Abstract
Ubiquitin-positive inclusion containing Fused in Sarcoma (FUS) defines a new subtype of frontotemporal lobar degeneration (FTLD). FTLD is characterized by progressive alteration in cognitions and it preferentially affects the superficial layers of frontotemporal cortex. Mutation of FUS is linked to amyotrophic lateral sclerosis and to motor neuron disease with FTLD. To examine FUS pathology in FTLD, we developed the first mammalian animal model expressing human FUS with pathogenic mutation and developing progressive loss of memory. In FUS transgenic rats, ubiquitin aggregation and FUS mislocalization were developed primarily in the entorhinal cortex of temporal lobe, particularly in the superficial layers of affected cortex. Overexpression of mutant FUS led to Golgi fragmentation and mitochondrion aggregation. Intriguingly, aggregated ubiquitin was not colocalized with either fragmented Golgi or aggregated mitochondria, and neurons with ubiquitin aggregates were deprived of endogenous TDP-43. Agonists of peroxisome proliferator-activated receptor gamma (PPAR-γ) possess anti-glial inflammation effects and are also shown to preserve the dendrite and dendritic spines of cortical neurons in culture. Here we show that rosiglitazone, a PPAR-γ agonist, rescued the dendrites and dendritic spines of neurons from FUS toxicity and preserved rats' spatial memory. Our FUS transgenic rats would be useful to the mechanistic study of cortical dementia in FTLD. As rosiglitazone is clinically used to treat diabetes, our results would encourage immediate application of PPAR-γ agonists in treating patients with cortical dementia.
INTRODUCTION
Frontotemporal lobar degeneration (FTLD) is a clinical syndrome that is characterized by progressive alteration in behavior, language and personality, with relative preservation of memory at early disease stages (1,2). About half FTLD cases show the pathology of tau-negative, ubiquitin-positive inclusions in the cytoplasm and neurites, particularly in the superficial layers of the frontotemporal cortex and in the dentate gyrus of the hippocampus (1–4). This subtype of FTLD with ubiquitin inclusions is now called FTLD-U. In most FTLD-U cases, ubiquitin inclusions entrap Tar DNA-binding protein 43 (TDP-43) (5), whereas a subtype of FTLD-U shows TDP-43-negative, ubiquitin-positive inclusion that is immunostained of Fused in Sarcoma (FUS) (6). Both TDP-43 and FUS are RNA- and DNA-binding proteins (7), but they define a distinct pathology of FTLD.
FUS can bind to both single- and double-stranded DNA sequences (8), promoting D-loop formation in DNA repair (9). FUS interacts with the p65 subunit of the nuclear factor κB to regulate gene transcription (10). FUS binds RNA molecules, particularly the RNA with GGUG sequence (11), and FUS is associated with the complex of splicing factors, regulating RNA splicing (12). In physiological conditions, FUS is mainly in the nucleus of neurons, but it shuttles between the nucleus and the cytoplasm to perform complex functions (13,14). In hippocampal pyramidal neurons, FUS is translocated to dendrites and accumulates in dendritic spines at excitatory post-synapses upon mGlR5 activation, participating in mRNA sorting to dendritic spines and regulating spine morphology (15,16). Expression of FUS is rapidly downregulated in the peripheral organs during postnatal development, but it remains at significant levels in neurons throughout the lifetime (17), suggesting that FUS may exert important functions in the central nerve system.
Mutation of FUS is linked to amyotrophic lateral sclerosis (ALS) (18–20), MND with dementia (21) and FTLD (22). Mutation in FUS changes the profile of its RNA binding (23), suggesting that pathogenic mutation in FUS gains toxic properties. Indeed, overexpression of mutant, but not the normal, FUS causes rapid paralysis in Caenorhabditis elegans and in rats (24–26). FUS deficiency causes early death in homogeneous C57/BL6 background (27), but causes only male infertility in heterogeneous genomic background (28). Studies in yeast and Drosophila suggest that cytoplasmic localization of mutant FUS is required for cytotoxicity (29,30). FUS and TDP-43 are two similar ribonucleoproteins and may interact with each other in a common pathway leading to neurodegeneration (31). In Drosophila and zebrafish (32,33), overexpression of FUS rescues the phenotypes of TDP-43 deficiency, but does not vice versa, suggesting that FUS may function downstream of TDP-43 in a pathway. Till now, the only mammalian animal model expressing human FUS develops rapid paralysis as the predominant phenotype, with limited loss of cortical neurons (25). How FUS pathology correlates with cortical dementia in FTLD remains to be determined.
To examine FUS in the pathogenesis of FTLD, we developed novel transgenic rats expressing mutant FUS restrictedly in neurons of the forebrain. This novel rat model reproduced the core phenotypes of FTLD: severe loss of neurites and dendritic spines; progressive loss of neurons particularly in the superficial layers of the cortex; and gradual development of FUS mislocalization and ubiquitin aggregates primarily in the entorhinal cortex of the temporal lobe. In this novel rat model, the spatial memory was well preserved when the neurites and dendritic spines were rescued by treatment with rosiglitazone.
RESULTS
Overexpression of mutant FUS in the forebrain caused progressive dementia in transgenic rats
Although mutation in the fus is causative of some familial ALS (18,19), FUS pathology defines a distinct subtype of FTLD that displays ubiquitin- and FUS-positive, but TDP-43 negative, inclusions (34,35). Ubiquitous expression of mutant human FUS in rats causes early-onset paralysis without a significant loss of motor neurons (25). To examine FUS pathobiology in FTLD, we attempted to overexpress mutant human FUS restrictedly in the forebrain and thus to induce cortical dementia in transgenic rats (Figs 1–3).
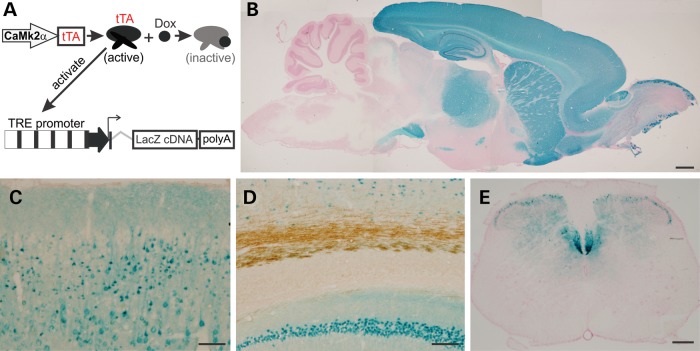
Figure 1.
Restricted expression of the reporter gene LacZ in the forebrain of transgenic rats. (A) The schematic shows a tetracycline-regulated gene expression system. The promoter of mouse Camk2a directs the synthesis of tTA, which is subject to the regulation by Dox. Free tTA binds to tetracycline-responsive elements (TRE) and activate the LacZ transgene. (B) A reconstructed photo shows the profile of LacZ expression on the sagittal section of a rat brain. Tissue sections were taken from a Camk2a-tTA/TRE-LacZ double-transgenic 4-week-old rat. Individual photos were assembled with Photoshop to demonstrate the profile of X-gal staining. (C and D) X-gal staining shows the expression patterns of the LacZ transgene in the cortex (C) and CA1 region (D). Tissue sections were first stained with X-gal and were further stained with an antibody to myelin basic protein. (E) A cross-section of cervical spinal cord shows that corticospinal tracks were stained with X-gal. Scale bars: (B) 1 mm; (C and D) 100 µm; (E) 200 µm.
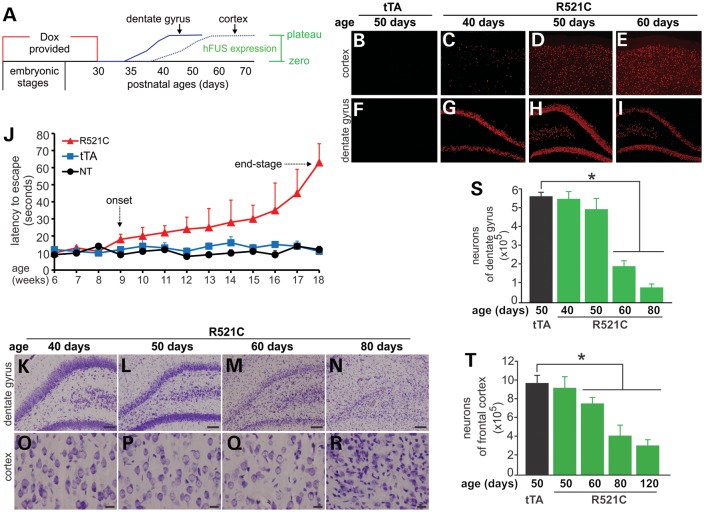
Figure 3.
