Abstract
The extract of UIC 10035, a strain obtained from a sample collected near the town of Homestead, south Florida, showed antiproliferative activity against MDA-MB-435 cells. Bioassay-guided fractionation led to the isolation of a series of cyclic lipodecapeptides, named minutissamides E - L (1 – 8). The planar structures were determined by analysis of HRESIMS, tandem MS, and 1D and 2D NMR data, and the stereoconfigurations were assigned by LC-MS analysis of the Marfey's derivatives after acid hydrolysis. Minutissamides E - L (1 – 8) exhibited antiproliferative activity against MDA-MB-435 cells with IC50 values ranging between 1 and 10 μM. The structures of minutissamides E - L (1 – 8) were closely related with those of the previously reported lipopeptides, puwainaphycins A - E and minutissamides A - D, characterized by the presence of a lipophilic -amino acid and three non-standard amino acids NMeAsn, OMeThr and Dhb (, -dehydro- -aminobutyric acid). The strain UIC 10035 was designated as cf. Anabaena sp. on the basis of morphological and 16S rRNA gene sequence analyses.
Keywords: Cyanobactreia, Anabaena, Cyclic lipopeptides, MDA-MB-435, Antiproliferative activity
1. Introduction
Cyclic lipopeptides represent a large subclass of non-ribosomal peptides, which often contain non-standard amino acids such as Dhb (, -dehydro- -aminobutyric acid), -hydroxylated amino acids, and N- and/or O-methylated amino acids.1, 2 This subset of non-ribosomal peptides has shown a wide range of biological activities including antibacterial and antifungal as well as cytotoxic activities, and some of these cyclic lipopeptides are regarded as promising candidates for clinical evaluation.3,4 Mode of action studies of the phytotoxin syringomycin from Pseudomonas syringae and the antibiotic daptomycin from Streptomyces roseosporus indicated that their lipophilic residues play key roles for the biological activities of these molecules.5,6 The presence of lipophilic residues allows these molecules to interact with cell membranes to form transmembrane pores, leading to rapid membrane depolarization by the leakage of intracellular ions, and eventually resulting in cell necrosis.
Cyanobacteria are well-known producers of cyclic lipopeptides.7 A variety of cyclic lipopeptides has been identified from both marine and freshwater cyanobacteria with a wide spectrum of biological activities, including antifungal, cardiotonic and cytotoxic activities.8-12 A recent study done by Hrouzek et al. revealed a possible mode of action of cytotoxic cyanobacterial lipopeptides.13 The elevated concentration of intracellular Ca2+ and subsequent cell necrosis were observed upon addition of puwainaphycins F/G, indicating that these cyanobacterial lipopeptides interact with cell membrane, causing membrane depolarization in a similar manner to the well-known pore-forming lipopeptides, such as daptomycin and syringomycin.
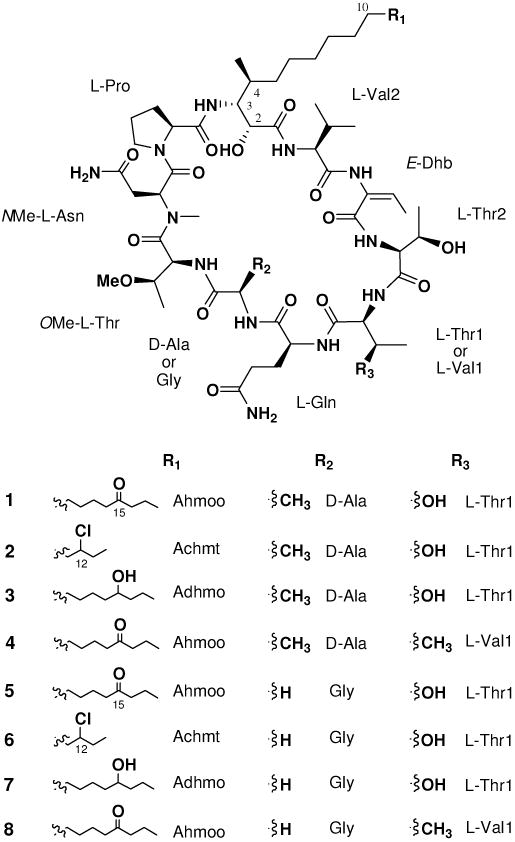
We recently reported four cyclic lipodecapeptides, named minutissamides A - D, possessing a ipophilic -amino acid residue and the nonstandard amino acids NMeAsn and Dhb, from the cultured cyanobacterium Anabaena minutissima UTEX 1613.14 Herein, we report the isolation, structure determination and biological activity of an additional set of cyclic lipodecapeptides, named minutissamides E - L (1 – 8) due to the structural similarity to the previously reported minutissamides A - D. The planar structures were determined by analysis of HRESIMS, tandem MS, and 1D and 2D NMR data, and the stereoconfigurations were assigned by advanced Marfey's analysis after acid hydrolysis. Minutissamides E - L (1 – 8) featured cyclic lipodecapeptide architectures characterized by the presence of one lipophilic -amino acid (octadecanoic or tetradecanoic acids) with 3-amino-2-hydroxy-4-methyl functionality, and three non-standard amino acids NMeAsn, OMeThr and Dhb.
2. Results and Discussion
Cf. Anabaena sp. UIC 10035 was isolated from a sample collected near the town of Homestead, South Florida, 2007, and cultured in an inorganic Z medium.15 The freeze-dried cells (7.5 g from 20 L) were extracted with a mixture of CH2Cl2 and MeOH (1:1, v/v), and dried in vacuo. The resulting extract (0.5 g) was fractionated using Diaion HP-20 resin with an increasing amount of iPrOH in H2O. Fractions eluting at 40 and 60 % aqueous iPrOH (v/v) exhibited antiproliferative activity against MD-MB-435 cells. LC-MS analysis of these fractions indicated the presence of a series of nitrogen-containing compounds with molecular weights ranging between 1150 and 1250 Da. These fractions were combined and subjected to HPLC purification using reversed-phase columns to yield eight cyclic lipodecapeptides, named minutissamides E - L (1 – 8).
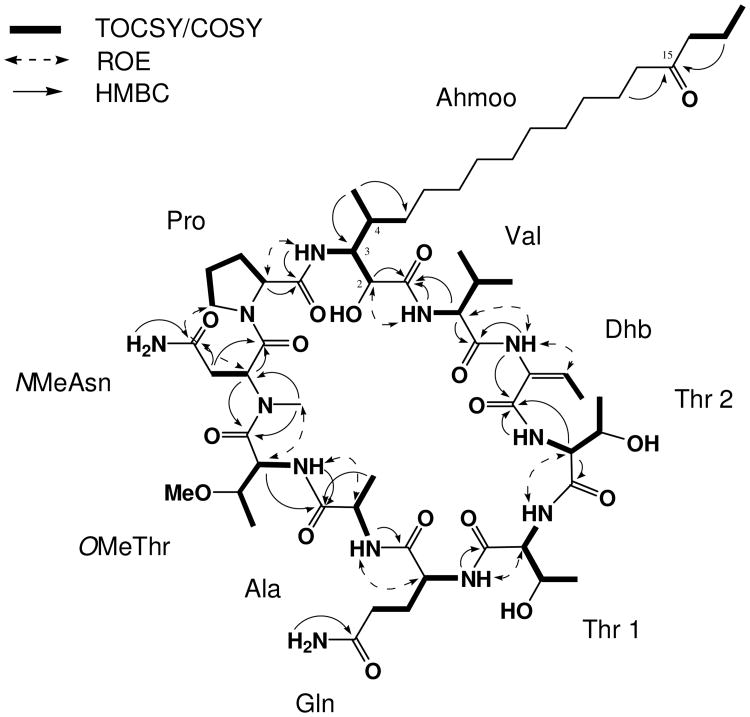
Minutissamide E (1) was obtained as a colorless, amorphous powder. The molecular formula was determined as C59H100N12O17 by HRESIMS analysis. The signal distribution pattern observed in the 1H NMR spectrum (amide NH, H 6.0 – 10.0; amino acid H, H 3.5 – 5.5; largely overlapped aliphatic CH2, H 1.25; aliphatic doublet CH3, H 0.5 – 2.0) suggested this compound to be a lipopeptide. The presence of ten amino acid residues were identified by 2D NMR analysis, including one lipophilic -amino acid, six standard amino acids and three non-standard amino acids. Analysis of the COSY and TOCSY spectra established the structures of six standard amino acids as Pro, Ala, Gln, Thr (2×) and Val. The structures of three non-standard amino acids were determined as NMeAsn, OMeThr and Dhb by interpretation of the COSY, HSQC and HMBC spectra (Figure 1). The structure of the -amino acid residue was also determined by analysis of 2D NMR data as described below. COSY correlations between NH (H 6.77) and H-3 (H 3.92), and between a doublet methyl (H 0.57) and H-4 (H 1.68), as well as sequential COSY correlations of H-2/H-3/H-4/H2-5, which in turn coupled with the overlapped methylene signals at H 1.25, indicated the presence of a 3-amino-2-hydroxy-4-methyl functionality coupled to a lipid portion (Figure 1). The isolated TOCSY correlation fragment of H2-16/H2-17/H3-18 and HMBC correlations from H2-17 (H 1.47) to a ketone carbon (C 210.6) indicated that a ketone was positioned at C-15, determining the structure of the -amino acid residue as Ahmoo (3-amino-2-hydroxy-4-methyl- 15-oxooctadecanoic acid).
