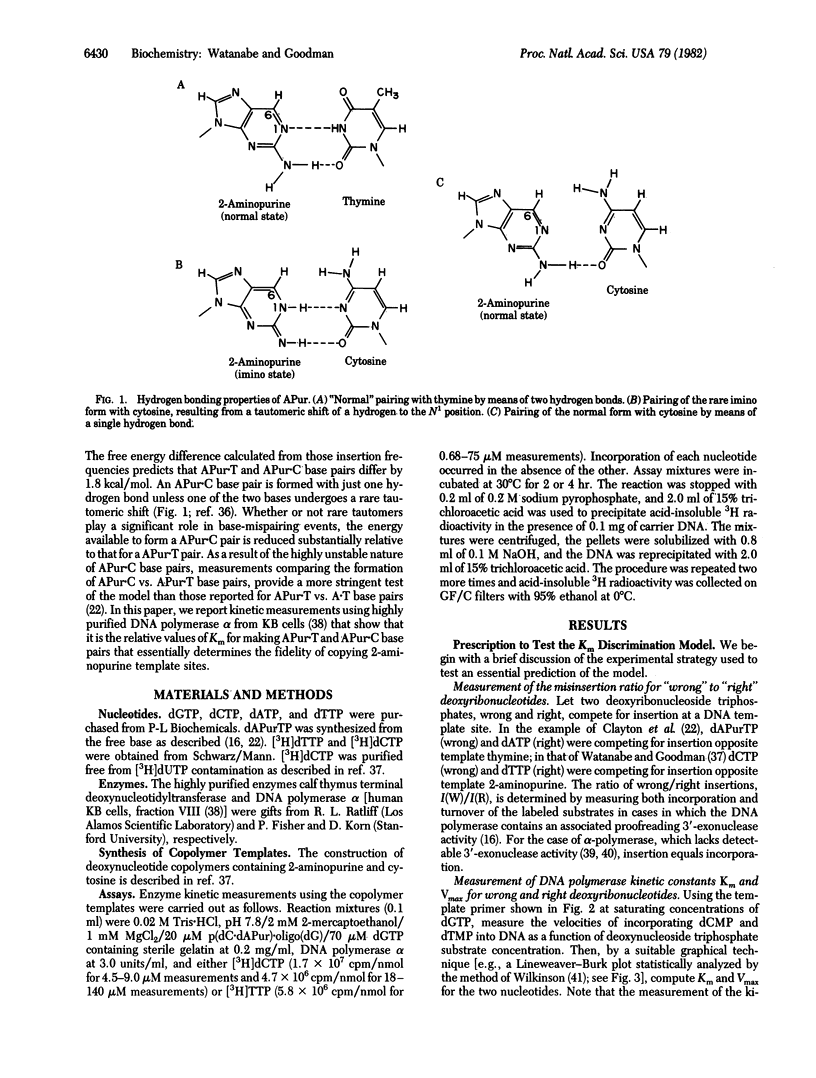
Abstract
Enzyme kinetic measurements are presented showing that Km rather than maximum velocity (Vmax) discrimination governs the frequency of forming 2-aminopurine X cytosine base mispairs by DNA polymerase alpha. An in vitro system is used in which incorporation of dTMP or dCMP occurs opposite a template 2-aminopurine, and values for Km and Vmax are obtained. Results from a previous study in which dTTP and dCTP were competing simultaneously for insertion opposite 2-aminopurine indicated that dTMP is inserted 22 times more frequently than dCMP. We now report that the ratio of Km values KCm/KTm = 25 +/- 6, which agrees quantitatively with the dTMP/dCMP incorporation ratio obtained previously. We also report that VCmax is indistinguishable from VTmax. These Km and Vmax data are consistent with predictions from a model, the Km discrimination model, in which replication fidelity is determined by free energy differences between matched and mismatched base pairs. Central to this model is the prediction that the ratio of Km values for insertion of correct and incorrect nucleotides specifies the insertion fidelity, and the maximum velocities of insertion are the same for both nucleotides.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bessman M. J., Muzyczka N., Goodman M. F., Schnaar R. L. Studies on the biochemical basis of spontaneous mutation. II. The incorporation of a base and its analogue into DNA by wild-type, mutator and antimutator DNA polymerases. J Mol Biol. 1974 Sep 15;88(2):409–421. doi: 10.1016/0022-2836(74)90491-4. [DOI] [PubMed] [Google Scholar]
- Brutlag D., Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. 36. A proofreading function for the 3' leads to 5' exonuclease activity in deoxyribonucleic acid polymerases. J Biol Chem. 1972 Jan 10;247(1):241–248. [PubMed] [Google Scholar]
- COMMONER B. DEOXYRIBONUCLEIC ACID AND THE MOLECULAR BASIS OF SELF-DUPLICATION. Nature. 1964 Aug 1;203:486–491. doi: 10.1038/203486a0. [DOI] [PubMed] [Google Scholar]
- Chang L. M. Low molecular weight deoxyribonucleic acid polymerase from calf thymus chromatin. II. Initiation and fidelity of homopolymer replication. J Biol Chem. 1973 Oct 25;248(20):6983–6992. [PubMed] [Google Scholar]
- Clayton L. K., Goodman M. F., Branscomb E. W., Galas D. J. Error induction and correction by mutant and wild type T4 DNA polymerases. Kinetic error discrimination mechanisms. J Biol Chem. 1979 Mar 25;254(6):1902–1912. [PubMed] [Google Scholar]
- Drake J. W., Allen E. F. Antimutagenic DNA polymerases of bacteriophage T4. Cold Spring Harb Symp Quant Biol. 1968;33:339–344. doi: 10.1101/sqb.1968.033.01.039. [DOI] [PubMed] [Google Scholar]
- Ehrenberg M., Blomberg C. Thermodynamic constraints on kinetic proofreading in biosynthetic pathways. Biophys J. 1980 Sep;31(3):333–358. doi: 10.1016/S0006-3495(80)85063-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fersht A. R. Fidelity of replication of phage phi X174 DNA by DNA polymerase III holoenzyme: spontaneous mutation by misincorporation. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4946–4950. doi: 10.1073/pnas.76.10.4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fersht A. R., Knill-Jones J. W. DNA polymerase accuracy and spontaneous mutation rates: frequencies of purine.purine, purine.pyrimidine, and pyrimidine.pyrimidine mismatches during DNA replication. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4251–4255. doi: 10.1073/pnas.78.7.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher P. A., Korn D. DNA polymerase-alpha. Purification and structural characterization of the near homogeneous enzyme from human KB cells. J Biol Chem. 1977 Sep 25;252(18):6528–6535. [PubMed] [Google Scholar]
