Abstract
The proline-, glutamate-, valine-, and lysine-rich (PEVK) domain of the giant muscle protein titin is thought to be an intrinsically unstructured random-coil segment. Various observations suggest, however, that the domain may not be completely devoid of internal interactions and structural features. To test the validity of random polymer models for PEVK, we determined the mean end-to-end distances of an 11- and a 21-residue synthetic PEVK peptide, calculated from the efficiency of the fluorescence resonance energy transfer (FRET) between an N-terminal intrinsic tryptophan donor and a synthetically added C-terminal IAEDANS acceptor obtained in steady-state and time-resolved experiments. We find that the contour-length scaling of mean end-to-end distance deviates from predictions of a purely statistical polymer chain. Furthermore, the addition of guanidine hydrochloride decreased, whereas the addition of salt increased the FRET efficiency, pointing at the disruption of structure-stabilizing interactions. Increasing temperature between 10 and 50°C increased the normalized FRET efficiency in both peptides but with different trajectories, indicating that their elasticity and conformational stability are different. Simulations suggest that whereas the short PEVK peptide displays an overall random structure, the long PEVK peptide retains residual, loose helical configurations. Transitions in the local structure and dynamics of the PEVK domain may play a role in the modulation of passive muscle mechanics.
Introduction
The elasticity of striated muscle is largely determined by the giant intrasarcomeric protein titin (1) (also called connectin (2)). The titin molecule spans half of the sarcomere; its N-terminus is part of the Z-disk and its C-terminus is anchored to the M-line (3,4). In the A-band, titin is an integral component of the thick filament (5). The extensible portion of titin in the sarcomere is restricted to the I-band section of the molecule (6), which displays large size variations among isoforms in different muscle types (7,8). Here titin is built of serially linked immunoglobulin (Ig)-like domains (proximal and distal tandem Ig segments) interspersed with unique sequences, most importantly the proline-, glutamate-, valine-, and lysine-rich (PEVK) domain (8). Upon stretching the sarcomere with physiological force levels, the tandem Ig segments together with the PEVK domain are responsible for the extension of the titin’s I-band section (9,10). Two distinct types of repeat motifs have been observed in the skeletal muscle PEVK segment (11)—PPAK and polyE. PPAK repeats (isoelectric point, pI ∼10) are 26–28-residue motifs that often begin with the amino acids PPAK. PolyE motifs (pI ∼4) display a preponderance of negatively charged glutamate residues. Recently it has been shown that truncation of PEVK leads to cardiomyopathy (12), and protein kinase C exerts its contraction modulatory effect via phosphorylating this domain (13). Interaction between the PEVK domain and the thin filament may regulate contractility by the generation of a variable internal viscous load (14–16).
It has been suggested that the PEVK region of titin is a random coil with little regular secondary structure, and that its elasticity can be described with the wormlike chain (WLC) model (17,18). Studies on titin PEVK peptides suggested that they exhibit most of the characteristics of intrinsically disordered proteins (19), probably owing to the preponderance of highly charged residues. However, other observations indicated that secondary structural features such as polyproline-type II helices may be present within the PEVK domain (20–22). Furthermore, electrostatic interactions between PPAK and polyE repeats may give rise to mechanical fatigue within titin (23,24). Finally, it has also been reported that PEVK elasticity is influenced to a great extent by hydrophobic interactions (25). Thus, PEVK may not be completely devoid of ordered structural features and intradomain interactions.
In this work, we examined the random coil nature of PEVK by testing the predictions of statistical polymer chain models. We used the WLC model, which envisions the polymer as an isotropic rod that is continuously flexible. The main parameters used in the model are the end-to-end distance (R), the contour length (L), and the persistence length of the chain (P) (26,27). The latter characterizes the bending rigidity, or stiffness of the polymer and it is defined as the length over which correlations in the direction of the tangent are lost. The wormlike chain model assumes that the chain segments do not interact with each other, as well as possessing a uniform persistence length distribution along the polymer. We used short PEVK peptides and measured the Förster’s resonance energy transfer (FRET) between the N-terminal tryptophan residue, as a donor, and the C-terminal IAEDANS fluorophore, as an acceptor.
Additionally, to gain an insight into the conformational dynamics of PEVK, we explored how external conditions, e.g., temperature, chemical denaturants, and ionic strength affect the FRET efficiency in the peptides. According to our observations, upon different physical-chemical conditions the PEVK peptides do not behave as purely isotropic rods, because the contour-length scaling of mean end-to-end distance deviates from the square-root law predicted for a statistical polymer chain, and the changes in temperature, ionic strength, and denaturant concentration cause distinct changes in FRET efficiency. Molecular dynamics simulations suggest that residual loose helical configurations may be present in the investigated peptides.
Materials and Methods
Preparation of peptide samples
Eleven- and twenty-one-residue-long PEVK peptides were produced by solid-state synthesis as described previously (28). The sequences of the peptides are WEEAYQEREV and WEEAYQEREVIQVQKEVYEE, and correspond to regions 17413–17442 and 17413–17472 (GenBank accession no. X90569 (version X90569.1)) within the titin sequence, respectively (8). These protein regions are encoded by exons 115 and 116 and fall within the polyE motif structure by sequence. Notably, the peptides are inclusive segments of the E115 peptide investigated by Duan et al. (19). During synthesis a cysteine was attached to the C-terminus of each peptide, resulting in 11- and 21-residue-long peptides, hereafter referred to as PEVK11 (WEEAYQEREVC) and PEVK21 (WEEAYQEREVIQVQKEVYEEC), respectively.