Figure 1.
Key 2D correlations of minutissamide E (1) used for the determination of the planar structure of 1.
The established ten amino acid residues were connected by combined analysis of the HMBC and ROESY spectra (Figure 1). Starting from the Ahmoo residue, HMBC correlations from Ahmoo NH to Pro C-1 (C 171.2) and from N-Me (H 2.93) to NMeAsn C-2 (C 49.7) and OMeThr C-1 (C 169.6), combined with NOE correlations between Ahmoo NH (H 6.77) and Pro H-2 (H 4.25), between Pro H2-5 (H 3.11 and 4.21) and NMeAsn H-2 (H 5.52), established a partial sequence of Ahmoo-Pro-NMeAsn-OMeThr. This sequence was further expanded into Ahmoo-Pro-NMeAsn-OMeThr-Ala-Gln-Thr1 by HMBC correlations from OMeThr NH (H 6.74) to Ala C-1 (C 171.9), from Ala NH (H 7.58) to Gln C-1 (C 171.1) and from Gln NH (H 7.25) to Thr1 C-1 (C 170.4), as well as NOE correlations between OMeThr NH (H 6.74) and Ala H-2 (H 4.19), between Ala NH (H 7.58) and Gln H-2 (H 4.08), and between Gln NH (H 7.25) and Thr1 H-2 (H 3.90). The complete sequence of Ahmoo-Pro-NMeAsn-OMeThr-Ala-Gln-Thr1-Thr2-Dhb-Val was established by HMBC correlations from Thr2 NH (H 8.37) to Dhb C-1 (H 163.9) and from Dhb NH to Val C-1 (C 168.8), together with NOE correlations between Thr1 NH (H 8.84) and Thr2 H-2 (H 5.02), and between Dhb NH (H 9.08) and Val H-2 (H 4.31). Lastly, HMBC correlations from Val NH (H 6.85) and Val H-2 (H 4.31) to Ahmoo C-1 (C 169.6), and a NOE correlation observed between Val NH and Ahmoo H-2, closed the ring, completing the cyclic lipodecapeptide structure of 1.
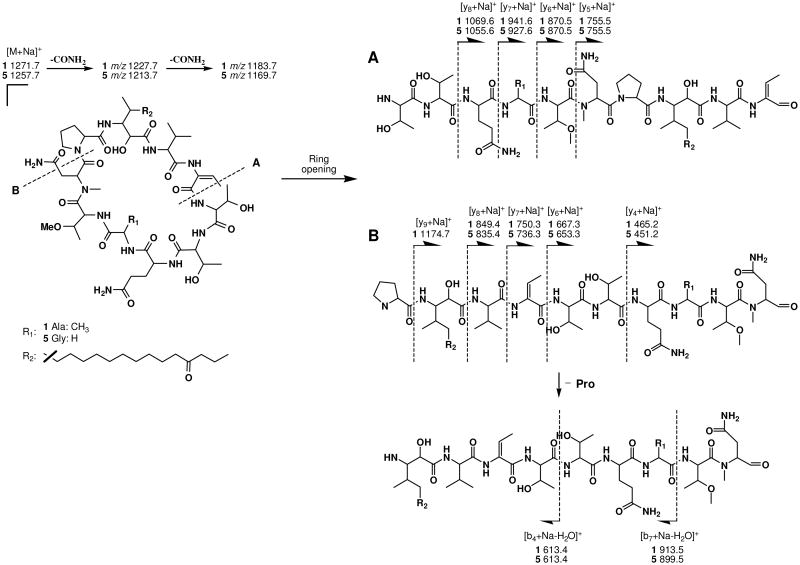
To confirm the amino acid sequence of 1, MS fragmentation analysis was carried out using quadrupole ion trap CID (collision-induced dissociation) MS/MS data (Figure 2). In the MS1 analysis, the most intense peak corresponded to the sodium adduct ion of 1 (m/z 1271.7). CID fragmentation of this sodiated precursor ion generated a set of y-type fragment ions instead of b-type, which are commonly observed in CID fragmentation of protonated precursor ions. This predominant formation of y-type fragments by CID of sodiated precursor ions has been previously reported. The two most intense ions at m/z 1227.7 and 1183.7, corresponding to [M + Na - CONH2]+ and [M + Na - 2CONH2]+, respectively, resulted from the sequential loss of the side chain amides from NMeAsn and Gln. Ring-opening at two different sites yielded two sets of y-type fragment ions (Figure 2). Ring-opening (A) between Dhb and Thr yielded ions at m/z 755.5, 870.5, 941.6 and 1069.6, corresponding to y5, y6, y7 and y8 fragments, respectively (Figure 2A). The second set of ions, resulted from ring-opening) between NMeAsn and Pro, included m/z 465.2, 667.3, 750.3, 849.4 and 1174.7, corresponding to y4, y6, y7, y8 and y9 fragments, respectively (Figure 2B). This fragmentation pattern agreed well with the sequence determined by 2D NMR analysis, thus further confirming the amino acid sequence of 1.
Figure 2.
ESI-MS/MS fragmentation of minutissamides E and I (1 and 5).
The stereoconfiguration of 1, including the geometric configuration of Dhb and the absolute configurations of - and -amino acids, was determined on the basis of NOE correlations and the advanced Marfey's method as well as comparison of 1H and 13C NMR chemical shifts. A strong NOE correlation, observed between Dhb NH and Dhb H-3, determined E-geometry for the Dhb residue. For the assignment of amino acid configurations, advance Marfey's analysis was carried out as previously described.17-19 LC-MS comparison between l-FDLA and dl-FDLA derivatives of the acid hydrolysate of 1 assigned l-configuration to Pro, Gln, and Val, and d-configuration to Ala. The l-FDLA derivative of the acid hydrolysate of 1 was further compared with those of authentic standards of Thr (l-Thr, d-Thr, l-allo-Thr and d-allo-Thr), NMeAsn (NMe-l-Asn and NMe-d-Asn) and OMeThr (OMe-l-Thr, OMe-d-Thr, OMe-l-allo-Thr and OMe-d-allo-Thr), assigning l-configuration to these amino acids. Minutissamide E (1) contained the same asymmetric centers (3-amino2-hydroxy-4-methyl) in the -amino acid residue as the previously described puwainaphycins and minutissamides A - D.9,14 These three consecutive stereocenters in Ahmoo showed nearly identical 1H (< 0.1 ppm) and 13C (< 1 ppm) NMR chemical shifts to those of the puwainaphycins and minutissamides A - D. In addition, a small 3JHH between H-2 and H-3 (4.8 Hz) and a large 3JHH between H-3 and H-4 (10.8 Hz) observed in Ahmoo of 1 were similar to those of the puwainaphycins (5.5 Hz and 11.3 Hz) and minutissamides A - D (5.6 Hz and 11.0 Hz). This suggested the relative configuration of Ahmoo in 1 to be the same as those found in the puwainaphycins and minutissamides A - D. The absolute configuration of Ahmoo was then determined using the advanced Marfey's method as previously described.14, 19 In LC-MS analysis, the dl-FDLA derivative of the acid hydrolysate of 1 exhibited two peaks at 67.0 and 72.2 min, corresponding to the molecular weight of the FDLA derivative of Ahmoo (m/z 638.3 [M + H]+), while the l-FDLA derivative resulted in one peak at 72.2 min. This result was consistent with those reported for the -amino acids Ahda (3-amino-2-hydroxydecanoic acid) in microginin19 and Hamd (3-amino-2-hydroxy-4-methyldodecanoic acid) in minutissamide A,14 suggesting the same absolute configuration (3R) in Ahmoo. Taken together, the absolute configuration of the three consecutive stereocenters (C-2, C-3 and C-4) in Ahmoo was assigned as 2R,3R,4S, completing the stereoconfiguration of 1.
Minutissamide F (2) was obtained as a colorless, amorphous powder. The molecular formula was determined as C55H93ClN12O16 by HRESIMS, which exhibited a molecular ion peak at m/z 1213.6645 ([M + H]+) and a M + 2 isotope peak. Comparison of the 1D and 2D NMR spectra of 2 and 1 revealed that compound 2 shared the same amino acid sequence as 1 and differed only in the -amino acid side chain. The four carbon differences in the molecular formula between 2 and 1 suggested that the -amino acid residue of 2 was composed of 14 carbons (tetradecanoic acid) instead of 18 carbons. The down-field proton signal of H-12 (H 3.99), as well as sequential COSY correlations from the terminal methyl H3-14 (H 0.96) to H-12 via one diastereotopic methylene H2-13 (H 1.64 and 1.78) and an HMBC correlation from H3-14 to C-12 (C 66.4), positioned the chlorine at C-12, establishing the structure of the -amino acid residue as Achmt (3-amino-12-chloro-2-hydroxy-4-methyltetradecanoic acid). Compound 2 showed nearly identical 1H and 13C NMR chemical shifts to those of 1 at all of the stereocenters, and the negative specific rotation ([ ] D -19) was also similar to that of 1 ([ ] D -29), thus indicating the stereoconfiguration of the stereocenters of 2 to be the same as those observed for 1. The absolute configuration of the additional chlorine-bearing stereocenter (C-12), located distant from other stereocenters in Achmt, was not assigned.
