Abstract
Resistin is a polypeptide hormone that was reported to be associated with insulin resistance, inflammation and risk of type 2 diabetes and cardiovascular disease. We conducted a genome-wide association (GWA) study on circulating resistin levels in individuals of European ancestry drawn from the two independent studies: the Nurses' Health Study (n = 1590) and the Health, Aging and Body Composition Study (n = 1658). Single-nucleotide polymorphisms (SNPs) identified in the GWA analysis were replicated in an independent cohort of Europeans: the Gargano Family Study (n = 659). We confirmed the association with a previously known locus, the RETN gene (19p13.2), and identified two novel loci near the TYW3/CRYZ gene (1p31) and the NDST4 gene (4q25), associated with resistin levels at a genome-wide significant level, best represented by SNP rs3931020 (P = 6.37 × 10–12) and SNP rs13144478 (P = 6.19 × 10−18), respectively. Gene expression quantitative trait loci analyses showed a significant cis association between the SNP rs3931020 and CRYZ gene expression levels (P = 3.68 × 10−7). We also found that both of these two SNPs were significantly associated with resistin gene (RETN) mRNA levels in white blood cells from 68 subjects with type 2 diabetes (both P = 0.02). In addition, the resistin-rising allele of the TYW3/CRYZ SNP rs3931020, but not the NDST4 SNP rs13144478, showed a consistent association with increased coronary heart disease risk [odds ratio = 1.18 (95% CI, 1.03–1.34); P = 0.01]. Our results suggest that genetic variants in TYW3/CRYZ and NDST4 loci may be involved in the regulation of circulating resistin levels. More studies are needed to verify the associations of the SNP rs13144478 with NDST4 gene expression and resistin-related disease.
INTRODUCTION
Resistin is a polypeptide hormone that belongs to a family of cysteine-rich proteins called resistin-like molecules (1). It was originally reported to link obesity to insulin resistance and diabetes as an adipocytokine in mice (2). Many subsequent human studies have yielded controversial results in the association of circulating resistin levels with insulin resistance, type 2 diabetes and cardiovascular disease (3–17). In mice, resistin is preferentially expressed in adipocytes, whereas in humans it is highly expressed in monocyte and macrophage and only modestly in adipose cells (18,19). Genetic factors may play important roles in the regulation of circulating resistin levels. It has been estimated that ∼70% of the variation of circulating resistin levels is heritable (20). Several single-nucleotide polymorphisms (SNPs) in the human resistin gene (RETN) have been associated with circulating resistin levels in previous candidate gene studies (12,20–23). A previous one-stage genome-wide association (GWA) study failed to indentify loci associated with circulating resisting levels at a genome-wide significant level (24). Therefore, we undertook a GWA analysis for circulating resistin levels in individuals of European ancestry from two discovery cohorts and an independent replication study.
RESULTS
The characteristics of participants in each cohort are shown in Table 1. In the discovery stage, we performed three GWA scans for resistin levels in a total of 3248 individuals of European ancestry from the Nurses' Health Study (NHS) type 2 diabetes controls, the NHS type 2 diabetes cases and the Health, Aging and Body Composition Study (Health ABC) separately. There was no evidence of systematic bias in the distribution of P-values for association tests [genomic inflation factors (λ) were 0.99, 0.99 and 1.02, respectively] (Supplementary Material, Fig. S1). The results from the three GWA scans were combined by a meta-analysis. A graphical summary of the meta-analysis results is displayed in Supplementary Material, Figure S2. No associations with resistin levels reached a genome-wide significance level (P = 5 × 10−8).
Table 1.
Characteristics of the participants in GWAS and replication samples
| Study | n | Age (years) | Females (%) | BMI (kg/m2) | Resistin (ng/ml) | Log resistin (ng/ml) |
|---|---|---|---|---|---|---|
| GWAS | ||||||
| NHS (control) | 892 | 57.2 ± 6.7 | 100 | 25.6 ± 4.9 | 18.2 ± 12.2 | 2.76 ± 0.50 |
| NHS (case) | 698 | 57.6 ± 6.7 | 100 | 30.0 ± 5.7 | 20.9 ± 17.0 | 2.88 ± 0.53 |
| Health ABC | 1658 | 73.6 ± 2.9 | 47.1 | 26.6 ± 4.1 | 19.7 ± 9.9 | 2.90 ± 0.38 |
| Replication | ||||||
| GFS | 659 | 37.5 ± 13.9 | 62.7 | 26.2 ± 4.7 | 5.9 ± 2.98 | 0.73 ± 0.19 |
Data are means ± SD or otherwise indicated.
We took four top SNPs representing independent loci having a stage 1 P < 5 × 10−5 for further replication in a replication sample of the Gargano Family Study (GFS, n = 659) (Table 2). SNPs were considered to be independent if the pair-wise linkage disequilibrium r2 < 0.1 and if they were apart at least 1 Mb from one another (25). The SNPs rs17372114, rs3931020 and rs6068258 were genotyped, and the SNP rs13144478 was imputed (MACH r2 = 0.9). The SNPs rs3931020 in the TYW3/CRYZ locus (1p31) and rs13144478 in the NDST4 locus (4q25–q26) showed directionally consistent associations with resistin levels in the GFS, and the P-values for the combined results of discovery and replication stages reached genome-wide significance (P = 6.4 × 10−12 and 6.2 × 10−18, respectively; Table 2). Regional association plots of SNPs in the TYW3/CRYZ and NDST4 loci are displayed in Figure 1. In addition, there were eight SNPs in a previously known locus (12,20–23), the RETN gene (from 7630 to 7650 kb, on chromosome 19p13.2), with association results available in our discovery data. Most of these SNPs were imputed with modest imputation quality (0.2 ≤ MACH r2 ≤ 0.7); and a previously reported SNP rs3745367 (IVS2 181G/A) (20) showed strongest association with resistin levels (P = 0.007) (Supplementary Material, Table S1). After combining our present result and previously reported data in the GFS (20), the SNP rs3745367 showed a suggestive association with circulating resistin levels (P = 2.66 × 10−6; Table 2).