In the case of acceptor-labeled peptides n-[[(iodoacetyl)amino]ethyl]-5-naphthylamine-1-sulfonate (IAEDANS) was attached covalently to the SH group of the extrinsic C-terminal cysteine during the solid-state synthesis process, thus achieving labeling efficiency and unbound fluorophore fraction approaching 100% and 0%, respectively. The purity of the synthetic peptides was >98%, as confirmed by analytical high-pressure liquid chromatography. The lyophilized peptide samples were dissolved in sodium phosphate buffer (25 mM NaH2PO4 pH 7.4, 0.2 M KCl, 4 mM MgCl2, 1 mM EGTA, 3 mM NaN3). Final concentrations were set to 0.25 mg/mL. Peptide concentration was determined by measuring the absorbance at 280 nm, using extinction coefficients obtained with the ProtParam tool (http://www.expasy.ch/tools/protparam.html).
Steady-state fluorescence measurements
Fluorescence spectra of PEVK peptides were measured using a Fluorolog-3 spectrofluorometer (Jobin Yvon, Longjumeau, France). Tryptophan was excited at 295 nm and emission was recorded between 305 and 700 nm. Excitation and emission slits were both set to 5 nm. Fluorescence intensities were corrected for the inner filter effect (29). Förster’s resonance energy transfer (FRET) efficiency (E) was calculated from steady-state fluorescence spectra as
| (1) |
where FDA and FD are the corrected donor fluorescence intensities integrated between 320 nm and 360 nm in the presence and absence of the acceptor, respectively. The distance between the donor and acceptor fluorophore (R) was calculated as
| (2) |
(30), where R0 is Förster’s critical distance at which transfer efficiency is 50%. The R0 value of the tryptophan-IAEDANS pair is 2.2 nm, as reported previously (31). In the case of temperature-dependent measurements, the f′ parameter (FRET efficiency normalized with the donor emission intensity) was calculated as
| (3) |
The temperature dependence of f′ characterizes the average flexibility of the protein matrix between the donor and acceptor (32).
Fluorescence lifetime experiments
Fluorescence decay of the donor fluorophore was acquired at 22°C with the time-correlated single photon-counting method by using a Chronos-BH spectrometer (ISS, Champaign, IL). For each curve, 3 × 108 excitations were summed at an excitation pulse rate of 1 MHz, and emission was collected at the magic-angle relative to the excitation polarization. Excitation of the Trp during the lifetime measurement did not cause detectable photobleaching. Data were evaluated by an iterative reconvolution method that employed a sum of single exponentials and distribution functions. Acceptable fits were obtained with 2–3 exponentials. Use of a continuous distribution of lifetimes did not improve the fitting statistics. The average lifetime () was calculated as
| (4) |
where αi and τi are the ith preexponential factor and lifetime component of tryptophan displaying a complex decay (33). FRET efficiency was calculated from the determined fluorescence lifetimes as
| (5) |
where and are the donor lifetimes in the presence and absence of the acceptor, respectively.
Polymer model calculations
The contour lengths (L) of the peptides were calculated as the number of residues in the peptide (11 or 21) × residue spacing (0.38 nm) + size of the IAEDANS acceptor molecule (0.87 nm) (34). Contour lengths obtained this way were 4.18 nm and 7.98 nm for PEVK11 and PEVK21, respectively. The average end-to-end length (R) of the peptides was determined from FRET efficiencies as described above. Apparent persistence length (P), measure of the bending rigidity of the polymer chains, was obtained numerically by using the formula for the mean-square end-to-end distance, assuming that the conformation of peptides is described by the WLC model (35,36):
| (6) |
The model assumes that the chain is free of interactions and homogeneous (persistence length is constant along its length). In the case when L ≫ P, the formula becomes 〈R2〉 = 2PL. However, this simplified formula cannot be used in our case, because the contour lengths of our peptides (4.18 nm and 7.98 nm) are comparable to persistence lengths previously measured for PEVK segments in single-molecule mechanics experiments (0.3–2.3 nm) (37–40). Persistence lengths were thus calculated by using Eq. 6.
Prediction of secondary structures of the peptides
The GOR4 (41), NetSurfP (42), Jpred3 (43), and PSIPRED (44) secondary structure prediction servers were applied to get an estimate for the possible secondary structures of the nonmodified peptides.
Molecular dynamics calculations
Both peptides were built with the aid of the TINKER and PCModel program packages and the backbone conformation of all amino acids were set to α-helical. The GROMOS 96 force field (45) was applied during the GROMACS (46) calculations. Force-field parameters of the EDANS-modified Cys were set on a similarity basis. Systems of the raw complexes, neutralizing counterions, and numerous water molecules of explicit single-point charge water model were placed into rectangular simulation boxes with a 9 Å distance between the peptide and the box walls. The systems were refined with steepest descent and BFGS molecular mechanics energy minimizations. The refined systems were subjected to 100-ns-long molecular dynamics (MD) simulations. A time step of 2 fs, constant temperature of 300 K, and LINCS bond constraints were applied. Temperature coupling was carried out using the Berendsen scheme. Long-range electrostatics was simulated using the particle-mesh Ewald method, and a van der Waals cutoff of 9 Å was used. Neighbor lists were used and updated every 10 fs. The resulting trajectory files were analyzed with programs of the GROMACS package and the DSSP program (47). Structural figures were prepared using the PyMol program of Dr. Warren L. DeLano.
Statistics
Mean ± standard deviation (44) are shown for data collected in four sets of experiments. Data were analyzed and displayed by using commercial software packages (Origin; OriginLab, Northampton, MA, and IgorPro; WaveMetrics, Lake Oswego, OR).
Results
Spectral properties of PEVK peptides
Absorption spectra of the labeled peptide samples were recorded to verify labeling and to determine the concentration of the peptides (Fig. 1). In the case of the IAEDANS-labeled peptides the absorption spectra displayed a significant peak at 336 nm, which corresponds to the absorption maximum of the acceptor. Comparison of the fluorescence emission spectra of the respective unlabeled and IAEDANS-labeled PEVK peptides confirmed the presence of energy transfer (Fig. 2). The intensity of the tryptophan donor emission in the presence of the acceptor (FDA) was much reduced compared with that in the absence of the acceptor (FD).
Figure 1.