Minutissamide G (3) was obtained as a colorless, amorphous powder. The HRESIMS spectrum of 3 displayed a molecular ion at m/z 1251.7564 ([M + H]+), suggesting the molecular formula as C59H102N12O17, which was two mass units higher than that of 1. The 1H NMR spectrum of 3 also closely resembled that of 1, except for in the -amino acid residue. The difference was the up-field shift of signals for H2-14 (H 1.26 and 1.30) and H2-16 (H 1.28) in the -amino acid residue as compared to those of 1. These two methylene signals were connected by the oxygenated methine proton H-15 (H 3.35) in the COSY spectrum, indicating the structure of the -amino acid in 3 to be Adhmo (3-amino-2,15-dihydroxy4-methyloctadecanoic acid). The position of a hydroxy group at C-15 was further supported by an HMBC correlation from the terminal methyl proton signal of H3-18 (H 0.86) to the down-fielded carbon signal of C-16 (C 39.9, the -carbon of a hydroxy group). The stereoconfiguration of 3 was suggested to be the same as that of 1, based on the nearly identical NMR chemical shifts, observed for all the stereocenters. In addition, compound 3 showed the negative specific rotation ([ ]D -22) similar to that of 1 ([ ] D -29). The assignment of the absolute configuration for the additional hydroxy-bearing carbinol stereocenter (C-15) in Adhmo was not attempted due to the limited amount of sample available.
Minutissamide H (4) was obtained as a colorless, amorphous powder. The molecular formula was deduced as C60H102N12O16 by HRESIMS analysis (m/z 1247.7622 [M + H]+). Detailed analysis of the COSY and TOCSY spectra suggested that the structure of 4 was similar to that of 1, but differed by one amino acid in the cyclic peptide core. In the TOCSY spectra, a new spin system, corresponding to the amino acid Val, replaced the spin system of Thr1, indicating the presence of Val1 instead of Thr1. The amino acid configurations of 4 were determined by advanced Marfey's analysis as described for 1. LC-MS comparison between the l- and dl-FDLA derivatives of the acid hydrolysate of 4 assigned the l-configuration to Val. The result of advanced Marfey's analysis also suggested that the absolute configurations of the rest of the other amino acids in 4 are the same as those found for 1.
Minutissamide I (5), a colorless, amorphous powder, displayed a molecular ion peak at m/z 1235.7328 ([M + H]+) in HRESIMS analysis, indicating a molecular formula of C58H98N12O17. The molecular weight of 5 was smaller than that of 1 by 14 mass units and identical to that of the previously reported cyclic lipodecapeptide, puwainaphycin A.9 Detailed analysis of the 2D NMR spectra revealed that the structure of 5 differs from that of 1 by the presence of Gly instead of Ala, thus sharing the same cyclicpeptide core as puwainaphycin A. The only difference found between 5 and puwainaphycin A was in the position of the ketone in the -amino acid residue Ahmoo. An isolated spin system, composed of the proton signals of H3-18 (H 0.83), H2-17 (H 1.46) and H2-16 (H 2.36), was found in the TOCSY spectrum. This, together with HMBC correlations from H3-18 and H2-17 to C-15 (C 210.9), indicated the ketone to be positioned at C-15 in 5 instead of at C-14 as found for puwainaphycin A. The amino acid sequence of 5 was further confirmed by analysis of tandem MS data, which showed a nearly identical fragmentation pattern to that of 1 (Figure 2). Lastly, advanced Marfey's analysis of the acid hydrolysate of 5, as well as 1H and 13C NMR data, suggested the absolute configurations of all of the amino acids to be the same as those reported for puwainaphycin A.
Minutissamide J (6) was also obtained as a colorless, amorphous powder. The HRESIMS spectrum of 6 displayed a molecular ion peak at m/z 1199.6477 ([M + H]+) as well as a M + 2 isotope peak, indicating the presence of a chlorine, thus the molecular formula of 6 was deduced as C54H91ClN12O16. Analysis of 1D and 2D NMR data of 6 revealed that the structure of 6 is similar to that of 5 except for the -amino acid residue. The 36 Da mass difference between 6 and 5 and the presence of a chlorine instead of an oxygen indicated the structure of the -amino acid residue in 6 to be Achmt, the same as found in 2. The chlorine was placed at C-12 by an HMBC correlation from the terminal methyl protons H3-14 (H 0.96) to the chlorinated carbon C-12 (C 66.4). The stereoconfiguration of 6 was considered to be the same as that found for 5 based on the nearly identical 1H and 13C NMR chemical shifts at all of the stereocenters and the negative specific rotation ([ ]D -19) similar to that of 5 ([ ]D -26). The absolute configuration of the additional chlorine-bearing stereocenter (C-12) in Achmt of 6 was not assigned.
The last two compounds, minutissamides K (7) and L (8), were obtained as a mixture in a ratio of 3:5 as determined by 1H NMR analysis. The HRESIMS spectrum of the mixture showed the two molecular ion peaks at m/z 1237.7381 (7, [M + H]+) and m/z 1233.7482 (8, [M + H]+), suggesting the molecular formulas as C58H100N12O17 and C59H100N12O16, respectively. Attempts to separate 7 and 8 using reversed-phase HPLC failed due to complete overlap of the two peaks, thus structure determination was carried out using the mixture. The majority of signals in the 1H NMR spectrum appeared to be identical between 7 and 8, except for one amino acid and the -amino acid residue. In the TOCSY spectrum, two doubled NH (H 9.03 and H 8.67) and -H (H 3.80 and H 3.82) signals, the sum of which showed the same integration as other signals, indicating thatthese signals belong to different spin systems, Thr1 and Val1. In addition, analysis of the COSY and HMBC spectra identified the presence of the two -amino acids, Adhmo and Ahmoo. The molecular weight and the integration ratio suggested the presence of Thr1 and Adhmo in 7, and Val1 and Ahmoo in 8. Advanced Marfey's analysis of the acid hydrolysate of the mixture assigned the l-configuration to all of the amino acids present in 7 and 8. Nearly identical 1H and 13C NMR data and of stereogenic centers (C2, C3 and C4) in the -amino acid residue to those of other compounds (1 - 6) as well as minutissamides A - D and the puwainaphycins and the negative specific rotation ([ ]D -26) suggested the absolute configuration of the -amino acid residue for both 7 and 8 to be the same (2R,3R,4S). The determination of the absolute configuration at C-15 in Adhmo was not attempted in this study due to the limited amount of sample available.
Cf. Anabaena sp. UIC 10035 produced several cyclic lipodecapeptides in a laboratory culture that showed close structural relationships with the previously reported cyclic lipodecapeptides, puwainaphycins A - E and minutissamides A - D isolated from Anabaena sp. BQ-16-1 and Anabaena minutissima UTEX 1613, respectively.9,14 However, minutissamides E - L showed higher structural diversity than the puwainaphycins and minutissamides A - D. This high structural diversity of cylic lipodecapeptides produced by the strain UIC 10035 is likely derived from its biosynthetic flexibility to incorporate two different amino acids (Ala/Gly and Thr1/Val1) by adenylation domains in two NRPS modules, and two different fatty acids (probably hexadecanoic acid/dodecanoic acid) by an acyl ligase in a PKS loading module followed by variable modifications including oxidation, chlorination and hydroxylation. As observed for the adenylation domain in anabaenapeptin biosynthesis,20 minutissamides E - L (1 - 8) produced by the strain UIC 10035 might also represent a good example of nature's strategy to diversify their structures by substrate promiscuity.
Minutissamides E - L (1 - 8) were evaluated for their antiproliferative activity against the MDA-MB-435 human melanoma cancer cell line. All of the compounds exhibited similar levels of activity with IC50 values ranging between 1 and 10 μM (1, 2.9 μM; 2, 1.2 μM; 3, 9.9 μM; 4, 1.1 μM; 5, 2.9 μM; 6, 2.6 μM; the mixture of 7 and 8, 2.9 μg/ml). Compounds 1 - 8 were also evaluated for antibacterial activity against Mycobacterium (M. smegmatis), Gram-positive (S. aureus, E. faecalis and S. pneumoniae) and Gram-negative (A. baumannii, E. coli and P. aeruginosa) bacteria, but showed no activity at the highest concentrations tested (10 μg/ml).
Taxonomic identification of UIC 10035 was established on the basis of morphological observation and phylogenetic analysis using a partial 16S rRNA gene sequence (1.2 Kb). The strain UIC 10035 was filamentous and developed heterocysts in nitrogen-deficient media, indicating it belonging to the order Nostocales. Long straight and yellow-green colored trichomes and the lack of mucilage indicated UIC 10035 potentially being of the genus Anabaena. However, the partial 16S rRNA gene sequence of UIC 10035 showed the highest sequence homology to those of Nostoc sp. PCC8112 (99.9 %), Nostoc sp. PCC8976 (98.5 %) and Nostoc elgonense TH3S05 (97.8 %). In the phylogenetic tree constructed using cyanobacterial 16S rRNA gene sequences (Figure 3), the monophyletic clade, containing the UIC 10035 strain and three Nostoc spp., was not clustered with any of the genus reference strains for either Nostoc or Anabaena, complicating the taxonomic assignment of the UIC 10035 strain. Pairwise distance analysis between clades indicated that this clade is the most closely related with the clade containing the strain Anabaena minutissima UTEX 1613 (0.03), the producer of minutissamides A - D, and the clade of the genus Anabaenopsis (0.03). Based on the morphological similarity and close phylogenetic relationship to the strain Anabaena minutissima UTEX 1613, the UIC 10035 strain was designated as a cf. Anabaena sp.
