Abstract
Chronic lymphoproliferative disorders of natural killer cells (CLPD-NKs) and T-cell large granular lymphocytic leukemias (T-LGLs) are clonal lymphoproliferations arising from either natural killer cells or cytotoxic T lymphocytes (CTLs). We have investigated for distribution and functional significance of mutations in 50 CLPD-NKs and 120 T-LGL patients by direct sequencing, allele-specific PCR, and microarray analysis. STAT3 gene mutations are present in both T and NK diseases: approximately one-third of patients with each type of disorder convey these mutations. Mutations were found in exons 21 and 20, encoding the Src homology 2 domain. Patients with mutations are characterized by symptomatic disease (75%), history of multiple treatments, and a specific pattern of STAT3 activation and gene deregulation, including increased expression of genes activated by STAT3. Many of these features are also found in patients with wild-type STAT3, indicating that other mechanisms of STAT3 activation can be operative in these chronic lymphoproliferative disorders. Treatment with STAT3 inhibitors, both in wild-type and mutant cases, resulted in accelerated apoptosis. STAT3 mutations are frequent in large granular lymphocytes suggesting a similar molecular dysregulation in malignant chronic expansions of NK and CTL origin. STAT3 mutations may distinguish truly malignant lymphoproliferations involving T and NK cells from reactive expansions.
Introduction
Since its original description, large granular lymphocyte leukemia (LGL) has been a subject of controversy, as to whether the disorder represents a lymphoid malignancy or an exaggerated reactive T-cell process. LGL is a chronic clonal lymphoproliferative disorder that can be phenotypically subdivided into T-cell LGL and natural killer LGL.1,2 Both subtypes, seemingly derived from distinct cell lineages, are morphologically similar, causing an accumulation of large granular lymphocytes that correspond to a mature cytotoxic effector type. The current World Health Organization (WHO) separates chronic proliferations of LGL according to their cell lineage into 2 entities: chronic lymphoproliferative disorders of NK cells (CLPD-NKs) and T-cell large granular lymphocytic leukemia (T-LGL), although no significant differences in clinical features or therapeutic approach are observed between them.3–6
Chronic expansions of LGLs primarily affects elderly persons and is probably underdiagnosed. It can be associated with severe cytopenias and other comorbidities, including autoimmune conditions and malignancies, complicating the diagnosis because of the overlap between reactive processes and clear malignant lymphoproliferation.7,8 While in T-LGL, objective molecular characterization relies on the detection of clonal expansions through the uniquely rearranged TCR, establishing the clonal nature of CLPD-NKs is difficult and often disputed.9,10 For both types, and distinct from typical polyclonal reactions, autoimmune phenomena are mediated by clonal, seemingly malignant immune cells. Recently, the presence of somatic mutations in STAT3 has been described in T-LGL, indicating that a significant proportion of cases represent a true malignant process.11
Because of pathogenetic and clinical similarities, we searched for mutations in STAT3 and other related members of the STAT signaling pathway in CLPD-NKs, T-LGL, and in closely related conditions with possibly reactive expansions.
Methods
Patients
Blood sample collection from patients was performed after informed consent, according to the protocols approved by the Institutional Review Board of Cleveland Clinic, Pennsylvania State University, and the Helsinki University Central Hospital and in accordance with the Declaration of Helsinki. Thirty-seven T-LGL patients included in a previous work are not incorporated in the present study, so the mutational distribution may not be coincident (eg, 7 D661V mutations were described in the previous report vs 3 such mutations reported herein). Based on World Health Organization guidelines, the following criteria were used to diagnose T-LGL leukemia: monoclonal TCRγ-chain rearrangement, an LGL count by peripheral blood smear of > 2000 cells/μL (not a critical criterion, patients who met all other criteria but with an LGL count < 2000 cells/μL were included); flow cytometric evidence of an abnormal CTL population characterized by expression of CD2, CD3, TCRαβ (or γδ), CD4 (in 2 cases), CD5dim, CD8, CD16/56, and CD57 with negativity of CD28. Each patient must have met at least 3 of these criteria to be included in the study. In addition, TCR Vβ expansions were detected and quantitated according to criteria described previously.12,13 Diagnosis of CLPD-NKs was established based on the following parameters: blood LGL count of > 700/μL and abnormal immunophenotype pattern, including the presence of CD56+CD16+CD2+CD3− and granzyme B-expressing cells. Persistence of the condition for more than 6 months was required. Cytopenias were classified as neutropenia (absolute neutrophil count, < 1.5 × 103/μL), anemia (hemoglobin, < 10 g/dL), and thrombocytopenia (platelet count, < 100 × 103/μL). Clinical responses were determined following the modified International Working Group criteria for myelodysplastic syndromes, as reported previously.14 Time-to-treatment failure was defined as the interval between the start of treatment and the need for initiating a second line of therapy and/or progressive disease (including relapse after remission).
ARMS-PCR
The presence of D661Y and Y640F mutations nondetectable by direct sequencing was determined by a DNA tetra-primer amplification refractory mutation system (ARMS) assay. The primer sequences for the D661Y assay were (5′-3′): forward inner primer (G allele), AAATCATCATGGGCTATAAGATCACGG; reverse inner primer (T allele), GGAGACACCAGGATATTGGTAGCGTA; forward outer primer, CCTAGCTGTAGGTTCCATGATCTTTCCT; and reverse outer primer, AAAATTAAATGCCAGGAACATGGAAAAT (product size: 2 outer primers, 290 bp; G allele, 182 bp; T allele, 161 bp). Primer sequences for the Y640F assay were (5′-3′): forward inner primer (A allele), ACCCAGATCCAGTCCGTGGAACCTTA; reverse inner primer (T allele), ACATGTTGTTCAGCTGCTGCTTTGAGA; forward outer primer, AAAAAATGGGCAGTTTTCTCTGAGATGACC; reverse outer primer, CCAGTGGAGACACCAGGATATTGGTAGC (product size: 2 outer primers, 197 bp; A allele, 119 bp; T allele, 131 bp)
Flow cytometry assays
KIR (killer cell immunoglobulin-like receptor) skewing in CLPD-NKs.
In 18 cases, paired healthy and patient NK cells were analyzed by 2-color flow cytometry using CyChrome-conjugated anti-CD56 (clone B159; BD Biosciences) or anti–CD16-FITC–conjugated antibodies in combination with the following PE-conjugated antibodies: anti-CD158a (KIR2DL1, KIR2DS1), anti-CD158b (KIR2DL2, KIR2DL3, KIR2DS2), anti-NKB1 (KIR3DL1), anti-KARp50 (KIR2DS4), and anti-NKG2A, which were all obtained from BD Biosciences. A purified antibody was used to stain NKG2D (0.5 μg; R&D Systems) and KIR2DL4 (0.5 μg; provided by Dr Eric Long, National Institute of Allergy and Infectious Diseases, Rockville, MD) followed by rat anti–mouse immunoglobulin (Ig)–conjugated PE secondary antibody. Six of these patients were included in a previous study.
