Abstract
There are currently at least 53 structures of components of nuclear transport in the Protein Databank. In addition to providing critical insights into molecular mechanisms of nuclear transport, these atomic resolution structures provide a large body of information that could guide biochemical and cell biological analyses involving nuclear transport proteins. This paper catalogs 53 crystal and NMR structures of nuclear transport proteins, with the emphasis on providing information useful for mutagenesis and overexpression of recombinant proteins.
1. Introduction
High resolution structures of macromolecular complexes are necessary to understand molecular mechanisms of cellular processes. The importance of structures is particularly evident in the cellular process of nucleocytoplasmic transport. The nuclear transport machinery consists of a large number of proteins that include components of the nuclear pore complex (nucleoporins), transport factors that recognize import or export substrates (Karyopherins/Importins/Exportins and TAP), Ran, its transporter NTF2 and its regulators, RanBP1, RanGAP and RanGEF. Macromolecular interactions in nuclear transport are complex. Each protein generally contacts multiple macromolecular ligands, binding to different partners in the cytoplasm versus the nucleus. Partner-switching in the different subcellular compartments is also frequently accompanied with large conformational changes in the proteins. High resolution structures of nuclear transport complexes have been crucial in revealing how a transport factor recognizes its ligands and how structural plasticity plays a central role in the different steps of nuclear import and export.
High resolution structures that have been determined in nuclear transport include those of Kapβs, Kapαs, Ran and its regulators RanGAP, RanGEF, RanBP1 and NTF2, mRNA export factor TAP and nucleoporins. The list of Kapβ structures includes nine Kapβ1/Impβ structures (unliganded, Ran-, substrate- and nucleoporin-complexes), two Kapβ2/Transportin structures, two Cse1 structures and a structure of a small Crm1 fragment. A large number of Kapα structures are available, including nine of mouse Kapα and five of the yeast homolog Kap60p, providing insight into the recognition of a variety of classical-NLSs and also nucleoporins such as Nup50 and Nup2p. Ran, its regulators RanBP1, NTF2, RanGAP and RanGEF as well as complexes involving these proteins are also well represented with a total of 12 structures. Structures in mRNA export include eight structures of TAP or its yeast homolog Mex67p, and finally, there are currently five structures of individual nucleoporin domains.
Other than their important roles in revealing molecular mechanisms of cellular processes, high resolution structures of macromolecular complexes also provide tremendous resources and tools for biochemical and cell biological experimental design. Structures could provide critical guidance in mutagenesis studies, especially when the aim is to disrupt specific interactions. Structure determination efforts, which require large amounts of proteins also provide very useful information about overexpression and purification of recombinant proteins.
This paper strives to catalog a comprehensive list of high resolution structures (mostly crystal structures) in nuclear transport in Tables 1–6 and Figs. 1–6. We have tabulated information about the identity of successful protein constructs for each structure as well as residues that are observed in those structures, to guide production of recombinant proteins. Many proteins involved in nuclear transport contain multiple modular and globular domains (such as TAP and nucleoporins), and knowledge of where individual domains begin and end obtained from the structures will allow design of protein constructs to optimize both folding and function of those domains. In contrast to the common globular proteins, both Kapα and Kapβ proteins contain multiple HEAT/ARM repeats such that these proteins are either elongated or spiral-shape. More importantly, every single HEAT repeat helix in these proteins contributes to the extended hydrophobic cores of the proteins. Thus, it is not trivial to generate deletion mutants involving HEAT/ARM repeats without interfering with the folding or solubility of the proteins. Structures of karyopherin fragments summarized here should provide information on the few deletion mutants that have been overexpressed successfully. Many of the structures tabulated in this paper are those of complexes of two or more proteins. These structures are very informative as they reveal the chemical and physical nature of the contact interfaces. We have also included contact residues in individual binding partners seen in structures of complexes, and also information on published interface mutants that disrupt specific protein–protein interactions. Such structural and biochemical data should aid significantly in mutagenesis analysis especially to disrupt specific functions in this large group of proteins.
Table 1.
Crystal structures of Kapβ1 complexes
| Structure | PDB-ID | Ref. | Organism | Resolution (Å) |
Protein constructs in crystals |
Residues in model |
Domains/motifs | Contact residues (molecule 1) |
Contact residues (molecule 2) |
Contact type | Disruptive interface mutants |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Kap95p-RanGTP | 2BKU | [1] | Kap95p: yeast | 2.7 | Kap95p: 1–861 | Kap95p: 1–861 | β1: | Kap95p: | Ran: | Ran: K37D/K152A binds | |
| HEAT repeats | I14 | L75 | HP* | Kap95p, but is unable to | |||||||