Restricted overexpression of mutant FUS in a rat forebrain causes progressive loss of neurons and memory. (A) The diagram shows a strategy to induce the expression of mutant human FUS (hFUS) (R521C) in Camk2a-tTA/TRE-FUSR521C transgenic rats. Breeding rats and their offspring were provided with Dox in drinking water and the transgenic offspring were deprived of Dox by the age of 30 days. After Dox was withdrawn, hFUS began to express from the zero level to the maximal levels (plateau). (B–I) Immunofluorescent staining revealed the gradual expression of mutant hFUS in Camk2a-tTA/TRE-FUSR521C double-transgenic rats (R521C), but not in Camk2a-tTA single-transgenic rats (tTA). (J) The Barnes maze revealed the reduction of spatial memory in mutant FUS transgenic rats (R521C), but not in Camk2a-tTA single-transgenic rats (tTA) or in non-transgenic rats (NT). Rat's spatial memory was measured with Banes maze once a week. Data are means + SEM (n = 8–10). (K–R) Cresyl violet staining revealed the progressive loss of neurons in Camk2a-tTA/TRE-FUSR521C double-transgenic rats (R521C). Scale bars: (K–N) 100µm; and (O–R) 20µm. (S and T) Stereological cell counting estimated the number of neurons in specified brain regions in the Camk2a-tTA/TRE-FUSR521C double-transgenic rats (R521C) and in the Camk2a-tTA single-transgenic rats (tTA). Data are means SEM (n = 6–7). *P < 0.05.
To express a transgene in the forebrain, we isolated the promoter (9 kb) of the mouse camk2a from a BAC clone and used it to drive tetracycline-responsive transactivator (tTA) in rats (Fig. 1A). We established one transgenic line that carries two copies of the tTA transgene as determined by quantitative PCR. To determine the profile of transgene expression, we crossed Camk2a-tTA transgenic rats with a LacZ reporter line (36). X-gal staining shows that expression of the reporter gene was restricted in the forebrain (Fig. 1B). X-gal-stained cells were observed in the hippocampus and cortex (Fig. 1C and D), but not in the corpus callosum (Fig. 1D). The corticospinal tracts in the spinal cord were also stained (Fig. 1E). Expression of the tTA in the rat forebrain did not affect neuron development and maturation and thus did not affect rats' spatial learning (Supplementary Material, Fig. S1).
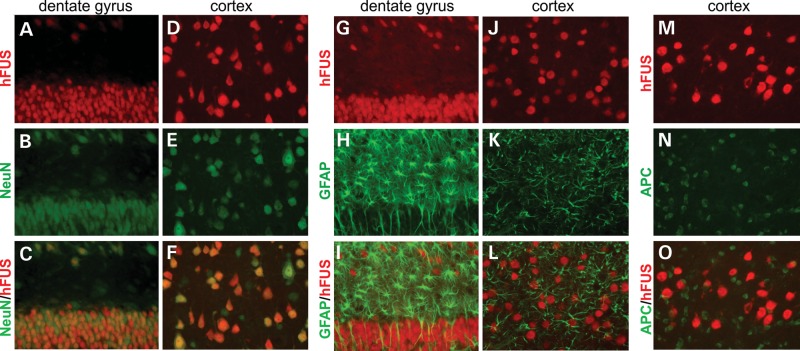
To induce expression of mutant human FUS in the rat forebrain, we crossed Camk2a-tTA transgenic line with mutant FUS transgenic rats (line 16) (25). In the double-transgenic rats, we observed expression of human FUS only in the neurons (Fig. 2A–F), but not in the glia (Fig. 2G–O). Profiles of transgene expression in Camk2a-tTA/TRE-FUS-R521C rats were consistent with that in Camk2a-tTA/TRE-LacZ rats (Figs 1 and 2), validating that the Camk2a promoter drives transgene expression restrictedly in the neurons of the rat forebrain.
Figure 2.
Camk2a-tTA activates human FUS transgene selectively in the neurons of a rat forebrain. (A–O) Double-labeling fluorescence staining reveals that human FUS (hFUS) is colocalized with the neuronal marker NeuN and is not colocalized with the glia markers (GFAP for astrocytes and APC for oligodendrocytes) in the dentate gyrus (A, B and G–I) and cortex (D–F and J–O) of Camk2a-tTA/TRE-FUSR521C double-transgenic rats. Transgenic rats were given Dox during embryonic and postnatal development and were deprived of Dox at 30 days of age. The hippocampus was taken from a 50-day-old rat (20 days off Dox), but the cortex was taken from a 60-day-old rat (30 days off Dox). All photos were taken at identical magnification.
We attempted to induce expression of mutant FUS in postnatal rats and thus to induce disease phenotypes in adult rats (Fig. 3). Expression of mutant FUS is subject to the regulation by doxycycline (Dox) (Fig. 1A). Dox was withdrawn from postnatal rats at the age of 30 days and expression of the FUS transgene was initiated gradually (Fig. 3A–I). After Dox withdrawal, the mutant FUS transgene was activated first in the hippocampus and then in the cortex (Fig. 3B–I). Overexpression of mutant FUS in the forebrain impaired rats' spatial memory, which was detected by Barnes maze assay (Fig. 3J). The rats began losing spatial memory (disease onset) about 5 weeks after Dox withdrawal, and by the age of 18 weeks most of the rats had difficulty in locating the escaping hole in the Barnes maze and reached disease end-stages (Fig. 3J). In the FUS transgenic rats, neurons were progressively lost in the hippocampus and cortex (Fig. 3K–R). Neuronal loss was quantified with stereological cell counting (Fig. 3S and T). The brain regions of neuronal loss were correlated with the initiation of transgene expression: expression of the mutant FUS transgene was detected earlier in the hippocampus than in the cortex (Fig. 3B–I) and thus the neuronal loss was first detected in the dentate gyrus (Fig. 3K–T). At the onset of the disease, neurons were lost severely in the dentate gyrus, but were only lost moderately in the cortex (Fig. 3). At disease end-stages, most of the neurons were lost in the frontal cortex (Fig. 3S and T). Our findings suggest that both the dentate gyrus and the frontal cortex contribute to spatial learning and spatial memory in rats.
Overexpression of mutant FUS decreased dendritic complexity
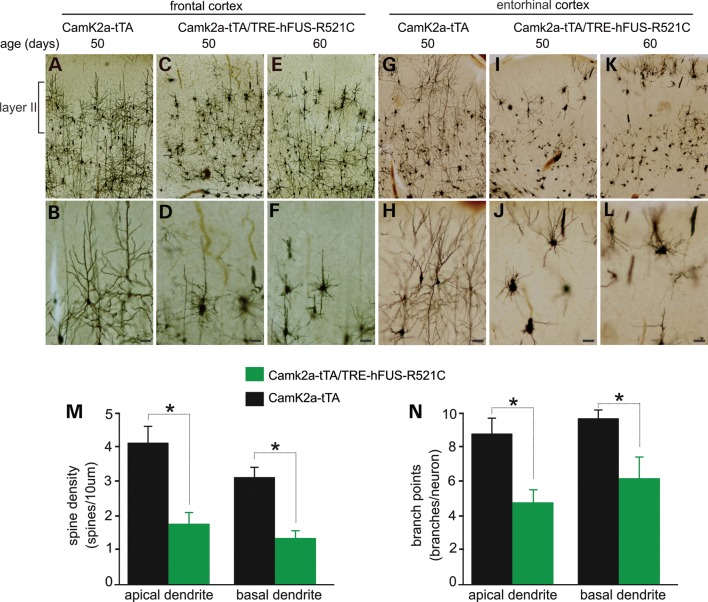
After the mutant FUS transgene was turned on in postnatal rats, the rats began losing spatial memory by the fifth week of transgene expression (Fig. 3J). At disease onset, no significant loss of neurons was detected in the cortex, though it was detected in the dentate gyrus (Fig. 3K–T). As neurites and spines play a critical role in neurotransmission, these structures might be destroyed and the neuronal functions might be disturbed before the neurons die. Using Golgi staining, we detected that the neurons in the frontal and entorhinal cortexes were severely deprived of neurites before memory loss (Fig. 4A–L). In the frontal cortex, neurons of layer II were first affected (Fig. 4A–F), displaying short and retracted neurites. Similarly, neurons in the entorhinal cortex lost neurites severely (Fig. 4G–L). Quantitative measures show that neurite branches and the spine density on both the apical and basal dendrites were markedly reduced in the transgenic rats (Fig. 4M and N). Neurite and dendritic spines are the primary targets of degeneration caused by mutation in FUS.
Figure 4.