In 7 patients, the NK phenotype was assessed using a 4-color approach with antibodies directed against CD2, CD3, CD57 (Immunotech), and CD158a (clone EB6; Beckman Coulter), CD158b/j (clone GL183; Beckman Coulter), CD158e (clone DX9; BD Biosciences), and CD158i (clone FES172; Immunotech), CD159a (cloneZ199, Immunotech), CD94 (clone Z199, IgG2b; Beckman Coulter), and NKG2D (clone 1D11, BioLegend)
TCR variable β-chain (Vβ) skewing T-LGL.
Fresh peripheral blood was stained for Vβ flow cytometry analysis to quantitate the percentage of each Vβ family in the CD4 and CD8 lymphocyte populations. The manufacturer's instructions (IOTest Beta Mark kit; Beckman Coulter) were modified as follows: 5 μL of PE cyanin (PC) 5-conjugated anti-CD4 (Beckman Coulter) and 5 μL of PC7 anti-CD8 (BD Biosciences) monoclonal antibodies were added. Anti-Vβ 6.7 FITC (Pierce Chemical), anti-CD3 FITC, anti-TCRαβ PC5, and anti-TCRγδ PE (Beckman Coulter) also were included in the panel. A 4-color acquisition protocol was used on an FC500 with CXP Version 2.2 software (Beckman Coulter). FCS Express Version 3.0 (De Novo Software) was used for analysis. The lymphocyte gate was set according to forward and side scatter. For Vβ family T-cell repertoire analysis, gates were set on CD4 (2 cases) and CD8 bright lymphocyte populations and then analyzed for Vβ distribution. Mean and SD values were provided by the manufacturer of the IOTest Beta Mark kit and are based on a control population of 85 volunteers, as described previously.15,16 In addition, a separate Vβ repertoire control group of 69 volunteers was analyzed that did not differ significantly from the Beckman group or previous publications.13 A significant clonal expansion was defined as an expansion that was greater than the mean ± 3 SD of healthy controls.
Apoptosis assay
To quantitate apoptosis, we performed 3-color flow cytometry with propidium iodide (Sigma-Aldrich), annexin-V allophycocyanin (Beckman Coulter), and CD3 FITC (Beckman Coulter). Forward and side scatter analyses were used to identify the lymphocyte population in peripheral blood samples from leukemic LGLs and normal PBMCs. This lymphocyte gate, then the CD3+ (T-LGL) or CD3− (CLPD-NKs) gate, was used to examine the population that stained positive for annexin-V allophycocyanin and propidium iodide in single-positive and double-positive quadrants, allowing for the calculation of percent early apoptotic, apoptotic, and dead cells. Mononuclear cells were separated from peripheral blood by density gradient centrifugation and then cultured in the presence or absence of 1μM or 10μM STA-21 (Enzo Life Sciences) in RPMI + 10% FCS for 48 hours.
Microarray analysis
Mononuclear cells were separated from peripheral blood by density gradient sedimentation (Mediatech). LGL cells were separated by flow cytometric sorting using anti-VB and CD8 mAb as described previously.17 Healthy, donor-derived CD8+CD57+ cells were isolated by flow cytometric sorting using CD3, CD8, and CD57 mAb. Total RNA was extracted from cells using Trizol (Invitrogen) Phase-Lock gel tubes (Eppendorf), cleaned RNAeasy columns (QIAGEN), and dissolved diethylpyrocarbonate water. Total cRNA was prepared using the in vitro–transcribed (Affymetrix) and hybridized to U133 arrays, according to the manufacturer's instructions (Affymetrix). All microarrays were examined for surface defects, grid placement, background intensity, housekeeping gene expression, and a 3:5 ratio of probe from genes of various lengths. Expression analysis was conducted using standard Affymetrix analysis software algorithms (Microarray Suite Version 5.0). Comparative analysis between expression profiles was carried out on GeneSpring Version 7.1 software (Agilent Technologies). Scanned images of Affymetrix chips were converted spreadsheet numbers using Affymetrix proprietary GeneChip Operating software. Gene expression data were normalized in 2 “per-gene normalization” and “per-sample normalization.” This approach been previously described in detail.18 Data for this microarray study are uploaded into the Gene Expression Omnibus under accession no. GSE39838.
Western immunoblotting
PBMCs from diagnosis were lysed in a buffer composed of 50mM Tris-Cl (pH 7.6), 5mM EDTA, 150mM NaCl, 0.5% NP-40, 0.5% Triton-X-100 (RIPA) containing 1 μg/mL leupeptin, aprotinin, and antipain; and 1 times concentration of phosphatase inhibitor cocktail 2 (Sigma-Aldrich P5726) containing sodium vanadate, sodium molybdate, and sodium tartrate (all from Sigma-Aldrich). Protein concentration was determined in all cell extracts using the BCA protein assay kit (Pierce Biotechnology). Unless otherwise indicated, 25 μg of total protein was loaded per lane in Laemmli SDS-PAGE sample loading buffer and boiled for 5 minutes before separation by 10% SDS-PAGE. A total of 25 μg of total protein was found to give ECL signals within the linear range for β-actin in titration experiments. The proteins were then transferred to a membrane for Western blot analysis. Antibodies were obtained from the following sources and used at the dilutions recommended by the manufacturer: anti-pSTAT3 (Tyr705), 1:1000 (#9131; Cell Signaling Technology), and anti–β-actin 1:25 000 (A3853 Sigma-Aldrich). Antibody detection was performed by standard ECL techniques as recommended by the manufacturer (GE Healthcare). Protein quantification was performed by densitometry using the ImageJ program (Rasband, W.S., ImageJ, National Institutes of Health, Bethesda, MD; http://imagej.nih.gov/ij/, 1997-2011.).
PCR direct sequencing assays
PCR primers were designed to amplify and sequence all coding exons in the Src homology 2 (SH2) domain of the STAT family genes (STAT1, STAT2, STAT4, STAT5a, STAT5b, and STAT6) and known mutational hotspot regions in other JAK/STAT pathway genes (gp130 exon 6, JAK2 V617F, JAK3 exons 13-17, RELA exon 5), and are available on request. For each PCR, 40 ng genomic DNA was used for PCR amplification followed by purification using Montage Cleanup kit (Millipore). Sequencing was performed using ABI 3730xl DNA analyzer (Applied Biosystems). All STAT3 mutations were detected by bidirectional sequencing and scored as pathogenic mutations on the basis of the observation that they were not detected in nonleukemic cells (CD4+ cells). All mutations were first compared with published SNP data (dbSNP; http://www.ncbi.nlm.nih.gov/projects/SNP).
Statistical analysis
Comparisons of proportions and ranks of variables between groups were performed by χ2 test, Fisher exact test, Student t test, or Mann-Whitney U test, as appropriate. We used the Kaplan-Meier and the Cox method to analyze overall survival and progression-free survival, with a 2-sided, P value ≤ .05 considered significant. In Cox models, examination of log (−log) survival plots and partial residuals was performed to assess that the underlying assumption of proportional hazards was met.
