teyton L. Crystal structures of mouse CD1d-iGb3 complex and its cognate Valpha14 T cell receptor suggest a model for dual recognition of foreign and self glycolipids. J Mol Biol. 2008;377:1104–1116. doi: 10.1016/j.jmb.2008.01.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mallevaey T, et al. A Molecular Basis for NKT Cell Recognition of CD1d-Self-Antigen. Immunity. 2011;34:315–326. doi: 10.1016/j.immuni.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Agea E, et al. Human CD1-restricted T cell recognition of lipids from pollens. J Exp Med. 2005;202:295–308. doi: 10.1084/jem.20050773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Russano AM, et al. Recognition of pollen-derived phosphatidyl-ethanolamine by human CD1d-restricted gamma delta T cells. J Allergy Clin Immunol. 2006;117:1178–1184. doi: 10.1016/j.jaci.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 74.Russano AM, et al. CD1-restricted recognition of exogenous and self-lipid antigens by duodenal gammadelta+ T lymphocytes. J Immunol. 2007;178:3620–3626. doi: 10.4049/jimmunol.178.6.3620. [DOI] [PubMed] [Google Scholar]
- 75.Godfrey DI, Macdonald HR, Kronenberg M, Smyth MJ, Van Kaer L. NKT cells: what’s in a name? Nat Rev Immunol. 2004;4:231–237. doi: 10.1038/nri1309. [DOI] [PubMed] [Google Scholar]
- 76.Wei DG, Curran SA, Savage PB, Teyton L, Bendelac A. Mechanisms imposing the Vbeta bias of Valpha14 natural killer T cells and consequences for microbial glycolipid recognition. J Exp Med. 2006;203:1197–1207. doi: 10.1084/jem.20060418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gapin L. iNKT cell autoreactivity: what is “self” and how is it recognized? Nat Rev Immunol. 2010;10:272–277. doi: 10.1038/nri2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cardell S, Tangri S, Chan S, Kronenberg M, Benoist C, Mathis D. CD1-restricted CD4+ T cells in major histocompatibility complex class II-deficient mice. J Exp Med. 1995;182:993–1004. doi: 10.1084/jem.182.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Park SH, Weiss A, Benlagha K, Kyin T, Teyton L, Bendelac A. The mouse CD1d-restricted repertoire is dominated by a few autoreactive T cell receptor families. J Exp Med. 2001;193:893–904. doi: 10.1084/jem.193.8.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Arrenberg P, Halder R, Dai Y, Maricic I, Kumar V. Oligoclonality and innate-like features in the TCR repertoire of type II NKT cells reactive to a beta-linked self-glycolipid. Proc Natl Acad Sci USA. 2010;107:10984–10989. doi: 10.1073/pnas.1000576107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Exley MA, et al. A major fraction of human bone marrow lymphocytes are Th2-like CD1d-reactive T cells that can suppress mixed lymphocyte responses. J Immunol. 2001;167:5531–5534. doi: 10.4049/jimmunol.167.10.5531. [DOI] [PubMed] [Google Scholar]
- 82.Exley MA, et al. Cutting edge: Compartmentalization of Th1-like noninvariant CD1d-reactive T cells in hepatitis C virus-infected liver. J Immunol. 2002;168:1519–1523. doi: 10.4049/jimmunol.168.4.1519. [DOI] [PubMed] [Google Scholar]
- 83.Durante-Mangoni E, et al. Hepatic CD1d expression in hepatitis C virus infection and recognition by resident proinflammatory CD1d-reactive T cells. J Immunol. 2004;173:2159–2166. doi: 10.4049/jimmunol.173.3.2159. [DOI] [PubMed] [Google Scholar]
- 84.Fuss IJ, et al. Nonclassical CD1d-restricted NK T cells that produce IL-13 characterize an atypical Th2 response in ulcerative colitis. J Clin Invest. 2004;113:1490–1497. doi: 10.1172/JCI19836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Engel I, et al. Co-receptor choice by V alpha14i NKT cells is driven by Th-POK expression rather than avoidance of CD8-mediated negative selection. J Exp Med. 2010;207:1015–1029. doi: 10.1084/jem.20090557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sidobre S, Kronenberg M. CD1 tetramers: a powerful tool for the analysis of glycolipid-reactive T cells. J Immunol Methods. 2002;268:107–121. doi: 10.1016/s0022-1759(02)00204-1. [DOI] [PubMed] [Google Scholar]