Neurites and spines are lost prior to neuron loss. (A–L) Representative photos of Golgi staining showing that affected neurons displayed contracted neurites particularly in the layer II of frontal and entorhinal cortexes in mutant FUS transgenic rats. All scale bars: 40 µm. (M and N) Spine densities and branch points were quantified for five neurons in the frontal cortex of individual rats. Data are means + SEM (n = 4). *P < 0.05.
Overexpression of mutant FUS led to fragmentation of Golgi complexes
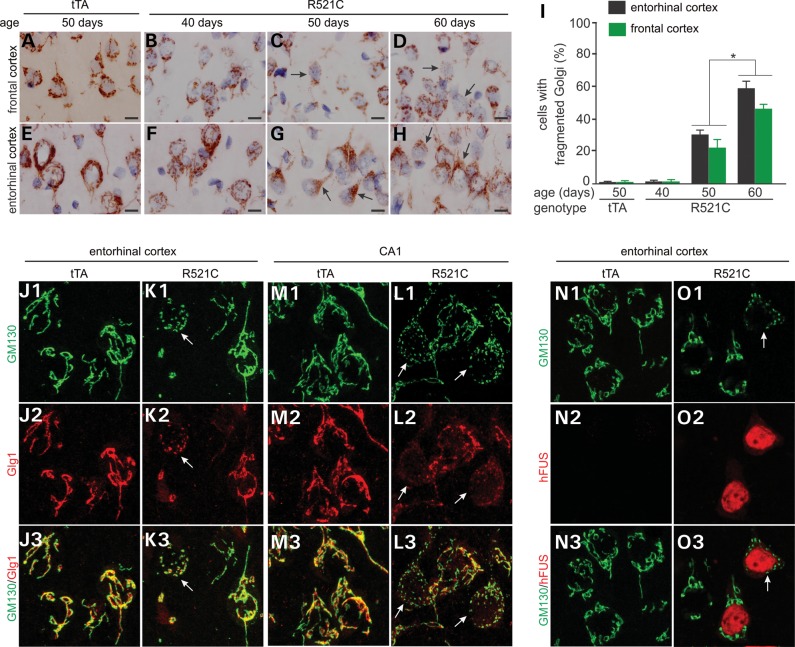
Mutation of FUS is linked to ALS and FTLD, and Golgi fragmentation is frequently observed in the motor neurons of ALS patients (18,19,37,38). We examined Golgi apparatus in mutant FUS transgenic rats and observed that both cis- and trans-Golgi complexes were fragmented in the neurons expressing mutant human FUS (Fig. 5, Supplementary Material, Figs S2 and S3). After the mutant FUS transgene was turned on, Golgi complexes were fragmented progressively in the rats at pre-symptomatic stages (Fig. 5A–I), suggesting that Golgi fragmentation is an early event in FUS pathology. Golgi fragmentation was observed in all of the brain regions examined, but appeared to be severe in the entorhinal cortex of the temporal lobe (Fig. 5). Fragmented Golgi complexes were restricted to the neurons expressing mutant human FUS (Fig. 5 and Supplementary Material, Fig. S3), suggesting that Golgi fragmentation is a result of the overexpression of mutant human FUS.
Figure 5.
Overexpression of mutant human FUS leads to Golgi fragmentation. (A–H) Immunostaining reveals fragmented Golgi apparatuses in the Camk2a-tTA/TRE-FUSR521C double-transgenic rat (R521C), but not in the Camk2a-tTA single-transgenic rat (tTA). Coronal sections (12 µm) of the forebrain were stained with an antibody to GM130. Scale bars: 10 µm. (I) The cells with fragmented Golgi were quantified. Ten images (×100) were examined for Golgi fragmentation in the frontal and entorhinal cortexes of individual rats. Data are means + SEM (n = 5). *P < 0.05. (J–L) Confocal microscopy reveals that the cis (stained with GM130) and trans (stained with Glg1) faces of Golgi were fragmented in affected cells. Arrows point to fragmented Golgi. (N and O) Confocal microscopy reveals that Golgi fragmentation occurred in neurons expressing mutant human FUS (R521). Coronal sections (12 µm) of the forebrain were stained with GM130 and with Glg1 or human FUS antibodies and Z-stacks of confocal images were projected to construct Golgi apparatuses (J–O).
Overexpression of mutant FUS led to ubiquitin aggregation and FUS mislocalization primarily in the entorhinal cortex
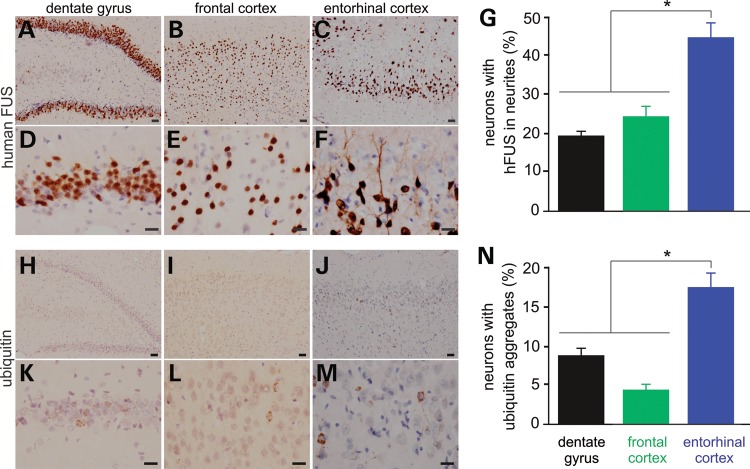
FUS shuttles between the nucleus and the cytoplasm in physiological conditions and its normal localization might be interrupted by pathogenic mutations (39,40). We examined FUS localization in mutant FUS transgenic rats at presymptomatic and symptomatic stages and we observed that mutant FUS accumulated in the cytoplasm and in the neurites (Fig. 6A–G and Supplementary Material, Fig. S4A–F). Compared with the other brain regions expressing mutant FUS, the entorhinal cortex was predisposed to FUS mislocalization (Fig. 6G). As the disease was progressing, mutant FUS progressively accumulated in the neurites and in the dendritic spines (Supplementary Material, Fig. S4). The entorhinal cortex is part of the temporal lobe that is primarily affected in FTLD. Our finding suggests that entorhinal cortical neurons are predisposed to mutation of FUS.
Figure 6.
FUS mislocalization and ubiquitin aggregation primarily develop in the entorhinal cortex of transgenic rats. (A–F) Immunostaining showing that mutant human FUS was mislocalized to the neurites of some neurons in the Camk2a-tTA/TRE-FUSR521C double-transgenic rats. (G) Neurons with human FUS in the neurites were quantified for selected brain regions. Data are means + SEM (n = 5). (H–M) Immunostaining showing that ubiquitin formed aggregates in some neurons. (N) Neurons with ubiquitin aggregates were quantified for selected brain regions. Data are means + SEM (n = 4). Neurons were examined for FUS mislocalization (human FUS in the neurites) and ubiquitin aggregation on six images (×40) of each selected brain region for individual rats at the age of 50 days. FUS mislocalization is defined as a neuron with human FUS in a neurite that is longer than the diameter of the cell body. *P < 0.05. Scale bars: (A–C and H–J) 40 µm; (D–F and K–M) 20 µm.
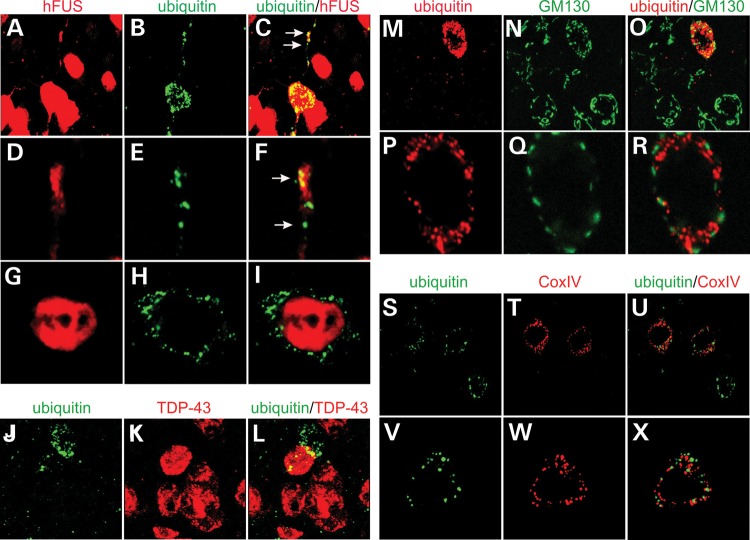
As the disease was progressing, ubiquitin was accumulated and aggregated in affected neurons particularly in the entorhinal cortex (Fig. 6H–N and Supplementary Material, Fig. S5). Similar to mislocalized FUS, aggregated ubiquitin was detected primarily in the entorhinal cortex (Fig. 6N). Aggregated ubiquitin and mislocalized human FUS were simultaneously detected in the neurites of the same affected neurons, but double-labeling fluorescence staining revealed that these two proteins were not physically colocalized in the neurites or the cytoplasm (Fig. 7A–I). Intriguingly, some neurons with ubiquitin aggregates were deprived of endogenous TDP-43 protein (Fig. 7J–L). Aggregated ubiquitin was not colocalized with fragmented Golgi complex (Fig. 7M–R). As mutant FUS was expressed, some neurons displayed an enhanced staining of Cox-IV indicative of mitochondrial clustering and damage (Supplementary Material, Fig. S5A–I). Aggregated ubiquitin also was not colocalized with damaged mitochondria (Fig. 7S–X). In mutant FUS transgenic rats, ubiquitin-positive inclusions entrap something other than fragmented Golgi complex and damaged mitochondria.