Results
Identification of STAT3 mutations
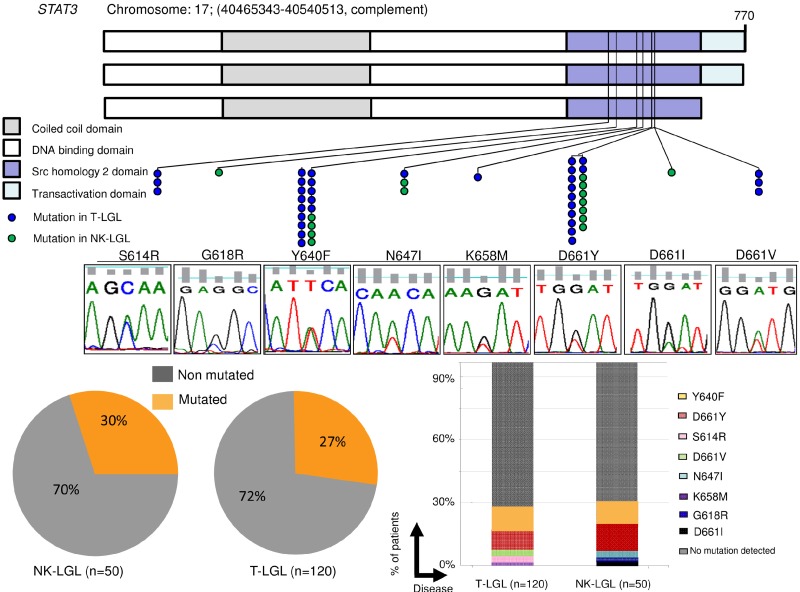
Using stringent diagnostic criteria, we have identified a large cohort of patients with CLPD-NKs and T-LGL that have been subjected to a molecular search for mutations associated with STAT3 signaling. We screened 170 cases and identified 49 STAT3 mutations in 48 cases (Figure 1). The cohort included 120 T-LGL and 50 CLPD-NKs (Table 1), but no significant differences in distribution of STAT3 mutations were found when comparing cell-lineage subsets. Of note is that no STAT3 mutation was identified in an independent cohort of 31 cases diagnosed with idiopathic neutropenia and identifiable LGLs in the blood smear but not fulfilling all diagnostic criteria for T-LGL or CLPD-NKs.
Figure 1.
Distribution of STAT3 mutations throughout gene domains and patient cohort. (A) STAT3 mutations (blue dots represent mutations in T-LGL; green dots, in CLPD-NKs) found in the SH2 domain, necessary for receptor association and tyrosine phosphodimer formation. The major domains of STAT3 are shown: coiled-coil domain, DNA-binding domain, SH2 domain, and transactivation domain. Lower panel: Corresponding representative Sanger sequence for each mutation found. (B) Percentage of patients with STAT3 mutations. Lesions were observed in 15 of 50 CLPD-NKs and 33 of 120 T-LGL patients when using Sanger and AS-PCR (7 cases not detected by Sanger). (C) Histograms showing the percentage of cases corresponding to each mutation. D661Y and Y640F accounted for ∼ 80% of all mutations found.
Table 1.
Clinical characteristics at baseline of the cohort and comparison according to the cell lineage
| Variable | Whole cohort (n = 170) | T-LGL (n = 120) | CLPD-NKs (n = 50) | P |
|---|---|---|---|---|
| Age | .74 | |||
| Median, y | 64 | 65 | 61 | |
| Range, y | 20-90 | 20-87 | 27-90 | |
| Sex, no. (%) | .9 | |||
| Male | 55 (94) | 55 (66) | 56 (28) | |
| Female | 45 (86) | 45 (54) | 44 (22) | |
| Symptoms at diagnosis, no. (%) | 53 (90) | 53 (63) | 54 (27) | .8 |
| Neutropenia, no. (%)* | 62 (105) | 64 (75) | 60 (30) | .7 |
| Lymphocytosis, no. (%)† | 54 (92) | 50 (60) | 64 (32) | .07 |
| Anemia, no. (%)‡ | 30 (52) | 31 (37) | 30 (15) | .9 |
| Thrombocytopenia, no. (%)§ | 14 (24) | 13 (16) | 16 (8) | .7 |
| LGL count in PB, × 109/L | .5 | |||
| Median | 3.4 | 3.1 | 3.7 | |
| Range | 0.2-22 | 0.3-22 | 0.9-13 | |
| Splenomegaly, no. (%) | 21 (37) | 25 (31) | 21 (6) | .4 |
| Presence of MGUS, no. (%) | 21 (36) | 22 (26) | 20 (10) | .9 |
| Associated autoimmune disease | ||||
| RA, no. (%) | 10 (17) | 11 (14) | 6 (3) | .10 |
| AIHA, no. (%) | 7 (12) | 5 (6) | 12 (6) | .15 |
| Associated BM disorder | ||||
| MDS, no. (%) | 5 (9) | 6 (7) | 4 (2) | .25 |
| PRCA, no. (%) | 5 (8) | 6 (7) | 2 (1) | .15 |
| Presence of B-cell malignancy, no. (%) | 9 (15) | 10 (12) | 6 (3) | .3 |
| Treatment lines | .3 | |||
| Median | 1.5 | 1.7 | 1.2 | |
| Patients with somatic mutation in STAT3-SH2 domain, no. (%) | 28 (48) | 27 (33) | 30 (15) | .6 |
PB indicates peripheral blood; MGUS, monoclonal gammopathy of undetermined significance; MDS, myelodysplastic syndrome; STAT3, signal transducer and activator of transcription 3; and SH2, Src homology 2 domain.
Absolute neutrophil count ≤ 1.5 × 109/L.
Absolute lymphocyte count ≥ 4 × 109/L.
Hemoglobin ≤ 10 g/dL.
Platelet count ≤ 100 × 109/L.
All mutations were located in the domain of STAT3 (residues 585-688) that shares homology with SH2 domains (Figure 1A). The STAT3 SH2 domain mediates STAT3 dimerization via binding of phosphotyrosine residue Y705.19 No additional somatic mutations were identified in the SH2 domain of other STAT family members (STAT1, STAT2, STAT4, STAT5a, STAT5b, and STAT6) and in other JAK/STAT pathway screened genes' hotspots (gp130 exon 6, JAK2 V617F, JAK3 exons 13-17, RELA exon 5) in a screening subcohort of 40 STAT3-negative cases (20 CLPD-NKs and 20 T LGLs). All mutations were heterozygous and proved to be somatic, as they were not observed in germline DNA. One patient harbored 2 STAT3 mutations (D661Y and Y640F). In a case with coexisting NK and CTL components, sorting revealed that the mutation originated in the NK-cell subset. Moreover, mutations were not only found in T-LGL with α/β restrictions but also in γ/δ rearranged cases: 1 of 2 γ/δ cases included in this study was mutated (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Two mutations, Y640F and D661Y, accounted for 80% of the somatic variations found, enabling the design of a more sensitive ARMS-PCR method for each of these alterations, which allowed us to find 7 new mutated cases after analyzing all Sanger-negative cases. Sequencing of cloned exon 21 STAT3 DNA products from these cases showed that the mutations were carried by one of the alleles in low proportion (< 10% of clones). Titration studies highlighted a higher sensitivity of ARMS-PCR assay compared with Sanger sequencing for detecting mutations, and follow-up analyses confirmed the value of this assay in detecting minimal residual disease throughout the course of the disease in an exemplary CLPD-NK patient (supplemental Figure 2). In general, ARMS-PCR may increase the diagnostic yield to clones representing < 10%.