- 87.Jahng A, Maricic I, Aguilera C, Cardell S, Halder RC, Kumar V. Prevention of autoimmunity by targeting a distinct, noninvariant CD1d-reactive T cell population reactive to sulfatide. J Exp Med. 2004;199:947–957. doi: 10.1084/jem.20031389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Halder RC, Aguilera C, Maricic I, Kumar V. Type II NKT cell-mediated anergy induction in type I NKT cells prevents inflammatory liver disease. J Clin Invest. 2007;117:2302–2312. doi: 10.1172/JCI31602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Arrenberg P, Maricic I, Kumar V. Sulfatide-mediated activation of type II natural killer T cells prevents hepatic ischemic reperfusion injury in mice. Gastroenterology. 2011;140:646–655. doi: 10.1053/j.gastro.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yang SH, et al. Sulfatide-reactive natural killer T cells abrogate ischemia-reperfusion injury. J Am Soc Nephrol. 2011;22:1305–1314. doi: 10.1681/ASN.2010080815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang B, Geng YB, Wang CR. CD1-restricted NK T cells protect nonobese diabetic mice from developing diabetes. J Exp Med. 2001;194:313–320. doi: 10.1084/jem.194.3.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kadri N, et al. CD4+ type II NKT cells mediate ICOS and programmed death-1-dependent regulation of Type 1 diabetes. J Immunol. 2012;188:3138–3149. doi: 10.4049/jimmunol.1101390. [DOI] [PubMed] [Google Scholar]
- 93.Baron JL, Gardiner L, Nishimura S, Shinkai K, Locksley R, Ganem D. Activation of a nonclassical NKT cell subset in a transgenic mouse model of hepatitis B virus infection. Immunity. 2002;16:583–594. doi: 10.1016/s1074-7613(02)00305-9. [DOI] [PubMed] [Google Scholar]
- 94.Terabe M, et al. A nonclassical non-Valpha14Jalpha18 CD1d-restricted (type II) NKT cell is sufficient for down-regulation of tumor immunosurveillance. J Exp Med. 2005;202:1627–1633. doi: 10.1084/jem.20051381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ambrosino E, et al. Cross-regulation between type I and type II NKT cells in regulating tumor immunity: a new immunoregulatory axis. J Immunol. 2007;179:5126–5136. doi: 10.4049/jimmunol.179.8.5126. [DOI] [PubMed] [Google Scholar]
- 96.Ambrosino E, Berzofsky JA, Terabe M. Regulation of tumor immunity: the role of NKT cells. Expert Opin Biol Ther. 2008;8:725–734. doi: 10.1517/14712598.8.6.725. [DOI] [PubMed] [Google Scholar]
- 97.Gadola SD, et al. Structure and binding kinetics of three different human CD1d-alpha-galactosylceramide-specific T cell receptors. J Exp Med. 2006;203:699–710. doi: 10.1084/jem.20052369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kjer-Nielsen L, et al. A structural basis for selection and cross-species reactivity of the semi-invariant NKT cell receptor in CD1d/glycolipid recognition. J Exp Med. 2006;203:661–673. doi: 10.1084/jem.20051777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Scott-Browne JP, et al. Germline-encoded recognition of diverse glycolipids by natural killer T cells. Nat Immunol. 2007;8:1105–1113. doi: 10.1038/ni1510. [DOI] [PubMed] [Google Scholar]
- 100.Florence WC, et al. Adaptability of the semi-invariant natural killer T-cell receptor towards structurally diverse CD1d-restricted ligands. EMBO J. 2009;28:3579–3590. doi: 10.1038/emboj.2009.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Borg NA, et al. CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature. 2007;448:44–49. doi: 10.1038/nature05907. [DOI] [PubMed] [Google Scholar]