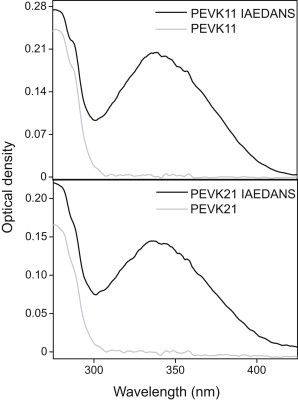
Absorption spectra of PEVK peptides in the absence (shaded line) and presence of the acceptor (solid line). Spectra were recorded at 0.2 M ionic strength, at 20°C.
Figure 2.
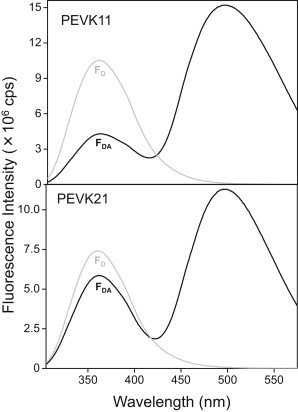
Fluorescence emission spectra of PEVK peptides in the absence (shaded line) and presence of the acceptor (solid line). Excitation wavelength was 295 nm, and optical slits were set to 5.0 nm on both the excitation and emission sides. Spectra were recorded at 0.2 M ionic strength, at 20°C.
Calculation of apparent persistence length
The donor-acceptor distance, corresponding to the end-to-end distance of the peptides, was calculated by measuring the decrease of donor fluorescence in the presence of the acceptor (Eq. 2). Under conditions corresponding most closely to the physiological (at 20°C and 0.2 M ionic strength), our experiments revealed average end-to-end distances (R) of 2.12 (±0.01) for PEVK11 and 2.69 (±0.05) nm for PEVK21. These values and the sequence-based contour lengths (4.18 and 7.98 nm, respectively) were used to calculate the apparent persistence length with Eq. 6. The calculated apparent persistence lengths (P) were 0.63 for PEVK11 and 0.48 nm for PEVK21.
Effect of chemical denaturants on PEVK peptides
To assess whether there are interactions within the PEVK peptides that might be disrupted with chemical denaturants, we studied the effect of guanidine hydrochloride (GuHCl) on the two peptides. Upon adding the chemical denaturant, FRET efficiency decreased gradually (Fig. 3). Increasing the GuHCl concentration from 0 to 6 M resulted in a decrease of transfer efficiency from 0.56 (±0.01) to 0.41 (±0.02) and from 0.23 (±0.02) to 0.16 (±0.003) in the case of PEVK21 and PEVK11, respectively. Considering that the R0 for the tryptophan-IAEDANS donor acceptor pair is known, the changes in transfer efficiency correspond to an increase of polymer-chain end-to-end distance (R) from 2.12 (±0.01) to 2.34 (±0.03) nm for PEVK11 and from 2.69 (±0.05) to 2.91 (±0.01) nm for PEVK21.
Figure 3.
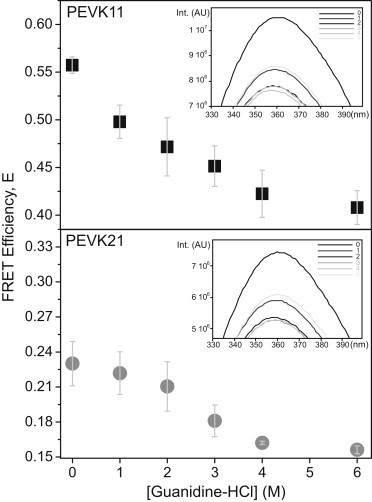
Effect of guanidine hydrochloride (GuHCl) on the transfer efficiency of PEVK11 (top) and PEVK21 (bottom) peptides. (Error bars) Standard deviation calculated from four independent experiments. (Insets) Donor (Trp) fluorescence emission spectra (with no acceptor) of the respective peptides, enlarged to show details of the changes.
The calculated apparent P-values increased by ∼0.18 nm for PEVK11 and ∼0.09 nm for PEVK21. Notably, in the case of PEVK21 the FRET efficiency decreased more steeply in the range of 2–4 M, than in the rest of the denaturant concentration range. In control experiments, we measured the changes in tryptophan fluorescence emission spectra without the acceptor as a function of GuHCl concentration (Fig. 3, insets). Fluorescence emission intensity was reduced by ∼25% upon GuHCl addition, and a 2-nm blue shift was observed. Interestingly, part of the emission intensity was recovered during the increase of GuHCl concentration from 4 M to 6 M.
Fluorescence lifetime measurements
To substantiate the transfer-efficiency results obtained in steady-state fluorescence experiments and to resolve structural or dynamical detail, time-domain fluorescence lifetime measurements were carried out at distinct denaturant concentrations. Representative time-domain fluorescence decay curves are shown in Fig. 4. Double-exponential fits gave the best results, suggesting that two populations contribute to the fluorescence behavior. Upon GuHCl addition, only the longer lifetime changed (shortened) significantly. Comparison of FRET efficiencies calculated from steady-state and average lifetime measurements is shown in Table 1. The transfer efficiencies obtained from average lifetime measurements are comparable to those obtained in steady-state measurements (within 4.2%).
Figure 4.
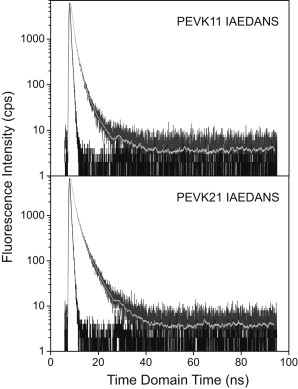
Fluorescence decay curves of PEVK11 and PEVK21 peptides. (Dark-shaded curve) Original data; (light-shaded line) model function; and (solid curve) instrument response function with the LED light source profile included.
Table 1.