Proliferation and survival signals in STAT3 mutant LGL cases
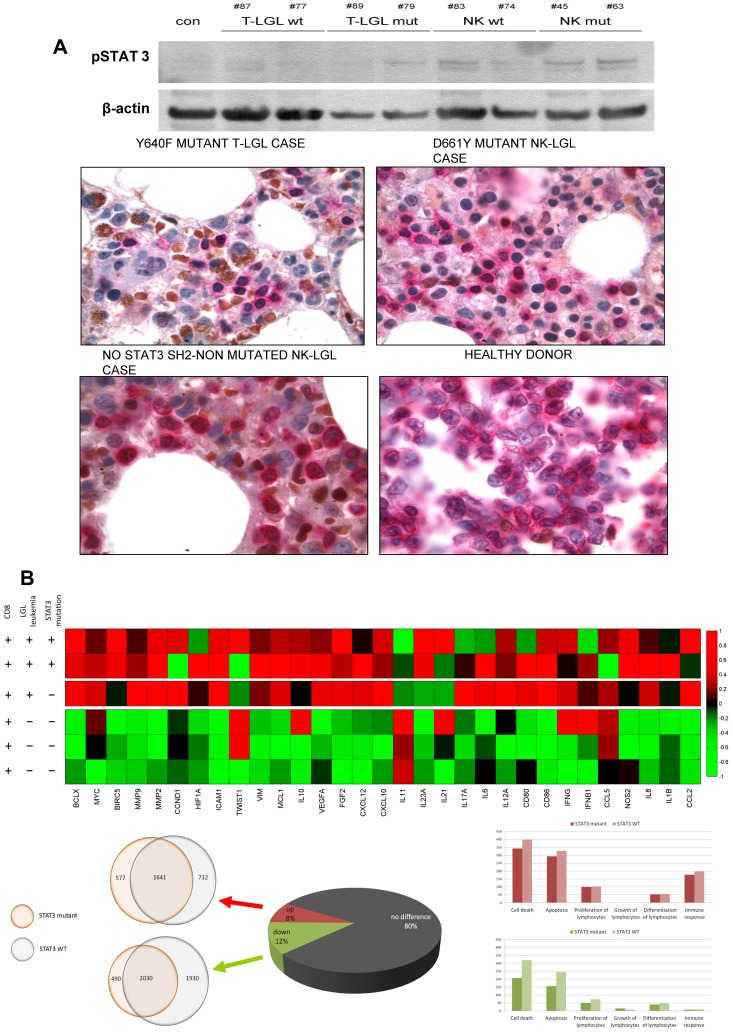
We then investigated whether mutant and wild-type cases show signs of activation along the STAT3 signaling pathway. Western blot analysis in leukemic LGLs from mutated or wild-type specimens and mononuclear cells in controls showed that phosphorylated-STAT3 (pSTAT3) protein was most abundant in mutated cases, irrespective of the cell lineage, but also expressed at higher levels in WTs compared with the amount in normal mononuclear cells, suggested constitutive activation in leukemic cells (Figure 2A top). Aberrant intracellular pSTAT3 signaling also was detected in STAT3 mutant cases of T and NK origin. Counterstaining with anticytoplasmic-CD3 identified the lymphoid derivation of this signal (Figure 2A bottom).
Figure 2.
Proliferation and survival signals in chronic LGL diseases. (A) Constitutive STAT3 activation in leukemic cells. (Ai) Western blot analysis in leukemic cells from 4 T-LGL patients (2 mutated and 2 wild-type), 4 CLPD-NK patients (2 mutated and 2 wild-type), and a control. (Aii) Aberrant intracellular pSTAT3 signal (brown) has been also detected in paraffin sections from bone marrow biopsy samples of STAT3-mutant and nonmutant cases of T and NK origin. Previous immunohistochemical staining with CD8, surface CD3, and CD2 defined the cell lineage of the lymphocyte infiltration. Positive double staining with anticytoplasmic CD3 (pink) and pSTAT3 (brown) showed aberrant pSTAT3 signal in the infiltrating lymphoid compartment. Finally, a healthy donor tonsil sample shows no brown nuclei in cCD3-positive cells. (B) STAT3 pathway-related genes deregulated. (Bi) Heat map reflecting color-coded expression levels from a set of genes known to be regulated by STAT3 (columns) in purified T-LGL cells from 3 patients and control samples (rows). (Bii) Pie chart depicting whole genome expression in the 3 T-LGL leukemia patients and overlapping circles showing a high degree of coincidences in deregulated genes in mutated and nonmutated patients. (Ciii) Histograms of whole genome expression levels separated by pathways, exposing a predominance of deregulation in apoptosis and cell death, both in mutated and nonmutated patients. Up-regulated pathway genes are shown in pink (top panel) and down-regulated pathway genes in green (bottom panel).
STAT3 signaling has been considered a major intrinsic pathway for cancer inflammation, capable of inducing a large number of downstream genes that are crucial for tumor promotion. Deregulation of the STAT3 pathway has also been specifically implicated in the pathogenesis of T-LGL. Consequently, we examined the gene expression pattern in T-LGL leukemias compared with controls, and the impact of the presence of STAT3 mutations on the transcriptional regulation of a specific set of genes previously described.20 For this analysis, a global expression microarray analysis was performed on LGL cells from 2 patients with a STAT3 mutation (D661Y and Y640F) and a patient with symptomatic T-LGL, showing a clearly dominant clone as determined by profound TCR Vβ chain usage, but without detectable STAT3 mutations. T-LGL cells were sorted according to the clonal expression of pathognomonic Vβ chain. As expected, sorted clonal cells showed high expression of CD57. Controls derived from flow-sorted CD8+CD57+ CTL effector cells from healthy donors. T-LGL cell populations discordantly expressed 7416 genes: 2966 that were up- and 4450 that were down-regulated (change > 2-fold; P = .02; Figure 2B bottom). Interestingly, mutated and WT disease samples shared ∼ 50% of deregulated genes, implying a role in both of deregulated apoptotic pathways. There was also a striking overexpression of 31 cancer inflammation-related genes described previously to be inducible by STAT3; interestingly, mutated and WT disease samples shared ∼ 50% of deregulated genes, showing both mutated and nonmutated patients a preference for deregulation of apoptotic pathways. Focusing on 31 cancer inflammation-related genes described previously to be inducible by STAT3, there was a striking pattern of overexpression of these genes; interestingly, mutant and nonmutant patients showed significant concordance (Figure 2B top), suggesting that, in addition to STAT3 mutations, other mechanisms or lesions can be responsible of deregulation of this pathway.