Figure 3.
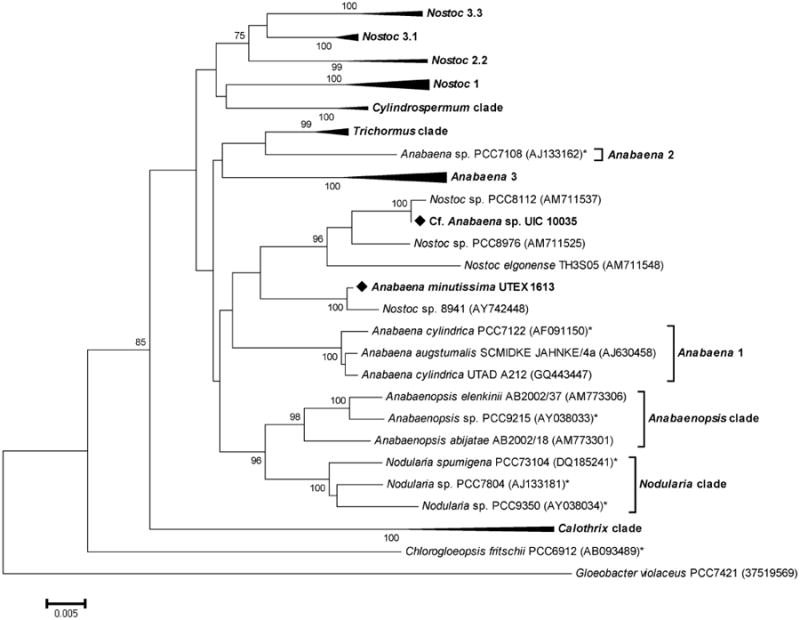
Phylogenetic relationships of 16S rRNA genes from cyanobacteria. Evolutionary distances were determined using the minimum evolution method with 1,000 replicate bootstrap re-samplings to construct the phylogenetic tree. Strains denoted with an asterisk (*) are “Bergey's” reference strains. Strains were obtained from NCBI with the accession number given in parentheses. Only bootstrap values greater than or equal to 75% are displayed. The complete phylogenetic tree can be found in the supplementary data.
3. Conclusion
Chemical investigation of the UIC 10035 strain, guided by antiproliferative assay against MDA-MB-435 cells, led to the isolation of eight antiproliferative cyclic lipodecapeptides, named minutissamides E - L (1 – 8). The planar structures were determined by analysis of the HRESIMS, tandem MS, and 1D and 2D NMR data, and the advanced Marfey's method was used to assign the stereoconfigurations. Minutissamides E - L (1 – 8) showed close structural relationships with the puwainaphycins and minutissamides A - D, characterized by the cyclic lipodecapeptide architectures containing the lipophilic -amino acid possessing 3-amino-2-hydroxy-4-methyl functionality, and three non-standard amino acids including NMeAsn, OMeThr and Dhb. Minutissamides E - L (1 – 8) exhibited antiproliferative activity against MDA-MB-435 cells with low micromolar IC50 values ranging between 1 and 10 μM. The strain UIC 10035 was designated as cf. Anabaena sp. on the basis of morphological and 16S rRNA gene sequence analyses.
4. Experimental Section
4.1 General Experimental Procedures
Optical rotations were measured using a Perkin-Elmer 241 polarimeter. UV and IR spectra were recorded on a Shimadzu UV-1700 spectrometer and a Thermo Nicolet 6700 FT-IR spectrometer, respectively. 1D and 2D NMR spectra were obtained on a Bruker Avance DRX 600 MHz NMR spectrometer with a 5 mm CPTXI Z-gradient probe and a Bruker Avance II 900 MHz NMR spectrometer with a 5 mm ATM CPTCI Z-gradient probe. 1H and 13C NMR chemical shifts were referenced to the DMSO-d6 solvent signals (H 2.50 and C 39.51, respectively). A mixing time of 80 ms was set for the TOCSY experiments and 200 ms for the T-ROESY experiments. The HMBC spectra were recorded with the 3JC-H set to 8 Hz, and the HSQC spectra were collected with the 1JC-H set to145 Hz. High-resolution ESI-MS and tandem MS spectra were acquired using Shimadzu IT-TOF and Thermo Finnigan LTQ FT mass spectrometers, respectively.
4.2 Biological Material
The strain UIC 10035 was isolated from the sample collected near the town of Homestead, South Florida, in 2007 (N 25°24.2′, W 80°33.4′). The strain was cultured in two 20 L glass flasks each containing 18 L of Z media15 under sterile aeration. Cultures were illuminated with fluorescent lamps at 1.03 klx with 18/6 light/dark cycle. The temperature of the culture was maintained at 22 °C. After 6 weeks, the biomass of cyanobacteria was harvested by centrifugation and freeze-dried.
4.3 Morphological and Phylogenetic Analyses for Taxonomic Identification
Morphological studies were performed using the cultivated cyanobacterium UIC 10035 (see Supplementary Data). Taxonomic identification of the cyanobacterial specimen was made in accordance with the modern taxonomic system.21 Genomic DNA of the strain UIC 10035 was extracted using the Wizard Genomic DNA purification kit (Promega) according to the manufacturer's protocol. PCR amplification and sequencing of the partial 16S rRNA gene sequence was carried out as previously described22 using the cyanobacterial specific primers 106F and 1509R.23 The resulting sequence was deposited in the NCBI GenBank under the accession number JX023442. Phylogenetic analysis was conducted using MEGA 5.0.23 The sequence of UIC 10035 was aligned with those of other cyanobacteria belonging to the order Nostocales using ClustalW with standard gap opening and extension penalties. The evolutionary history was inferred using the neighbor-joining, minimum evolution and maximum parsimony methods, which showed nearly identical topology with similar bootstrap values.
4.4 Extraction and Isolation
The freeze-dried biomass (7.5 g) from 36 L (2×18 L) of culture was harvested and extracted with CH2Cl2-MeOH (1:1, v/v) to yield an extract (458.7 mg). The resulting extract was fractionated using Diaion HP-20 with increasing amounts of iPrOH in H2O to generate eight sub-fractions (0, 20, 40, 60, 70, 80, 90, 100 % iPrOH in H2O, v/v). Fractions eluting at 40 and 60 % iPrOH (v/v) exhibited antiproliferative activity against MDA-MB-435 cells (95 % and 100% at 25 μg/ml, respectively). LC- MS dereplication of these fractions indicated the presence of a series of nitrogen-containing compounds with molecular weights, ranging between 1,150 and 1,250 Da. Reversed-phase HPLC of these fractions using C8 column (Varian 10 mm × 250 mm, 3 mL/min) with a linear gradient from 60 to 80% aqueous MeOH (v/v) for 50 min yielded three sub-fractions and two pure compounds, minutissamides F (2, 1.1 mg) and I (5, 1.7 mg). The resulting sub-fractions were re-purified by reversed-phase HPLC using C18 column with the same gradient condition as described above to yield minutissamides E (1, 1.3 mg), G (3, 0.5 mg), H (4, 0.6 mg) and J (6, 0.7 mg), and the mixture (1.1 mg) of minutissamides K (7) and L (8).
4.4.1 Minutissamide E (1)
Colorless, amorphous powder; [ ]25D -29 (c 0.09, MeOH); UV (MeOH) max (log) 203 (4.52), 234 (3.88) nm; IR (neat) 3325 (br), 2930, 2855, 1662 (br), 1535 cm−1; 1H and 13C NMR (see Table 1); HRESIMS m/z 1249.7411 [M + H]+ (calcd for C59H101N12O17, 1249.7408)
Table 1. NMR Spectroscopic data for minutissamides E -H (1 – 4) in DMSO-d6.