Figure 7.
Ubiquitin aggregation and Golgi fragmentation co-exist in affected neurons. (A–I) Confocal microscopy reveals that aggregated ubiquitin was not physically colocalized with human FUS in the entorhinal cortical neurons of transgenic rats. Z-stacks images were projected to show the profile of ubiquitin aggregation and human FUS mislocalization (A–C). Single-scanned images (thickness: 2 µm) showed the localization of ubiquitin aggregates and human FUS. (J–L) Confocal microscopy reveals that neurons with ubiquitin aggregates were depleted of rat endogenous TDP-43. (M–R) Confocal microscopy reveals that ubiquitin aggregates coexisted, but were not colocalized, with Golgi fragments in the same affected neurons. Z-stack images were projected to show the profile of ubiquitin aggregation and Golgi fragmentation (M–O). Single-scanned images (2 µm) showed the localization of ubiquitin aggregates and Golgi fragments (P–R). (S–X) Confocal microscopy reveals that ubiquitin aggregates were not colocalized with damaged mitochondria immunostained of Cox-IV. Lateral entorhinal cortex was taken from a Camk2a-tTA/TRE-FUSR521C transgenic rat at the age of 60 days.
Astrocytes and microglia became reactive in response to neurodegeneration in FUS transgenic rats
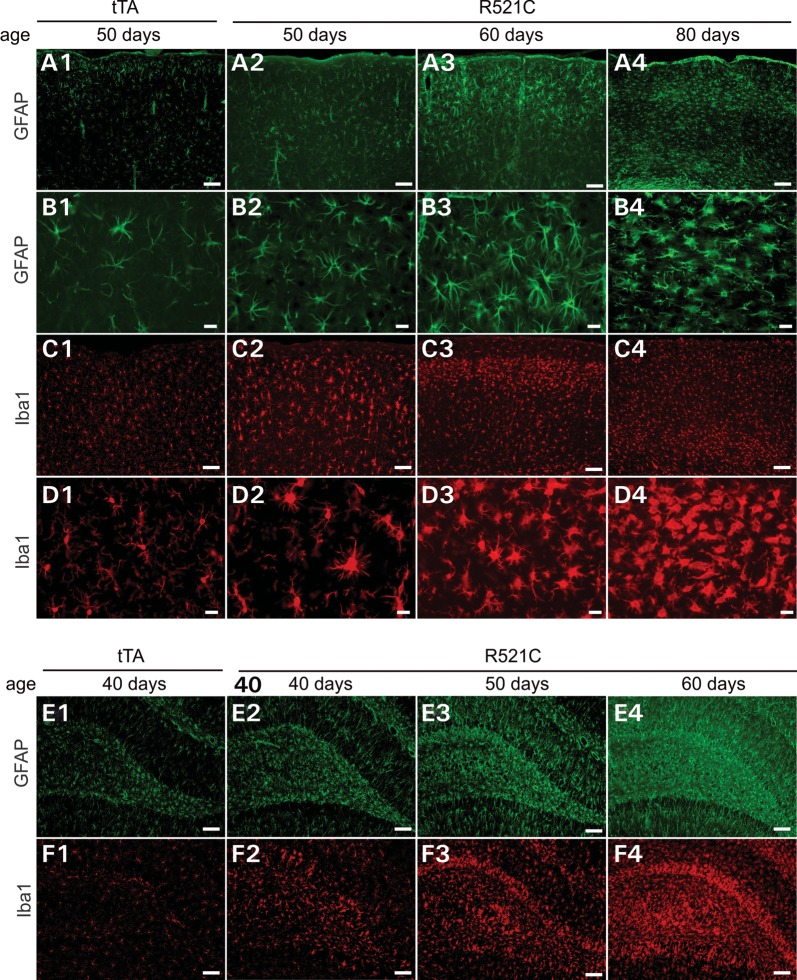
Glial reaction is a common feature of neurodegenerative diseases, including FTLD. We examined astrocytic and microglial reaction in mutant FUS transgenic rats and observed that both microglia and astrocytes were activated in all of the brain regions examined (Fig. 8). Glial reaction was closely associated with neuron death. As neuronal loss occurred earlier in the hippocampus than in the cortex, reactive astrocytes and microglia were detected first in the hippocampus (Fig. 8E and F) and then in the cortex (Fig. 8A–D). In the frontal cortex, neurons of layers II and III were first affected (Fig. 4). Accordingly, reactive microglia and astrocytes were first detected in the layers II and III of frontal cortex (Fig. 8A3 and C3). Glial reaction is related to neuronal death.
Figure 8.
Astrocytes and microglia are overtly activated in mutant FUS transgenic rats. (A–F) Immunostaining for GFAP (a marker of astrocyte) and Iba1 (a marker of microglia) revealed that astrocytes and microglia were activated in the cortex (A–D) and dentate gyrus (E and F) of Camk2a-tTA/TRE-FUSR521C double-transgenic rat (R521C) when compared with Camk2a-tTA single-transgenic rat (tTA). The glial cells were first activated in the layers II and III of cortex (A3 and C2–3). Scale bars: (A1–4, C1–4, E1–4 and F1–4): 100 µm; (B1–4 and D1–4): 20 µm.
Rosiglitazone rescued dendritic spine loss in FUS transgenic rats
In mutant FUS transgenic rats, neurites and dendritic spines were the primary targets of neurodegeneration, and glial cells were remarkably activated in response to the neuronal death (Figs 4 and 8). Rosiglitazone is a potent agonist of peroxisome proliferator-activated receptor gamma (PPAR-γ) and possesses anti-inflammation effects on glial reaction (41,42). Recent studies showed that rosiglitazone rescues dendritic spine loss in cultured primary neurons (43). We examined the neuroprotective effects of rosiglitazone in mutant FUS transgenic rats.