Inhibition of STAT3 results in LGL apoptosis
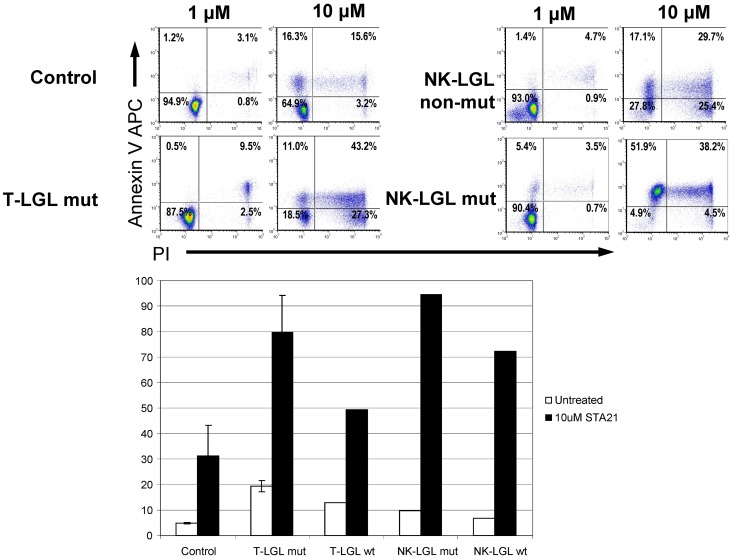
We then examined the effects of STAT3 inhibition in CLPD-NKs and T-LGL cells using STA-21, a novel synthetic inhibitor of STAT3 dimerization, DNA binding, and STAT3-dependent luciferase reporter activity.20 STA-21 alone induced an increase in annexin-V–allophycocyanin binding in leukemic LGLs (both NK and T; Figure 3). Increasing doses of STA-21 induced a dose-dependent increment in the percentage of apoptotic cells in leukemic LGLs but had little effect on normal lymphocytes from the same sample and also from controls. Both leukemic NK and CTL cells showed a much higher sensitivity than their nonmalignant counterparts. However, the effect of inhibitor was not specific to STAT3 mutant cells because patients without STAT3 mutations showed similar response to STAT3 inhibition. These results indicate that only the leukemic cells (both mutant and WT), and not nonleukemic cells, are most sensitive to the apoptosis-inducing effects of STA-21.
Figure 3.
Effect of STA-21 on apoptosis of malignant LGLs. Leukemic and control cells were harvested after 48 hours of STA-21 treatment and analyzed with propidium iodide and annexin V staining assays. (Top) Dose-dependent increase in apoptosis. (Bottom) Histograms depicting percentage of cells undergoing apoptosis after treatment compared with untreated cells.
Clinical correlates of STAT3 mutations
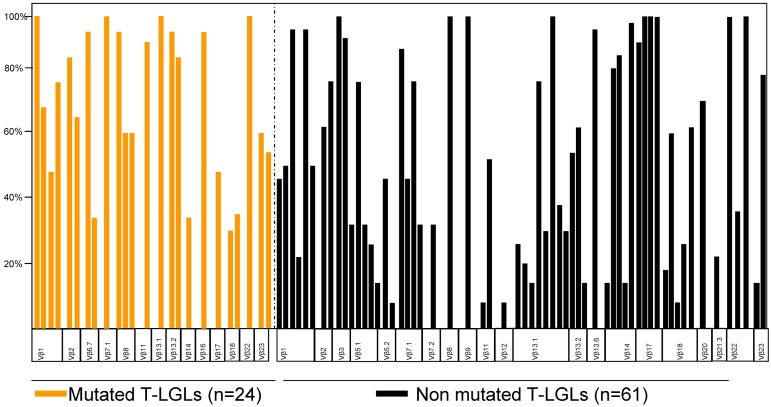
We first studied the correlation between the detection of STAT3 and clonal size as determined by morphologic enumeration of LGLs and Vβ TCR repertoire analysis. No correlation was found between the presence of a STAT3 mutation and the absolute LGL count (Table 2). While some patients with otherwise typical cells and extreme clonal expansions did not have STAT3 mutations, overall the STAT3 mutant subset of T-LGLs had more dominant clones, with a remarkable expansion in the panel: in all mutated cases, the immunodominant clone size exceeded 25% of the whole CD8 compartment, whereas in the nonmutated cohort, 23% of cases did not reach the 25% threshold (Figure 4). In addition, no specific restricted Vβ subset was observed to be characteristic of each group. We performed a KIR expression analysis in half of our CLPD-NK cohort. All STAT3-mutated patients7 were included among the 23 of 25 cases that showed an abnormal pattern of expression (Table 3; supplemental Figure 4).
Table 2.
Clinical differences at baseline between the STAT 3 SH2 domain mutated and nonmutated LGL leukemia patients
| Variable | STAT 3 SH2 domain mutated patients (48) | STAT 3 SH2 domain nonmutated (122) | P |
|---|---|---|---|
| Age | .96 | ||
| Median, y | 63 | 64 | |
| Range, y | 35-80 | 20-90 | |
| Sex, no. (%) | .06 | ||
| Male | 58 (28) | 52 (66) | |
| Female | 42 (20) | 48 (54) | |
| Symptoms at diagnosis, no. (%) | 75 (36) | 45 (54) | < .001 |
| Neutropenia, no. (%)* | 70 (34) | 59 (71) | .14 |
| Lymphocytosis, no. (%)† | 56 (27) | 52 (65) | .7 |
| Anemia, no. (%)‡ | 50 (24) | 34 (28) | .06 |
| Thrombocytopenia, no. (%)§ | 13 (6) | 15 (18) | .7 |
| LGL count in PB, × 109/L | 3.9 | 3.3 | .45 |
| Median | 0.4-20 | 0.5-9 | |
| Splenomegaly, no. (%) | 33 (16) | 26 (32) | .5 |
| Presence of MGUS, no. (%) | 14 (8) | 20 (28) | .3 |
| Associated autoimmune disease, no. (%) | 22 | 24 | .052 |
| RA, no. (%) | 18 (9) | 7 (8) | .039 |
| AIHA, no. (%) | 14 (7) | 4 (5) | |
| Associated BM disorder | |||
| MDS, no. (%) | 8 (4) | 5 (6) | .437 |
| PRCA, no. (%) | 0 | 6 (8) | .027 |
| Presence of B-cell malignancy, no. (%) | 4 (2) | 10 (13) | .162 |
| Treatment lines | .03 | ||
| Median | 2.4 | 1.2 | |
| Lineage, no. (%) | .6 | ||
| T-cell | 28 (33) | 72 (89) | |
| NK-cell | 30 (15) | 70 (35) |
PB indicates peripheral blood; MGUS, monoclonal gammopathy of undetermined significance; MDS, myelodysplastic syndrome; STAT3, signal transducer and activator of transcription 3; and SH2, Src homology 2 domain.
Absolute neutrophil count ≤ 1.5 × 109/L.
Absolute lymphocyte count ≥ 4 × 109/L.
Hemoglobin ≤ 10 g/dL.
Platelet count ≤ 100 × 109/L.
Figure 4.
Marked immunodominant Vβ expansions can be seen both in STAT3 SH2 domain-mutated (orange bars) and nonmutated patients (black bars).
Table 3.
KIR expression and STAT3 mutational status in CLPD-NK patients
| Patient code no. | Mutation status | Abnormal KIR expression | Reference |
|---|---|---|---|
| 2-color flow cytometry | |||
| 3 | No | Yes | Present study |
| 5 | No | Yes | 23 |
| 6 | No | Yes | Present study |
| 10 | N647I | Yes | Present study |
| 11 | No | Yes | Present study |
| 12 | No | Yes | Present study |
| 13 | Y640F | Yes | 23 |
| 15 | No | Yes | Present study |
| 17 | No | Yes | Present study |
| 18 | No | Yes | Present study |
| 19 | No | Yes | Present study |
| 20 | No | Yes | 23 |
| 22 | No | Yes | 23 |
| 23 | D661I | Yes | Present study |
| 28 | No | Yes | 23 |
| 34 | D661Y | Yes | Present study |
| 36 | Y640F | Yes | 23 |
| 38 | No | Yes | Present study |
| 4-color flow cytometry | |||
| 63 | Y640F | Yes | Present study |
| 83 | No | Yes | Present study |
| 47 | D661Y | Yes | Present study |
| 57 | No | Yes | Present study |
| 61 | No | No | Present study |
| 62 | No | No | Present study |
| 74 | No | Yes | Present study |
Abnormal KIR expression included homogeneous reactivity to a single KIR receptor and/or absence of reactivity to 1 or more KIR receptors.