Comparison of FRET efficiencies
| Peptide | FRET efficiency (%) |
||
|---|---|---|---|
| [GuHCl] (M) | Steady state | Lifetime-based | |
| PEVK11 | 0 | 55.7 | 59.8 |
| 3 | 45.1 | 49.2 | |
| 6 | 40.8 | 45.0 | |
| PEVK21 | 0 | 23.0 | 23.3 |
| 3 | 18.1 | 17.9 | |
| 6 | 15.6 | 16.2 | |
Fluorescence resonance energy transfer efficiency values determined based on steady-state fluorescence or fluorescence lifetime-based measurements for the two peptides at different concentrations of GuHCl.
Effect of ionic strength on the conformation of PEVK peptides
To investigate the effect of ionic strength on the conformation of peptides, we recorded fluorescence emission spectra at varying KCl concentrations and calculated the FRET efficiencies (Fig. 5). Energy transfer efficiency increased by 6% in the case of PEVK11. In the case of PEVK21, a greater increase in transfer efficiency was observed (10.3%) upon increasing the ionic strength. Notably, in the case of PEVK21, most of the FRET efficiency increase occurred between KCl concentration of 0.2 and 1.0 M. Further increase in ionic strength resulted in a small increase of FRET efficiency. In control experiments, we measured the changes in tryptophan fluorescence emission spectra without the acceptor as a function of KCl concentration (Fig. 5, insets). Fluorescence emission intensity was reduced by ∼15% upon KCl addition, and a 2-nm blue shift was observed.
Figure 5.
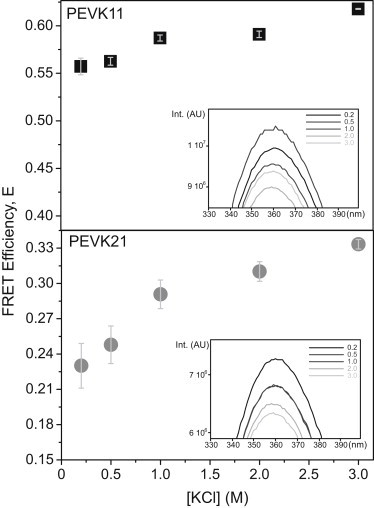
Effect of ionic strength (0.2–3 M KCl) on the transfer efficiency of PEVK11 (top) and PEVK21 (bottom) peptides. (Error bars) Standard deviation calculated from four independent experiments. (Insets) Donor (Trp) fluorescence emission spectra (with no acceptor) of the respective peptides enlarged to show details of the changes.
Effect of temperature on the conformational dynamics of PEVK peptides
Increasing the temperature from 10 to 50°C led to an increase in FRET efficiency (from 20.7 to 24.2%) for PEVK21, but only small changes for PEVK11. Considering that increasing temperatures may be used to perturb molecular structure via increase in molecular fluctuations, the structural dynamics of the PEVK peptides was characterized by measuring the normalized FRET efficiency (relative f′, Eq. 3) (32) across the temperature range of 10–50°C. Upon increasing temperature, the relative f′ increases by 30% and 70% for PEVK11 and PEVK21, respectively (Fig. 6). The trajectories of the f′ change were different in the case of the two peptides: whereas for PEVK11 the f′ function leveled out, in the case of PEVK21 a steep increase was observed followed by a departure from the original trend at 40°C.
Figure 6.
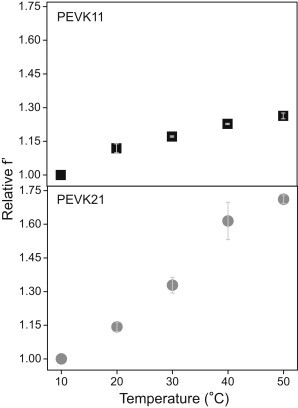
Normalized f′ parameter of PEVK11 and PEVK21 peptides as a function of temperature. (Error bars) Standard deviation calculated from four independent experiments.
Structural calculations
To investigate whether residual structural features persist in the PEVK peptides, we carried out 100-ns-long molecular dynamics simulations for each peptide (see the Supporting Material). For PEVK21, all servers predicted an α-helical structure for the core residues, whereas for PEVK11 either helical or random coil structure or their combinations were assigned by different secondary structure prediction servers (details of the predictions are listed in the Supporting Material). Thus, for both peptides, α-helical secondary structures were assumed as input for the molecular-mechanics energy minimizations and the subsequent 100-ns MD equilibrations. During the first 10 ns of the MD simulation, the initial α-helical structure (Fig. 7) disappeared partially (PEVK21) or completely (PEVK11), and loose π-helical, bend, or turn motifs appeared. After the energy minimization, the distances between the aromatic side chains of the N-terminal Trp1 and the C-terminal EDANS-modified Cys residues were 29 and 22 Å, in the case of PEVK21 and PEVK11, respectively.
Figure 7.
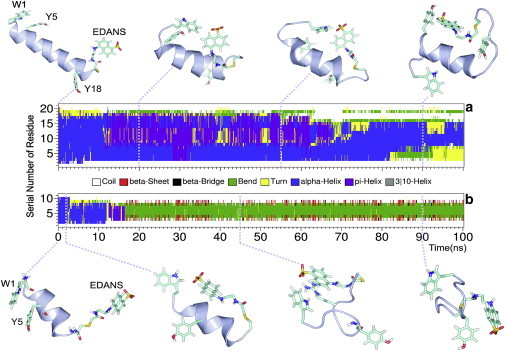
Results of the 100-ns MD simulations of PEVK21 (A) and PEVK11 (B) peptides. Color bands represent the change of secondary structure at each amino-acid residue during simulation time. Conformations of both peptides featuring important changes are also shown.
During the first 20 ns of the MD simulation of PEVK21, the initial distance dropped to ≤10 Å (see the Supporting Material) and remained there for a long, ∼35-ns period showing that the interaction between the above two terminal aromatic side chains is relatively stable. Notably, the helical (turn) conformation of the central residues survived during this period. In the case of PEVK11, the terminal aromatic side chains contacted each other soon after 2 ns with a distance ≤10 Å. At this time, the helical backbone conformation was still stable, and therefore the contact was temporary. However, following the above-mentioned collapse of the helical structure the bent backbone stabilized the aromatic contact after ∼40 ns for a longer period. The Tyr5 residues at both peptides could also approach the C-terminal EDANS-bound Cys residues (Fig. 7) corresponding to a 20 Å distance between the terminal rings.