| Ran: dog | Ran: 1–176 | Ran: 9–176 | K66 | D77 | Polar | displace IBB | |||||
| N67 | D77 | Polar | |||||||||
| E164 | R110 | Polar | |||||||||
| E288 | R140 | Polar | |||||||||
| E295 | R140 | Polar | |||||||||
| E295 | K141 | Polar | |||||||||
| W345 | R140 | HP* | |||||||||
| N515 | N156 | Polar | |||||||||
| Q570 | R29 | Polar | |||||||||
| E615 | K37 | Polar | |||||||||
| D616 | K37 | Polar | |||||||||
| D617 | K152 | Polar | |||||||||
| Q650 | K37 | Polar | |||||||||
| Kapβ1-RanGppNHp | 1IBR | [2] | β1: human | 2.3 | β1: 1–462 | β1 (chain B): | β1: | β1: | Ran: | ||
| 2–459 | HEAT repeats | L59 | V111 | HP* | |||||||
| Ran: human | Ran: 1–216 | K62 | D77 | Polar | |||||||
| β1 (chain D): | K68 | D107 | Polar | ||||||||
| 2–439 | D160 | R110 | Polar | ||||||||
| R232 | E113 | Polar | |||||||||
| Ran: 9–176 | E281 | R140 | Polar | ||||||||
| E281 | K141 | Polar | |||||||||
| D288 | R140 | Polar | |||||||||
| D338 | R166 | Polar | |||||||||
| Kapβ1 | 1GCJ | [3] | Mouse | 2.6 | 1–449 | 1–449 | HEAT repeats | ||||
| Kapβ1-IBBKapα | 1QGK | [4] | β1: human | 2.5 | β1: 1–867 | β1: 1–867 | β1: | β1: | α: | β1: | |
| HEAT repeats | E281 | R13 | Polar | W864 (~35-fold) | |||||||
| α: 11–54 | α: 11–54 | α: IBB | D288 | R13 | Polar | ||||||
| D339 | K20 | Polar | W864/W342 | ||||||||
| D340 | K20 | Polar | W864/W430 | ||||||||
| W342 | R13 | HP* | W864/W472 | ||||||||
| W342 | L14 | HP* | (~400-fold) | ||||||||
| K346 | F17 | HP* | |||||||||
| V350 | R13 | HP* | W342/W430/W864 | ||||||||
| M388 | K18 | HP* | W342/W472/W864 | ||||||||
| D426 | K18 | Polar | W430/W472/W864 | ||||||||
| T427 | K18 | Polar | (~950-fold) | ||||||||
| W430 | K18 | HP* | |||||||||
| 1QGR | 2.3 | β1: 1–867 | β1: 1–614, | N469 | N19 | Polar | [5] | ||||
| 621–867 | W472 | N19 | HP* | ||||||||
| α: 11–54 | W472 | K22 | HP* | ||||||||
| α: 27–54 | E530 | R31 | Polar | ||||||||
| R593 | N35 | Polar | |||||||||
| D627 | R28 | Polar | |||||||||
| D627 | R31 | Polar | |||||||||
| M630 | I32 | HP* | |||||||||
| D676 | R39 | Polar | |||||||||
| E767 | K43 | Polar | |||||||||
| D824 | R51 | Polar | |||||||||
| W864 | R51 | HP* | |||||||||
| W864 | V53 | HP* | |||||||||
| Kapβ1-PTHrP | 1M5N | [6] | Human | 2.9 | β1: 1–485 | β1: 1–485 | β1: | β1: | PTHrP: | ||
| HEAT repeats | I295 | K93 | HP* | ||||||||
| PTHrP: 67–94 | PTHrP: 67–94 | W430 | P86 | HP* | |||||||
| W472 | K89 | HP* | |||||||||
| Kapβ1-SREBP-2 | 1UKL | [7] | β1: mouse | 3.0 | β1: 1–876 | β1: 1–876 | β1: | β1: | SREBP-2: | ||
| HEAT repeats | I295 | Y379 (D) | HP* | ||||||||
| SREBP-2: human | SREBP-2: 343–403 | SREBP-2: 343–403 | F752 | Y376 (C) | HP* | ||||||
| SREBP-2: | F752 | Y379 (C) | HP* | ||||||||
| basic helix | D753 | K372 (C) | Polar | ||||||||
| –loop-helix | D756 | R371 (C) | Polar | ||||||||
| leucine zipper | D812 | K378 (C) | Polar | ||||||||
| Kapβ1- | 1F59 | [8] | β1: human | 2.8 | β1: 1–442 | β1: 1–440 | β1: | β1: | FxFG: | β1: I178D | |
| Nsp1p I | HEAT repeats | L174 | F16 | HP* | |||||||
| Nsp1p: yeast | Nsp1p: | FxFG: Nsp1p 8–20 and 41–48 | I178 | F16 | HP* | ||||||
| 497–608 | F217 | F14 | HP* | ||||||||
| F217 | F16 | HP* | |||||||||
| I218 | F16 | HP* | |||||||||
| Y255 | F46 | HP* | |||||||||
| 1O6O | 2.8 | β1: 1–442 | β1: 2–440 | ||||||||
| Nsp1p: | FxFG: 9–14 | ||||||||||
| 497–608 | |||||||||||
| Kapβ1-GLFG | 1O6P | [9] | β1: human | 2.8 | β1: 1–442 | β1: 1–440 | β1: | β1: | GLFG: | β1: I178D | |
| HEAT repeats | L174 | F6 | HP* | ||||||||
| GLFG: | GLFG peptide: | GLFG: 4–7 | T175 | L5 | HP* | ||||||
| Synthetic peptide | 1DSGGLFGSK9 | I178 | F6 | HP* | |||||||
| F217 | F6 | HP* | |||||||||
| I218 | F6 | HP* | |||||||||
| Kap95p-nup1p | 2BPT | [10] | Kap95p: yeast | 2.0 | Kap95p: | Kap95p: | β1: | Kap95p: | nup1p: | nup1p: | |
| 1–861 | 1–861 | HEAT repeats | L181 | F1008 | HP* | F977D/F987D | |||||
| nup1p: yeast | I182 | P1004 | HP* | ||||||||
| nup1p: | nup1p: | I182 | F1008 | HP* | |||||||
| 963–1076 | 974–1012 | V185 | F1008 | HP* | |||||||
| Y224 | I1007 | HP* | |||||||||
| Y224 | F1008 | HP* | |||||||||
| M226 | F987 | HP* | |||||||||
| Q227 | N986 | Polar | |||||||||
| Q227 | F987 | HP* | |||||||||
| C230 | I985 | HP* | |||||||||
| C230 | F987 | HP* | |||||||||
| Y262 | F987 | HP* | |||||||||
| M263 | F977 | HP* | |||||||||
| Y268 | P979 | HP* | |||||||||
| L270 | P983 | HP* | |||||||||
| L270 | I985 | HP* | |||||||||
| F317 | F977 | HP* | |||||||||
HP, Hydrophobic contact.
Table 6.
Structures of nucleoporins
| Structure | PDB-ID | Ref. | Organism | Method | Resolution (Å) | Protein constructs in crystals |
Residues in model | Domains/motifs |
|---|---|---|---|---|---|---|---|---|
| Nup35 | 1WWH | [51] | Mouse | Crystal | 2.70 | 149–267 | 169–249 | Mppn domain |
| c-Nup98 autoprotease domain |
1KO6 | [52] | Human | Crystal | 3.00 | 710–870 | 712–870 | Auto-proteolytic/ pore-targeting |
| c-Nup116p | 2AIV | [53] | Yeast | NMR | 967–1113 | 967–1113 | Pore-targeting | |
| n-Nup133 | 1XKS | [54] | Human | Crystal | 2.35 | 67–514 | 75–162, 170–201, 206–251, 270–177 |
7-Bladed β-propeller |
| n-Nup159p | 1XIP | [55] | Yeast | Crystal | 2.5 | 2–387 | 2–347, 362–381 | 7-Bladed β-propeller |
Fig. 1.
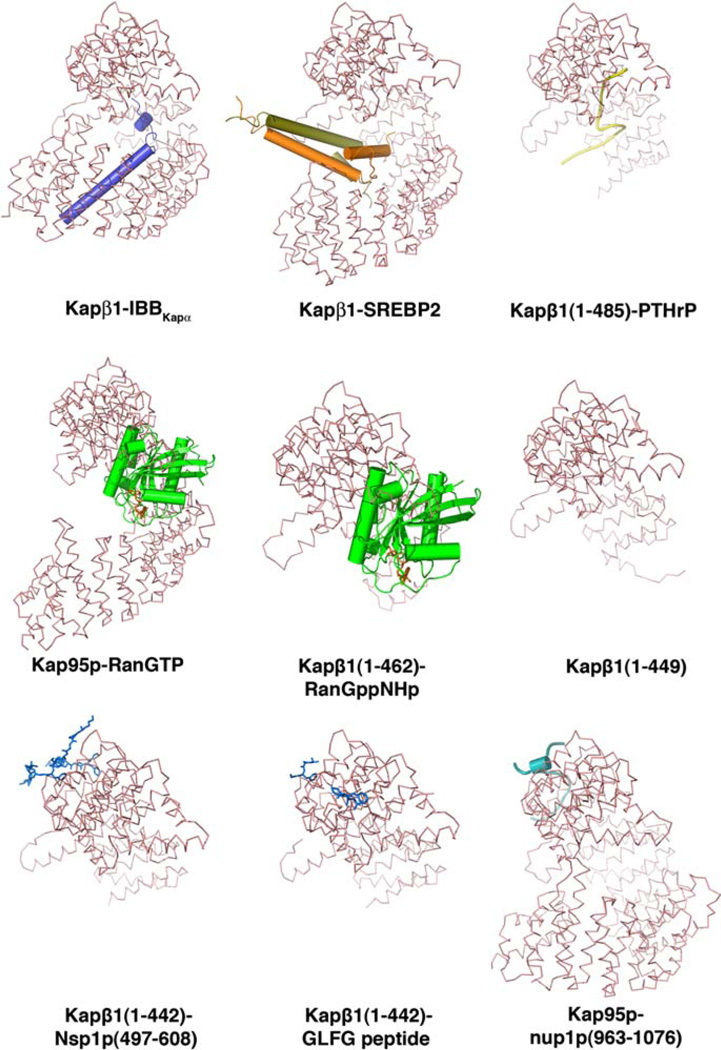
Structures of Kapβ1/Impβ complexes. Kapβ1 proteins are shown as red ribbons. Substrates IBB (blue) and SREBP2 (orange) are drawn as cylinders and ribbons, PTHrp as a yellow ribbon. Ran is shown in green and nucleoporin peptides in blue (Nsp1p and the GLFG peptide drawn as stick figure and nup1p as cylinder/ribbon). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this paper.)
Fig. 6.
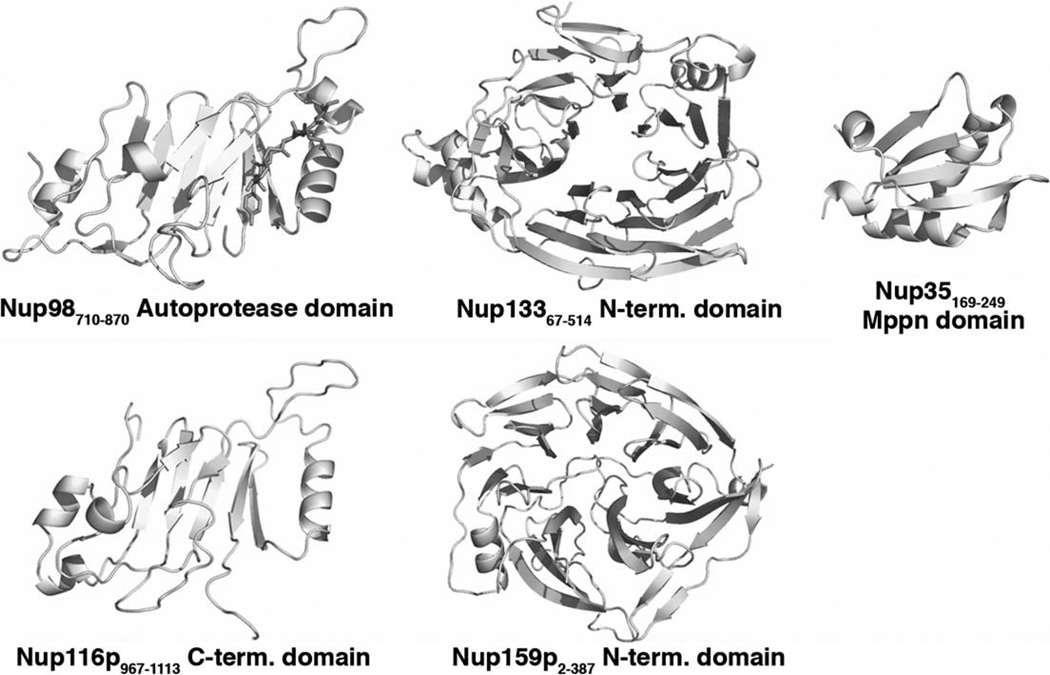
Structures of nucleoporin domains.