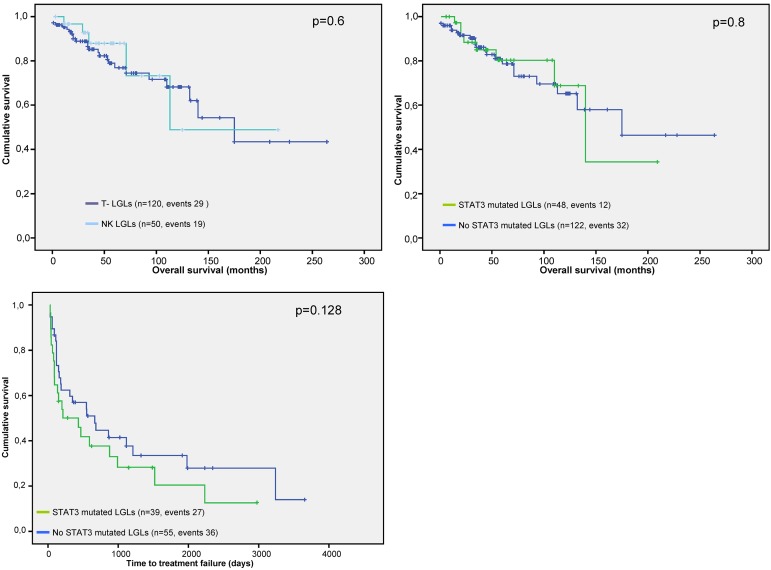
Patients with a somatic mutation in STAT3 were characterized by a higher frequency of symptomatic disease at baseline (75% vs 40%; P < .001) and the requirement of more lines of therapy through the course of their disease (2.4 vs 1.2; P = .03; Table 2). Indolent cases (as determined based on “need to treat” bases) were predominantly grouped within the nonmutated patients: 59 wild-type versus 6 mutant cases. No significant differences were found when analyzing overall survival between CLPD-NKs and T-LGL (Figure 5A) or between the mutated and nonmutated subsets (Figure 5B). The STAT3-mutated cohort had a shorter time-to-treatment failure compared with those without mutations (Figure 5C), although no statistical significance was reached (P = .128). Focusing on comorbidities, both rheumatoid arthritis and autoimmune hemolytic anemia were present in significantly higher frequencies in mutated cases, whereas, surprisingly, pure red cell aplasia (PRCA) cases were invariably related to non-STAT3 SH2 mutant cases (Table 2).
Figure 5.
Survival outcomes and time-to-treatment failure in patients with CLPD-NKs and T-LGL. P values presented correspond to the Cox regression between the groups indicated. (A) Comparison of survival outcomes according to the leukemic cell lineage. (B) Comparison of survival outcomes depending on the STAT3 SH2 domain mutational status. (C) Differences in the time-to-treatment failure in patients with or without STAT3 SH2 domain mutation. Time-to-treatment failure was defined as the interval between the start of treatment and the need for initiating a second line of therapy and/or progressive disease (including relapse after remission).
Discussion
Leukemic LGLs can arise from either CTLs or NK cells and represent a probably underdiagnosed, often indolent, clonal lymphoproliferation that overlaps with reactive processes via the provisional entity of monoclonal T-cell clonopathy of unclear significance.7 Although prognosis is generally good, many patients have chronic morbidities because of cytopenias and the disease is, essentially, not curable. Our discovery of somatic mutations in the SH2 domain of STAT3 in T-LGL, and now also in CLPD-NKs, displays its main value both in the diagnostic setting: (1) STAT3 mutations may be a useful tool to discriminate malignant NK lymphoproliferations from reactive expansions, in particular to establish clonality using STAT3 mutation as a clonal marker; and (2) the shared altered pathway and clinical behavior strongly support the suitability of merging both entities in 1 disease category; and in pathogenesis: we show that enhanced, and reversible apoptosis, is a hallmark of STAT3 lymphocytes.
Despite a similar cytotoxic effector function, NK cells, unlike T cells, mediate non-MHC–restricted cytotoxicity and do not express the surface CD3/TCR complex or go through TCR gene rearrangement.2 These evident differences led to segregation of chronic LGLs in hematologic tumor classifications according to their cell lineage, although no significant disparity in clinical behavior has been established.4–6 Here we show that T-LGL and CLPD-NKs are associated with somatic STAT3 mutations in a significant proportion of cases, a finding that demonstrates a shared altered signaling pathway mediating aberrant survival of the clonal cells of T- or NK-cell origin. A skewed NK receptor expression on NK cells has been described in patients with the NK lymphoproliferative disease of granular lymphocytes.21 In particular, differences in the repertoire of KIR receptors expressed on NK cells from patients with CLPD-NKs in contrast to those of healthy persons can assist in the difficult process of assessing clonality in this disorder.10,22 In our cohort, all of our mutated CLPD-NK patients showed some degree of KIR expression restriction, supporting its value as a marker of malignant clonality.
The presence of the same acquired molecular lesion in T-LGL and CLPD-NKs raises the question of whether those lesions might distinguish truly malignant, transformed leukemias from reactive processes because of a continuum of cellular immune responses spanning polyclonal, oligoclonal, and strict clonal lymphoproliferations. The concerns and difficulties involved in separating, clinically and pathogenetically, those entities have been highlighted by us and others.2,7,23 We show herein higher expansions of the Vβ immunodominant clones among mutated patients, corresponding with a higher frequency of symptomatic disease at baseline. However, cases without typical STAT3 mutations are characterized by a great deal of heterogeneity, with indolent and symptomatic patients or cases with either extreme or very modest clonal expansions. One could speculate that some symptomatic patients with clear expansions might harbor other somatic mutations affecting the JAK/STAT3 pathway. That hypothesis led us to screen patients negative for a STAT3 exon 21 mutation by a more sensitive method, to expand the search to the SH2 domain of the rest of the STAT family genes and to genes with reported mutations involved in JAK/STAT3 signaling. Twenty percent of the mutations reported here were found by this search, including an additional 7 cases by allele-specific PCR in exon 21 and 3 cases with a somatic mutation in exon 20 of STAT3. It is therefore likely that increased sensitivity may result in detection of subliminal mutated clones in a higher proportion of cases. Targeted screening of other potentially involved genes did not yield positive results, but it appears that STAT3 mutant-negative cases may also display dependence on STAT3-mediated signals with a stimulatory input, potentially at a more proximal level. Alternatively, this pathway has been demonstrated to be persistently deregulated in cancers, including lymphomas, by mechanisms distinct from acquired alterations in the genetic code. These include mechanisms, such as microenvironmental paracrine activation or epigenetic modulation.24–26 While acknowledging the cautiousness in the data interpretation because of the relatively scarce number of patients arrayed, our downstream deregulation and activation studies suggest that STAT3 can be activated independent of key oncogenic driver mutations in its SH2 domain in a subset of patients with LGL disorders.