- Freese E. B., Freese E. On the specificity of DNA polymerase. Proc Natl Acad Sci U S A. 1967 Mar;57(3):650–657. doi: 10.1073/pnas.57.3.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas D. J., Branscomb E. W. Enzymatic determinants of DNA polymerase accuracy. Theory of coliphage T4 polymerase mechanisms. J Mol Biol. 1978 Oct 5;124(4):653–687. doi: 10.1016/0022-2836(78)90176-6. [DOI] [PubMed] [Google Scholar]
- Gillin F. D., Nossal N. G. Control of mutation frequency by bacteriophage T4 DNA polymerase. I. The CB120 antimutator DNA polymerase is defective in strand displacement. J Biol Chem. 1976 Sep 10;251(17):5219–5224. [PubMed] [Google Scholar]
- Goodman M. F., Hopkins R., Gore W. C. 2-Aminopurine-induced mutagenesis in T4 bacteriophage: a model relating mutation frequency to 2-aminopurine incorporation in DNA. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4806–4810. doi: 10.1073/pnas.74.11.4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall Z. W., Lehman I. R. An in vitro transversion by a mutationally altered T4-induced DNA polymerase. J Mol Biol. 1968 Sep 28;36(3):321–333. doi: 10.1016/0022-2836(68)90158-7. [DOI] [PubMed] [Google Scholar]
- Hershfield M. S. On the role of deoxyribonucleic acid polymerase in determining mutation rates. Characterization of the defect in the T4 deoxyribonucleic acid polymerase caused by the ts L88 mutation. J Biol Chem. 1973 Feb 25;248(4):1417–1423. [PubMed] [Google Scholar]
- Hibner U., Alberts B. M. Fidelity of DNA replication catalysed in vitro on a natural DNA template by the T4 bacteriophage multi-enzyme complex. Nature. 1980 May 29;285(5763):300–305. doi: 10.1038/285300a0. [DOI] [PubMed] [Google Scholar]
- Hopfield J. J. Kinetic proofreading: a new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc Natl Acad Sci U S A. 1974 Oct;71(10):4135–4139. doi: 10.1073/pnas.71.10.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfield J. J. The energy relay: a proofreading scheme based on dynamic cooperativity and lacking all characteristic symptoms of kinetic proofreading in DNA replication and protein synthesis. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5248–5252. doi: 10.1073/pnas.77.9.5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch A. L., Miller C. A mechanism for keeping mutations in check. J Theor Biol. 1965 Jan;8(1):71–80. doi: 10.1016/0022-5193(65)90093-7. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Loeb L. A. Fidelity of mammalian DNA polymerases. Science. 1981 Aug 14;213(4509):765–767. doi: 10.1126/science.6454965. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Loeb L. A. On the fidelity of DNA replication. The accuracy of Escherichia coli DNA polymerase I in copying natural DNA in vitro. J Biol Chem. 1980 Oct 25;255(20):9961–9966. [PubMed] [Google Scholar]
- Liu C. C., Burke R. L., Hibner U., Barry J., Alberts B. Probing DNA replication mechanisms with the T4 bacteriophage in vitro system. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):469–487. doi: 10.1101/sqb.1979.043.01.053. [DOI] [PubMed] [Google Scholar]