2. Concluding remarks
We have summarized 53 structures of Kapβs, Kapαs, Ran and its regulators RanGAP, RanGEF, RanBP1 and NTF2, mRNA export factor TAP and nucleoporins. The information provided should be useful for both overexpression of recombinant proteins as well as for analysis and experimental manipulation of specific interactions of the proteins. We look forward to many more structure of nuclear transport proteins in the future, especially to complexes of different Kapβs with their substrates to understand NLS/NES specificity in the different pathways and also to structures of nucleoporin subcomplexes to understand the assembly of the nuclear pore complex and mechanism of translocation through the pore.
Fig. 2.
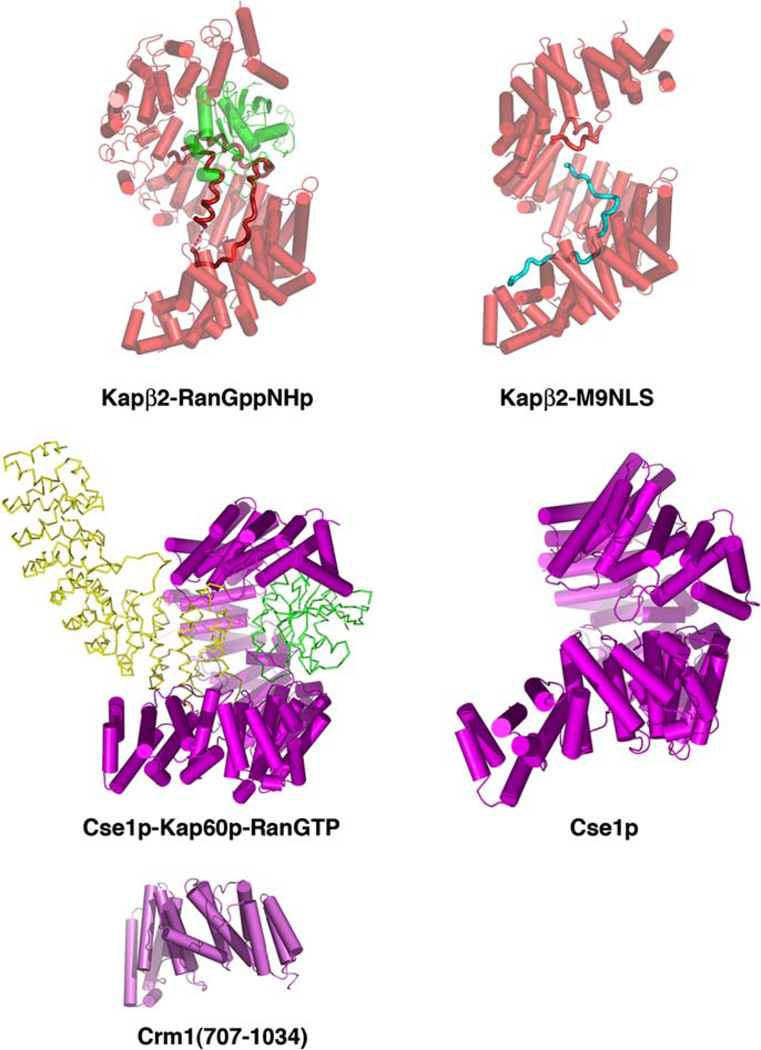
Structures of Kapβ2/Transportin complexes, Cse1p complexes and a Crm1 fragment. In the two Kapβ2 complexes, both Kapβ2 (red) and Ran (green) are shown as cylinders/ribbons and substrate M9NLS is shown as a cyan ribbon. Cse1p and Crm1 are both shown as purple cylinders/ribbons, and Ran (green) and Kapα (yellow) in the Cse1p-Ran-Kapα complex as ribbons. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this paper.)
Fig. 3.
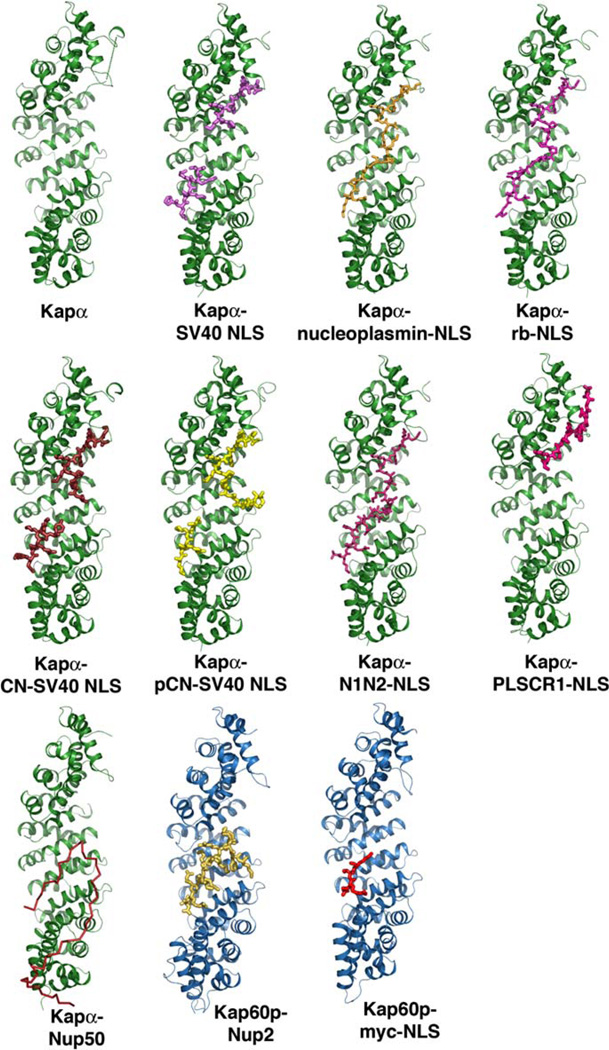
Structures of mouse Kapα and yeast Kap60p complexes. Mouse Kapαs are all drawn as green ribbons and yeast Kap60p as blue ribbons. With the exception of Nup50p (red ribbon), all other ligands are shown as stick figures. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this paper.)
Fig. 4.
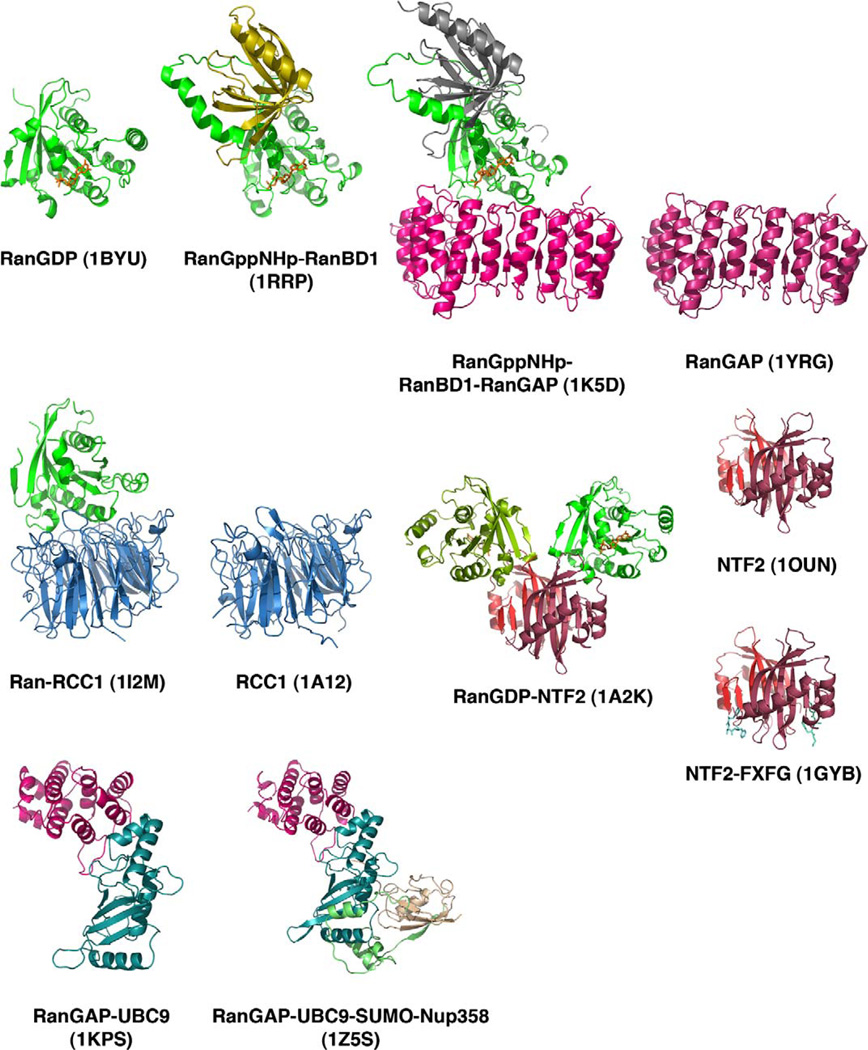
Structures of complexes involving Ran, NTF2, RanGAP and RanGEF. All 11 structures are shown as ribbon diagrams, with Ran in green, RanGAP in magenta, RCC1 RanGEF in blue, NTF2 in red, and RanBD1 or RanBP1-domain in gold (RanGppNHp-RBD1; 1RRP) and grey (RanGppNHp-RanBD1-RanGAP; 1K5D). RanGAP ligands, UBC9 is in aquamarine, SUMO is light brown and a fragment of Nup358 is in light green.