| minutissamide E (1)e | minutissamide F (2) | minutissamide G (3) | minutissamide H (4) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C a | H b | mult. (J in Hz) | C a | H b | mult. (J in Hz) | C d | H c | mult. (J in Hz) | C d | H c | mult. (J in Hz) | |||
| Ahmoo, Achmt or Adhmo | 1 | 169.6 | 169.6 | 169.6 | 169.5 | |||||||||
| 2 | 69.6 | 4.17 | d (4.8) | 69.6 | 4.17 | d (4.8) | 69.6 | 4.17 | d (4.8) | 69.6 | 4.16 | d (4.8) | ||
| 3 | 56.0 | 3.92 | td (10.8, 4.8) | 55.9 | 3.94 | td (10.8, 4.8) | 56.0 | 3.94 | td (10.8, 4.8) | 56.0 | 3.94 | td (10.8, 4.8) | ||
| 4 | 32.2 | 1.68 | m | 32.2 | 1.68 | m | 32.2 | 1.68 | m | 32.2 | 1.68 | m | ||
| 5 | 33.4 | 1.17 | m | 33.4 | 1.17 | m | 33.4 | 1.17 | m | 33.4 | 1.17 | m | ||
| 1.62 | m | 1.62 | m | 1.61 | m | 1.62 | m | |||||||
| 6 | 25.5 | 1.17 | m | 25.5 | 1.17 | m | 25.5 | 1.17 | m | 25.5 | 1.17 | m | ||
| 1.25 | m | 1.26 | m | 1.25 | m | 1.25 | m | |||||||
| 7 | 29.7 | 1.25 | m | 29.7 | 1.25 | m | 29.7 | 1.25 | m | 29.7 | 1.25 | m | ||
| 8 | 29.3 | 1.25 | m | 29.1 | 1.25 | m | 29.3 | 1.25 | m | 29.3 | 1.25 | m | ||
| 9 | 29.1 | 1.25 | m | 28.6 | 1.25 | m | 29.1 | 1.25 | m | 29.1 | 1.25 | m | ||
| 10 | 29.0 | 1.25 | m | 26.1 | 1.36 | m | 29.0 | 1.25 | m | 29.0 | 1.25 | m | ||
| 1.46 | m | |||||||||||||
| 11 | 28.9 | 1.25 | m | 37.5 | 1.63 | m | 28.9 | 1.25 | m | 28.9 | 1.25 | m | ||
| 1.73 | m | |||||||||||||
| 12 | 28.6 | 1.25 | m | 66.4 | 3.99 | m | 28.6 | 1.25 | m | 28.6 | 1.25 | m | ||
| 13 | 23.3 | 1.44 | m | 30.9 | 1.64 | m | 30.8 | 1.25 | m | 23.3 | 1.44 | m | ||
| 1.78 | m | |||||||||||||
| 14 | 41.9 | 2.37 | t (7.2) | 10.8 | 0.96 | t (7.2) | 37.7 | 1.26 | m | 41.9 | 2.37 | t (7.2) | ||
| 1.30 | m | |||||||||||||
| 15 | 210.6 | 69.2 | 3.35 | m | 210.6 | |||||||||
| 16 | 43.8 | 2.36 | t (7.2) | 39.9 | 1.28 | m | 43.8 | 2.36 | t (7.2) | |||||
| 17 | 16.7 | 1.47 | sf (7.2) | 18.9 | 1.26 | m | 16.7 | 1.47 | sf (7.2) | |||||
| 18 | 13.6 | 0.83 | t (7.2) | 14.5 | 0.86 | t (7.2) | 13.6 | 0.83 | t (7.2) | |||||
| 3-NH | 6.77 | d (10.8) | 6.77 | d (10.8) | 6.78 | d (10.2) | 6.76 | d (10.7) | ||||||
| 4-Me | 16.0 | 0.57 | d (6.6) | 16.0 | 0.58 | d (6.6) | 16.0 | 0.58 | d (6.6) | 16.0 | 0.57 | d (6.6) | ||
| Pro | 1 | 171.2 | 171.2 | 171.2 | 171.2 | |||||||||
| 2 | 59.9 | 4.25 | dd (8.4, 2.4) | 56.0 | 4.25 | dd (8.4, 2.4) | 56.0 | 4.26 | dd (7.8, 2.4) | 59.8 | 4.26 | dd (7.8) | ||
| 3 | 30.1 | 1.94 | m | 30.1 | 1.94 | m | 30.1 | 1.94 | m | 30.1 | 1.94 | m | ||
| 1.99 | m | 1.99 | m | 1.99 | m | 1.99 | m | |||||||
| 4 | 23.4 | 1.71 | m | 23.4 | 1.71 | m | 23.4 | 1.71 | m | 23.3 | 1.70 | m | ||
| 1.84 | m | 1.84 | m | 1.84 | m | 1.85 | m | |||||||
| 5 | 46.7 | 3.11 | m | 46.7 | 3.10 | m | 46.7 | 3.11 | m | 46.7 | 3.10 | m | ||
| 4.21 | m | 4.20 | m | 4.21 | m | 4.21 | m | |||||||
| NMeAsn | 1 | 167.4 | 167.5 | 167.4 | 167.4 | |||||||||
| 2 | 49.7 | 5.52 | dd (11.4, 3.6) | 49.7 | 5.52 | dd (12.0, 3.0) | 49.7 | 5.53 | dd (12.0, 3.0) | 49.7 | 5.52 | dd (11.4, 3.6) | ||
| 3 | 33.7 | 1.97 | overlapped | 33.7 | 1.97 | overlapped | 33.7 | 1.97 | overlapped | 33.7 | 1.97 | overlapped | ||
| 3.00 | dd (15.6, 12.0) | 3.00 | dd (15.6, 12.0) | 3.00 | dd (5.6. 12.0) | 2.99 | dd (16.0, 11.9) | |||||||
| 4 | 171.5 | 171.5 | 171.5 | 171.5 | ||||||||||
| N-Me | 30.5 | 2.93 | s | 30.5 | 2.93 | s | 30.5 | 2.93 | s | 30.5 | 2.92 | s | ||
| NH2 | 5.99 | s | 6.00 | s | 6.01 | s | 6.02 | s | ||||||
| 7.49 | s | 7.49 | s | 7.50 | s | 7.49 | s | |||||||
| OMeThr | 1 | 169.6 | 169.6 | 169.6 | 169.5 | |||||||||
| 2 | 52.6 | 4.79 | dd (9.0, 1.8) | 52.6 | 4.79 | dd (9.6, 2.4) | 52.6 | 4.80 | dd (9.6, 1.8) | 52.6 | 4.80 | dd (9.2, 1.4) | ||
| 3 | 75.1 | 3.71 | qd (6.0, 1.8) | 75.1 | 3.71 | qd (6.0, 2.4) | 75.1 | 3.72 | qd (6.0, 1.8) | 75.0 | 3.72 | qd (6.0, 1.4) | ||
| 4 | 14.7 | 0.95 | d (6.0) | 14.7 | 0.95 | d (6.0) | 14.7 | 0.96 | d (6.0) | 14.6 | 0.95 | d (6.0) | ||
| O-Me | 55.6 | 3.13 | s | 55.6 | 3.13 | s | 55.6 | 3.14 | s | 55.6 | 3.13 | s | ||
| NH | 6.74 | d (9.0) | 6.74 | d (9.6) | 6.75 | d (9.6) | 6.78 | overlapped | ||||||
| Ala | 1 | 171.9 | 171.9 | 171.9 | 171.8 | |||||||||
| 2 | 49.1 | 4.19 | p (7.8) | 49.1 | 4.19 | p (7.8) | 49.1 | 4.20 | p (7.8) | 49.1 | 4.20 | p (7.8) | ||
| 3 | 16.3 | 1.29 | d (7.8) | 16.3 | 1.29 | d (7.8) | 16.3 | 1.29 | d (7.8) | 16.3 | 1.29 | d (7.8) | ||
| NH | 7.58 | d (7.8) | 7.58 | d (7.8) | 7.59 | d (7.8) | 7.64 | d (7.8) | ||||||
| Gln | 1 | 171.1 | 171.1 | 171.1 | 170.6 | |||||||||
| 2 | 52.9 | 4.08 | m | 52.9 | 4.08 | m | 52.9 | 4.08 | m | 53.2 | 4.07 | m | ||
| 3 | 26.5 | 1.78 | m | 26.5 | 1.77 | m | 26.5 | 1.78 | m | 26.1 | 1.81 | m | ||
| 2.03 | m | 2.03 | m | 2.03 | m | 2.02 | m | |||||||
| 4 | 31.8 | 2.15 | m | 31.8 | 2.14 | m | 31.8 | 2.15 | m | 31.7 | 2.15 | m | ||
| 2.19 | m | |||||||||||||
| 5 | 173.9 | 173.9 | 173.9 | 173.6 | ||||||||||
| NH | 7.25 | d (9.0) | 7.24 | 7.26 | d (7.8) | 7.41 | d (8.0) | |||||||
| NH2 | 6.80 | s | 6.79 | s | 6.80 | s | 6.77 | s | ||||||
| 7.27 | s | 7.27 | s | 7.28 | s | 7.34 | s | |||||||
| Thr1 or Val1 | 1 | 170.4 | 170.5 | 170.4 | NAd | |||||||||
| 2 | 61.3 | 3.90 | m | 61.3 | 3.90 | dd (4.2, 3.0) | 61.3 | 3.91 | m | 60.5 | 3.91 | t (3.9) | ||
| 3 | 65.2 | 4.17 | overlapped | 65.2 | 4.18 | overlapped | 65.2 | 4.17 | overlapped | 28.8 | 2.20 | m | ||
| 4 | 20.6 | 1.25 | overlapped | 20.6 | 1.25 | d (6.0) | 20.6 | 1.25 | overlapped | 17.6 | 0.98 | d (6.8) | ||
| 4′ | 19.0 | 1.03 | d (6.8) | |||||||||||
| NH | 8.84 | brs | 8.96 | brs | 8.83 | brs | 8.54 | brs | ||||||
| Thr2 | 1 | 174.2 | 174.2 | 174.2 | 174.4 | |||||||||
| 2 | 56.6 | 5.02 | dd (10.2, 2.4) | 56.6 | 5.02 | dd (9.6, 2.4) | 56.6 | 5.03 | brd (11.4) | 56.6 | 4.99 | dd (9.7, 1.9) | ||
| 3 | 70.3 | 4.57 | brm | 70.3 | 14.58 | brm | 70.3 | 4.58 | brm | 70.2 | 4.55 | brm | ||
| 4 | 19.0 | 1.25 | overlapped | 19.0 | 1.25 | d (6.0) | 19.0 | 1.25 | overlapped | 19.1 | 1.25 | overlapped | ||
| NH | 8.37 | d (10.2) | 8.37 | d (9.6) | 8.38 | d (10.2) | 8.42 | d (9.7) | ||||||
| Dhb | 1 | 163.9 | 163.9 | 163.9 | 163.9 | |||||||||
| 2 | 132.4 | 132.4 | 132.4 | 132.4 | ||||||||||
| 3 | 117.3 | 5.38 | q (7.2) | 117.4 | 5.38 | q (7.8) | 117.4 | 5.39 | q (7.2) | 117.3 | 5.39 | q (7.2) | ||
| 4 | 13.2 | 1.75 | d (7.2)T | 13.2 | 1.75 | d (7.8) | 13.2 | 1.75 | d (7.2) | 13.2 | 1.74 | d (7.2) | ||
| NH | 9.08 | s | 9.08 | s | 9.09 | s | 9.10 | s | ||||||
| Val2 | 1 | 168.8 | 168.8 | 168.8 | 168.6 | |||||||||
| 2 | 55.5 | 4.31 | dd (9.0, 6.0) | 55.5 | 4.31 | dd (9.0, 6.6) | 55.5 | 4.32 | dd (9.0, 6.6) | 55.5 | 4.32 | dd (9.0, 6.3) | ||
| 3 | 32.7 | 1.81 | m | 32.7 | 1.81 | m | 32.7 | 1.81 | m | 32.5 | 1.81 | m | ||
| 4 | 18.4 | 0.83 | d (6.0) | 18.4 | 0.83 | d (6.6) | 18.4 | 0.83 | d (6.6) | 18.3 | 0.82 | d (6.8) | ||
| 4′ | 19.0 | 0.89 | d (6.0) | 19.0 | 0.89 | d (6.6) | 19.0 | 0.89 | d (6.6) | 19.0 | 0.87 | d (6.8) | ||
| NH | 6.85 | d (9.0) | 6.85 | d (9.0) | 6.86 | d (9.6) | 6.87 | d (9.2) | ||||||
Aassigned from the DEPT-Q spectrum recorded at 226 MHz,
Measured at 600 MHz,
Assigned from the HSQC and HMBC spectra.