Discussion
The PEVK domain of titin has been assumed to lack any higher-order structure and to behave as a random coil or a permanently unfolded polypeptide (8,17,18). The configuration and mechanical force-response of PEVK has been described by the WLC model (26), which assumes that different segments of the chain do not interact with each other, and the persistence length (the measure of the chain’s bending rigidity) is constant along the length of the polymer. Some studies, however, indicated that higher-order structural elements may be present within the PEVK (21,22) and that entropic mechanisms alone cannot account for the observed elasticity of this region (18).
To investigate whether the random polymer chain model is applicable to short fragments from the N-terminal third of PEVK (largest, m. soleus isoform), we studied the configuration of 2-, 11-, and 21-residue-long synthetic PEVK peptides by determining their end-to-end lengths using FRET measurements. The peptides contained an N-terminal tryptophan intrinsically present within the PEVK sequence, as a donor, and a C-terminally placed IAEDANS molecule as an acceptor. Under these circumstances the calculated contour length of the longer peptide was <10 nm, making it suitable for fluorescence energy transfer studies. End-to-end distances were calculated from the measured energy transfer efficiencies and were used to determine apparent persistence lengths of the peptides using Eq. 6, by assuming random-chain behavior according to the WLC model. FRET efficiencies were determined by steady-state measurements and were substantiated by calculations of the average fluorescence lifetime from time-domain measurements time-correlated single-photon counting (Table 1).
The apparent persistence lengths, under nondenaturing conditions, of the PEVK11 and PEVK21 peptides were 0.63 (±0.01) and 0.48 (±0.02) nm, respectively. Notably, these values are only apparent persistence lengths calculated under the assumption that the peptides possess random-coil structure, the average shape of which can be approximated with the WLC model. Previous nanomechanical studies on larger PEVK segments reported a broad distribution of persistence lengths. Values found for the constitutively expressed PEVK region were ranging between 0.4 and 2.5 nm (37) or 0.1 and 2.5 nm (40). The broad distributions were attributed to different extents of proline isomerization (37), or to a varying number of manipulated molecules, with a value of 1.4 nm corresponding to the single molecule (40).
Studies on the spatial variability of persistence lengths along PEVK showed that segments expressed from different PEVK exons had similar average persistence lengths (39), whereas the three-thirds of the human soleus PEVK fragment exhibited different persistence lengths, between 0.7 and 1.4 nm. This latter variability has been suggested to provide a mechanism for a hierarchical extensibility of PEVK (38). Our values are in the lower regime of the above ranges, and are similar to persistence lengths reported for unfolded polypeptide chains (0.4–0.44 nm (48–50)). The lower value obtained for PEVK21 indicates that there might be factors that modulate the flexibility of this peptide, resulting in a smaller apparent persistence length. The considerably different values observed in case of the two peptides suggest that the peptides do not behave as ideal polymer chains, and the assumptions of the WLC model are likely not valid in their case. This means that conformational variability (varying persistence length) and/or intrachain interactions may be present within the PEVK peptides.
Chemical denaturation caused a progressive decrease in the FRET efficiencies of the peptides (Fig. 3), but with slightly different trajectories. In the case of PEVK21, the drop in FRET efficiency became steep when increasing the GuHCl from 2 M to 4 M. The progressive decrease in FRET efficiency indicates a gradual increase in the end-to-end distance of the chains, caused most likely by the rupture of chain-shortening intramolecular interactions. The effect may be due to the weakening of hydrophobic interactions (51). Approximately 40% of the component amino acids of both peptides have hydrophobic side chains. Therefore, an internal hydrophobic interaction is indeed likely to stabilize the peptide structure, resulting in chain shortening. Upon chemical denaturation, the hydrophobic interactions are weakened and the end-to-end distance of the peptides increases.
The steplike transition seen in the case of PEVK21 upon increasing the GuHCl concentration from 2 M to 4 M may be caused by the rupture of a more stable configuration. Interestingly, the addition of GuHCl results in a small blue shift in the absorption spectrum of Trp, indicating that the local environment of the residue became more apolar. Conceivably, the Trp side chain screens the side chains of the internal hydrophobic residues released from their structural interactions upon the addition of GuHCl.
Upon increasing the ionic strength by adding KCl to the peptides in the concentration range of 0.2–3 M, FRET efficiency increased (Fig. 5). Increase in FRET efficiency corresponds to a decrease in mean end-to-end distance of the peptides. The change was more or less monotonic in the case of PEVK11, but it was biphasic and more pronounced in the case of PEVK21: a steep change is seen upon increasing KCl concentration from 0.2 M to 1.0 M, then the change became more gradual upon increasing KCl concentration further. There are two possible mechanisms that may explain the changes evoked by increasing ionic strength: electrostatic screening and amplification of the hydrophobic effect. Because the majority of charged residues in both peptides are glutamates, electrostatic screening may result in the weakening of intramolecular repulsion, leading to chain contraction. Additionally, the increase of ionic strength may cause the amplification of hydrophobic interaction (52), leading to chain contraction and collapse. The biphasic nature of the FRET efficiency increase seen in the case of PEVK21 suggests that this peptide has a more stable structure than PEVK11. Changes in PEVK properties caused by changes in ionic strength have been observed before. Decreasing the ionic strength caused an increase in titin-based myofibril stiffness (18), or in the persistence length of a human fetal titin PEVK domain (24). Thus, local variations in ionic strength may fine-tune muscle viscoelasticity by altering intramolecular interactions within the PEVK domain.