Fig. 5.
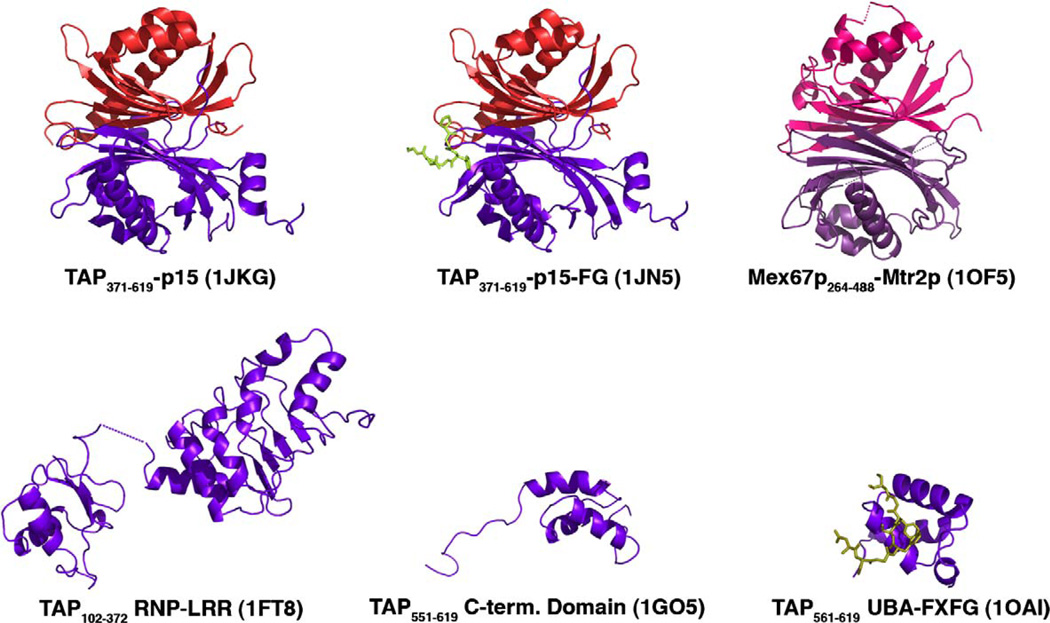
Structures of mRNA export factor TAP. TAP, Mex67p, p15 and MTR2p are all drawn as ribbons, and FG nucleoporin peptides are shown as stick figures. Domains of TAP and its yeast homolog Mex67p are in purple, p15 and its homolog Mtr2p are in red, and nucleoporin peptides in light green. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this paper.)
Table 2.
Crystal structures of Kapβ2, Csel complexes and a Crml fragment
| Structure | PDB-ID | Ref. | Organism | Resolution (Å) | Protein constructs in crystals |
Residues in model |
Domains/ motifs |
Contact residues (mol. 1) |
Contact residues (mol. 2) |
Contact residues (mol. 3) |
Contact type |
Disruptive interface mutants |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Kapβ2- | 1QBK | [11] | β2: human | 3.0 | β2: 1–890 | β2: | β2: | β2: | Ran: | |||
| RanGppnhp | Ran: human | Ran: 1–216 | 3–152, 158–166, | HEAT repeats | I114 | R110 | HP* | |||||
| 170–890 | S165 | R110 | Polar | |||||||||
| E275 | K141 | Polar | ||||||||||
| Ran: | E278 | K141 | Polar | |||||||||
| 8–197 | I315 | P172 | HP* | |||||||||
| E332 | N154 | Polar | ||||||||||
| R336 | D148 | Polar | ||||||||||
| P337 | Y155 | HP* | ||||||||||
| W373 | N143 | Polar | ||||||||||
| E682 | K127 | Polar | ||||||||||
| Kapβ2-M9NLS | 2H4M | [12] | β2: human | 3.1 | β2: 1–336- | β2: | β2: | β2: | M9NLS: | β2: | ||
| GGSGGSG- | 6–36, | HEAT repeats | R805 | N268 | Polar | W460A/ | ||||||
| 368–890 | 44–77, | K765 | Q269 | Polar | W730A | |||||||
| 57–73, | E809,I773 | F273 | HP* | |||||||||
| M9NLS: from | M9NLS: 257–305 | 80–319, | M9NLS: | W730 | P275 | HP* | ||||||
| hnRNP Al, human | 368–890 | extended | T766 | M276 | HP* | |||||||
| T547 | N280 | Polar | M9NLS: | |||||||||
| M9NLS: | F584,E588, | F281 | HP* | G274A/ | ||||||||
| 257–305 | V643 | HP* | P288A/ | |||||||||
| A505,T506, | R284 | Polar | Y289A | |||||||||
| E509,D543, | Polar | |||||||||||
| T547 | Polar | |||||||||||
| L419,I457, | P288 | HP* | ||||||||||
| W460 | HP* | |||||||||||
| A380,A381, | Y289 | HP* | ||||||||||
| D384,L419, | HP* | |||||||||||
| R464 | Polar | |||||||||||
| Csel | 1Z3H | [13] | Yeast | 3.1 | 1–959 | Chain A: 2–529, | HEAT repeats | |||||
| 544–870, 892–959 | ||||||||||||
| Chain B: 2–156, | ||||||||||||
| 162–525, 545– | ||||||||||||
| 875, 896–959 | ||||||||||||
| Csel-Kap60p- | IWA5 | [14] | Csel: yeast | 2.0 | Csel: 1–960 | Csel: | Csel: | Csel: | Kap60p: | Ran: | Ran: | |
| Ran | 1–193, 196–248, | HEAT repeats | R159 | E42 | Polar | R76A/ | ||||||
| 255–371, 376– | F164 | V41 | HP* | D77A | ||||||||
| 413, 418–880, | F164 | L43 | HP* | |||||||||
| 886–958 | E166 | R17 | Polar | |||||||||
| Kap60p: yeast | Kap60p: | Kap60p: 12–19, | Kap60p: IBB, | I167 | L43 | HP* | ||||||
| 1–530 | 31–58, | armadillo | D220 | R44 | Polar | K37D/ | ||||||
| 87–513 | domain | D227 | K45 | Polar | K152A | |||||||
| D79 | R44 | Polar | ||||||||||
| Ran: canine | Ran: 1–176 | Ran: | R321 | D435 | Polar | |||||||
| 7–176 | E387 | K431 | Polar | R95D/ | ||||||||
| R579 | E514 | Polar | K99A/ | |||||||||
| K62 | R76 | Polar | K130A/ | |||||||||
| N63 | D77 | Polar | K134A | |||||||||
| N609 | K152 | Polar | ||||||||||
| D653 | K37 | Polar | ||||||||||
| E656 | K152 | Polar | ||||||||||
| H894 | T93 | Polar | ||||||||||
| H894 | D128 | Polar | ||||||||||
| E484 | KI34 | Polar | ||||||||||
| Q490 | K95 | Polar | ||||||||||
| N492 | K99 | Polar | ||||||||||
| Y498 | K130 | Polar | ||||||||||
| E506 | K130 | Polar | ||||||||||
| E511 | K132 | Polar | ||||||||||
| Crml | 1W9C | [15] | Human | 2.3 | 707–1034 | 707–1027 | HEAT repeats | |||||
Table 3.