Not assigned due to the signal missing.
Complete NMR table for minutissamide E including 2D NMR data can be found in the Supplementary data.
Sextet
4.4.2 Minutissamide F (2)
Colorless, amorphous powder; [ ]25D - 19 (c 0.05, MeOH); UV (MeOH) max (log) 202 (4.39), 233 (3.69) nm; IR (neat) 3324 (br), 2926, 2855, 1630 (br), 1539 cm−1; 1H and 13C NMR (see Table 1); HRESIMS m/z 1213.6645 [M + H]+ (calcd for C55H94ClN12O16, 1213.6599)
4.4.3 Minutissamide G (3)
Colorless, amorphous powder; [ ]25D - 22 (c 0.02, MeOH); UV (MeOH) max (log) 202 (4.24), 233 (3.54) nm; IR (neat) 3338 (br), 2928, 2852, 1676 (br), 1540 cm−1; 1H and 13C NMR (see Table 1); HRESIMS m/z 1251.7608 [M + H]+ (calcd for C59H103N12O17, 1251.7564)
4.4.4 Minutissamide H (4)
Colorless, amorphous powder; [ ]25D - 30 (c 0.02, MeOH); UV (MeOH) max (log) 201 (4.16), 233 (3.42) nm; IR (neat) 3320, 2920, 2851, 1634 (br), 1535 cm−1; 1H and 13C NMR (see Table 1); HRESIMS m/z 1247.7622 [M + H]+ (calcd for C60H103N12O16, 1247.7615)
4.4.5 Minutissamide I (5)
Colorless, amorphous powder; [ ]25D - 26 (c 0.13, MeOH); UV (MeOH) max (log) 203 (4.46), 233 (3.81) nm; IR (neat) 3322 (br), 2929, 2855, 1658 (br), 1643 (br), 1537 cm−1; 1H and 13C NMR (see Table 2); HRESIMS m/z 1235.7238 [M + H]+ (calcd for C58H99N12O17, 1235.7251)
Table 2. NMR Spectroscopic data for minutissamides I - L (5 – 8) in DMSO-d6.
| minutissamide I (5) | minutissamide J (6) | mixture | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| minutissamide K (7) | minutissamide L (8) | |||||||||||||
| C a | H b | mult. (J in Hz) | C c | H b | mult. (J in Hz) | C c | H b | mult. (J in Hz) | C c | H b | mult. (J in Hz) | |||
| Ahmoo, Achmt or Adhmo | 1 | 169.8 | 169.9 | 169.8 | 169.8 | |||||||||
| 2 | 69.6 | 4.16 | d (5.1) | 69.7 | 4.16 | d (5.1) | 69.5 | 4.16 | d (4.7) | 69.5 | 4.16 | d (4.7) | ||
| 3 | 56.2 | 3.93 | td (10.6, 5.1) | 56.0 | 3.93 | td (10.3, 5.1) | 56.2 | 3.92 | brt (10.6) | 56.2 | 3.92 | brt (10.6) | ||
| 4 | 32.3 | 1.70 | m | 32.3 | 1.69 | m | 32.3 | 1.68 | m | 32.3 | 1.68 | m | ||
| 5 | 33.5 | 1.17 | m | 33.4 | 1.17 | m | 33.5 | 1.16 | m | 33.5 | 1.16 | m | ||
| 1.60 | m | 1.61 | m | 1.61 | m | 1.61 | m | |||||||
| 6 | 25.6 | 1.17 | m | 25.5 | 1.16 | m | 25.6 | 1.16 | m | 25.6 | 1.16 | m | ||
| 1.25 | m | 1.25 | m | 1.23 | m | 1.23 | m | |||||||
| 7 | 29.8 | 1.25 | m | 29.7 | 1.25 | m | 29.7 | 1.25 | m | 29.7 | 1.25 | m | ||
| 8 | 29.3 | 1.25 | m | 29.1 | 1.25 | m | 29.3 | 1.25 | m | 29.3 | 1.25 | m | ||
| 9 | 29.1 | 1.25 | m | 28.6 | 1.25 | m | 29.1 | 1.25 | m | 29.1 | 1.25 | m | ||
| 10 | 29.1 | 1.25 | m | 26.1 | 1.35 | m | 29.0 | 1.25 | m | 29.0 | 1.25 | m | ||
| 1.45 | m | |||||||||||||
| 11 | 29.0 | 1.25 | m | 37.5 | 1.63 | m | 28.9 | 1.25 | m | 28.9 | 1.25 | m | ||
| 1.72 | m | |||||||||||||
| 12 | 28.7 | 1.25 | m | 66.4 | 3.98 | m | 28.6 | 1.25 | m | 28.6 | 1.25 | m | ||
| 13 | 23.4 | 1.44 | m | 30.8 | 1.63 | m | 30.8 | 1.25 | m | 23.3 | 1.43 | m | ||
| 1.78 | m | |||||||||||||
| 14 | 42.0 | 2.37 | t (7.2) | 10.7 | 0.96 | t (7.2) | 37.3 | 1.26 | m | 42.2 | 2.37 | t (7.2) | ||
| 15 | 210.9 | 69.2 | 3.35 | m | 211.0 | |||||||||
| 16 | 43.8 | 2.36 | t (7.2) | 39.7 | 1.26 | m | 43.7 | 2.36 | t (7.2) | |||||
| 17 | 16.8 | 1.46 | sd (7.2) | 18.9 | 1.26 | m | 16.7 | 1.45 | sd (7.2) | |||||
| 1.38 | m | |||||||||||||
| 18 | 13.7 | 0.83 | t (7.2) | 14.2 | 0.84 | overlapped | 13.7 | 0.82 | t (7.2) | |||||
| 3-NH | 6.83 | d (10.3) | 6.83 | d (10.5) | 6.86 | overlapped | 6.86 | overlapped | ||||||
| 4-Me | 16.1 | 0.57 | d (6.7) | 16.1 | 0.57 | d (6.7) | 16.1 | 0.56 | d (6.7) | 16.1 | 0.56 | d (6.7) | ||
| Pro | 1 | 171.3 | 171.3 | 171.3 | 171.3 | |||||||||
| 2 | 59.9 | 4.26 | dd (8.0, 1.8) | 60.0 | 4.26 | dd (8.0, 1.8) | 60.0 | 4.26 | dd (8.0, 1.8) | 60.0 | 4.26 | dd (8.0, 1.8) | ||
| 3 | 30.2 | 1.92 | m | 30. 1 | 1.92 | m | 30.2 | 1.92 | m | 30.2 | 1.92 | m | ||
| 1.99 | m | 1.98 | m | 1.99 | m | 1.99 | m | |||||||
| 4 | 23.4 | 1.69 | m | 23.4 | 1.68 | m | 23.4 | 1.68 | m | 23.4 | 1.68 | m | ||
| 1.84 | m | 1.83 | m | 1.83 | m | 1.83 | m | |||||||
| 5 | 47.0 | 3.15 | m | 47.0 | 3.15 | m | 47.0 | 3.16 | m | 47.0 | 3.16 | m | ||
| 4.18 | m | 4.18 | m | 4.18 | m | 4.18 | m | |||||||