Reversal of the antiapoptotic phenotype in response to upstream STAT3 inhibitors occurs in a proportion of patients with T-LGL.27 We hypothesized that this effect could account for those cases without a STAT3 mutation. A downstream STAT3 selective inhibitor used in our study induced apoptosis of CLPD-NKs or T-LGL irrespective of mutational status, and it appears that leukemic cells are more sensitive than their normal counterparts. In this respect, the association of STAT3 mutations with the need for more lines of therapy shown here suggests that targeting the STAT3 mutation may be a viable therapeutic strategy. Of note, dominant negative mutations in the DNA-binding domain of STAT3 can cause Job syndrome, a primary immunodeficiency with predisposition to B-cell lymphomas; thus, systemic suppression of STAT3 function may have adverse consequences.28–31 Further studies will determine whether there are distinct STAT3 signaling patterns among mutated and nonmutated samples that can measure the response to different Stat3 inhibitors.
We show here that coexistence of RA or AIHA is more frequent in mutant CLPD-NKs or T-LGL patients than in wild-type cases and that we could not detect mutations in patients with concomitant PRCA. RA is the most common autoimmune disease associated with these disorders,32 and both entities share essential pathogenic features, including expansion of CTLs, constitutive overexpression of cytotoxic molecules, and apoptosis resistance.33–35 Our results strengthen the notion of this pathogenic association, which appears closer in mutant cases. For instance, STAT3 promotes survival of RA synovial fibroblasts and, in the therapeutic setting, widely used sulfur-containing gold compounds have been shown recently to depend on the blockade of JAK1/STAT3 signaling.36 Mutant cases also had a higher prevalence of concurrent AIHA; again, STAT3 constitutive activation might be responsible for apoptotic-resistant autoreactive T-cell clones.37 In contrast, no mutation was found in 7 patients with CLPD-NKs or T-LGL and PRCA, where a similar line of argument involving autoreactive apoptotic-resistant immunity could be followed. Further studies are needed to address this apparent controversy; nevertheless, STAT3 SH2 domain-mutated LGLs clones are unlikely to be a major contributor to the development of PRCA. No significant differences between mutant and nonmutant cases were found when considering other reported associations, such as the presence of B-cell processes or concomitant myelodysplastic syndrome.
In conclusion, the present study of CLPD-NKs or T-LGL shows that STAT3 SH2 somatic mutations can be found in a similar percentage either in NK or T cell entities. Our results strongly suggest a common specific clinical and pathogenic pattern driven by a shared genetic lesion in those cases irrespective of the cell lineage.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (CA98472, T.P.L.; and 2K24HL077522, J.P.M.), Fundacion CajaMadrid (A.J.), the Finnish government (special subsidy for health sciences, research, and training), and the Academy of Finland, the Finnish Cancer Societies, the Sigrid Juselius Foundation, the Finnish Association of Hematology, the National Clinical Graduate School, and the Signe and Ane Gyllenberg Foundation.
Footnotes
There is an Inside Blood commentary on this article in this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: A.J., S.M., T.P.L., and J.P.M. were responsible for overall design, data collection, data analysis, data interpretation, statistical analysis, manuscript preparation, and writing, completion, and final approval of the manuscript; M.J.C., H.M., H.K., F.L., K.P.N., T.O., B.P., M.A., I.G.-S., K.G., L.D., E.D.H., K.M., D.Z., M.W.W., K.P., M.A.S., and A.L. gathered data and edited and approved the final manuscript; and all authors approved the final version of the manuscript and the submission.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jaroslaw P. Maciejewski, Taussig Cancer Institution/R40, 9500 Euclid Ave, Cleveland, OH 44195; e-mail: maciejj@ccf.org.
References
- 1.Loughran TP, Jr, Kadin ME, Starkebaum G, et al. Leukemia of large granular lymphocytes: association with clonal chromosomal abnormalities and autoimmune neutropenia, thrombocytopenia, and hemolytic anemia. Ann Intern Med. 1985;102(2):169–175. doi: 10.7326/0003-4819-102-2-169. [DOI] [PubMed] [Google Scholar]
- 2.Loughran TP., Jr Clonal diseases of large granular lymphocytes. Blood. 1993;82(1):1–14. [PubMed] [Google Scholar]
- 3.Swerdlow SH. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. 4th Ed. Lyon, France: International Agency for Research on Cancer; 2008. International Agency for Research on Cancer, World Health Organization. [Google Scholar]
- 4.Swerdlow SH, Harris NL, editors. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. 4th Ed. Lyon, France: International Agency for Research on Cancer; 2008. [Google Scholar]
- 5.Bareau B, Rey J, Hamidou M, et al. Analysis of a French cohort of patients with large granular lymphocyte leukemia: a report on 229 cases. Haematologica. 2010;95(9):1534–1541. doi: 10.3324/haematol.2009.018481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Epling-Burnette PK, Sokol L, Chen X, et al. Clinical improvement by farnesyltransferase inhibition in NK large granular lymphocyte leukemia associated with imbalanced NK receptor signaling. Blood. 2008;112(12):4694–4698. doi: 10.1182/blood-2008-02-136382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Posnett DN, Sinha R, Kabak S, Russo C. Clonal populations of T cells in normal elderly humans: the T cell equivalent to “benign monoclonal gammapathy.”. J Exp Med. 1994;179(2):609–618. doi: 10.1084/jem.179.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bigouret V, Hoffmann T, Arlettaz L, et al. Monoclonal T-cell expansions in asymptomatic individuals and in patients with large granular leukemia consist of cytotoxic effector T cells expressing the activating CD94:NKG2C/E and NKD2D killer cell receptors. Blood. 2003;101(8):3198–3204. doi: 10.1182/blood-2002-08-2408. [DOI] [PubMed] [Google Scholar]
- 9.Ryan DK, Alexander HD, Morris TC. Routine diagnosis of large granular lymphocytic leukaemia by Southern blot and polymerase chain reaction analysis of clonal T cell receptor gene rearrangement. Mol Pathol. 1997;50(2):77–81. doi: 10.1136/mp.50.2.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zambello R, Falco M, Della Chiesa M, et al. Expression and function of KIR and natural cytotoxicity receptors in NK-type lymphoproliferative diseases of granular lymphocytes. Blood. 2003;102(5):1797–1805. doi: 10.1182/blood-2002-12-3898. [DOI] [PubMed] [Google Scholar]