Crystal structures of Kapα complexes
| Structure | PDB-ID | Ref. | Organism | Resolution (Å) | Protein constructs in crystals |
Residues in model |
Domains/ motifs |
Contact residues (mol. 1) |
Contact residues (mol. 2) |
Contact type |
Disruptive interface mutants |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Kapα | 1IAL | [16] | Mouse | 2.5 | 1–529 | 44–54, 70–496 | Armadillo domain (ARM), | ARM | IBB | ||
| IBB | T151 | K49 | Polar | ||||||||
| T155 | K49 | Polar | |||||||||
| D192 | K49 | Polar | |||||||||
| D270 | Q46 | Polar | |||||||||
| Kapα-SV40 | 1EJL | [17] | Kapα: mouse | 2.8 | α: 70–529 | α: 72–497 | α: ARM | α: | SV40: | ||
| monopartite | W142 | K131 | HP* | ||||||||
| NLS | SV40: 126–132 | sv40: 126–132 | T155 | K128 | Polar | ||||||
| D192 | K128 | Polar | |||||||||
| T328 | K128 | Polar | |||||||||
| E354 | K131 | Polar | |||||||||
| S360 | K129 | Polar | |||||||||
| E396 | K129 | Polar | |||||||||
| W399 | K129 | HP* | |||||||||
| Kapα- | 1Q1T | [18] | Kapα: mouse | 2.5 | α: 70–529 | α: 70–496 | α: ARM | α: | SV40: | ||
| SV40 CN | T155 | K128 | Polar | ||||||||
| SV40: 110–132 | SV40: | D192 | K128 | Polar | |||||||
| 123–133 | E396 | R130 | Polar | ||||||||
| 127–134 | |||||||||||
| Kapα-phospho- | 1Q1S | [18] | Kapα: | 2.3 | α: 70–529 | α: 64–497 | α: ARM | α: | SV40: | ||
| SV40CN | Mouse | T155 | K128 | Polar | |||||||
| SV40: 110–132 | SV40: | D192 | K128 | Polar | |||||||
| 119–133 | E396 | R130 | Polar | ||||||||
| 128–132 | |||||||||||
| Kapα- PLSCR1 | 1Y2A | [19] | Kapα: mouse | 2.2 | α: 70–529 | α: 70–529 | α: ARM | α: | NLS: | ||
| monopartite | |||||||||||
| NLS | E107 | H262 | Polar | ||||||||
| PLSCR1: | PLSCR1: | W142 | K261 | HP* | |||||||
| 257–266 | 257–266 | W142 | W263 | HP* | |||||||
| T155 | K258 | Polar | |||||||||
| W184 | I299 | HP* | |||||||||
| W184 | K261 | HP* | |||||||||
| D192 | K258 | Polar | |||||||||
| Kapα- | 1EJY | [17] | Kapα: mouse | 2.9 | α: 70–529 | α: 72–497 | α: ARM | α: | NLS: | ||
| nucleoplasmin | T155 | K167 | Polar | ||||||||
| bipartite NLS | Nucleoplasmin: 155–170 | Nucleoplasmin: 155–170 | D192 | K167 | Polar | ||||||
| W231 | K168 | HP* | |||||||||
| S360 | R156 | Polar | |||||||||
| E396 | R156 | Polar | |||||||||
| Kapα-N1N2 | 1PJN | [20] | Kapα: mouse | 2.5 | α: 70–529 | α: 72–496 | α: ARM | α: | N1N2: | SV40: | |
| bipartite NLS | W142 | K554 | HP* | K128 | |||||||
| N1N2: Xenopus, peptide | N1N2: 533–556 | N1N2: 535–555 | N146 | S553 | Polar | [21–23] | |||||
| T155 | K551 | Polar | |||||||||
| Q181 | K554 | Polar | |||||||||
| S276 | K547 | Polar | |||||||||
| Y277 | K547 | HP* | |||||||||
| T322 | K539 | Polar | |||||||||
| T328 | K537 | Polar | |||||||||
| S360 | R538 | Polar | |||||||||
| E396 | R538 | Polar | |||||||||
| Kapα-RB bipartite | 1PJM | [20] | Kapα: mouse | 2.5 | α: 70–529 | α: 71–497 | α: ARM | α: | RB: | ||
| NLS | T155 | K874 | Polar | ||||||||
| RB: human peptide | RB: 859–879 | RB: 859–878 | Q181 | R877 | Polar | ||||||
| D192 | K874 | Polar | |||||||||
| Y277 | K871 | Polar | |||||||||
| T328 | K861 | Polar | |||||||||
| E396 | R862 | Polar | |||||||||
| Kapα-nup50 | 2C1M | [24] | Kapα: mouse | 2.2 | α: 70–529 | α: 75–498 | α: ARM | α: | Nup50: | Nup50p: R38A/ | |
| K240 | E9 | Polar | R45D | ||||||||
| Nup50: mouse | Nup50: 1–109 | Nup50: 1–46 | L307 | W15 | HP* | ||||||
| T328 | K3 | Polar | |||||||||
| K348 | D16 | Polar | |||||||||
| S360 | R4 | Polar | |||||||||
| K392 | E7 | Polar | |||||||||
| E396 | R4 | Polar | |||||||||
| D471 | R45 | Polar | |||||||||
| E474 | R45 | Polar | |||||||||
| E493 | K41 | Polar | |||||||||
| Kap60p | 1BK5 | [25] | Kap60p: yeast | 2.2 | Kap60p: 88–530 | Kap60p: 89–509 | Kap60p: ARM | ||||
| Kap60p-SV40 | 1BK6 | [25] | Kap60p: yeast | 2.8 | Kap60p: | Kap60p: | Kap60p: ARM | Kap60p: | SV40: | Kap60p: N157A/ | |
| monopartite | 88–530 | 89–509 | W153 | K131 | HP* | D203K | |||||
| NLS | T166 | K128 | Polar | ||||||||
| SV40: 125–133 | SV40: 127–132, | Q192 | K131 | Polar | |||||||
| 127–131 | W195 | K129 | HP* | ||||||||
| D203 | K128 | Polar | |||||||||
| W237 | K127 | HP* | |||||||||
| E272 | K129 | Polar | |||||||||
| D276 | K127 | Polar | |||||||||
| D331 | K127 | Polar | |||||||||
| E402 | K128 | Polar | |||||||||
| Kap60 p-c-myc | 1EE4 | [26] | Kap60p: yeast | 2.1 | Kap60p: | Kap60p: | Kap60p: ARM | Kap60p: | NLS: | ||
| monopartite | 88–530 | 87–509 | T166 | K323 | Polar | ||||||
| NLS | c-myc: human peptide | W195 | K326 | HP* | |||||||
| c-myc: 319–329 | c-myc: | D203 | K323 | Polar | |||||||
| 320–328 | D276 | R324 | Polar | ||||||||
| 323–326 | T334 | K323 | Polar | ||||||||
| S366 | R324 | Polar | |||||||||
| E402 | R324 | Polar | |||||||||
| Kap60p- | 1EE5 | [26] | Kap60p: yeast | 2.4 | Kap60p: | Kap60p: | Kap60p: | Kap60p: | NLS: | ||
| Nucleopalsmin | 88–530 | 90–506 | ARM | W195 | K170 | HP* | |||||
| bipartite NLS | D203 | K167 | Polar | ||||||||
| Nucleoplasmin: | Nucleoplasmin: | Nucleoplasmin: | W237 | K168 | HP* | ||||||
| Xenopus peptide | 154–172 | 153–171 | W279 | K162 | HP* | ||||||
| T334 | K155 | Polar | |||||||||
| S366 | R156 | Polar | |||||||||
| E402 | R156 | Polar | |||||||||
| Kap60p | 1UN0 | [27] | Kap60p: yeast | 2.6 | Kap60p: | Kap60p: | Kap60p: | Kap60p: | Nup1p: | ||
| -nup2p | (2C1T) | [24] | 88–530 | 88–526 | ARM | S240 | R47 | Polar | |||
| nup1p: yeast | N241 | R47 | Polar | ||||||||
| Nup1p: 1–51 | Nup1p: 36–51 | Nup1p: | D276 | R47 | Polar | ||||||
| N-terminal Kap60p | W279 | K45 | HP* | ||||||||
| Binding domain and NPC | W363 | K40 | HP* | ||||||||
| targeting domain | E402 | R38 | Polar | ||||||||
HP, Hydrophobic contact.