| NMeAsn | 1 | 167.7 | 167.8 | 167.7 | 167.7 | |||||||||
| 2 | 49.6 | 5.56 | dd (11.5, 2.7) | 49.7 | 5.56 | dd (11.5, 2.7) | 49.7 | 5.55 | dd (11.5, 2.7) | 49.7 | 5.55 | dd (11.5, 2.7) | ||
| 3 | 34.0 | 1.96 | overlapped | 34.0 | 1.96 | overlapped | 33.9 | 1.95 | overlapped | 33.9 | 1.95 | overlapped | ||
| 3.02 | dd (15.5, 11.5) | 3.02 | dd (15.5, 11.5) | 3.01 | dd (15.5, 11.5) | 3.01 | dd (15.5, 11.5 | |||||||
| 4 | 171.7 | 171.7 | 171.7 | 171.7 | ||||||||||
| N-Me | 30.4 | 2.94 | s | 30.4 | 2.94 | s | 30.6 | 2.93 | brs | 30.6 | 2.93 | brs | ||
| NH2 | 5.99 | s | 5.98 | s | 6.11 | s | 6.11 | s | ||||||
| 7.52 | s | 7.52 | s | 7.53 | s | 7.53 | s | |||||||
| OMeThr | 1 | 169.8 | 169.9 | 169.9 | 169.9 | |||||||||
| 2 | 53.2 | 4.71 | dd (9.2, 2.7) | 53.2 | 4.71 | dd (9.2, 3.2) | 53.1 | 4.71 | brd (9.2) | 53.1 | 4.72 | brd (9.2) | ||
| 3 | 74.9 | 3.72 | qd (6.3, 2.7) | 74.9 | 3.72 | qd (6.3, 3.2) | 75.0 | 3.72 | qd (6.6, 2.4) | 75.0 | 3.72 | qd (6.6, 2.4) | ||
| 4 | 15.2 | 0.99 | d (6.3) | 15.2 | 0.99 | d (6.3) | 15.2 | 0.98 | d (6.6) | 15.2 | 0.98 | d (6.6) | ||
| O-Me | 55.9 | 3.16 | s | 55.9 | 3.16 | s | 55.8 | 3.15 | s | 55.8 | 3.15 | s | ||
| NH | 6.80 | overlapped | 6.80 | overlapped | 6.80 | overlapped | 6.80 | overlapped | ||||||
| Gly | 1 | 169.0 | 169.0 | 169.1 | 169.1 | |||||||||
| 2 | 3.21 | dd (17.1, 5.1) | 3.21 | dd (17.1, 5.1) | 3.20 | dd (17.1, 5.1) | 3.20 | dd (17.1, 5.1) | ||||||
| 2′ | 42.6 | 3.98 | dd (17.1, 7.4) | 42.5 | 3.98 | dd (17.1, 7.4) | 42.6 | 3.97 | dd (17.1, 7.3) | 42.6 | 3.97 | dd (17.1, 7.3) | ||
| NH | 7.95 | brt (6.0) | 7.95 | brt (6.0) | 7.97 | overlapped | 7.97 | overlapped | ||||||
| Gln | 1 | 172.0 | 172.0 | 172.0 | 171.6 | |||||||||
| 2 | 53.1 | 4.09 | m | 53.0 | 4.09 | m | 53.1 | 4.08 | m | 53.4 | 4.08 | m | ||
| 3 | 26.5 | 1.81 | m | 26.4 | 1.81 | m | 26.3 | 1.83 | m | 26.5 | 1.83 | m | ||
| 2.03 | m | 2.03 | m | 2.00 | m | 2.00 | m | |||||||
| 4 | 31.8 | 2.17 | m | 31.9 | 2.17 | m | 31.9 | 2.16 | m | 31.9 | 2.16 | m | ||
| 5 | 174.2 | 174.1 | 174.2 | 174.2 | ||||||||||
| NH | 7.36 | brd | 7.37 | brd | 7.35 | brd | 7.51 | brd | ||||||
| NH2 | 6.80 | s | 6.81 | s | 6.86 | s | 6.83 | s | ||||||
| 7.29 | s | 7.29 | s | 7.33 | s | 7.39 | s | |||||||
| Thr1 or Val1 | 1 | 170.8 | 170.8 | 170.8 | 171.2 | |||||||||
| 2 | 62.1 | 3.81 | t (3.8) | 62.1 | 3.82 | t (4.0) | 62.0 | 3.80 | t (3.8) | 61.4 | 3.82 | t (4.0) | ||
| 3 | 65.5 | 4.11 | m | 65.5 | 4.11 | M | 65.5 | 4.10 | m | 28.6 | 2.14 | m | ||
| 4 | 20.7 | 1.23 | overlapped | 20.8 | 1.23 | overlapped | 20.7 | 1.23 | overlapped | 18.1 | 0.99 | d (7.0) | ||
| 4′ | 19.2 | 1.01 | d (7.0) | |||||||||||
| NH | 8.97 | brs | 8.84 | brs | 9.03 | brs | 8.67 | brs | ||||||
| Thr2 | 1 | 174.2 | 174.2 | 174.2 | 173.6 | |||||||||
| 2 | 56.9 | 4.88 | dd (9.1, 2.3) | 56.9 | 4.88 | dd (9.4, 2.5) | 57.0 | 4.87 | brd (9.4) | 57.0 | 4.83 | brd (9.1) | ||
| 3 | 70.0 | 4.60 | brm | 70.0 | 4.60 | brm | 70.0 | 4.60 | brm | 70.0 | 4.60 | brm | ||
| 4 | 19.0 | 1.24 | overlapped | 19.0 | 1.24 | overlapped | 19.0 | 1.23 | overlapped | 19.1 | 1.23 | overlapped | ||
| NH | 8.35 | d (9.3) | 8.35 | d (9.3) | 8.39 | d (9.1) | 8.42 | d (9.6) | ||||||
| Dhb | 1 | 164.2 | 164.2 | 164.2 | 164.6 | |||||||||
| 2 | 132.4 | 132.5 | 132.5 | 132.5 | ||||||||||
| 3 | 117.4 | 5.35 | q (7.3) | 117.3 | 5.35 | q (7.3) | 117.7 | 5.35 | q (7.3) | 117.7 | 5.35 | q (7.3) | ||
| 4 | 12.4 | 1.73 | d (7.3) | 12.4 | 1.73 | d (7.3) | 12.5 | 1.72 | d (7.3) | 12.5 | 1.72 | d (7.3) | ||
| NH | 9.08 | s | 9.08 | s | 9.14 | s | 9.14 | s | ||||||
| Val2 | 1 | 168.9 | 168.8 | 168.9 | 168.9 | |||||||||
| 2 | 55.9 | 4.29 | dd (8.8, 6.5) | 55.8 | 4.29 | dd (8.8, 6.5) | 55.9 | 4.28 | dd (8.8, 6.1) | 55.9 | 4.28 | dd (8.8, 6.1) | ||
| 3 | 32.8 | 1.81 | m | 32.7 | 1.80 | m | 32.7 | 1.79 | m | 32.7 | 1.79 | m | ||
| 4 | 18.5 | 0.82 | d (6.8) | 18.5 | 0.82 | d (6.8) | 18.5 | 0.81 | brd | 18.5 | 0.81 | brd | ||
| 4′ | 18.1 | 0.88 | d (6.8) | 19.1 | 0.88 | d (6.8) | 19.1 | 0.87 | brd | 19.1 | 0.87 | brd | ||
| NH | 6.87 | d (8.9) | 6.87 | d (9.2) | 6.89 | overlapped | 6.89 | overlapped | ||||||
Aassigned from the DEPT-Q spectrum recorded at 226 MHz,
Measured at 600 MHz,
Assigned from the HSQC and HMBC spectra.