Table 2.
Associations of SNPs with resistin levels
| SNP | Chr | Near gene | Effect/reference allelea | Discovery stage |
P-value for heterogeneity | Replication |
Combined |
P-value for heterogeneity | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EAF | Beta | SE | P-value | EAF | Beta | SE | P-value | n | Beta | SE | P-value | ||||||
| Novel loci | |||||||||||||||||
| rs17372114 | 1 | C1orf168 | T/C | 0.59 | 0.05 | 0.01 | 5.86E − 06 | 0.77 | 0.58 | 0.03 | 0.01 | 0.01 | 3903 | 0.04 | 0.01 | 5.27E − 07 | 0.49 |
| rs3931020 | 1 | TYW3/CRYZ | C/T | 0.69 | 0.05 | 0.01 | 3.29E − 05 | 0.06 | 0.60 | 0.06 | 0.01 | 3.19E − 08 | 3892 | 0.05 | 0.01 | 6.37E − 12 | 0.09 |
| rs13144478 | 4 | NDST4 | T/A | 0.09 | 0.12 | 0.03 | 7.03E − 06 | 0.38 | 0.10 | 0.13 | 0.02 | 1.70E − 13 | 3898 | 0.13 | 0.02 | 6.19E − 18 | 0.58 |
| rs6068258 | 20 | ZFP64 | G/T | 0.32 | 0.05 | 0.01 | 2.25E − 06 | 0.43 | 0.32 | 0.02 | 0.01 | 0.15 | 3894 | 0.04 | 0.01 | 1.11E − 05 | 0.08 |
| Known locus | |||||||||||||||||
| rs3745367 | 19 | RETN | A/G | 0.23 | 0.04 | 0.01 | 0.007 | 0.45 | 0.22 | 0.05 | 0.02 | 0.03 | 3847 | 0.04 | 0.01 | 2.66E − 06 | 0.78 |
aEffect alleles represent the alleles associated with resistin levels.
Figure 1.
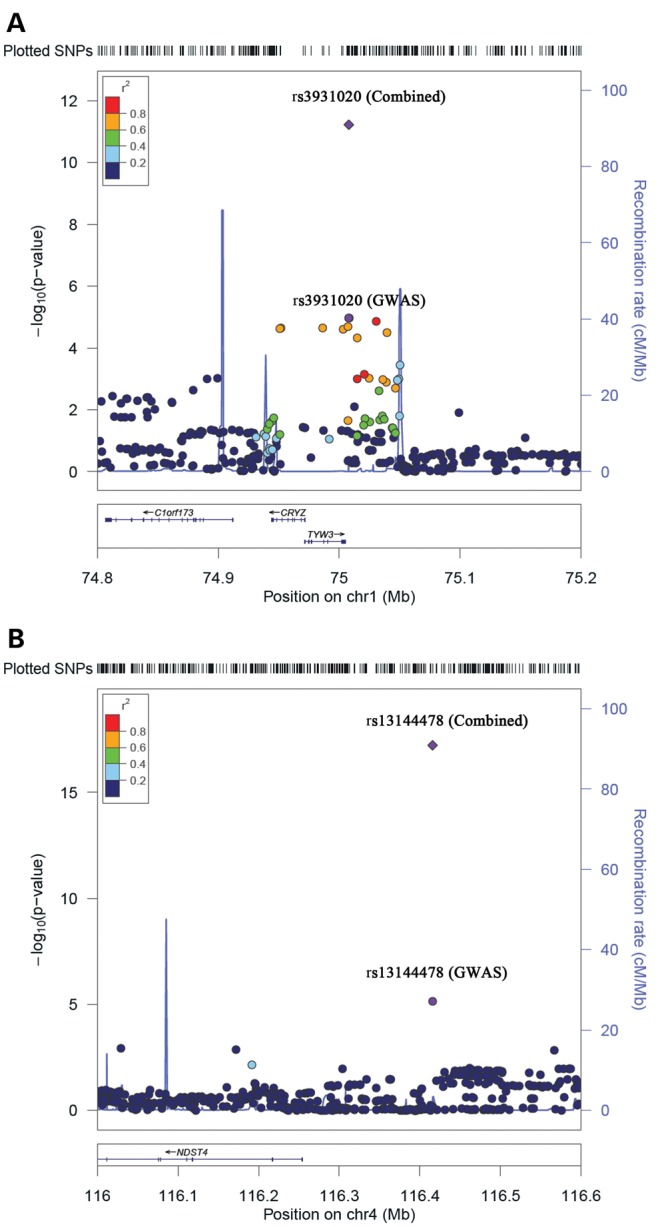
Regional association plot of SNPs with circulating resistin levels. (A) This panel included 342 SNPs within the region from 74.8 to 75.2 Mb on chromosome 1. (B) This panel included 509 SNPs within the region from 116 to 116.6 Mb on chromosome 4. The vertical axis representing the –log10 P-values. Recombination rates in this region are plotted in the background in blue.
To further examine the positional candidate genes of the identified loci, gene expression quantitative trait loci (eQTL) analyses were performed in the Gene Expression Omnibus (GEO) database with expression data from human lymphoblastoid cell lines (26). The SNP rs3931020 showed a significant cis association with CRYZ gene expression levels (P = 3.68 × 10−7), and another SNP rs277369 in linkage disequilibrium with rs3931020 (r2 = 0.711) showed the most significant association (P = 6.81 × 10−24). The SNP rs3931020 also showed a significant but weaker cis association with TYW3 gene expression (P = 2.7 × 10−4), compared with the CRYZ gene. Unfortunately, the SNP rs13144478 or correlated SNPs near the NDST4 gene were not available in the gene expression database.