Table 4.
Structures of Ran and NTF2 complexes
| Structure | PDB-ID | Ref. | Organism | Method | Resolution (Å) |
Protein constructs in crystals |
Residues in model |
Domains/ motifs |
Contact residues (mol. 1) |
Contact residues (mol. 2) |
Contact residues (mol. 3) |
Contact residues (mol. 4) |
Contact type |
Disruptive interface mutants |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RanGDP | 1BYU | [28] | Canine | Crystal | 2.15 | 1–216 | A: 2–216 | G-domain | ||||||
| B: 5–208 | ||||||||||||||
| RanGDP Q69L | 3RAN | [28] | Canine | Crystal | 2.3 | 1–216 | 5–205 | G-domain | ||||||
| RCC1 | 1A12 | [29] | Human | Crystal | 1.7 | 1–421 | 21–421 | 7-Bladed propeller | ||||||
| Ran-RCC1 | 1I2M | [30] | Ran: human | Crystal | 1.8 | Ran: | Ran: | Ran: | Ran: | RCC1: | Ran: G19V [31] | |||
| 1–216 | 8–31, 37–177 | G-domain | K71 | H304 | Polar | |||||||||
| RCC1: human | RCC1: | RCC1: | RCC1: | R76 | E334 | Polar | RCC1: D128A, | |||||||
| D182A, H304A | ||||||||||||||
| [32,33] | ||||||||||||||
| 1–421 | 24–232, | 7-Bladed propeller | S94 | R147 | Polar | |||||||||
| 239–417 | R95 | D95 | Polar | |||||||||||
| K99 | D128 | Polar | ||||||||||||
| N100 | Q303 | Polar | ||||||||||||
| R106 | D384 | Polar | ||||||||||||
| D107 | R320 | Polar | ||||||||||||
| R110 | Y323 | Polar | ||||||||||||
| K134 | D95 | Polar | ||||||||||||
| R140 | D44 | Polar | ||||||||||||
| RanGAP (rna1p) | 1YRG | [34] | S. pombe | Crystal | 2.66 | 1–387 | 2–344 | LRR | ||||||
| RanBD2 of RanBP2 | 1XKE | [35] | human | NMR | 2028–2154 | 2028–2154 | PH | |||||||
| (RanBP2) | (RanBP2) | |||||||||||||
| Ran-GppNHp-RanBD1 | 1RRP | [36] | Ran: human | Crystal | 2.9 | Ran: 1–216 | Ran: (A) 8–211 | RanBD1: PH | Ran: | RanBD1: | ||||
| F11 | V21 | HP* | ||||||||||||
| RanBD1 of | RanBD1 of | (C): 8–187 | Ran: G-domain | F11 | V22 | HP* | ||||||||
| Ran BP2: human | Ran BP2: | R29 | E59 | Polar | ||||||||||
| 1155–1321 (1–167) | RanBD1 of | K38 | E56 | Polar | ||||||||||
| Ran BP2: 17–150 | ||||||||||||||
| E158 | Q84 | Polar | ||||||||||||
| W163 | F18 | HP* | ||||||||||||
| K167 | F18 | HP* | ||||||||||||
| L168 | V21 | HP* | ||||||||||||
| I169 | V22 | HP* | ||||||||||||
| E186 | K75 | Polar | ||||||||||||
| D211 | K46 | Polar | ||||||||||||
| Ran-GppNHp-RanBP1- | 1K5D | [37] | Ran: human | Crystal | 2.7 | Ran: 1–216 | Ran: 8–213 | RanGAP: LRR | Ran: | RanGAP: | RanBP1: | |||
| RanGAP | D18 | R168 | Polar | |||||||||||
| RanBP1: | RanBP1: | RanBP1: | L43 | I79 | HP* | |||||||||
| human | ||||||||||||||
| 1–201 | 22–167 | E46 | K76 | Polar | ||||||||||
| RanGAP: | K71 | D103 | Polar | |||||||||||
| S.pombe | RanGAP: | RanGAP: | D128 | R200 | Polar | |||||||||
| 1–386 | 2–345 | K130 | D225 | Polar | ||||||||||
| K130 | L255 | HP* | ||||||||||||
| V9 | L33 | HP* | ||||||||||||
| R29 | E69 | Polar | ||||||||||||
| N55 | E35 | Polar | ||||||||||||
| W163 | F27 | HP* | ||||||||||||
| K167 | D24 | Polar | ||||||||||||
| K167 | Q26 | Polar | ||||||||||||
| L168 | I30 | HP* | ||||||||||||
| I169 | V31 | HP* | ||||||||||||
| I169 | L33 | HP* | ||||||||||||
| V177 | I38 | HP* | ||||||||||||
| L201 | K111 | HP* | ||||||||||||
| L201 | V121 | HP* | ||||||||||||
| D213 | K54 | Polar | ||||||||||||
| D213 | R141 | Polar | ||||||||||||
| Ran-GDP-RanBP1-RanGAP | 1K5G | [37] | Ran: human | Crystal | 3.1 | Ran: 1–216 | Ran: 8–213 | RanGAP: LRR | Ran: | RanGAP: | RanBP1: | |||
| D18 | R168 | Polar | ||||||||||||
| RanBP1: human | RanBP1: 1–201 | RanBP1: 22–167 | L43 | I79 | HP* | |||||||||
| RanGAP: Yeast (S. pombe) | D128 | R200 | Polar | |||||||||||
| K130 | D225 | Polar | ||||||||||||
| RanGAP: 1–386 | RanGAP: 2–345 | K130 | L255 | HP* | ||||||||||
| V9 | L33 | HP* | ||||||||||||
| R29 | E69 | Polar | ||||||||||||
| N55 | E35 | Polar | ||||||||||||
| R56 | E35 | Polar | ||||||||||||
| W163 | F27 | HP* | ||||||||||||
| K167 | D24 | Polar | ||||||||||||
| K167 | Q26 | Polar | ||||||||||||
| L168 | I30 | HP* | ||||||||||||
| I169 | V31 | HP* | ||||||||||||
| I169 | L33 | HP* | ||||||||||||
| V177 | I38 | HP* | ||||||||||||
| L201 | K111 | HP* | ||||||||||||
| L201 | V121 | HP* | ||||||||||||
| D213 | K54 | Polar | ||||||||||||
| D213 | R141 | Polar | ||||||||||||
| Ubc9-RanGAP1 | 1KPS | [38] | Ubc9: human | Crystal | 2.5 | Ubc9: 1–159 | Ubc9: 3–158 | Ubc9: | RanGAP1: | |||||
| K74 | E528 | Polar | ||||||||||||
| RanGAP1: mouse | Y87 | E528 | HP* | |||||||||||
| RanGAP1: 420–589 | RanGAP1: 434–589 | C93 | K526 | Polar | ||||||||||
| D127 | K526 | Polar | ||||||||||||
| E132 | N512 | Polar | ||||||||||||
| SUMO-1- | 1Z5S | [39] | Human | Crystal | 3.0 | SUMO-1: 18–97 | SUMO-1: 20–97 | RanBP2: | SUMO-1: | RanGAP1: | Ubc9: | Nup358: | ||
| RanGAP1- | E3 ligase domain | R63 | R104 | Polar | ||||||||||
| Ubc9- | RanGAP1: 419–587 | RanGAP1: 432–587 | Q92 | E122 | Polar | |||||||||
| Nup358/ | K39 | D2631 | Polar | |||||||||||
| RanBP2 | Ubc9: 1–158 | Ubc9: | R54 | E2637 | Polar | |||||||||
| 2–157 | E526 | K74 | Polar | |||||||||||
| R13 | E2675 | Polar | ||||||||||||
| Nup358: 2631–2695 | Nup358: 2631–2693 | R13 | D2673 | Polar | ||||||||||
| K30 | D2676 | Polar | ||||||||||||
| NTF2 | 1OUN | [40] | Rat | Crystal | 1.6 | 1–127 | (A): 2–126 | |||||||
| (B): 4–124 | ||||||||||||||
| NTF2-RanGDP | 1A2K | [41] | NTF2: rat | Crystal | 2.5 | NTF2: 1–127 | NTF2: | Ran: G-protein domain | NTF2: | Ran: | NTF2: E42K, | |||
| D92/94N [42] | ||||||||||||||
| 4–127 | W41 | F72 | HP* | |||||||||||
| Ran: canine | E42 | R76 | Polar | Ran: | ||||||||||
| Ran: 1–216 | Ran: | F61 | F72 | HP* | Q69L [28] | |||||||||
| (C) 8–206 | I64 | F72 | HP* | |||||||||||
| (D) 8–204 | L89 | F72 | HP* | |||||||||||
| D92 | K71 | Polar | ||||||||||||
| D94 | K71 | Polar | ||||||||||||
| F119 | F72 | HP* | ||||||||||||
| NTF2 N77Y-FxFG | 1GYB | [43] | Yeast | Crystal | 1.9 | NTF2: 1–125 | NTF2: | NTF2 | FxFG | NTF2: | ||||
| (A): | (A): | |||||||||||||
| 3–124 | Q43/45D | |||||||||||||
| (A and B) | F5 | F43 | HP* | |||||||||||
| 5–124 | P73 | F43 | HP* | W7A [44] | ||||||||||
| (C and D) | P73 | F45 | HP* | |||||||||||
| NTF2(B): | FxFg(B): | |||||||||||||
| FxFG from Nsp1p: | FxFG: | E34 | F45 | HP* | ||||||||||
| 40–48 | 42–46 | M36 | F43 | HP* | ||||||||||
| M36 | F45 | HP* | ||||||||||||
| Q43 | F43 | HP* | ||||||||||||
| F115 | F45 | HP* | ||||||||||||
HP, Hydrophobic contact.