- 11.Koskela HL, Eldfors S, Ellonen P, et al. Somatic STAT3 mutations in large granular lymphocytic leukemia. N Engl J Med. 2012;366(20):1905–1913. doi: 10.1056/NEJMoa1114885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Semenzato G, Zambello R, Starkebaum G, Oshimi K, Loughran TP., Jr The lymphoproliferative disease of granular lymphocytes: updated criteria for diagnosis. Blood. 1997;89(1):256–260. [PubMed] [Google Scholar]
- 13.Mohan SR, Clemente MJ, Afable M, et al. Therapeutic implications of variable expression of CD52 on clonal cytotoxic T cells in CD8+ large granular lymphocyte leukemia. Haematologica. 2009;94(10):1407–1414. doi: 10.3324/haematol.2009.009191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheson BD, Greenberg PL, Bennett JM, et al. Clinical application and proposal for modification of the International Working Group (IWG) response criteria in myelodysplasia. Blood. 2006;108(2):419–425. doi: 10.1182/blood-2005-10-4149. [DOI] [PubMed] [Google Scholar]
- 15.Langerak AW, van Den Beemd R, Wolvers-Tettero IL, et al. Molecular and flow cytometric analysis of the Vbeta repertoire for clonality assessment in mature TCRalphabeta T-cell proliferations. Blood. 2001;98(1):165–173. doi: 10.1182/blood.v98.1.165. [DOI] [PubMed] [Google Scholar]
- 16.Morice WG, Kimlinger T, Katzmann JA, et al. Flow cytometric assessment of TCR-Vbeta expression in the evaluation of peripheral blood involvement by T-cell lymphoproliferative disorders: a comparison with conventional T-cell immunophenotyping and molecular genetic techniques. Am J Clin Pathol. 2004;121(3):373–383. doi: 10.1309/3A32-DTVM-H640-M2QA. [DOI] [PubMed] [Google Scholar]
- 17.O'Keefe CL, Plasilova M, Wlodarski M, et al. Molecular analysis of TCR clonotypes in LGL: a clonal model for polyclonal responses. J Immunol. 2004;172(3):1960–1969. doi: 10.4049/jimmunol.172.3.1960. [DOI] [PubMed] [Google Scholar]
- 18.Zeng W, Miyazato A, Chen G, Kajigaya S, Young NS, Maciejewski JP. Interferon-gamma-induced gene expression in CD34 cells: identification of pathologic cytokine-specific signature profiles. Blood. 2006;107(1):167–175. doi: 10.1182/blood-2005-05-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wen Z, Zhong Z, Darnell JE., Jr Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell. 1995;82(2):241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 20.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song H, Wang R, Wang S, Lin J. A low-molecular-weight compound discovered through virtual database screening inhibits Stat3 function in breast cancer cells. Proc Natl Acad Sci U S A. 2005;102(13):4700–4705. doi: 10.1073/pnas.0409894102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zambello R, Trentin L, Ciccone E, et al. Phenotypic diversity of natural killer (NK) populations in patients with NK-type lymphoproliferative disease of granular lymphocytes. Blood. 1993;81(9):2381–2385. [PubMed] [Google Scholar]
- 23.Epling-Burnette PK, Painter JS, Chaurasia P, et al. Dysregulated NK receptor expression in patients with lymphoproliferative disease of granular lymphocytes. Blood. 2004;103(9):3431–3439. doi: 10.1182/blood-2003-02-0400. [DOI] [PubMed] [Google Scholar]
- 24.Clemente MJ, Wlodarski MW, Makishima H, et al. Clonal drift demonstrates unexpected dynamics of the T-cell repertoire in T-large granular lymphocyte leukemia. Blood. 2011;118(16):4384–4393. doi: 10.1182/blood-2011-02-338517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grivennikov S, Karin M. Autocrine IL-6 signaling: a key event in tumorigenesis? Cancer Cell. 2008;13(1):7–9. doi: 10.1016/j.ccr.2007.12.020. [DOI] [PubMed] [Google Scholar]
- 26.Sriuranpong V, Park JI, Amornphimoltham P, Patel V, Nelkin BD, Gutkind JS. Epidermal growth factor receptor-independent constitutive activation of STAT3 in head and neck squamous cell carcinoma is mediated by the autocrine/paracrine stimulation of the interleukin 6/gp130 cytokine system. Cancer Res. 2003;63(11):2948–2956. [PubMed] [Google Scholar]
- 27.Yoshikawa H, Matsubara K, Qian GS, et al. SOCS-1, a negative regulator of the JAK/STAT pathway, is silenced by methylation in human hepatocellular carcinoma and shows growth-suppression activity. Nat Genet. 2001;28(1):29–35. doi: 10.1038/ng0501-29. [DOI] [PubMed] [Google Scholar]
- 28.Epling-Burnette PK, Liu JH, Catlett-Falcone R, et al. Inhibition of STAT3 signaling leads to apoptosis of leukemic large granular lymphocytes and decreased Mcl-1 expression. J Clin Invest. 2001;107(3):351–362. doi: 10.1172/JCI9940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holland SM, DeLeo FR, Elloumi HZ, et al. STAT3 mutations in the hyper-IgE syndrome. N Engl J Med. 2007;357(16):1608–1619. doi: 10.1056/NEJMoa073687. [DOI] [PubMed] [Google Scholar]
- 30.Minegishi Y, Saito M, Tsuchiya S, et al. Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature. 2007;448(7157):1058–1062. doi: 10.1038/nature06096. [DOI] [PubMed] [Google Scholar]
- 31.Kumanovics A, Perkins SL, Gilbert H, Cessna MH, Augustine NH, Hill HR. Diffuse large B cell lymphoma in hyper-IgE syndrome due to STAT3 mutation. J Clin Immunol. 2010;30(6):886–893. doi: 10.1007/s10875-010-9452-z. [DOI] [PubMed] [Google Scholar]
- 32.Wallet N, Ghez D, Delarue R, et al. Diffuse large B-cell lymphoma in hyperimmunoglobulinemia E syndrome. Clin Lymphoma Myeloma. 2007;7(6):425–427. doi: 10.3816/CLM.2007.n.022. [DOI] [PubMed] [Google Scholar]
- 33.Starkebaum G. Leukemia of large granular lymphocytes and rheumatoid arthritis. Am J Med. 2000;108(9):744–745. doi: 10.1016/s0002-9343(00)00386-7. [DOI] [PubMed] [Google Scholar]
- 34.Namekawa T, Snyder MR, Yen JH, et al. Killer cell activating receptors function as costimulatory molecules on CD4+CD28null T cells clonally expanded in rheumatoid arthritis. J Immunol. 2000;165(2):1138–1145. doi: 10.4049/jimmunol.165.2.1138. [DOI] [PubMed] [Google Scholar]
- 35.Gulan G, Ravlic-Gulan J, Strbo N, et al. Systemic and local expression of perforin in lymphocyte subsets in acute and chronic rheumatoid arthritis. J Rheumatol. 2003;30(4):660–670. [PubMed] [Google Scholar]
- 36.Korb A, Pavenstadt H, Pap T. Cell death in rheumatoid arthritis. Apoptosis. 2009;14(4):447–454. doi: 10.1007/s10495-009-0317-y. [DOI] [PubMed] [Google Scholar]
- 37.Kim NH, Lee MY, Park SJ, Choi JS, Oh MK, Kim IS. Auranofin blocks interleukin-6 signalling by inhibiting phosphorylation of JAK1 and STAT3. Immunology. 2007;122(4):607–614. doi: 10.1111/j.1365-2567.2007.02679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ward FJ, Hall AM, Cairns LS, et al. Clonal regulatory T cells specific for a red blood cell autoantigen in human autoimmune hemolytic anemia. Blood. 2008;111(2):680–687. doi: 10.1182/blood-2007-07-101345. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.