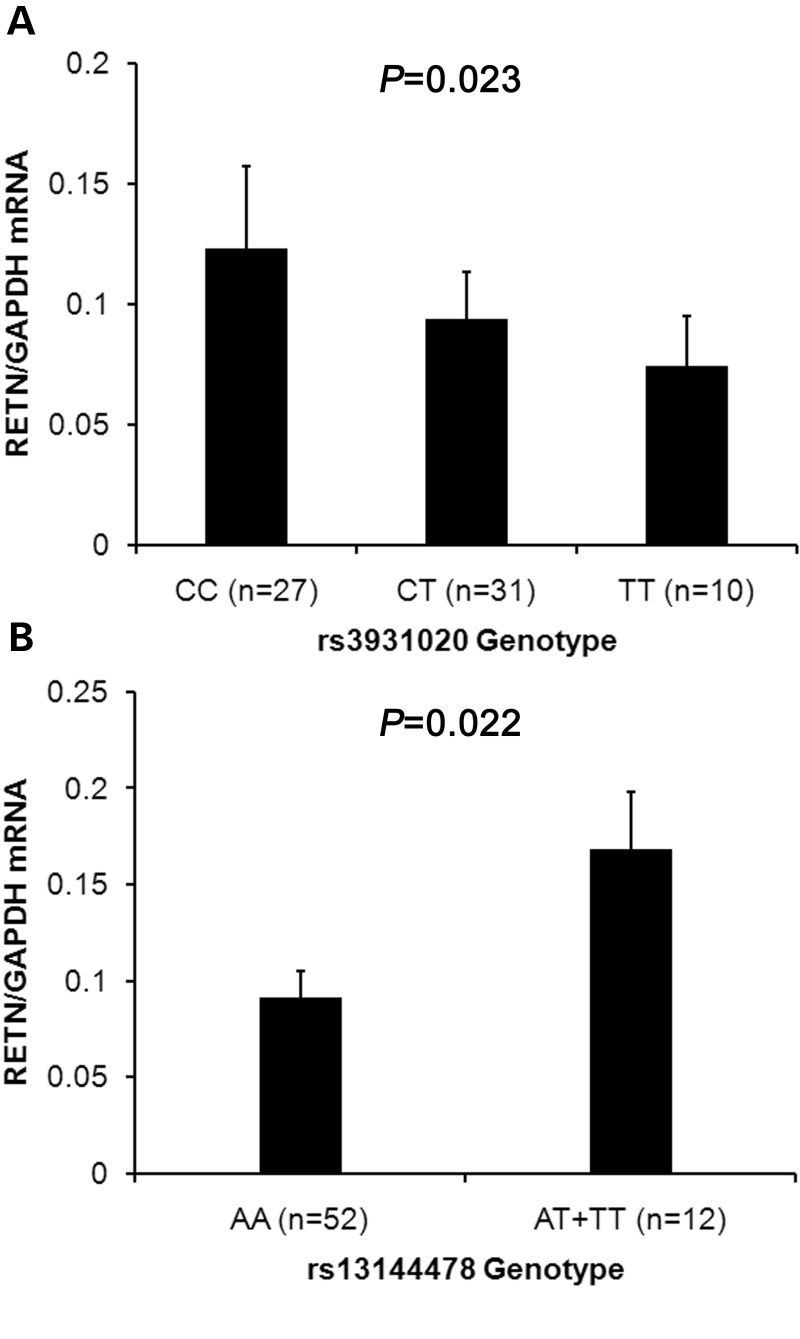
We also examined the association between the two newly identified SNPs and RETN expression levels in white blood cells. Consistent with the associations with circulating resistin levels, the rs3931020 C allele was significantly associated with higher RETN mRNA levels (P = 0.023 under an additive inheritance model, Fig. 2A), and RETN mRNA was significantly higher in carriers of the SNP rs13144478 T allele than that in non-T allele carriers (P = 0.022 under a dominant inheritance model since only one homozygous individual for the T allele was observed, Fig. 2B). Adjustments for age and sex did not change the observed association between the SNPs and RETN mRNA expression.
Figure 2.
RETN mRNA levels by the rs3931020 and rs13144478 genotypes. (A) Data are means + SE of RETN mRNA (normalized by using GAPDH) by the rs3931020 genotypes; P-value was calculated under an additive model. (B) Data are means + SE of RETN mRNA (normalized by using GAPDH) by the rs13144478 genotypes. Data from AT (n = 11) and TT (n = 1) individuals are pooled and showed as combined; P-value was calculated under a dominant model.
We further tested the associations of the newly identified resistin-associated SNPs with risk of type 2 diabetes (2591 cases; 3052 controls) and coronary heart disease (CHD) (776 cases; 1677 controls) in case–control samples from the NHS and Health Professional Follow-up Study (HPFS) (Table 3). In the pooled analysis, after adjustment for age and BMI, the SNP rs3931020 was associated with an increased CHD risk [odds ratio (OR) = 1.18, 95% CI 1.03–1.34; P = 0.01] but not type 2 diabetes risk, toward the same direction observed with resistin level. The association between the SNP rs3931020 and risk of CHD did not change after further adjustment for type 2 diabetes status. No association was observed between the SNP rs13144478 and type 2 diabetes or CHD risk. In addition, we further examined the association between the SNP rs3931020 and risk of CHD in the CARDIoGRAM (27), a consortium of GWA studies for CHD, and found that there was a marginal association between this SNP and risk of CHD (P = 0.09).