Table 5.
Structures of mRNA export complexes
| Structure | PDB-ID | Ref. | Organism | Method | Resolution (Å) | Protein constructs in crystals |
Residues in model | Domains/motifs | Contact residues (mol. 1) |
Contact residues (mol. 2) |
Contact type |
Disruptive interface mutants |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TAP-RBD (LRR) | 1FO1 | [45] | Human | Crystal | 2.9 | 102–372 | 123–191, 203–362 | Partially disordered | ||||
| RNP domain and LRR | ||||||||||||
| domains | ||||||||||||
| TAP-RBD (RNP) | 1FT8 | [45] | Human | Crystal | 3.15 | 102–372 | 119–198, 205–362 | RNP domains and | ||||
| LRR domains | ||||||||||||
| TAP-novel RBD | 1KOH | [46] | Human | Crystal | 3.8 | 96–372 | A: 105–362 | RNP and LRR | ||||
| (native) | B: 201–367 | domains | ||||||||||
| C: 105–362 | ||||||||||||
| D: 201–372 | ||||||||||||
| Tap/NXF1-C- term |
1GO5 | [47] | Human | NMR | 551–619 | 551–619 | UBA-like domain | |||||
| TAP-UBA-FXFG | 1OAI | [48] | Human | Crystal | 1 | TAP: | TAP: | TAP: UBA-like | TAP: | FG: | TAP: C588 | |
| domain | ||||||||||||
| 561–619 | 561–619 | M580 | F15 | HP* | ||||||||
| FXFG: | FxFG: 10–18 | W584 | F15 | HP* | ||||||||
| DSGFSFGSK | K587 | F15 | HP* | |||||||||
| C588 | F15 | HP* | ||||||||||
| A602 | F15 | HP* | ||||||||||
| L606 | F15 | HP* | ||||||||||
| E611 | F13 | HP* | ||||||||||
| P613 | F13 | HP* | ||||||||||
| TAP-p15 | 1JKG | [49] | TAP: human | Crystal | 1.9 | TAP: | TAP: | TAP: UBA-like | TAP: | p15: | TAP: | |
| domain | ||||||||||||
| 371–619 | 370–423, 431–555 | NTF2-like domain | R440 | D76 | Polar | D482R | ||||||
| K446 | E18 | Polar | I518R | |||||||||
| p15: human | p15: 2–140 | p15: 1–240 | p15: | D482 | R134 | Polar | ||||||
| NTF2-like domain | 1518 | V80 | HP* | |||||||||
| F535 | C96 | HP* | ||||||||||
| TAP-p15-FG | 1JN5 | [49] | TAP: human | Crystal | 2.8 | TAP: | TAP: | TAP: UBA-like | TAP: | FG: | TAP: | |
| domain, | ||||||||||||
| 371–619 | 371–422, 430–555 | NTF2-like domain | L383 | F1810 | HP* | L383R, L386R, | ||||||
| L386 | F1810 | HP* | W594A | |||||||||
| p15: 2–140 | p15: 2–140 | p15: | P521 | F1810 | HP* | |||||||
| p15: human | NTF2-like domain | L527 | F1810 | HP* | ||||||||
| FG: | ||||||||||||
| FG: | 1808–1812 | |||||||||||
| 1805–1816 | ||||||||||||
| Of Nup214 | ||||||||||||
| Mex67-Mtr2 | 1OF5 | [50] | Mex67: | Crystal | 2.8 | Mex67: | Mex67: | NTF2-like domain | Mex67: | Mtr2: | ||
| Saccharomyces | 264–407, 436–488 | 268–315, 354–107, | D446 | S148 | Polar | |||||||
| cerevisiae | 436–488 | T466 | N99 | Polar | ||||||||
| Mtr2: S. cerevisiae | Mtr2: 1–184 | Mtr2: | E385 | N170 | Polar | |||||||
| 14–106, | ||||||||||||
| 141–177 | ||||||||||||
HP, Hydrophobic contact.
References
- 1.Lee SJ, Matsuura Y, Liu SM, Stewart M. Nature. 2005;435:693–696. doi: 10.1038/nature03578. [DOI] [PubMed] [Google Scholar]
- 2.Vetter IR, Arndt A, Kutay U, Gorlich D, Wittinghofer A. Cell. 1999;97:635–646. doi: 10.1016/s0092-8674(00)80774-6. [DOI] [PubMed] [Google Scholar]
- 3.Lee SJ, Imamoto N, Sakai H, Nakagawa A, Kose S, Koike M, Yamamoto M, Kumasaka T, Yoneda Y, Tsukihara T. J. Mol. Biol. 2000;302:251–264. doi: 10.1006/jmbi.2000.4055. [DOI] [PubMed] [Google Scholar]
- 4.Cingolani G, Petosa C, Weis K, Muller CW. Nature. 1999;399:221–229. doi: 10.1038/20367. [DOI] [PubMed] [Google Scholar]
- 5.Koerner C, Guan T, Gerace L, Cingolani G. J. Biol. Chem. 2003;278:16216–16221. doi: 10.1074/jbc.M301137200. [DOI] [PubMed] [Google Scholar]
- 6.Cingolani G, Bednenko J, Gillespie MT, Gerace L. Mol. Cell. 2002;10:1345–1353. doi: 10.1016/s1097-2765(02)00727-x. [DOI] [PubMed] [Google Scholar]
- 7.Lee SJ, Sekimoto T, Yamashita E, Nagoshi E, Nakagawa A, Imamoto N, Yoshimura M, Sakai H, Chong KT, Tsukihara T, Yoneda Y. Science. 2003;302:1571–1575. doi: 10.1126/science.1088372. [DOI] [PubMed] [Google Scholar]
- 8.Bayliss R, Littlewood T, Stewart M. Cell. 2000;102:99–108. doi: 10.1016/s0092-8674(00)00014-3. [DOI] [PubMed] [Google Scholar]
- 9.Bayliss R, Littlewood T, Strawn LA, Wente SR, Stewart M. J. Biol. Chem. 2002;277:50597–50606. doi: 10.1074/jbc.M209037200. [DOI] [PubMed] [Google Scholar]
- 10.Liu SM, Stewart M. J. Mol. Biol. 2005;349:515–525. doi: 10.1016/j.jmb.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 11.Chook YM, Blobel G. Nature. 1999;399:230–237. doi: 10.1038/20375. [DOI] [PubMed] [Google Scholar]
- 12.Lee BJ, Cansizoglu AE, Süel KE, Louis TH, Zhang Z, Chook YM. Cell. 2006;126:543–558. doi: 10.1016/j.cell.2006.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cook A, Fernandez E, Lindner D, Ebert J, Schlenstedt G, Conti E. Mol. Cell. 2005;18:355–367. doi: 10.1016/j.molcel.2005.03.021. [DOI] [PubMed] [Google Scholar]
- 14.Matsuura Y, Stewart M. Nature. 2004;432:872–877. doi: 10.1038/nature03144. [DOI] [PubMed] [Google Scholar]
- 15.Petosa C, Schoehn G, Askjaer P, Bauer U, Moulin M, Steuerwald U, Soler-Lopez M, Baudin F, Mattaj IW, Muller CW. Mol. Cell. 2004;16:761–775. doi: 10.1016/j.molcel.2004.11.018. [DOI] [PubMed] [Google Scholar]
- 16.Kobe B. Nat. Struct. Biol. 1999;6:388–397. doi: 10.1038/7625. [DOI] [PubMed] [Google Scholar]
- 17.Fontes MR, Teh T, Kobe B. J. Mol. Biol. 2000;297:1183–1194. doi: 10.1006/jmbi.2000.3642. [DOI] [PubMed] [Google Scholar]
- 18.Fontes MR, Teh T, Toth G, John A, Pavo I, Jans DA, Kobe B. Biochem. J. 2003;375:339–349. doi: 10.1042/BJ20030510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen MH, Ben-Efraim I, Mitrousis G, Walker-Kopp N, Sims PJ, Cingolani G. J. Biol. Chem. 2005;280:10599–10606. doi: 10.1074/jbc.M413194200. [DOI] [PubMed] [Google Scholar]