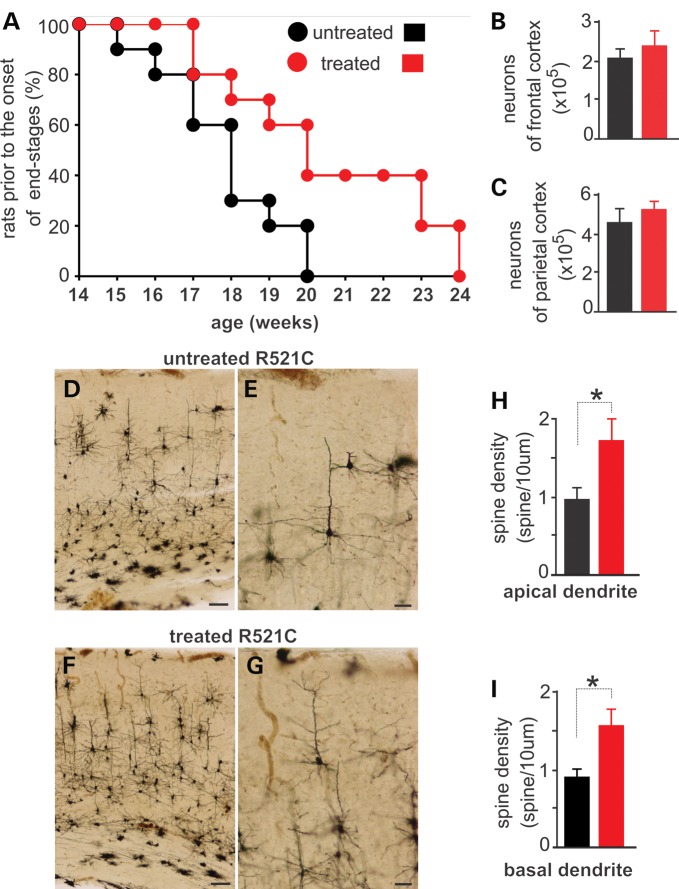
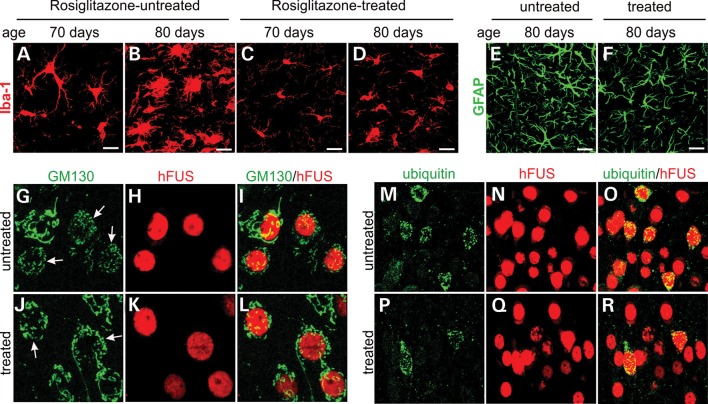
Mutant rats were treated with rosiglitazone or vehicle at the onset of memory loss (Fig. 9). By the time of disease onset, neurons in the dentate gyrus were lost severely and glial cells were overtly reactive (Figs 3 and 8). Treatment with rosiglitazone slowed down the progression of cortical dementia in FUS transgenic rats (Fig. 9A). We examined neuronal loss in rosiglitazone-treated rats by stereological cell counting and found that neuronal loss was not rescued by rosiglitazone (Fig. 9B and C). We further examined dendritic spines by Golgi staining and observed that rosiglitazone rescued the loss of dendritic spines caused by mutation of FUS (Fig. 9D–I). Rosiglitazone is known for its effects on glial reaction. Indeed, rosiglitazone mitigated astroglial and microglial reaction (Fig. 10A–F), with a dramatic effect on microglial activation. Moreover, treatment with rosiglitazone mitigated Golgi fragmentation and ubiquitin aggregation in the neurons expressing mutant FUS (Fig. 10G–R). Consistent with previous findings in culture (43), our data show that rosiglitazone prevented cortical dementia from progression by preserving dendritic spines.
Figure 9.
Rosiglitazone treatment preserves neurites and memory in mutant FUS transgenic rats. (A) The graph shows disease progression in Camk2a-tTA/TRE-FUSR521C double-transgenic rats that were treated with (treated) or without (untreated) rosiglitazone. Rats' spatial memory was weekly determined in a Barnes maze and disease stages were determined accordingly. Disease onset was defined as unrecoverable increase in the latency to find an escaping hole in a Barnes maze, and disease end-stages were defined as the escaping latency >60 s. Rats that reached the disease end-stages were counted and terminated. Rats received rosiglitazone treatment from disease onset onwards. Data are means + SEM (n = 15). (B and C) Stereological cell counting reveals no difference in the number of neurons between rosiglitazone-treated and untreated FUS transgenic rats. Data are means + SEM (n = 5). (D–G) Golgi staining reveals that neurites were preserved in rosiglitazone-treated rats (F and G) when compared with the untreated rats (D and E). Scale bars: (D and F) 100 µm; (E and G) 40 µm. (H and I) Spine density was quantified for five neurons per rat. Data are means + SEM (n = 4). *P < 0.05.
Figure 10.
Rosiglitazone mitigates glial reaction and ubiquitin aggregation. (A–F) Immunofluorescent staining reveals microglial (Iba-1: A–D) and astroglial (GFAP: E and F) reaction in rosiglitazone-treated (C, D and F) and -untreated (A, B and E) rats. Scale bars: 20 µm. (G–R) Confocal microscopy reveals that rosiglitazone moderately mitigated Golgi fragmentation (G–L) and ubiquitin aggregation (M–R) in rosiglitazone-treated rats (J–L and P–R) compared with untreated rats (G–I and M–O). Camk2a-tTA/TRE-FUSR521C double-transgenic rats received rosiglitazone treatment from disease onset onwards and were terminated at the age of 70 days (A and C) or 80 days (B, D and E–R). Rat entorhinal cortices were analyzed for Golgi fragmentation (GM130) and ubiquitin aggregation. Neurons expressing mutant FUS were identified by immunostaining for human FUS (hFUS). Arrows point to fragmented Golgi apparatus (G and J).
DISCUSSION
Overexpression of mutated human FUS in the rat forebrain initiated a dying-back process of neurodegeneration that was accompanied by gradual development of ubiquitin aggregates and FUS mislocalization. Similar to the pathology of FTLD, the pathological changes observed in FUS transgenic rats were predominant in the temporal lobe, and the neuronal death was preceded by remarkable Golgi fragmentation. In FUS transgenic rats, anti-glial inflammation treatment prevented the loss of neurites and dendritic spines and preserved spatial memory.
FUS pathology in our rat model preferentially affected the temporal lobe that is primarily affected in FTLD. Although mutant FUS was expressed in the whole forebrain, including the frontal, parietal and temporal cortexes and the hippocampus, FUS mislocalization and ubiquitin aggregates were primarily developed in the entorhinal cortex that is part of the temporal lobe. The pathological changes preceded the neuronal loss and possibly are related to the neurodegeneration in FUS transgenic rats. Consistent with the findings in patients with FTLD (1–4), neurodegeneration in our FUS transgenic rats was a dying-back process and preferentially affected the superficial layers of the cortex at early disease stages. Although aggregated ubiquitin and mislocalized mutant FUS were simultaneously found in the same neurons, ubiquitin immunostaining was not physically colocalized with the staining of mutant FUS, differing from clinical observation that reveals the colocalization of ubiquitin with FUS in the same inclusions (6). This difference in FUS pathology might be caused by a relatively faster progression of disease phenotypes in rats than that in patients or might result from the distinction of species. FUS and TDP-43 are two similar ribonucleoproteins and are both involved in ALS and FTLD. Viral delivery of TDP-43 induces cytoplasmic mislocalization of TDP-43 in monkey, but expresses TDP-43 only in the nucleus in rats (44). Mitochondrion and Golgi apparatus are involved in neurodegeneration (45–47). Expression of human FUS with pathogenic mutation induced Golgi fragmentation in the cortical neurons, which preceded neuronal death (Fig. 5). Damaged mitochondria form aggregates that can be labeled of Cox-IV (48). Expression of mutant FUS in rats induced aggregation of mitochondria (Fig. 7). Intriguingly, ubiquitin-positive inclusions did not contain damaged Golgi or mitochondria (Fig. 7). What ubiquitin-positive inclusions entrap in FUS transgenic rats remains to be determined. FUS may function in the downstream of TDP-43 in a common pathway as FUS rescues TDP-43-deficient phenotypes in invertebrate animal models (32,33). Unexpectedly, few cells with ubiquitin-positive inclusion were deprived of TDP-43 in FUS transgenic rats (Fig. 7). It remains to determine how TDP-43 depletion is related to FUS pathology. Our FUS transgenic rat is the first mammalian animal model recapitulating the core phenotypes of FTLD and would be useful to the mechanistic study of FUS pathogenesis.
Astroglia and microglia are commonly reactive to neurodegeneration in patients and in the animal models of neurodegenerative diseases (49–53). In response to neurodegeneration in FUS transgenic rats, astrocytes and microglia were overtly activated and their activation was closely related to neuronal death (Fig. 8). Reactive glial cells produce neurotoxic cytokines and the other unidentified mediators that induce neuroinflammation and neurotoxicity (51,54–57). Activation of PPAR-γ suppresses the secretion of cytokines from reactive glia and neuronal response to glial activation (58,59). Rosiglitazone is a potent PPAR-γ agonist (60), preserves memory in transgenic mouse models of Alzheimer's disease (41,61) and improves the cognition in patients with Alzheimer's disease (62,63). Recent study showed that rosiglitazone preserves the neurites and dendritic spines of cortical neurons in culture (43). As the mouse models tested of PPAR-γ agonists show cognitive deficits without significant loss of neurons (41,61), it is not known whether rosiglitazone can rescue both neurons and neurites in an animal model of cortical dementia. In our FUS transgenic rats, treatment with rosiglitazone preserved the spatial memory by rescuing neurites and dendritic spines rather than neurons (Fig. 9). Activation of PPAR-γ receptor exerts neuroprotective effects selectively on neuronal terminals rather than on the cell body. Rosiglitazone has a long history in clinical use and is worth testing of its therapeutic effects in FTLD patients.
MATERIALS AND METHODS
Ethics statement
Animal use followed NIH guidelines and was approved by the Institutional Animal Care and Use Committee (IACUC) at Thomas Jefferson University.