Table 3.
Associations of SNPs with type 2 diabetes and CHD risk in the NHS and HPFSa
| Type 2 diabetes |
CHD |
|||||
|---|---|---|---|---|---|---|
| Cases/controls | OR (95% CI) | P-value | Cases/controls | OR (95% CI) | P-value | |
| rs3931020 | ||||||
| NHS | 1459/1739 | 1.01 (0.90–1.13) | 0.89 | 341/800 | 1.05 (0.86–1.27) | 0.66 |
| HPFS | 1119/1294 | 1.03 (0.90–1.18) | 0.65 | 435/877 | 1.30 (1.09–1.55) | 0.004 |
| Combinedb | 1.02 (0.93–1.11) | 0.70 | 1.18 (1.03–1.34) | 0.01 | ||
| rs13144478 | ||||||
| NHS | 1459/1741 | 0.97 (0.78–1.21) | 0.79 | 341/800 | 0.73 (0.50–1.05) | 0.09 |
| HPFS | 1119/1293 | 1.19 (0.93–1.52) | 0.17 | 435/877 | 1.11 (0.80–1.53) | 0.53 |
| Combinedb | 1.06 (0.90–1.25) | 0.47 | 0.92 (0.72–1.18) | 0.51 | ||
aORs and P-values were estimated in an additive genetic model (resistin-rising allele was coded as effect allele) adjusted for age and BMI.
bResults were combined between men (HPFS) and women (NHS) using inverse variance weights under fixed model, as there was no heterogeneity between men and women (all P-values for heterogeneity >0.10).
DISCUSSION
In the first two-stage GWA analysis of circulating resistin levels in individuals of European ancestry, we identified two novel loci near the TYW3/CRYZ gene (1p31) and the NDST4 gene (4q25), associated with resistin levels at a genome-wide significant level, best represented by SNP rs3931020 and SNP rs13144478, respectively. Our in silico analysis indicates that the SNP rs3931020 strongly regulate the downstream located CRYZ gene. The CRYZ gene encodes zeta-crystallin, a NADPH-dependent quinone reductase which is expressed in lenses and many other tissues (28). This protein specifically binds to adenine–uracil-rich elements in 3′-UTR of mRNA, and it has been reported to act as trans-acting factors in the regulation of certain mRNAs (29,30). We also found that the SNP rs3931020 was associated with resistin mRNA levels in human white blood cells, with the consistent direction of genetic effect on circulating resistin levels. Although entirely speculative, our data allow one to speculate that the association of the SNP rs3931020 with circulating and mRNA resistin levels might be mediated through its effect in increasing CRYZ expression, which in turn, by stabilizing RETN mRNA, increases circulating resistin levels. The SNP rs3931020 also regulate the upstream TYW3 gene, which is involved in wybutosine synthesis. Wybutosine is a hyper-modified guanosine with a tricyclic nucleoside found at the 3′-position adjacent to the anticodon of eukaryotic phenylalanine tRNA (31). Whether a potential role of the TYW3 gene in tRNA modification may underlie the observed association between rs3931020 and resistin mRNA and circulating levels is presently unknown. Further mapping studies of this locus as well as functional studies are needed to address this possibility. Another novel finding of this study is that the resistin-rising allele (C-allele) of TYW3/CRYZ SNP rs3931020 was associated with increased CHD risk, but not with type 2 diabetes risk. Our result is in line with the fact that most of previous studies reported consistent associations between elevated circulating resistin levels and increased risk of cardiovascular disease (8–11,13,15–17), although the results in the association between resistin and type 2 diabetes were controversial (3,5,7,11,14).
The SNP rs13144478 is located downstream of the NDST4 gene, encoding heparin sulfate N-deacetylase/N-sulfotransferase 4 (32), which is not known to be involved in the regulation of circulating resistin levels. The newly observed association between the NDST4 gene and resistin needs to be interpreted with caution, since cis eQTL data of the NDST4 SNP rs13144478 was not available and no other SNPs in or near this locus showed strong association with resistin. More studies are needed to verify the results and to clarify the underlying mechanisms.
The major strengths of our study included large and well-defined cohorts, high-quality genotype data, minimal population stratification and consistent evidence in expression analysis (RETN mRNA). Several limitations need to be acknowledged. Resistin levels were lower in the replication sample than those in discovery cohorts, which might be due to difference in laboratory methods in resistin measurements and various characteristics of study populations. However, these differences may not affect the genetic associations, as the association analyses were conducted separately in each cohort and then combined by a meta-analysis, and there was no significant heterogeneity in genetic associations among these different study cohorts. We confirmed the association for one previously reported SNP rs3745367 in the RETN locus (20), but the association did not reach a GWA significance level. Previously reported SNPs in the 3′ region of RETN (23) were not significantly associated with resistin levels in our study, which might be due to the poor imputation quality (MACH r2 ≈ 0.2) of these SNPs in the discovery data. Owing to the relative small sample size of our replication cohort, we selected only four top independent signals identified from the discovery stage for replication. Studies with more replication samples and a greater number of the top-ranked GWAS hits might result in the identification of additional genetic variants. In addition, our study samples exclusively consist of Caucasians with European ancestry. Therefore, these findings may not be generalizable to other populations (33).
In conclusion, we found two novel loci at TYW3/CRYZ and NDST4 gene regions associated with circulating resistin levels and provided consistent data on the association for RETN mRNA levels. We observed that the newly identified SNP rs3931020 at the TYW3/CRYZ locus, but not the SNP rs13144478 at the NDST4 locus, was associated with an increased risk of CHD. More studies are warranted to verify the associations of the NDST4 locus with resistin and related disease. Further studies are needed to identify the molecular mechanisms that link these genomic loci to circulating resistin levels and to examine their effects on resistin-related disease risk.