- Lo K. Y., Bessman M. J. An antimutator deoxyribonucleic acid polymerase. I. Purification and properties of the enzyme. J Biol Chem. 1976 Apr 25;251(8):2475–2479. [PubMed] [Google Scholar]
- Loeb L. A., Dube D. K., Beckman R. A., Koplitz M., Gopinathan K. P. On the fidelity of DNA replication. Nucleoside monophosphate generation during polymerization. J Biol Chem. 1981 Apr 25;256(8):3978–3987. [PubMed] [Google Scholar]
- Loeb L. A., Springgate C. F., Battula N. Errors in DNA replication as a basis of malignant changes. Cancer Res. 1974 Sep;34(9):2311–2321. [PubMed] [Google Scholar]
- Loeb L. A., Weymouth L. A., Kunkel T. A., Gopinathan K. P., Beckman R. A., Dube D. K. On the fidelity of DNA replication. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):921–927. doi: 10.1101/sqb.1979.043.01.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mildvan A. S. Mechanism of enzyme action. Annu Rev Biochem. 1974;43(0):357–399. doi: 10.1146/annurev.bi.43.070174.002041. [DOI] [PubMed] [Google Scholar]
- Muzyczka N., Poland R. L., Bessman M. J. Studies on the biochemical basis of spontaneous mutation. I. A comparison of the deoxyribonucleic acid polymerases of mutator, antimutator, and wild type strains of bacteriophage T4. J Biol Chem. 1972 Nov 25;247(22):7116–7122. [PubMed] [Google Scholar]
- Ninio J. Kinetic amplification of enzyme discrimination. Biochimie. 1975;57(5):587–595. doi: 10.1016/s0300-9084(75)80139-8. [DOI] [PubMed] [Google Scholar]
- Reha-Krantz L. J., Bessman M. J. Studies on the biochemical basis of mutation VI. Selection and characterization of a new bacteriophage T4 mutator DNA polymerase. J Mol Biol. 1981 Feb 5;145(4):677–695. doi: 10.1016/0022-2836(81)90309-0. [DOI] [PubMed] [Google Scholar]
- Ripley L. S. Influence of diverse gene 43 DNA polymerases on the incorporation and replication in vivo of 2-aminopurine at A X T base-pairs in bacteriophage T4. J Mol Biol. 1981 Aug 5;150(2):197–216. doi: 10.1016/0022-2836(81)90449-6. [DOI] [PubMed] [Google Scholar]
- Sedwick W. D., Shu-Fong Wang T., Korn D. "Cytoplasmic" deoxyribonucleic acid polymerase. Structure and properties of the highly purified enzyme from human KB cells. J Biol Chem. 1975 Sep 10;250(17):7045–7056. [PubMed] [Google Scholar]
- Speyer J. F., Karam J. D., Lenny A. B. On the role of DNA polymerase in base selection. Cold Spring Harb Symp Quant Biol. 1966;31:693–697. doi: 10.1101/sqb.1966.031.01.088. [DOI] [PubMed] [Google Scholar]
- Topal M. D., DiGuiseppi S. R., Sinha N. K. Molecular basis for substitution mutations. Effect of primer terminal and template residues on nucleotide selection by phage T4 DNA polymerase in vitro. J Biol Chem. 1980 Dec 25;255(24):11717–11724. [PubMed] [Google Scholar]
- WILKINSON G. N. Statistical estimations in enzyme kinetics. Biochem J. 1961 Aug;80:324–332. doi: 10.1042/bj0800324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S. M., Goodman M. F. On the molecular basis of transition mutations: frequencies of forming 2-aminopurine.cytosine and adenine.cytosine base mispairs in vitro. Proc Natl Acad Sci U S A. 1981 May;78(5):2864–2868. doi: 10.1073/pnas.78.5.2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissbach A. Eukaryotic DNA polymerases. Annu Rev Biochem. 1977;46:25–47. doi: 10.1146/annurev.bi.46.070177.000325. [DOI] [PubMed] [Google Scholar]