When temperature was increased from 10°C to 50°C, the normalized FRET efficiency (f′) increased in the case of both peptides, although along different trajectories (Fig. 6). Whereas the changes were small and nearly leveled out in the case of PEVK11, a biphasic response was observed in the case of PEVK21: f′ increased with increasing steepness between 10°C and 40°C, then the slope became much smaller upon further increase of the temperature. It has previously been shown both theoretically (32) and experimentally (53) that the normalized FRET efficiency parameter is sensitive to local changes in protein conformation and matrix elasticity. Increasing the temperature is used to perturb protein structure by evoking conformational fluctuations. Monotonic f′ versus temperature trajectories correspond to elastic protein matrix: the greater the slope, the greater the protein matrix flexibility (32,53).
Distinct change in the trajectory is commonly interpreted as being caused by a conformational change (53). The greater overall slope of the f′ versus temperature function seen in the case of PEVK21 indicates that this peptide has a greater overall protein matrix elasticity than PEVK11. The observation corresponds well with the calculation of apparent persistence length of the peptides under nondenaturing conditions. The 0.48 nm (±0.02) persistence length of PEVK21 indicates that it is more flexible (i.e., has a smaller bending rigidity) than PEVK11, the apparent persistence length of which was calculated to be 0.63 nm (±0.01). Importantly, the properties monitored by either the temperature-dependent f′ or the apparent persistence length are conformational fluctuations. Thus, the most plausible explanation of the temperature-evoked effect is that the peptides go through transitions between conformational states separated by small energy barriers. The effect is much more pronounced in the case of PEVK21, in the case of which steep temperature-dependence and biphasic-f′-versus-temperature response were observed. Altogether, the f′-versus-temperature measurements support the notion that PEVK21 possesses a more stable structure than PEVK11.
We also examined the effect of calcium on the conformation of the peptides. Interestingly, upon increasing the calcium concentration from pCa 9 to pCa 2, we observed no significant change in FRET efficiency (<2% increase) and the corresponding end-to-end distances (<0.03 nm) (data not shown). These results suggest that calcium has only negligible effect on the conformation of the examined PEVK peptides. This is in contrast with previous findings reporting calcium-induced conformational changes that reduced the bending rigidity of poly-E PEVK fragments (54). The peptides studied in that work by Linke et al. (10) had lengths >100 nm, more than an order-of-magnitude greater than that of the peptides investigated here. Conceivably, calcium effects depend on long-range interactions along the PEVK structure, which are precluded in the case of short peptides such as those used in this article.
The MD simulations (Fig. 7) indicate that PEVK21 tends to adopt a loose helical core, whereas PEVK11 fluctuates without well-defined secondary structure. In agreement with our results, Duan et al. (19) also predicted a high α-helical content of PEVK peptides (of which the E115 peptide’s C-terminus is identical to PEVK21, excluding the last amino acid), although these predictions were not supported by circular dichroism measurements. The assumption of an approximately two-turn-long α-helical segment within PEVK21, as predicted by the simulations, alters the contribution of this segment to the contour length of the peptide. The seven-residue-long random coil segment (7 × 0.38 nm = 2.66 nm) would be replaced by the length of a two-turn-long helix (2 × 0.54 nm = 1.08 nm). This modifies the predicted contour length from 7.98 nm to 6.4 nm and the calculated apparent persistence length from 0.48 nm to 0.62 nm. In both peptides, the contact between the terminal aromatic side chains is possible for considerable time intervals.
It is conceivable that the random, structureless unit of PEVK is ∼10 amino-acids-long, but transient interactions such as electrostatic and, importantly, hydrophobic interactions, may arise in longer segments. Such weak interactions arise as enthalpic contributions to the elasticity of PEVK, which has been suggested previously in Linke et al. (18). Considering that these weak interactions are modulated by external, solvent factors, titin elasticity may dynamically vary its apparent elasticity in response to rapid environmental changes. The two peptides we used belong to the polyE sequence motifs, which represent only a smaller fraction of the whole PEVK segment. However, because there is no major difference in the amino-acid composition of our peptides and that of the overall PEVK, each containing amino acids that may participate in electrostatic or hydrophobic interactions, we find possible that our results may be extrapolated to other PEVK fragments or even the whole PEVK region.
Conclusion
Our results indicate that titin’s PEVK domain is probably not a completely unstructured, random protein module. The variability observed in the persistence length of short PEVK peptides indicate that transient structural features and intrachain interactions are possibly present within the peptides. These interactions are mostly hydrophobic in nature, and, as suggested previously (18,25), they may provide an additional enthalpic contribution to PEVK elasticity, in addition to the purely entropic term based on WLC behavior. Most importantly, we observed these effects at the level of short peptides, whereas most previous studies were performed on larger PEVK segments or myofibrils. Although in short peptides the ionic-strength-dependent changes are subtle, in the full-length domain the effect may be large enough to provide a mechanism for the fine-tuning of local elasticity.
Acknowledgments
We thank András Lukács for discussion and inner filter effect calculation.
This work was supported by grants from the Hungarian Science Foundation (OTKA grant Nos. F49514, K73256, and K84133), the Hungarian National Office of Research and Technology (grant Nos. NANOAMI KFKT-1-2006-0021 and OMFB-380/2006), the Hungarian Medical Research Council (grant No. ETT-229/09), and the Hungarian National Development Agency (grant No. TAMOP-4.2.1.B-09/1/KMR-2010-0001). C.H. is thankful to the National Information Infrastructure Development Institute, Hungary for computational infrastructure and time provided by the NIIF supercomputing facility.