- 102.Pellicci DG, et al. Differential recognition of CD1d-alpha-galactosyl ceramide by the V beta 8.2 and V beta 7 semi-invariant NKT T cell receptors. Immunity. 2009;31:47–59. doi: 10.1016/j.immuni.2009.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Patel O, et al. Vβ2 natural killer T cell antigen receptor-mediated recognition of CD1d-glycolipid antigen. Proc Natl Acad Sci USA. 2011;108:19007–19012. doi: 10.1073/pnas.1109066108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Li Y, Girardi E, Wang J, Yu ED, Painter GF, Kronenberg M, Zajonc DM. The Vα14 invariant natural killer T cell TCR forces microbial glycolipids and CD1d into a conserved binding mode. J Exp Med. 2010;207:2383–2393. doi: 10.1084/jem.20101335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Aspeslagh S, et al. Galactose-modified iNKT cell agonists stabilized by an induced fit of CD1d prevent tumour metastasis. EMBO J. 2011;30:8413–8421. doi: 10.1038/emboj.2011.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pellicci DG, et al. Recognition of β-linked self glycolipids mediated by natural killer T cell antigen receptors. Nat Immunol. 2011;12:827–834. doi: 10.1038/ni.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Girardi E, et al. Unique Interplay between Sugar and Lipid in Determining the Antigenic Potency of Bacterial Antigens for NKT Cells. PLoS Biol. 2011;9:e1001189. doi: 10.1371/journal.pbio.1001189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yu ED, Girardi E, Wang J, Zajonc DM. Cutting edge: structural basis for the recognition of β-linked glycolipid antigens by invariant NKT cells. J Immunol. 2011;187:2079–2083. doi: 10.4049/jimmunol.1101636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Patel O, et al. NKT TCR Recognition of CD1d-α-C-Galactosylceramide. J Immunol. 2011;187:827–833. doi: 10.4049/jimmunol.1100794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kerzerho J, et al. Structural and Functional Characterization of a Novel Nonglycosidic Type I NKT Agonist with Immunomodulatory Properties. J Immunol. 2012;188:2254–2265. doi: 10.4049/jimmunol.1103049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.López-Sagaseta J, Sibener LV, Kung JE, Gumperz J, Adams EJ. Lysophospholipid presentation by CD1d and recognition by a human natural killer T-cell receptor. EMBO J. 2012;24:1–13. doi: 10.1038/emboj.2012.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rudolph MG, Stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu Rev Immunol. 2006;24:419–466. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- 113.Brossay L, et al. CD1d-mediated recognition of an alpha-galactosylceramide by natural killer T cells is highly conserved through mammalian evolution. J Exp Med. 1998;188:1521–1528. doi: 10.1084/jem.188.8.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sidobre S, et al. The T cell antigen receptor expressed by Valpha14i NKT cells has a unique mode of glycosphingolipid antigen recognition. Proc Natl Acad Sci USA. 2004;101:12254–12259. doi: 10.1073/pnas.0404632101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wun KS, et al. A molecular basis for the exquisite CD1d-restricted antigen specificity and functional responses of natural killer T cells. Immunity. 2011;34:327–339. doi: 10.1016/j.immuni.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tynan FE, et al. A T cell receptor flattens a bulged antigenic peptide presented by a major histocompatibility complex class I molecule. Nat Immunol. 2007;8:268–276. doi: 10.1038/ni1432. [DOI] [PubMed] [Google Scholar]
- 117.Zhou D, et al. Lysosomal glycosphingolipid recognition by NKT cells. Science. 2004;306:1786–1789. doi: 10.1126/science.1103440. [DOI] [PubMed] [Google Scholar]
- 118.Wun KS, et al. A minimal binding footprint on CD1d-glycolipid is a basis for selection of the unique human NKT TCR. J Exp Med. 2008;205:939–949. doi: 10.1084/jem.20072141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bozna BL, et al. Binding strength and dynamics of invariant natural killer cell T cell receptor/CD1d-glycosphingolipid interaction on living cells by single molecule force spectroscopy. J Biol Chem. 2011;286:15973–15979. doi: 10.1074/jbc.M110.192674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sidobre S, Naidenko OV, Sim B-C, Gascoigne NRJ, Garcia KC, Kronenberg M. The V alpha 14 NKT cell TCR exhibits high-affinity binding to a glycolipid/CD1d complex. J Immunol. 2002;169:1340–1348. doi: 10.4049/jimmunol.169.3.1340. [DOI] [PubMed] [Google Scholar]