MATERIALS AND METHODS
Study populations
The present study is a two-stage GWA study of circulating resistin levels following similar study design of our previous GWA analyses of biomarkers (34–37). The discovery cohorts consisted of 3248 individuals from the NHS (n = 1590) and the Health ABC (n = 1658). The NHS was established in 1976 when 121 700 female registered nurses aged 30–55 years residing in 11 large US states completed a mailed questionnaire on their medical history and lifestyle (38). A total of 32 826 women provided blood samples between 1989 and 1990. NHS participants for the current GWA scans were a subset of women who had data on resistin levels (n = 1590) included in a nested case–control study of type 2 diabetes [3221 women (1467 cases, 1754 controls)] (39). For the analysis of associations between SNPs and risk of type 2 diabetes, we included the total nested case–control samples from the NHS (3221 women) and 2422 men (1124 cases, 1298 controls) from the HPFS (39). In addition, in order to test the associations with risk of CHD, we included nested case–control samples [1141 women (341 cases, 800 controls) and 1312 men (435 cases, 877 controls)] from the NHS and HPFS (40). Detailed information about these nested case–control studies has been described elsewhere (39,40). Plasma resistin was assayed by ELISA (Linco Research, Inc.). The minimum detectable range of this assay is 0.16 ng/ml for a sample size of 10 μl (diluted 1:10), with an intra-assay CV% of 3.2–7.0%. Genotyping was done using the Affymetrix Genome-Wide Human 6.0 Array (Santa Clara, CA, USA). Genotypic data were checked for quality as described in detail elsewhere (39). Briefly, all samples used in the present study achieved a call rate of ≥98%. Individual SNPs were excluded if they were monomorphic, had a missing call rate of ≥2%, more than one discordance, a Hardy–Weinberg equilibrium (HWE) P-value of <1 × 10−4 or a minor allele frequency (MAF) of <0.02. We used MACH (version 1.0.16; http://www.sph.umich.edu/csg/abecasis/MACH) to impute SNPs on chromosome 1–22 with NCBI build 36 of Phase II HapMap CEU data (release 22) as the reference panel.
The Health ABC study is a longitudinal cohort study designed to investigate relationships among health conditions, body composition, social and behavioral factors and functional decline. The study population included 3075 well-functioning black and white men and women aged 70–79 (48% men, 42% Blacks) from Pittsburgh, Pennsylvania and Memphis, Tennessee. Baseline interview and clinic-based examination occurred between April 1997 and June 1998. For this study, only white participants with a resistin measurement (n = 1658) were included. Resistin was measured on EDTA plasma by the Human Resistin ELISA Kit (Linco Research, Inc.) at the University of Pennsylvania. Intra- and inter-assay CV% for this assay were 4.5 and 7.4%, respectively. The lowest 5% extreme values were repeated. Genotyping was performed by the Center for Inherited Disease Research using the Illumina Human1M-Duo BeadChip system. Samples were excluded from the data set for the reasons of sample failure, genotypic sex mismatch and first-degree relative of an included individual based on genotype data. Genotyping was successful for 1 151 215 SNPs in 2802 unrelated individuals (1663 Caucasians and 1139 African Americans). SNPs with an MAF ≥1%, call rate ≥97% and HWE P ≥ 10−6 were used for imputation. The MACH software (version 1.0.16) was used to impute SNPs on chromosome 1–22 with NCBI build 36 of Phase II HapMap CEU data (release 22) as the reference panel. A total of 2 543 887 SNPs are available for the current analysis.
In the replication sample of the GFS, a total of 659 non-diabetic individuals (247men and 412 women) from 235 families were recruited in the Gargano area in center-east Italy and examined as previously described (20,41). Serum resistin concentrations were measured by a commercial ELISA kit (BioVendor, Brno Czech Republic). Inter- and intra-assay CV% were 3.2–4 and 6.3–7.2%, respectively. It has been shown that resistin levels measured by BioVendor ELISA are lower than those measured by Linco ELISA, but these two methods have a high correlation coefficient of 0.92 according to the manufactory protocol of Linco ELISA. Replication SNPs were genotyped by TaqMan SNP allelic discrimination technique, by means of an ABI 7000 (Applied Biosystems, CA, USA). Call rate and concordance rate were ≥98% and >99%, respectively. All the SNPs were in HWE (P > 0.05).
Statistical analysis
In the discovery stage, following the analytical strategy widely used in the high-quality GWA consortia (25,42), we performed GWA analysis separately in each study sample and then combined by a meta-analysis. In the NHS, GWA analysis of resistin levels across ∼2.5 million SNPs (imputed data were expressed as allele dosage) was performed separately in control and diabetic case samples by using linear regression under an additive genetic model adjusting for age and BMI in the ProbABEL package (43). In Health ABC, linear regression modeling under an additive genetic model was used with adjustment for age, gender and study sites. Meta-analysis of these three GWAS scans was conducted using inverse variance weights under a fixed-effect model in METAL (http://www.sph.umich.edu/csg/abecasis/Metal). Associations between resistin levels and each SNP in the GFS were tested by a linear mixed effects model implemented in SOLAR which accounts for within-family correlations with adjustment for age and gender. Each SNP was included in a model as a fixed effect with additive coding. Similarly, fixed-effect models were used to combine the results from discovery and replication analyses in METAL. Resistin levels were log-transformed before analysis. The associations between SNPs and the risk of type 2 diabetes and CHD in the NHS and HPFS were analyzed using the logistic regression model, adjusted for age and BMI.