Animal experiments
Creation of FUS and LacZ transgenic rats was reported (25,36). Transgenic rats expressing tTA were created using a similar procedure described previously (36). The promoter of the mouse calcium/calmodulin-dependent protein kinase type II subunit alpha (Camk2a) gene was isolated from a BAC clone (CHORI: RP24-243J21) and was used to drive the tTA transgene. Linearized transgenic DNA was purified from agar gel and was injected into the pronuclei of fertilized eggs of Sprague–Dawley rats to produce transgenic founder rats (36). Transgenes were maintained on the SD genomic background and the Camk2a-tTA transgene was identified by PCR analysis of the rat tail DNA with the following primers: 5′-TGAAAGGCAGGCAGGTGTTG-3′ (forward) and 5′-TCCAAGGCAGAGTTGATGAC-3′ (reverse).
Rat's spatial learning and memory were examined with a Barnes maze (Med Associates) as described previously (25). Mutant FUS transgenic rats were trained to locate the escaping hole on a Barnes maze three times per day, for 3 consecutive days. After the training sections, the rats were examined of spatial memory once a week for determining the progression of disease phenotypes: disease onset was defined as unrecoverable increase in the time of locating the escaping hole on a Barnes maze, and disease end-stage was defined as the inability to locate the escaping hole on a Barnes maze within 60 s.
Rosiglitazone was dissolved in sterilized water and was administered to rats by gavage. From the disease onset, rats were given rosiglitazone at 10 mg/kg body weight, once per day until they reached the disease end-stages.
Histology and immunostaining
As described previously (25,36), rats were deeply anesthetized and were transcardially perfused with 4% paraformaldehyde dissolved in 1× PBS buffer. After perfusion, rat tissues were dissected and were dehydrated as described previously (25,36). Tissue sections of 12 μm were immunostained with the following primary antibodies described previously (25,36): rabbit polyclonal antibody to human FUS (made in-house) (25), chicken antibody to ubiquitin (Sigma), mouse monoclonal antibodies against Iba-1 (Wako Chemical) or GFAP (Millipore), mouse monoclonal anti-APC (Calbiochem), rabbit polyclonal antibody to TDP-43 (Proteintech), mouse monoclonal antibody to GM130 (BD Bioscience), rabbit polyclonal antibody to GLG1 (Abjent), rabbit polyclonal antibody to Cox-IV (Cell Signaling) and mouse monoclonal antibody to NeuN (Millipore). For histochemistry, immunostained sections were visualized with an ABC kit in combination with diaminobenzidine (Vector) and counterstained with hematoxylin to display nuclei. For immunofluorescent staining, tissue sections were incubated with specific primary antibodies and then with secondary antibodies labeled with fluorescent dyes (Jackson ImmunoResearch). The primary antibodies were incubated overnight at 4°C and the secondary antibodies were incubated for 2 h at room temperature. In determining the colocalization of two proteins, fluorescent staining was documented by confocal microscopy (Imaging Facility of Kimmel Cancer Center at Jefferson) and the single-layer image was scanned with a Zeiss LSM510 META confocal system. To reveal the integrity of Golgi trans and cis complexes, Z-stacks of confocal images (at 1 µm of intervals) were projected to reconstruct Golgi structure.
Stereological cell counting
The total number of neurons was estimated with stereological cell counting for the following brain regions in one hemisphere: the frontal cortex (from the apical forebrain to the first occurrence of corpus callosum), the parietal cortex (from the first occurrence of corpus callosum to the first occurrence of hippocampus) and dentate gyrus. Rat forebrains were cut into serial coronal sections (20 μm) and every 12th section (a total of 15 to 18 sections) was counted for neurons in the defined brain regions. Tissue sections were stained with Cresyl violet and mounted in sequential order (rostral–caudal). The number of targeted neurons was estimated using a procedure described previously (25,36).
Golgi staining
Dendrites and dendritic spines were visualized by Golgi impregnation method following the manufacturer's instruction (FD NeuroTechnologies). Five neurons in each selected brain region of individual rats were examined for dendrite branches and dendritic spine density (64). Dendritic spine density was calculated by dividing the number of spines with the length of dendrites. Both apical and basal dendrites were examined for spine density.
Statistical analysis
The number of neurons in the defined region was statistically compared between groups of transgenic rats, and comparison among experimental groups was performed by one-way ANOVA followed by Tukey's post hoc test. The null hypothesis was rejected at the level of 0.05.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
ACKNOWLEDGEMENTS
We thank Ms Xiao-Tao Wei for technical assistance.
Conflict of Interest statement. None declared.
FUNDING
This work was supported by the National Institutes of Health (NS072696 and NS072113 to X.-G.X and NS073829 to H.Z.).
REFERENCES
- 1.Galimberti D., Scarpini E. Genetics and biology of Alzheimer's disease and frontotemporal lobar degeneration. Int. J. Clin. Exp. Med. 2010;3:129–143. [PMC free article] [PubMed] [Google Scholar]
- 2.Hu W.T., Trojanowski J.Q., Shaw L.M. Biomarkers in frontotemporal lobar degenerations—progress and challenges. Prog. Neurobiol. 2011;95:636–648. doi: 10.1016/j.pneurobio.2011.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mackenzie I.R., Foti D., Woulfe J., Hurwitz T.A. Atypical frontotemporal lobar degeneration with ubiquitin-positive, TDP-43-negative neuronal inclusions. Brain. 2008;131:1282–1293. doi: 10.1093/brain/awn061. [DOI] [PubMed] [Google Scholar]
- 4.Mackenzie I.R. The neuropathology and clinical phenotype of FTD with progranulin mutations. Acta Neuropathol. 2007;114:49–54. doi: 10.1007/s00401-007-0223-8. [DOI] [PubMed] [Google Scholar]
- 5.Neumann M., Sampathu D.M., Kwong L.K., Truax A.C., Micsenyi M.C., Chou T.T., Bruce J., Schuck T., Grossman M., Clark C.M., et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 6.Neumann M., Rademakers R., Roeber S., Baker M., Kretzschmar H.A., Mackenzie I.R.A. A new subtype of frontotemporal lobar degeneration with FUS pathology. Brain. 2009;132:2922–2931. doi: 10.1093/brain/awp214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sephton C.F., Cenik C., Kucukural A., Dammer E.B., Cenik B., Han Y., Dewey C.M., Roth F.P., Herz J., Peng J., et al. Identification of neuronal RNA targets of TDP-43-containing ribonucleoprotein complexes. J. Biol. Chem. 2011;286:1204–1215. doi: 10.1074/jbc.M110.190884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan A.Y., Riley T.R., Coady T., Bussemaker H.J., Manley J.L. TLS/FUS (translocated in liposarcoma/fused in sarcoma) regulates target gene transcription via single-stranded DNA response elements. Proc. Natl Acad. Sci. USA. 2012;109:6030–6035. doi: 10.1073/pnas.1203028109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baechtold H., Kuroda M., Sok J., Ron D., Lopez B.S., Akhmedov A.T. Human 75-kDa DNA-pairing protein is identical to the pro-oncoprotein TLS/FUS and is able to promote D-loop formation. J. Biol. Chem. 1999;274:34337–34342. doi: 10.1074/jbc.274.48.34337. [DOI] [PubMed] [Google Scholar]
- 10.Uranishi H., Tetsuka T., Yamashita M., Asamitsu K., Shimizu M., Itoh M., Okamoto T. Involvement of the pro-oncoprotein TLS (translocated in liposarcoma) in nuclear factor-kappa B p65-mediated transcription as a coactivator. J. Biol. Chem. 2001;276:13395–13401. doi: 10.1074/jbc.M011176200. [DOI] [PubMed] [Google Scholar]
- 11.Lerga A., Hallier M., Delva L., Orvain C., Gallais I., Marie J., Moreau-Gachelin F. Identification of an RNA binding specificity for the potential splicing factor TLS. J. Biol. Chem. 2001;276:6807–6816. doi: 10.1074/jbc.M008304200. [DOI] [PubMed] [Google Scholar]