- 121.Stanic A, et al. Another view of T cell antigen recognition: cooperative engagement of glycolipid antigens by Va14Ja18 natural TCR. J Immunol. 2003;171:4539–4551. doi: 10.4049/jimmunol.171.9.4539. [DOI] [PubMed] [Google Scholar]
- 122.Wu LC, Tuot DS, Lyons DS, Garcia KC, Davis MM. Two-step binding mechanism for T-cell receptor recognition of peptide MHC. Nature. 2002;418:552–556. doi: 10.1038/nature00920. [DOI] [PubMed] [Google Scholar]
- 123.Matsuda JL, Gapin L, Fazilleau N, Warren K, Naidenko OV, Kronenberg M. Natural killer T cells reactive to a single glycolipid exhibit a highly diverse T cell receptor beta repertoire and small clone size. Proc Natl Acad Sci USA. 2001;98:12636–12641. doi: 10.1073/pnas.221445298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gui M, Li J, Wen L-J, Hardy RR, Hayakawa K. TCR beta chain influences but does not solely control autoreactivity of Valpha14J281T cells. J Immunol. 2001;167:6239–6246. doi: 10.4049/jimmunol.167.11.6239. [DOI] [PubMed] [Google Scholar]
- 125.Ronet C, Mempel M, Thieblemont N, Lehuen A, Kourilsky P, Gachelin G. Role of the complementarity-determining region 3 (CDR3) of the TCR-beta chains associated with the Valpha14 semi-invariant TCR alpha-chain in the selection of CD4+ NK T Cells. J Immunol. 2001;166:1755–1762. doi: 10.4049/jimmunol.166.3.1755. [DOI] [PubMed] [Google Scholar]
- 126.Exley M, Garcia J, Balk SP, Porcelli S. Requirements for CD1d recognition by human invariant Valpha24+ CD4-CD8- T cells. J Exp Med. 1997;186:109–120. doi: 10.1084/jem.186.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Porcelli S, Yockey C, Brenner M, Balk S. Antigen receptor (TCR) expression by human peripheral blood CD4-8-cr T cells demonstrates preferential use of several V beta genes and an invariant TCR alpha chain. J Exp Med. 1993;178:1–16. doi: 10.1084/jem.178.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Matulis G, et al. Innate-like control of human iNKT cell autoreactivity via the hypervariable CDR3beta loop. PLoS Biol. 2010;8:e1000402. doi: 10.1371/journal.pbio.1000402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Benlagha K, Weiss A, Beavis A, Teyton L, Bendelac A. In vivo identification of glycolipid antigen-specific T cells using fluorescent CD1d tetramers. J Exp Med. 2000;191:1895–1903. doi: 10.1084/jem.191.11.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kobayashi E, Motoki K, Uchida T, Fukushima H, Koezuka Y. KRN7000, a novel immunomodulator, and its antitumor activities. Oncol Res. 1995;7:529–534. [PubMed] [Google Scholar]
- 131.Yamaguchi Y, et al. Enhancing effects of (2S,3S,4R)-1-O-(alpha-D-galactopyranosyl)-2-(N-hexacosanoylamino)-1,3,4-octadecanetriol (KRN7000) on antigen-presenting function of antigen-presenting cells and antimetastatic activity of KRN7000-pretreated antigen-presenting cells. Oncol Res. 1996;8:399–407. [PubMed] [Google Scholar]
- 132.Burdin N, et al. Selective ability of mouse CD1 to present glycolipids: alpha-galactosylceramide specifically stimulates V alpha 14+ NK T lymphocytes. J Immunol. 1998;161:3271–3281. [PubMed] [Google Scholar]
- 133.Hayakawa Y, Takeda K, Yagita H, Kakuta S, Kaer LV, Saiki I, Okumura K. Critical contribution of IFN-gamma and NK cells, but not perforin-mediated cytotoxicity, to anti-metastatic effect of alpha-galactosylceramide. Eur J Immunol. 2001;31:1720–1727. [PubMed] [Google Scholar]
- 134.Smyth MJ. Sequential production of interferon-gamma by NK1.1+ T cells and natural killer cells is essential for the antimetastatic effect of alpha -galactosylceramide. Blood. 2002;99:1259–1266. doi: 10.1182/blood.v99.4.1259. [DOI] [PubMed] [Google Scholar]
- 135.Toura I, Kawano T, Akutsu Y, Nakayama T, Ochiai T, Taniguchi M. Cutting edge: inhibition of experimental tumor metastasis by dendritic cells pulsed with alpha-galactosylceramide. J Immunol. 1999;163:2387–2391. [PubMed] [Google Scholar]
- 136.Cui J, et al. Requirement for Valpha14 NKT cells in IL-12-mediated rejection of tumors. Science. 1997;278:1623–1626. doi: 10.1126/science.278.5343.1623. [DOI] [PubMed] [Google Scholar]