eQTL and RETN mRNA expression analyses
We performed eQTL analyses in the GEO database, in which data on 408 273 SNPs and eQTL from measurements of 54 675 transcripts representing 20 599 genes in Epstein–Barr virus-transformed lymphoblastoid cell lines from 400 children were available (26). In the RETN mRNA expression analysis, RETN RNA were extracted by using the PAXgene Blood RNA System (PreAnalytiX, GmbH, Germany) from white blood cells of 68 patients with type 2 diabetes (38 male and 30 female, mean age 65.1 ± 7.0 years), consecutively recruited at the IRCCS ‘Casa Sollievo della Sofferenza’ in San Giovanni Rotondo (Gargano, Italy). RETN mRNA levels were measured by quantitative real-time PCR in triplicates and normalized using the GAPDH housekeeping gene. The difference in mRNA levels by genotypes was estimated by ANCOVA models (SPSS 15.0, SPSS, Inc., Chicago, IL, USA), according to additive and dominant models of inheritance. Log-transformed values were used to account for the skewed distribution of the RETN/GAPDH ratios.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
ACKNOWLEDGEMENTS
We are indebted to the staff and participants of the NHS, HPFS, Health ABC, GFS and CARDIoGRAM consortium for their dedication and contributions.
Conflict of Interest statement. None declared.
FUNDING
This research in the NHS and HPFS is supported by grants DK091718, HL071981, U01HG004399, HL71981, HL073168, U01HG004399, DK58845 and DK46200. L.Q. is a recipient of the American Heart Association Scientist Development Award (0730094N). The genotyping of the HPFS and NHS CHD GWAS was supported by an unrestricted grant from Merck Research Laboratories, North Wales, PA, USA. This research in the Health ABC was supported by NIH/NIA contracts N01AG62101, N01AG62103 and N01AG62106. The genome-wide association study in the Health ABC was funded by NIA grant 1R01AG032098-01A1 to Wake Forest University Health Sciences, and genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, contract number HHSN268200782096C. This research was supported in part by the Intramural Research Program of the NIH, National Institute on Aging. Part of the research described in this paper was supported by Accordo Programma Quadro in Materia di Ricerca Scientifica nella Regione Puglia-PST 2006, an Italian Ministry of Health grant (RC2011 and RC2012), an EFSD/Pfizer grant (C.M.).
REFERENCES
- 1.Steppan C.M., Brown E.J., Wright C.M., Bhat S., Banerjee R.R., Dai C.Y., Enders G.H., Silberg D.G., Wen X., Wu G.D., et al. A family of tissue-specific resistin-like molecules. Proc. Natl Acad. Sci. USA. 2001;98:502–506. doi: 10.1073/pnas.98.2.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steppan C.M., Bailey S.T., Bhat S., Brown E.J., Banerjee R.R., Wright C.M., Patel H.R., Ahima R.S., Lazar M.A. The hormone resistin links obesity to diabetes. Nature. 2001;409:307–312. doi: 10.1038/35053000. [DOI] [PubMed] [Google Scholar]
- 3.Lee J.H., Chan J.L., Yiannakouris N., Kontogianni M., Estrada E., Seip R., Orlova C., Mantzoros C.S. Circulating resistin levels are not associated with obesity or insulin resistance in humans and are not regulated by fasting or leptin administration: cross-sectional and interventional studies in normal, insulin-resistant, and diabetic subjects. J. Clin. Endocrinol. Metab. 2003;88:4848–4856. doi: 10.1210/jc.2003-030519. [DOI] [PubMed] [Google Scholar]
- 4.McTernan P.G., Fisher F.M., Valsamakis G., Chetty R., Harte A., McTernan C.L., Clark P.M.S., Smith S.A., Barnett A.H., Kumar S. Resistin and type 2 diabetes: regulation of resistin expression by insulin and rosiglitazone and the effects of recombinant resistin on lipid and glucose metabolism in human differentiated adipocytes. J. Clin. Endocrinol. Metab. 2003;88:6098–6106. doi: 10.1210/jc.2003-030898. [DOI] [PubMed] [Google Scholar]
- 5.Fujinami A., Obayashi H., Ohta K., Ichimura T., Nishimura M., Matsui H., Kawahara Y., Yamazaki M., Ogata M., Hasegawa G., et al. Enzyme-linked immunosorbent assay for circulating human resistin: resistin concentrations in normal subjects and patients with type 2 diabetes. Clin. Chim. Acta. 2004;339:57–63. doi: 10.1016/j.cccn.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 6.Vozarova de Courten B., Degawa-Yamauchi M., Considine R.V., Tataranni P.A. High serum resistin is associated with an increase in adiposity but not a worsening of insulin resistance in Pima Indians. Diabetes. 2004;53:1279–1284. doi: 10.2337/diabetes.53.5.1279. [DOI] [PubMed] [Google Scholar]
- 7.Youn B.S., Yu K.Y., Park H.J., Lee N.S., Min S.S., Youn M.Y., Cho Y.M., Park Y.J., Kim S.Y., Lee H.K., et al. Plasma resistin concentrations measured by enzyme-linked immunosorbent assay using a newly developed monoclonal antibody are elevated in individuals with type 2 diabetes mellitus. J. Clin. Endocrinol. Metab. 2004;89:150–156. doi: 10.1210/jc.2003-031121. [DOI] [PubMed] [Google Scholar]
- 8.Pischon T., Bamberger C.M., Kratzsch J., Zyriax B.-C., Algenstaedt P., Boeing H., Windler E. Association of plasma resistin levels with coronary heart disease in women. Obesity. 2005;13:1764–1771. doi: 10.1038/oby.2005.215. [DOI] [PubMed] [Google Scholar]