Supporting Material
References
- 1.Wang K., McClure J., Tu A. Titin: major myofibrillar components of striated muscle. Proc. Natl. Acad. Sci. USA. 1979;76:3698–3702. doi: 10.1073/pnas.76.8.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maruyama K. Connectin/titin, giant elastic protein of muscle. FASEB J. 1997;11:341–345. doi: 10.1096/fasebj.11.5.9141500. [DOI] [PubMed] [Google Scholar]
- 3.Fürst D.O., Osborn M., Weber K. The organization of titin filaments in the half-sarcomere revealed by monoclonal antibodies in immunoelectron microscopy: a map of ten nonrepetitive epitopes starting at the Z line extends close to the M line. J. Cell Biol. 1988;106:1563–1572. doi: 10.1083/jcb.106.5.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trinick J., Tskhovrebova L. Titin: a molecular control freak. Trends Cell Biol. 1999;9:377–380. doi: 10.1016/s0962-8924(99)01641-4. [DOI] [PubMed] [Google Scholar]
- 5.Cazorla O., Freiburg A., Granzier H. Differential expression of cardiac titin isoforms and modulation of cellular stiffness. Circ. Res. 2000;86:59–67. doi: 10.1161/01.res.86.1.59. [DOI] [PubMed] [Google Scholar]
- 6.Tskhovrebova L., Trinick J. Role of titin in vertebrate striated muscle. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2002;357:199–206. doi: 10.1098/rstb.2001.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freiburg A., Trombitas K., Labeit S. Series of exon-skipping events in the elastic spring region of titin as the structural basis for myofibrillar elastic diversity. Circ. Res. 2000;86:1114–1121. doi: 10.1161/01.res.86.11.1114. [DOI] [PubMed] [Google Scholar]
- 8.Labeit S., Kolmerer B. Titins: giant proteins in charge of muscle ultrastructure and elasticity. Science. 1995;270:293–296. doi: 10.1126/science.270.5234.293. [DOI] [PubMed] [Google Scholar]
- 9.Gautel M., Goulding D. A molecular map of titin/connectin elasticity reveals two different mechanisms acting in series. FEBS Lett. 1996;385:11–14. doi: 10.1016/0014-5793(96)00338-9. [DOI] [PubMed] [Google Scholar]
- 10.Linke W.A., Ivemeyer M., Labeit S. Towards a molecular understanding of the elasticity of titin. J. Mol. Biol. 1996;261:62–71. doi: 10.1006/jmbi.1996.0441. [DOI] [PubMed] [Google Scholar]
- 11.Greaser M. Identification of new repeating motifs in titin. Proteins. 2001;43:145–149. doi: 10.1002/1097-0134(20010501)43:2<145::aid-prot1026>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 12.Granzier H.L., Radke M.H., Gotthardt M. Truncation of titin’s elastic PEVK region leads to cardiomyopathy with diastolic dysfunction. Circ. Res. 2009;105:557–564. doi: 10.1161/CIRCRESAHA.109.200964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hidalgo C., Hudson B., Granzier H. PKC phosphorylation of titin’s PEVK element: a novel and conserved pathway for modulating myocardial stiffness. Circ. Res. 2009;105:631–638. doi: 10.1161/CIRCRESAHA.109.198465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chung C.S., Bogomolovas J., Granzier H.L. Titin-actin interaction: PEVK-actin-based viscosity in a large animal. J. Biomed. Biotechnol. 2011;2011:310791. doi: 10.1155/2011/310791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kulke M., Fujita-Becker S., Linke W.A. Interaction between PEVK-titin and actin filaments: origin of a viscous force component in cardiac myofibrils. Circ. Res. 2001;89:874–881. doi: 10.1161/hh2201.099453. [DOI] [PubMed] [Google Scholar]
- 16.Nagy A., Cacciafesta P., Kellermayer M.S. Differential actin binding along the PEVK domain of skeletal muscle titin. J. Cell Sci. 2004;117:5781–5789. doi: 10.1242/jcs.01501. [DOI] [PubMed] [Google Scholar]
- 17.Labeit S., Kolmerer B., Linke W.A. The giant protein titin. Emerging roles in physiology and pathophysiology. Circ. Res. 1997;80:290–294. doi: 10.1161/01.res.80.2.290. [DOI] [PubMed] [Google Scholar]
- 18.Linke W.A., Ivemeyer M., Kolmerer B. Nature of PEVK-titin elasticity in skeletal muscle. Proc. Natl. Acad. Sci. USA. 1998;95:8052–8057. doi: 10.1073/pnas.95.14.8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duan Y., DeKeyser J.G., Greaser M.L. Studies on titin PEVK peptides and their interaction. Arch. Biochem. Biophys. 2006;454:16–25. doi: 10.1016/j.abb.2006.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gutierrez-Cruz G., Van Heerden A.H., Wang K. Modular motif, structural folds and affinity profiles of the PEVK segment of human fetal skeletal muscle titin. J. Biol. Chem. 2001;276:7442–7449. doi: 10.1074/jbc.M008851200. [DOI] [PubMed] [Google Scholar]
- 21.Ma K., Kan L., Wang K. Polyproline II helix is a key structural motif of the elastic PEVK segment of titin. Biochemistry. 2001;40:3427–3438. doi: 10.1021/bi0022792. [DOI] [PubMed] [Google Scholar]
- 22.Ma K., Wang K. Malleable conformation of the elastic PEVK segment of titin: non-cooperative interconversion of polyproline II helix, β-turn and unordered structures. Biochem. J. 2003;374:687–695. doi: 10.1042/BJ20030702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kellermayer M.S., Smith S.B., Granzier H.L. Mechanical fatigue in repetitively stretched single molecules of titin. Biophys. J. 2001;80:852–863. doi: 10.1016/S0006-3495(01)76064-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Forbes J.G., Jin A.J., Wang K. Titin PEVK segment: charge-driven elasticity of the open and flexible polyampholyte. J. Muscle Res. Cell Motil. 2005;26:291–301. doi: 10.1007/s10974-005-9035-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walther K.A., Gräter F., Fernandez J.M. Signatures of hydrophobic collapse in extended proteins captured with force spectroscopy. Proc. Natl. Acad. Sci. USA. 2007;104:7916–7921. doi: 10.1073/pnas.0702179104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bustamante C., Marko J.F., Smith S. Entropic elasticity of λ-phage DNA. Science. 1994;265:1599–1600. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- 27.Kellermayer M.S., Smith S.B., Bustamante C. Folding-unfolding transitions in single titin molecules characterized with laser tweezers. Science. 1997;276:1112–1116. doi: 10.1126/science.276.5315.1112. [DOI] [PubMed] [Google Scholar]