- 137.Baxevanis CN, Gritzapis AD, Papamichail M. In vivo antitumor activity of NKT cells activated by the combination of IL-12 and IL-18. J Immunol. 2003;171:2953. doi: 10.4049/jimmunol.171.6.2953. [DOI] [PubMed] [Google Scholar]
- 138.Parekh VV, et al. Glycolipid antigen induces long-term natural killer T cell anergy in mice. J Clin Invest. 2005;115:2572–2583. doi: 10.1172/JCI24762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Tahir SM, et al. Loss of IFN-gamma production by invariant NK T cells in advanced cancer. J Immunol. 2001;167:4046–4050. doi: 10.4049/jimmunol.167.7.4046. [DOI] [PubMed] [Google Scholar]
- 140.Dhodapkar MV, et al. A reversible defect in natural killer T cell function characterizes the progression of premalignant to malignant multiple myeloma. J Exp Med. 2003;197:1667–1676. doi: 10.1084/jem.20021650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Giaccone G, et al. A phase I study of the natural killer T-cell ligand alpha-galactosylceramide (KRN7000) in patients with solid tumors. Clin Cancer Res. 2002;8:3702–3709. [PubMed] [Google Scholar]
- 142.Miyamoto K, Miyake S, Yamamura T. A synthetic glycolipid prevents autoimmune encephalomyelitis by inducing TH2 bias of natural killer T cells. Nature. 2001;413:531–534. doi: 10.1038/35097097. [DOI] [PubMed] [Google Scholar]
- 143.Schmieg J, Yang G, Franck RW, Tsuji M. Superior protection against malaria and melanoma metastases by a C-glycoside analogue of the natural killer T cell ligand alpha-Galactosylceramide. J Exp Med. 2003;198:1631–1641. doi: 10.1084/jem.20031192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Im JS, et al. Kinetics and cellular site of glycolipid loading control the outcome of natural killer T cell activation. Immunity. 2009;30:888–898. doi: 10.1016/j.immuni.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Bai L, et al. Lysosomal recycling terminates CD1d-mediated presentation of short and polyunsaturated variants of the NKT cell lipid antigen alphaGalCer. Proc Natl Acad Sci USA. 2009;106:10254–10259. doi: 10.1073/pnas.0901228106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Bai L, et al. Distinct APCs explain the cytokine bias of α-galactosylceramide variants in vivo. J Immunol. 2012;188:3053–3061. doi: 10.4049/jimmunol.1102414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Venkataswamy MM, Porcelli SA. Lipid and glycolipid antigens of CD1d-restricted natural killer T cells. Semin Immunol. 2010;22:68–78. doi: 10.1016/j.smim.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Silk JD, et al. Cutting edge: nonglycosidic CD1d lipid ligands activate human and murine invariant NKT cells. J Immunol. 2008;180:6452–6456. doi: 10.4049/jimmunol.180.10.6452. [DOI] [PubMed] [Google Scholar]
- 149.Cerundolo V, Barral P, Batista FD. Synthetic iNKT cell-agonists as vaccine adjuvants--finding the balance. Curr Opin Immunol. 2010;22:417–424. doi: 10.1016/j.coi.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 150.Li X, et al. Design of a potent CD1d-binding NKT cell ligand as a vaccine adjuvant. Proc Natl Acad Sci USA. 2010;107:13010–13015. doi: 10.1073/pnas.1006662107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Padte NN, Li X, Tsuji M, Vasan S. Clinical development of a novel CD1d-binding NKT cell ligand as a vaccine adjuvant. Clin immunol. 2011;140:142–151. doi: 10.1016/j.clim.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Blomqvist M, et al. Multiple tissue-specific isoforms of sulfatide activate CD1d-restricted type II NKT cells. Eur J Immunol. 2009;39:1726–1735. doi: 10.1002/eji.200839001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Girardi E, et al. Type II Natural Killer T cells recognize sulfatide self-antigens using features of both innate-like and conventional T cells. Nat Immunol. 2012 doi: 10.1038/ni.2371. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Adams JJ, et al. T cell receptor signaling is limited by docking geometry to peptide-major histocompatibility complex. Immunity. 2011;35:681–693. doi: 10.1016/j.immuni.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]