- 9.Reilly M.P., Lehrke M., Wolfe M.L., Rohatgi A., Lazar M.A., Rader D.J. Resistin is an inflammatory marker of atherosclerosis in humans. Circulation. 2005;111:932–939. doi: 10.1161/01.CIR.0000155620.10387.43. [DOI] [PubMed] [Google Scholar]
- 10.Burnett M.S., Devaney J.M., Adenika R.J., Lindsay R., Howard B.V. Cross-sectional associations of resistin, coronary heart disease, and insulin resistance. J. Clin. Endocrinol. Metab. 2006;91:64–68. doi: 10.1210/jc.2005-1653. [DOI] [PubMed] [Google Scholar]
- 11.Yaturu S., Daberry R.P., Rains J., Jain S. Resistin and adiponectin levels in subjects with coronary artery disease and type 2 diabetes. Cytokine. 2006;34:219–223. doi: 10.1016/j.cyto.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 12.Osawa H., Tabara Y., Kawamoto R., Ohashi J., Ochi M., Onuma H., Nishida W., Yamada K., Nakura J., Kohara K., et al. Plasma resistin, associated with single nucleotide polymorphism-420, is correlated with insulin resistance, lower HDL Cholesterol, and high-sensitivity C-reactive protein in the Japanese general population. Diabetes Care. 2007;30:1501–1506. doi: 10.2337/dc06-1936. [DOI] [PubMed] [Google Scholar]
- 13.Hoefle G., Saely C.H., Risch L., Koch L., Schmid F., Rein P., Aczél S., Berchtold S., Drexel H. Relationship between the adipose-tissue hormone resistin and coronary artery disease. Clin. Chim. Acta. 2007;386:1–6. doi: 10.1016/j.cca.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 14.Qi Q., Wang J., Li H., Yu Z., Ye X., Hu F.B., Franco O.H., Pan A., Liu Y., Lin X. Associations of resistin with inflammatory and fibrinolytic markers, insulin resistance, and metabolic syndrome in middle-aged and older Chinese. Eur. J. Endocrinol. 2008;159:585–593. doi: 10.1530/EJE-08-0427. [DOI] [PubMed] [Google Scholar]
- 15.Butler J., Kalogeropoulos A., Georgiopoulou V., de Rekeneire N., Rodondi N., Smith A.L., Hoffmann U., Kanaya A., Newman A.B., Kritchevsky S.B., et al. Serum resistin concentrations and risk of new onset heart failure in older persons: the Health, Aging, and Body Composition (Health ABC) Study. Arterioscler. Thromb. Vasc. Biol. 2009;29:1144–1149. doi: 10.1161/ATVBAHA.109.186783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frankel D.S., Vasan R.S., D'Agostino R.B., Sr, Benjamin E.J., Levy D., Wang T.J., Meigs J.B. Resistin, adiponectin, and risk of heart failure: the Framingham Offspring Study. J. Am. Coll. Cardiol. 2009;53:754–762. doi: 10.1016/j.jacc.2008.07.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang M.H., Na B., Schiller N.B., Whooley M.A. Association of resistin with heart failure and mortality in patients with stable coronary heart disease: data from the Heart and Soul Study. J. Card. Fail. 2010;17:24–30. doi: 10.1016/j.cardfail.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 18.Patel L., Buckels A.C., Kinghorn I.J., Murdock P.R., Holbrook J.D., Plumpton C., Macphee C.H., Smith S.A. Resistin is expressed in human macrophages and directly regulated by PPAR gamma activators. Biochem. Biophys. Res. Commun. 2003;300:472–476. doi: 10.1016/s0006-291x(02)02841-3. [DOI] [PubMed] [Google Scholar]
- 19.Yang R.Z., Huang Q., Xu A., McLenithan J.C., Eison J.A., Shuldiner A.R., Alkan S., Gong D.-W. Comparative studies of resistin expression and phylogenomics in human and mouse. Biochem. Biophys. Res. Commun. 2003;310:927–935. doi: 10.1016/j.bbrc.2003.09.093. [DOI] [PubMed] [Google Scholar]
- 20.Menzaghi C., Coco A., Salvemini L., Thompson R., De Cosmo S., Doria A., Trischitta V. Heritability of serum resistin and its genetic correlation with insulin resistance-related features in nondiabetic Caucasians. J. Clin. Endocrinol. Metab. 2006;91:2792–2795. doi: 10.1210/jc.2005-2715. [DOI] [PubMed] [Google Scholar]
- 21.Cho Y., Youn B.S., Chung S., Kim K., Lee H., Yu K.Y., Park H., Shin H., Park K. Common genetic polymorphisms in the promoter of resistin gene are major determinants of plasma resistin concentrations in humans. Diabetologia. 2004;47:559–565. doi: 10.1007/s00125-003-1319-x. [DOI] [PubMed] [Google Scholar]
- 22.Osawa H., Onuma H., Ochi M., Murakami A., Yamauchi J., Takasuka T., Tanabe F., Shimizu I., Kato K., Nishida W., et al. Resistin SNP-420 determines its monocyte mRNA and serum levels inducing type 2 diabetes. Biochem. Biophys. Res. Commun. 2005;335:596–602. doi: 10.1016/j.bbrc.2005.07.122. [DOI] [PubMed] [Google Scholar]