- 20.Fontes MR, Teh T, Jans D, Brinkworth RI, Kobe B. J. Biol. Chem. 2003;278:27981–27987. doi: 10.1074/jbc.M303275200. [DOI] [PubMed] [Google Scholar]
- 21.Lanford RE, Butel JS. Cell. 1984;37:801–813. doi: 10.1016/0092-8674(84)90415-x. [DOI] [PubMed] [Google Scholar]
- 22.Kalderon D, Richardson WD, Markham AF, Smith AE. Nature. 1984;311:33–38. doi: 10.1038/311033a0. [DOI] [PubMed] [Google Scholar]
- 23.Colledge WH, Richardson WD, Edge MD, mith AE. Mol. Cell. Biol. 1986;6:4136–4139. doi: 10.1128/mcb.6.11.4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuura Y, Stewart M. EMBO J. 2005;24:3681–3689. doi: 10.1038/sj.emboj.7600843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Conti E, Uy M, Leighton L, Blobel G, Kuriyan J. Cell. 1998;94:193–204. doi: 10.1016/s0092-8674(00)81419-1. [DOI] [PubMed] [Google Scholar]
- 26.Conti E, Kuriyan J. Struct. Fold Des. 2000;8:329–338. doi: 10.1016/s0969-2126(00)00107-6. [DOI] [PubMed] [Google Scholar]
- 27.Matsuura Y, Lange A, Harreman MT, Corbett AH, Stewart M. EMBO J. 2003;22:5358–5369. doi: 10.1093/emboj/cdg538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stewart M, Kent HM, McCoy AJ. J. Mol. Biol. 1998;284:1517–1527. doi: 10.1006/jmbi.1998.2204. [DOI] [PubMed] [Google Scholar]
- 29.Renault L, Nassar N, Vetter I, Becker J, Klebe C, Roth M, Wittinghofer A. Nature. 1998;392:97–101. doi: 10.1038/32204. [DOI] [PubMed] [Google Scholar]
- 30.Renault L, Kuhlmann J, Henkel A, Wittinghofer A. Cell. 2001;105:245–255. doi: 10.1016/s0092-8674(01)00315-4. [DOI] [PubMed] [Google Scholar]
- 31.Lounsbury KM, Richards SA, Carey KL, Macara IG. J. Biol. Chem. 1996;271:32834–32841. doi: 10.1074/jbc.271.51.32834. [DOI] [PubMed] [Google Scholar]
- 32.Azuma Y, Seino H, Seki T, Uzawa S, Klebe C, Ohba T, Wittinghofer A, Hayashi N, Nishimoto T. J. Biochem. (Tokyo) 1996;120:82–91. doi: 10.1093/oxfordjournals.jbchem.a021397. [DOI] [PubMed] [Google Scholar]
- 33.Azuma Y, Renault L, Garcia-Ranea JA, Valencia A, Nishimoto T, Wittinghofer A. J. Mol. Biol. 1999;289:1119–1130. doi: 10.1006/jmbi.1999.2820. [DOI] [PubMed] [Google Scholar]
- 34.Hillig RC, Renault L, Vetter IR, Drell Tt, Wittinghofer A, Becker J. Mol. Cell. 1999;3:781–791. doi: 10.1016/s1097-2765(01)80010-1. [DOI] [PubMed] [Google Scholar]
- 35.Geyer JP, Doker R, Kremer W, Zhao X, Kuhlmann J, Kalbitzer HR. J. Mol. Biol. 2005;348:711–725. doi: 10.1016/j.jmb.2005.02.033. [DOI] [PubMed] [Google Scholar]
- 36.Vetter IR, Nowak C, Nishimoto T, Kuhlmann J, Wittinghofer A. Nature. 1999;398:39–46. doi: 10.1038/17969. [DOI] [PubMed] [Google Scholar]
- 37.Seewald MJ, Korner C, Wittinghofer A, Vetter IR. Nature. 2002;415:662–666. doi: 10.1038/415662a. [DOI] [PubMed] [Google Scholar]
- 38.Bernier-Villamor V, Sampson DA, Matunis MJ, Lima CD. Cell. 2002;108:345–356. doi: 10.1016/s0092-8674(02)00630-x. [DOI] [PubMed] [Google Scholar]
- 39.Reverter D, Lima CD. Nature. 2005;435:687–692. doi: 10.1038/nature03588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bullock TL, Clarkson WD, Kent HM, Stewart M. J. Mol. Biol. 1996;260:422–431. doi: 10.1006/jmbi.1996.0411. [DOI] [PubMed] [Google Scholar]
- 41.Stewart M, Kent HM, McCoy AJ. J. Mol. Biol. 1998;277:635–646. doi: 10.1006/jmbi.1997.1602. [DOI] [PubMed] [Google Scholar]
- 42.Clarkson WD, Corbett AH, Paschal BM, Kent HM, McCoy AJ, Gerace L, Silver PA, Stewart M. J. Mol. Biol. 1997;272:716–730. doi: 10.1006/jmbi.1997.1255. [DOI] [PubMed] [Google Scholar]
- 43.Bayliss R, Leung SW, Baker RP, Quimby BB, Corbett AH, Stewart M. EMBO J. 2002;21:2843–2853. doi: 10.1093/emboj/cdf305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bayliss R, Ribbeck K, Akin D, Kent HM, Feldherr CM, Gorlich D, Stewart M. J. Mol. Biol. 1999;293:579–593. doi: 10.1006/jmbi.1999.3166. [DOI] [PubMed] [Google Scholar]
- 45.Liker E, Fernandez E, Izaurralde E, Conti E. EMBO J. 2000;19:5587–5598. doi: 10.1093/emboj/19.21.5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ho DN, Coburn GA, Kang Y, Cullen BR, Georgiadis MM. Proc. Natl. Acad. Sci. USA. 2002;99:1888–1893. doi: 10.1073/pnas.042698599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grant RP, Hurt E, Neuhaus D, Stewart M. Nat. Struct. Biol. 2002;9:247–251. doi: 10.1038/nsb773. [DOI] [PubMed] [Google Scholar]
- 48.Grant RP, Neuhaus D, Stewart M. J. Mol. Biol. 2003;326:849–858. doi: 10.1016/s0022-2836(02)01474-2. [DOI] [PubMed] [Google Scholar]
- 49.Fribourg S, Braun IC, Izaurralde E, Conti E. Mol. Cell. 2001;8:645–656. doi: 10.1016/s1097-2765(01)00348-3. [DOI] [PubMed] [Google Scholar]
- 50.Fribourg S, Conti E. EMBO Rep. 4:699–703. doi: 10.1038/sj.embor.embor883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Handa N, Murayama K, Kukimoto M, Hamana H, Uchikubo C, Takemoto T, Terada T, Shirouzu M, Yokoyama S. to be published. [Google Scholar]
- 52.Hodel AE, Hodel MR, Griffis ER, Hennig KA, Ratner GA, Xu S, Powers MA. Mol. Cell. 2002;10:347–358. doi: 10.1016/s1097-2765(02)00589-0. [DOI] [PubMed] [Google Scholar]
- 53.Robinson MA, Park S, Sun ZY, Silver PA, Wagner G, Hogle JM. J. Biol. Chem. 2005;280:35723–35732. doi: 10.1074/jbc.M505068200. [DOI] [PubMed] [Google Scholar]
- 54.Berke IC, Boehmer T, Blobel G, Schwartz TU. J. Cell Biol. 2004;167:591–597. doi: 10.1083/jcb.200408109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weirich CS, Erzberger JP, Berger JM, Weis K. Mol. Cell. 2004;16:749–760. doi: 10.1016/j.molcel.2004.10.032. [DOI] [PubMed] [Google Scholar]