- 12.Meissner M., Lopato S., Gotzmann J., Sauermann G., Barta A. Proto-oncoprotein TLS/FUS is associated to the nuclear matrix and complexed with splicing factors PTB, SRm160 and SR proteins. Exp. Cell Res. 2003;283:184–195. doi: 10.1016/s0014-4827(02)00046-0. [DOI] [PubMed] [Google Scholar]
- 13.Wang X., Arai S., Song X., Reichart D., Du K., Pascual G., Tempst P., Rosenfeld M.G., Glass C.K., Kurokawa R. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tradewell M.L., Yu Z., Tibshirani M., Boulanger M.C., Durham H.D., Richard S. Arginine methylation by PRMT1 regulates nuclear-cytoplasmic localization and toxicity of FUS/TLS harbouring ALS-linked mutations. Hum. Mol. Genet. 2012;21:136–149. doi: 10.1093/hmg/ddr448. [DOI] [PubMed] [Google Scholar]
- 15.Fujii R. TLS facilitates transport of mRNA encoding an actin-stabilizing protein to dendritic spines. J. Cell Sci. 2005;118:5755–5765. doi: 10.1242/jcs.02692. [DOI] [PubMed] [Google Scholar]
- 16.Fujii R., Okabe S., Urushido T., Inoue K., Yoshimura A., Tachibana T., Nishikawa T., Hicks G.G., Takumi T. The RNA binding protein TLS is translocated to dendritic spines by mGluR5 activation and regulates spine morphology. Curr. Biol. 2005;15:587–593. doi: 10.1016/j.cub.2005.01.058. [DOI] [PubMed] [Google Scholar]
- 17.Huang C., Xia P.Y., Zhou H. Sustained expression of TDP-43 and FUS in motor neurons in rodent's lifetime. Int. J. Biol. Sci. 2010;6:396–406. doi: 10.7150/ijbs.6.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwiatkowski T.J., Jr, Bosco D.A., Leclerc A.L., Tamrazian E., Vanderburg C.R., Russ C., Davis A., Gilchrist J., Kasarskis E.J., Munsat T., et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 19.Vance C., Rogelj B., Hortobagyi T., De Vos K.J., Nishimura A.L., Sreedharan J., Hu X., Smith B., Ruddy D., Wright P., et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Couthouis J., Hart M.P., Shorter J., DeJesus-Hernandez M., Erion R., Oristano R., Liu A.X., Ramos D., Jethava N., Hosangadi D., et al. A yeast functional screen predicts new candidate ALS disease genes. Proc. Natl Acad. Sci. USA. 2011;108:20881–20890. doi: 10.1073/pnas.1109434108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan J., Deng H.X., Siddique N., Fecto F., Chen W., Yang Y., Liu E., Donkervoort S., Zheng J.G., Shi Y., et al. Frameshift and novel mutations in FUS in familial amyotrophic lateral sclerosis and ALS/dementia. Neurology. 2010;75:807–814. doi: 10.1212/WNL.0b013e3181f07e0c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Langenhove T., van der Zee J., Sleegers K., Engelborghs S., Vandenberghe R., Gijselinck I., Van den Broeck M., Mattheijssens M., Peeters K., De Deyn P.P., et al. Genetic contribution of FUS to frontotemporal lobar degeneration. Neurology. 2010;74:366–371. doi: 10.1212/WNL.0b013e3181ccc732. [DOI] [PubMed] [Google Scholar]
- 23.Hoell J.I., Larsson E., Runge S., Nusbaum J.D., Duggimpudi S., Farazi T.A., Hafner M., Borkhardt A., Sander C., Tuschl T. RNA targets of wild-type and mutant FET family proteins. Nat. Struct. Mol. Biol. 2011;18:1428–1431. doi: 10.1038/nsmb.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murakami T., Yang S.P., Xie L., Kawano T., Fu D., Mukai A., Bohm C., Chen F., Robertson J., Suzuki H., et al. ALS mutations in FUS cause neuronal dysfunction and death in Caenorhabditis elegans by a dominant gain-of-function mechanism. Hum. Mol. Genet. 2011;21:1–9. doi: 10.1093/hmg/ddr417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang C., Zhou H., Tong J., Chen H., Liu Y.-J., Wang D., Wei X., Xia X.G. FUS transgenic rats develop the phenotypes of amyotrophic lateral sclerosis and frontotemporal lobar degeneration. PLOS Genet. 2011;7:e1002011. doi: 10.1371/journal.pgen.1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vaccaro A., Tauffenberger A., Aggad D., Rouleau G., Drapeau P., Parker J.A. Mutant TDP-43 and FUS cause age-dependent paralysis and neurodegeneration in C. elegans. PLoS ONE. 2012;7:e31321. doi: 10.1371/journal.pone.0031321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hicks G.G., Singh N., Nashabi A., Mai S., Bozek G., Klewes L., Arapovic D., White E.K., Koury M.J., Oltz E.M., et al. FUS deficiency in mice results in defective B-lymphocyte development and activation, high levels of chromosomal instability and perinatal death. Nat. Genet. 2000;24:175–179. doi: 10.1038/72842. [DOI] [PubMed] [Google Scholar]
- 28.Kuroda M., Sok J., Webb L., Baechtold H., Urano F., Yin Y., Chung P., de Rooij D.G., Akhmedov A., Ashley T., et al. Male sterility and enhanced radiation sensitivity in TLS(−/−) mice. EMBO J. 2000;19:453–462. doi: 10.1093/emboj/19.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lanson N.A., Jr, Maltare A., King H., Smith R., Kim J.H., Taylor J.P., Lloyd T.E., Pandey U.B. A Drosophila model of FUS-related neurodegeneration reveals genetic interaction between FUS and TDP-43. Hum. Mol. Genet. 2011;20:2510–2523. doi: 10.1093/hmg/ddr150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ju S., Tardiff D.F., Han H., Divya K., Zhong Q., Maquat L.E., Bosco D.A., Hayward L.J., Brown R.H., Jr, Lindquist S., et al. A yeast model of FUS/TLS-dependent cytotoxicity. PLoS Biol. 2011;9:e1001052. doi: 10.1371/journal.pbio.1001052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ling S.C., Albuquerque C.P., Han J.S., Lagier-Tourenne C., Tokunaga S., Zhou H., Cleveland D.W. ALS-associated mutations in TDP-43 increase its stability and promote TDP-43 complexes with FUS/TLS. Proc. Natl Acad. Sci. USA. 2010;107:13318–13323. doi: 10.1073/pnas.1008227107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang J.W., Brent J.R., Tomlinson A., Shneider N.A., McCabe B.D. The ALS-associated proteins FUS and TDP-43 function together to affect Drosophila locomotion and life span. J. Clin. Invest. 2011;121:4118–4126. doi: 10.1172/JCI57883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kabashi E., Bercier V., Lissouba A., Liao M., Brustein E., Rouleau G.A., Drapeau P. FUS and TARDBP but not SOD1 interact in genetic models of amyotrophic lateral sclerosis. PLoS Genet. 2011;7:e1002214. doi: 10.1371/journal.pgen.1002214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seelaar H., Klijnsma K.Y., Koning I., Lugt A., Chiu W.Z., Azmani A., Rozemuller A.J.M., Swieten J.C. Frequency of ubiquitin and FUS-positive, TDP-43-negative frontotemporal lobar degeneration. J. Neurol. 2009;257:747–753. doi: 10.1007/s00415-009-5404-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Urwin H., Josephs K.A., Rohrer J.D., Mackenzie I.R., Neumann M., Authier A., Seelaar H., Swieten J.C., Brown J.M., Johannsen P., et al. FUS pathology defines the majority of tau- and TDP-43-negative frontotemporal lobar degeneration. Acta Neuropathol. 2010;120:33–41. doi: 10.1007/s00401-010-0698-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang C., Tong J., Bi F., Zhou H., Xia X.G. Mutant TDP-43 in motor neurons promotes the onset and progression of ALS in rats. J. Clin. Invest. 2012;122:107–118. doi: 10.1172/JCI59130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fujita Y., Okamoto K. Golgi apparatus of the motor neurons in patients with amyotrophic lateral sclerosis and in mice models of amyotrophic lateral sclerosis. Neuropathol. 2005;25:388–394. doi: 10.1111/j.1440-1789.2005.00616.x. [DOI] [PubMed] [Google Scholar]