- 23.Hivert M.F., Manning A.K., McAteer J.B., Dupuis J.E., Fox C.S., Cupples L.A., Meigs J.B., Florez J.C. Association of variants in RETN with plasma resistin levels and diabetes-related traits in the Framingham Offspring Study. Diabetes. 2009;58:750–756. doi: 10.2337/db08-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Melzer D., Perry J.R.B., Hernandez D., Corsi A.-M., Stevens K., Rafferty I., Lauretani F., Murray A., Gibbs J.R., Paolisso G., et al. A Genome-wide association study identifies protein quantitative trait loci (pQTLs) PLoS Genet. 2008;4:e1000072. doi: 10.1371/journal.pgen.1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Speliotes E.K., Willer C.J., Berndt S.I., Monda K.L., Thorleifsson G., Jackson A.U., Allen H.L., Lindgren C.M., Luan J.A., Magi R., et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat. Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dixon A.L., Liang L., Moffatt M.F., Chen W., Heath S., Wong K.C.C., Taylor J., Burnett E., Gut I., Farrall M., et al. A genome-wide association study of global gene expression. Nat. Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 27.Schunkert H., Konig I.R., Kathiresan S., Reilly M.P., Assimes T.L., Holm H., Preuss M., Stewart A.F.R., Barbalic M., Gieger C., et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat. Genet. 2011;43:333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gonzalez P., Rao P.V., Zigler J.S. Organization of the human zeta-crystallin/quinone reductase gene (CRYZ) Genomics. 1994;21:317–324. doi: 10.1006/geno.1994.1272. [DOI] [PubMed] [Google Scholar]
- 29.Lapucci A., Lulli M., Amedei A., Papucci L., Witort E., Di Gesualdo F., Bertolini F., Brewer G., Nicolin A., Bevilacqua A., et al. zeta-Crystallin is a bcl-2 mRNA binding protein involved in bcl-2 overexpression in T-cell acute lymphocytic leukemia. FASEB J. 2010;24:1852–1865. doi: 10.1096/fj.09-140459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fernández M., Porté S., Crosas E., Barberà N., Farrés J., Biosca J., Parés X. Human and yeast zeta-crystallins bind AU-rich elements in RNA. Cell. Mol. Life Sci. 2007;64:1419–1427. doi: 10.1007/s00018-007-7091-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noma A., Kirino Y., Ikeuchi Y., Suzuki T. Biosynthesis of wybutosine, a hyper-modified nucleoside in eukaryotic phenylalanine tRNA. EMBO J. 2006;25:2142–2154. doi: 10.1038/sj.emboj.7601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aikawa J., Grobe K., Tsujimoto M., Esko J.D. Multiple isozymes of heparan sulfate/heparin GlcNAc N-deacetylase/GlcN N-sulfotransferase. J. Biol. Chem. 2001;276:5876–5882. doi: 10.1074/jbc.M009606200. [DOI] [PubMed] [Google Scholar]
- 33.Menzaghi C., Trischitta V. Genetics of serum resistin: a paradigm of population-specific regulation? Diabetologia. 2009;53:226–228. doi: 10.1007/s00125-009-1589-z. [DOI] [PubMed] [Google Scholar]
- 34.Qi L., Cornelis M.C., Kraft P., Jensen M., van Dam R.M., Sun Q., Girman C.J., Laurie C.C., Mirel D.B., Hunter D.J., et al. Genetic variants in ABO blood group region, plasma soluble E-selectin levels and risk of type 2 diabetes. Hum. Mol. Genet. 2010;19:1856–1862. doi: 10.1093/hmg/ddq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun Q., Cornelis M.C., Kraft P., Qi L., van Dam R.M., Girman C.J., Laurie C.C., Mirel D.B., Gong H., Sheu C.-C., et al. Genome-wide association study identifies polymorphisms in LEPR as determinants of plasma soluble leptin receptor levels. Hum. Mol. Genet. 2010;19:1846–1855. doi: 10.1093/hmg/ddq056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He M., Cornelis M.C., Kraft P., van Dam R.M., Sun Q., Laurie C.C., Mirel D.B., Chasman D.I., Ridker P.M., Hunter D.J., et al. Genome-wide association study identifies variants at the IL18-BCO2 locus associated with interleukin-18 levels. Arterioscler. Thromb. Vasc. Biol. 2010;30:885–890. doi: 10.1161/ATVBAHA.109.199422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qi L., Menzaghi C., Salvemini L., De Bonis C., Trischitta V., Hu F.B. Novel locus FER is associated with serum HMW adiponectin levels. Diabetes. 2011;60:2197–2201. doi: 10.2337/db10-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Colditz G.A., Manson J.E., Hankinson S.E. The Nurses' Health Study: 20-year contribution to the understanding of health among women. J. Womens Health. 1997;6:49–62. doi: 10.1089/jwh.1997.6.49. [DOI] [PubMed] [Google Scholar]
- 39.Qi L., Cornelis M.C., Kraft P., Stanya K.J., Linda Kao W.H., Pankow J.S., Dupuis J.E., Florez J.C., Fox C.S., Paré G., et al. Genetic variants at 2q24 are associated with susceptibility to type 2 diabetes. Hum. Mol. Genet. 2010;19:2706–2715. doi: 10.1093/hmg/ddq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jensen M.K., Pers T.H., Dworzynski P., Girman C.J., Brunak S.R., Rimm E.B. Protein interaction-based genome-wide analysis of incident coronary heart disease. Circ. Cardiovasc. Genet. 2011;4:549–556. doi: 10.1161/CIRCGENETICS.111.960393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Menzaghi C., Salvemini L., Paroni G., De Bonis C., Mangiacotti D., Fini G., Doria A., Di Paola R., Trischitta V. Circulating high molecular weight adiponectin isoform is heritable and shares a common genetic background with insulin resistance in nondiabetic white Caucasians from Italy: evidence from a family-based study. J. Intern. Med. 2009;267:287–294. doi: 10.1111/j.1365-2796.2009.02141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dupuis J., Langenberg C., Prokopenko I., Saxena R., Soranzo N., Jackson A.U., Wheeler E., Glazer N.L., Bouatia-Naji N., Gloyn A.L., et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat. Genet. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aulchenko Y., Struchalin M., van Duijn C. ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics. 2010;11:134. doi: 10.1186/1471-2105-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.