- 28.Zarándi M., Soós K., Penke B. Synthesis of Aβ[1-42] and its derivatives with improved efficiency. J. Pept. Sci. 2007;13:94–99. doi: 10.1002/psc.801. [DOI] [PubMed] [Google Scholar]
- 29.Lakowicz J. Springer; New York: 2006. Principles of Fluorescence Spectroscopy. [Google Scholar]
- 30.Lakowicz J.R. Principles of frequency-domain fluorescence spectroscopy and applications to cell membranes. Subcell. Biochem. 1988;13:89–126. doi: 10.1007/978-1-4613-9359-7_3. [DOI] [PubMed] [Google Scholar]
- 31.Fairclough R.H., Cantor C.R. The use of singlet-singlet energy transfer to study macromolecular assemblies. Methods Enzymol. 1978;48:347–379. doi: 10.1016/s0076-6879(78)48019-x. [DOI] [PubMed] [Google Scholar]
- 32.Somogyi B., Matkó J., Damjanovich S. Förster-type energy transfer as a probe for changes in local fluctuations of the protein matrix. Biochemistry. 1984;23:3403–3411. doi: 10.1021/bi00310a004. [DOI] [PubMed] [Google Scholar]
- 33.Lakowicz J.R. Principles of Fluorescence Spectroscopy. Springer Science+Business Media; New York: 2006. Time-domain lifetime measurements; pp. 141–143. [Google Scholar]
- 34.Hammarström P., Owenius R., Lindgren M. High-resolution probing of local conformational changes in proteins by the use of multiple labeling: unfolding and self-assembly of human carbonic anhydrase II monitored by spin, fluorescent, and chemical reactivity probes. Biophys. J. 2001;80:2867–2885. doi: 10.1016/S0006-3495(01)76253-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flory P.J. Hanser Publishers; Munich, Vienna, New York: 1989. Statistical Mechanics of Chain Molecules. [Google Scholar]
- 36.Rivetti C., Guthold M., Bustamante C. Scanning force microscopy of DNA deposited onto mica: equilibration versus kinetic trapping studied by statistical polymer chain analysis. J. Mol. Biol. 1996;264:919–932. doi: 10.1006/jmbi.1996.0687. [DOI] [PubMed] [Google Scholar]
- 37.Li H., Oberhauser A.F., Fernandez J.M. Multiple conformations of PEVK proteins detected by single-molecule techniques. Proc. Natl. Acad. Sci. USA. 2001;98:10682–10686. doi: 10.1073/pnas.191189098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagy A., Grama L., Kellermayer M.S. Hierarchical extensibility in the PEVK domain of skeletal-muscle titin. Biophys. J. 2005;89:329–336. doi: 10.1529/biophysj.104.057737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sarkar A., Caamano S., Fernandez J.M. The elasticity of individual titin PEVK exons measured by single molecule atomic force microscopy. J. Biol. Chem. 2005;280:6261–6264. doi: 10.1074/jbc.C400573200. [DOI] [PubMed] [Google Scholar]
- 40.Watanabe K., Nair P., Granzier H. Molecular mechanics of cardiac titin’s PEVK and N2B spring elements. J. Biol. Chem. 2002;277:11549–11558. doi: 10.1074/jbc.M200356200. [DOI] [PubMed] [Google Scholar]
- 41.Garnier J., Gibrat J.F., Robson B. GOR method for predicting protein secondary structure from amino acid sequence. Methods Enzymol. 1996;266:540–553. doi: 10.1016/s0076-6879(96)66034-0. [DOI] [PubMed] [Google Scholar]
- 42.Petersen B., Petersen T.N., Lundegaard C. A generic method for assignment of reliability scores applied to solvent accessibility predictions. BMC Struct. Biol. 2009;9:51. doi: 10.1186/1472-6807-9-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cole C., Barber J.D., Barton G.J. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36(Web Server issue):W197–W201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bryson K., McGuffin L.J., Jones D.T. Protein structure prediction servers at University College London. Nucleic Acids Res. 2005;33(Web Server issue):W36–W38. doi: 10.1093/nar/gki410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oostenbrink C., Villa A., van Gunsteren W.F. A biomolecular force field based on the free enthalpy of hydration and solvation: the GROMOS force-field parameter sets 53A5 and 53A6. J. Comput. Chem. 2004;25:1656–1676. doi: 10.1002/jcc.20090. [DOI] [PubMed] [Google Scholar]
- 46.van der Spoel D., Lindahl E., Berendsen H.J. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 47.Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 48.Buscaglia M., Lapidus L.J., Hofrichter J. Effects of denaturants on the dynamics of loop formation in polypeptides. Biophys. J. 2006;91:276–288. doi: 10.1529/biophysj.105.071167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oberhauser A.F., Marszalek P.E., Fernandez J.M. The molecular elasticity of the extracellular matrix protein tenascin. Nature. 1998;393:181–185. doi: 10.1038/30270. [DOI] [PubMed] [Google Scholar]
- 50.Rief M., Gautel M., Gaub H.E. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 51.Zangi R., Zhou R., Berne B.J. Urea’s action on hydrophobic interactions. J. Am. Chem. Soc. 2009;131:1535–1541. doi: 10.1021/ja807887g. [DOI] [PubMed] [Google Scholar]
- 52.Zangi R., Hagen M., Berne B.J. Effect of ions on the hydrophobic interaction between two plates. J. Am. Chem. Soc. 2007;129:4678–4686. doi: 10.1021/ja068305m. [DOI] [PubMed] [Google Scholar]
- 53.Nyitrai M., Hild G., Somogyi B. Effect of Ca2+-Mg2+ exchange on the flexibility and/or conformation of the small domain in monomeric actin. Biophys. J. 1998;74:2474–2481. doi: 10.1016/S0006-3495(98)77955-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Labeit D., Watanabe K., Granzier H. Calcium-dependent molecular spring elements in the giant protein titin. Proc. Natl. Acad. Sci. USA. 2003;100:13716–13721. doi: 10.1073/pnas.2235652100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