- 38.Xu G., Karch C., Li N., Lin N., Fromholt D., Gonzales V., Borchelt D.R. Receptor-associated protein (RAP) plays a central role in modulating Abeta deposition in APP/PS1 transgenic mice. PLoS ONE. 2008;3:e3159. doi: 10.1371/journal.pone.0003159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dormann D., Rodde R., Edbauer D., Bentmann E., Fischer I., Hruscha A., Than M.E., Mackenzie I.R., Capell A., Schmid B., et al. ALS-associated fused in sarcoma (FUS) mutations disrupt transportin-mediated nuclear import. EMBO J. 2010;29:2841–2857. doi: 10.1038/emboj.2010.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bosco D.A., Lemay N., Ko H.K., Zhou H., Burke C., Kwiatkowski T.J., Jr, Sapp P., McKenna-Yasek D., Brown R.H., Jr., Hayward L.J. Mutant FUS proteins that cause amyotrophic lateral sclerosis incorporate into stress granules. Hum. Mol. Genet. 2010;19:4160–4175. doi: 10.1093/hmg/ddq335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Escribano L., Simón A.-M., Pérez-Mediavilla A., Salazar-Colocho P., Río J.D., Frechilla D. Rosiglitazone reverses memory decline and hippocampal glucocorticoid receptor down-regulation in an Alzheimer's disease mouse model. Biochem. Biophys. Res. Commun. 2009;379:406–410. doi: 10.1016/j.bbrc.2008.12.071. [DOI] [PubMed] [Google Scholar]
- 42.Yi J.-H., Park S.-W., Brooks N., Lang B.T., Vemuganti R. PPARγ agonist rosiglitazone is neuroprotective after traumatic brain injury via anti-inflammatory and anti-oxidative mechanisms. Brain Res. 2008;1244:164–172. doi: 10.1016/j.brainres.2008.09.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brodbeck J., Balestra M.E., Saunders A.M., Roses A.D., Mahley R.W., Huang Y. Rosiglitazone increases dendritic spine density and rescues spine loss caused by apolipoprotein E4 in primary cortical neurons. Proc. Natl Acad. Sci. USA. 2008;105:1343–1346. doi: 10.1073/pnas.0709906104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uchida A., Sasaguri H., Kimura N., Tajiri M., Ohkubo T., Ono F., Sakaue F., Kanai K., Hirai T., Sano T., et al. Non-human primate model of amyotrophic lateral sclerosis with cytoplasmic mislocalization of TDP-43. Brain. 2012;135:833–846. doi: 10.1093/brain/awr348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Manfredi G., Xu Z. Mitochondrial dysfunction and its role in motor neuron degeneration in ALS. Mitochondrion. 2005;5:77–87. doi: 10.1016/j.mito.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 46.Gonatas N.K., Stieber A., Gonatas J.O. Fragmentation of the Golgi apparatus in neurodegenerative diseases and cell death. J. Neurol. Sci. 2006;246:21–30. doi: 10.1016/j.jns.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 47.Farg M.A., Soo K.Y., Walker A.K., Pham H., Orian J., Horne M.K., Warraich S.T., Williams K.L., Blair I.P., Atkin J.D. Mutant FUS induces endoplasmic reticulum stress in amyotrophic lateral sclerosis and interacts with protein disulfide-isomerase. Neurobiol. Aging. 2012 doi: 10.1016/j.neurobiolaging.2012.02.009. [Epub ahead of print, available online 28 March 2012] [DOI] [PubMed] [Google Scholar]
- 48.Xu Y.F., Gendron T.F., Zhang Y.J., Lin W.L., D'Alton S., Sheng H., Casey M.C., Tong J., Knight J., Yu X., et al. Wild-type human TDP-43 expression causes TDP-43 phosphorylation, mitochondrial aggregation, motor deficits and early mortality in transgenic mice. J. Neurosci. 2010;30:10851–10859. doi: 10.1523/JNEUROSCI.1630-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Igaz L.M., Kwong L.K., Lee E.B., Chen-Plotkin A., Swanson E., Unger T., Malunda J., Xu Y., Winton M.J., Trojanowski J.Q., et al. Dysregulation of the ALS-associated gene TDP-43 leads to neuronal death and degeneration in mice. J. Clin. Invest. 2011;121:726–738. doi: 10.1172/JCI44867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Swarup V., Phaneuf D., Bareil C., Robertson J., Rouleau G.A., Kriz J., Julien J.P. Pathological hallmarks of amyotrophic lateral sclerosis/frontotemporal lobar degeneration in transgenic mice produced with TDP-43 genomic fragments. Brain. 2011;134:2610–2626. doi: 10.1093/brain/awr159. [DOI] [PubMed] [Google Scholar]
- 51.Swarup V., Phaneuf D., Dupre N., Petri S., Strong M., Kriz J., Julien J.P. Deregulation of TDP-43 in amyotrophic lateral sclerosis triggers nuclear factor kappab-mediated pathogenic pathways. J. Exp. Med. 2011;208:2429–2447. doi: 10.1084/jem.20111313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang L., Gutmann D.H., Roos R.P. Astrocyte loss of mutant SOD1 delays ALS disease onset and progression in G85R transgenic mice. Hum. Mol. Genet. 2011;20:286–293. doi: 10.1093/hmg/ddq463. [DOI] [PubMed] [Google Scholar]
- 53.Bento-Abreu A., Van Damme P., Van Den Bosch L., Robberecht W. The neurobiology of amyotrophic lateral sclerosis. Eur. J. Neurosci. 2010;31:2247–2265. doi: 10.1111/j.1460-9568.2010.07260.x. [DOI] [PubMed] [Google Scholar]
- 54.Papadeas S.T., Kraig S.E., O'Banion C., Lepore A.C., Maragakis N.J. Astrocytes carrying the superoxide dismutase 1 (SOD1G93A) mutation induce wild-type motor neuron degeneration in vivo. Proc. Natl Acad. Sci. USA. 2011;108:17803–17808. doi: 10.1073/pnas.1103141108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Diaz-Amarilla P., Olivera-Bravo S., Trias E., Cragnolini A., Martinez-Palma L., Cassina P., Beckman J., Barbeito L. Phenotypically aberrant astrocytes that promote motoneuron damage in a model of inherited amyotrophic lateral sclerosis. Proc. Natl Acad. Sci. USA. 2011;108:18126–18131. doi: 10.1073/pnas.1110689108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nagai M., Re D.B., Nagata T., Chalazonitis A., Jessell T.M., Wichterle H., Przedborski S. Astrocytes expressing ALS-linked mutated SOD1 release factors selectively toxic to motor neurons. Nat. Neurosci. 2007;10:615–622. doi: 10.1038/nn1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamanaka K., Chun S.J., Boillee S., Fujimori-Tonou N., Yamashita H., Gutmann D.H., Takahashi R., Misawa H., Cleveland D.W. Astrocytes as determinants of disease progression in inherited amyotrophic lateral sclerosis. Nat. Neurosci. 2008;11:251–253. doi: 10.1038/nn2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luna-Medina R. Regulation of inflammatory response in neural cells in vitro by thiadiazolidinones derivatives through peroxisome proliferator-activated receptor activation. J. Biol. Chem. 2005;280:21453–21462. doi: 10.1074/jbc.M414390200. [DOI] [PubMed] [Google Scholar]
- 59.Luna-Medina R., Cortes-Canteli M., Alonso M., Santos A., Martinez A., Perez-Castillo A. Regulation of inflammatory response in neural cells in vitro by thiadiazolidinones derivatives through peroxisome proliferator-activated receptor gamma activation. J. Biol. Chem. 2005;280:21453–21462. doi: 10.1074/jbc.M414390200. [DOI] [PubMed] [Google Scholar]
- 60.Xu J., Drew P.D. Peroxisome proliferator-activated receptor-gamma agonists suppress the production of IL-12 family cytokines by activated glia. J. Immunol. 2007;178:1904–1913. doi: 10.4049/jimmunol.178.3.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pedersen W., McMillan P., Kulstad J., Leverenz J., Craft S., Haynatzki G. Rosiglitazone attenuates learning and memory deficits in Tg2576 Alzheimer mice. Exp. Neurol. 2006;199:265–273. doi: 10.1016/j.expneurol.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 62.Watson G.S., Cholerton B.A., Reger M.A., Baker L.D., Plymate S.R., Asthana S., Fishel M.A., Kulstad J.J., Green P.S., Cook D.G., et al. Preserved cognition in patients with early Alzheimer disease and amnestic mild cognitive impairment during treatment with rosiglitazone: a preliminary study. Am. J. Geriatr. Psychiatry. 2005;13:950–958. doi: 10.1176/appi.ajgp.13.11.950. [DOI] [PubMed] [Google Scholar]
- 63.Risner M.E., Saunders A.M., Altman J.F., Ormandy G.C., Craft S., Foley I.M., Zvartau-Hind M.E., Hosford D.A., Roses A.D. Efficacy of rosiglitazone in a genetically defined population with mild-to-moderate Alzheimer's disease. Pharmacogenomics J. 2006;6:246–254. doi: 10.1038/sj.tpj.6500369. [DOI] [PubMed] [Google Scholar]
- 64.Woolley C.S., Gould E., Frankfurt M., McEwen B.S. Naturally occurring fluctuation in dendritic spine density on adult hippocampal pyramidal neurons. J. Neurosci. 1990;10:4035–4039. doi: 10.1523/JNEUROSCI.10-12-04035.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.