Sextet
4.4.6 Minutissamide J (6)
Colorless, amorphous powder; [ ]25D - 19 (c 0.05, MeOH); UV (MeOH) (log) 202 (4.29), 233 (3.62) nm; IR (neat) 3294 (br), 2931, 2857, 1674 (br), 1540 cm−1; 1H and 13C NMR (see Table 2); HRESIMS m/z 1199.6477 [M + H]+ (calcd for C54H92ClN12O16, 1199.6443)
4.4.7 Minutissamides K (7) and L (8)
Colorless, amorphous powder; [ ]25D - 26 (c 0.04, MeOH); UV (MeOH) max (log) 202 (4.28), 233 (3.59) nm; IR (neat) 3291 (br), 2932, 2855, 1679 (br), 1541 cm−1; 1H and 13C NMR (see Table 2); HRESIMS for 7 m/z 1237.7381 [M + H]+ (calcd for C58H101N12O17, 1237.7408); HRESIMS for 8 m/z 1233.7482 [M + H]+ (calcd for C59H101N12O16, 1233.7459)
4.5 Advanced Marfey's analysis
Approximately 0.3 mg of 1 was hydrolyzed with 6 N HCl (500 μL) for 16 h at 110 °C. The resulting acid hydrolysate was separated into two equal portions for derivatization with either l-FDLA or dl-FDLA. Each portion was dissolved in H2O (50 μL), and mixed with 1N NaHCO3 (20 μL) and l-FDLA or dl-FDLA (20 μL, 10 mg/mL in acetone). Then, acetone was added to a final volume of 200 μL, and the reaction mixtures were heated to 40 °C and stirred for 1 h. After cooling to rt, the reaction was neutralized with 1N HCl (20 μL), and the resulting reaction mixtures were air-dried and re-dissolved in CH3CN (300 μL). LC-MS analysis was carried out using a reversed-phase column (Alltima C18, 250 × 4.6 mm, 5 μm, 1.0 mL/min) with a linear gradient from 20 % to 65 % aqueous CH3CN containing 0.1% formic acid for 50 min. The selective ion chromatograms of l-FDLA and dl-FDLA for each amino acid were compared for the assignment of amino acid configurations. The dl-FDLA derivative exhibited two peaks corresponding to the l- and d-FDLA derivatives of each amino acid: Ahmoo 67.0 (S, C-3) and 72.4 (R, C-3) min; Pro 30.4 (L) and 34.1 (D) min; NMeAsn 24.0 (D) and 26.1 (L) min; Ala 30.9 (L) and 35.4 (D) min; Gln 27.2 (L) and 28.7 (D) min; Thr 24.5 (L) and 30.6 (d) min; Val 35.8 (l) and 43.8 (d) min. The l-FDLA derivative gave one peak for each amino acid at 72.4 (Ahmoo), 30.5 (Pro), 26.3 (NMeAsn), 35.6 (Ala), 27.4 (Gln), 24.7 (Thr) and 35.9 (Val) min, confirming the R configuration at C-3 for Ahmoo, the d-configuration for Ala and the l-configuration for Pro, Ala, Gln and Val. The presence of NMe-l-Asn and l-Thr was confirmed by chromatographic com parison of the l-FDLA derivative with those of the amino acid standards NMe-l-Asn (26.1 min), NMe-d-Asn (24.1 min), l-Thr (24.5), d-Thr (30.6 min), l-allo-Thr (25.7 min) and d-allo-Thr (27.9 min). Advanced Marfey's analysis of 4, 5 and the mixture of 7 and 8 was also carried out as described above.
4.6 Preparation of OMeThr standards
Prior to methylation, the amino group of l-Thr was protected according to the previously described protocol.25 Briefly, Boc2O (2.5 eq, 3 mL of 2M solution in THF) and NaHCO3 (3 eq) were added to a solution of l-Thr (200 mg) in H2O (3 mL), and stirred overnight at rt. The turbid solution was extracted with Et2O twice, and the aqueous layer was carefully acidified to pH 3 at 0 °C using saturated citric acid. The resulting acidic solution was extracted with CH2Cl2 three times. The CH2Cl2 layer was dried using Na2SO4 and evaporated to yield NBoc-l-Thr (143 mg). For methylation, 2,6-di-tert-butylpyridine (0.5 mL) and methyl trifluoromethansulfonate (methyl triflate, 1 mL) were sequentially added to a solution of NBoc-l-Thr (50 mg) in CH2Cl2 (2 mL) at 0 °C. The mixture was warmed to rt and stirred overnight. The resulting yellowish solution was cooled to 0 °C, and saturated NaHCO3 was added. The organic phase was washed with H2O and purified by silica gel column chromatography eluting with n-hexane:EtOAc (99:1, v/v) and EtOAc. The EtOAc fraction was dried in vacuo to give NBoc-OMe-l-Thr methyl ester (40 mg) as light yellowish oil. NBoc-OMe-l-Thr methyl ester (5 mg) was hydrolyzed using 6 N HCl (1 mL) for 16 h at 110 °C to yield the final product OMe-l-Thr. OMe-d- Thr, OMe-l-allo-Thr and OMe-d-allo-Thr standards were prepared in the same way using d-Thr and dl-allo-Thr as starting compounds. The prepared OMeThr standards were derivatized with l-FDLA as described above and analyzed by LC-MS using a reversed-phase column (Alltima C18, 250 × 4.6 mm, 5 μm, 1.0 mL/min) with a linear gradient from 30 % to 50 % aqueous CH3CN (0.1% formic acid) for 50 min. The l-FDLA derivatives of OMe-l-Thr, OMe-l-allo-Thr, OMe-d-Thr and OMe-d-allo-Thr standards were eluted at 33.0, 32.6, 44.1 and 47.6 min, respectively. The l-FDLA derivative of the acid hydrolysate of 1 gave a peak at 33.0 min corresponding to OMeThr, indicating the presence of OMe-l-Thr. The l-FDLA peak was used as an internal standard to calibrate the retention time difference.
4.7 Antiproliferative Assay against MDA-MB-435 cells
Human melanoma MDA-MB-43526 cells were purchased from the American Type Culture Collection, (Manassas, VA). The cells were propagated at 37 °C in 5% CO2 in RPMI 1640 medium supplemented with fetal bovine serum (10%), penicillin (100 units/ml) and streptomycin (100 μg/ml). Cells in log phase growth were harvested by trypsinization. A total of 5,000 cells were seeded per well of a 96-well plate and incubated overnight at 37 °C in 5% CO2. Samples dissolved in DMSO were then sequentially diluted and added to the appropriate wells (total volume 100 μL). Each compound was tested at the following concentrations (μg/ml): 25, 5.0, 1.0, 0.2 and 0.04. The cells were incubated in the presence of test substance for ninety-six hours at 37 °C and evaluated for viability with a commercial absorbance assay (CellTiter 96® AQueous One Solution Cell Proliferation Assay, Promega Corp, Madison, WI). Activity was expressed as the percentage of viable cells present relative to the negative (solvent) control. The positive control was vinblastine tested at 1 ng/mL, which had 49% viable cells after treatment.
Supplementary Material
Acknowledgments
This research was supported by PO1 CA125066 from NCI/NIH. We thank Dr. B. Ramirez from Center for Structural Biology at UIC for providing an access to NMR instruments. We also thank Dr. G. Chlipala for help with taxonomic identification. LC-MS analysis was performed at UIC Research Resource Center (RRC) Mass Spectrometry Facility. We thank Carrie Crot from the Mass Spectrometry Facility at UIC for acquiring LTQ FT tandem MS data.
Footnotes
Supplementary data: Supplementary data associated with this article can be found, in the oline version, at http://
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and notes
- 1.Sieber SA, Marahiel MA. Chem Rev. 2005;105:715–738. doi: 10.1021/cr0301191. [DOI] [PubMed] [Google Scholar]
- 2.Strieker M, Marahiel MA. ChemBioChem. 2009;10:607–616. doi: 10.1002/cbic.200800546. [DOI] [PubMed] [Google Scholar]
- 3.Baltz RH, Miao V, Wrigley SK. Nat Prod Rep. 2005;22:717–741. doi: 10.1039/b416648p. [DOI] [PubMed] [Google Scholar]
- 4.Barrett D. Biochim Biophys Acta. 2002;1587:224–233. doi: 10.1016/s0925-4439(02)00085-6. [DOI] [PubMed] [Google Scholar]
- 5.Takemoto JY, Zhang L, Taguchi N, Tachikawa T, Miyakawa T. J Gen Microbiol. 1991;137:653–659. [Google Scholar]
- 6.Straus SK, Hancock REW. Biochim Biophys Acta. 2006;1758:1215–1223. doi: 10.1016/j.bbamem.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 7.Welker M, Döhren H. FEMS Microbiol Rev. 2006;30:530–563. doi: 10.1111/j.1574-6976.2006.00022.x. [DOI] [PubMed] [Google Scholar]
- 8.Moore RE, Bornemann V, Niemczura WP, Gregson JM, Chen JL, Norton TR, Patterson GML, Helms GL. J Am Chem Soc. 1989;111:6128–6132. [Google Scholar]
- 9.Gregson JM, Chen JL, Patterson GML, Moore RE. Tetrahedron. 1992;48:3727–3734. [Google Scholar]
- 10.MacMillan JB, Ernst-Russell MA, Ropp JS, Molinski TF. J Org Chem. 2002;67:8210–8215. doi: 10.1021/jo0261909. [DOI] [PubMed] [Google Scholar]
- 11.Bonnard I, Rolland M, Salmon JM, Debiton E, Barthomeuf C, Banaigs B. J Med Chem. 2007;50:1266–1279. doi: 10.1021/jm061307x. [DOI] [PubMed] [Google Scholar]
- 12.Plaza A, Bewley CA. J Org Chem. 2006;71:6898–6907. doi: 10.1021/jo061044e. [DOI] [PubMed] [Google Scholar]
- 13.Hrouzek P, Kuzma M, erný J, Novák P, Fi er R, imek P, Luke ová A, Kopecký J. Chem Res Toxicol. 2012;25:1203–1211. doi: 10.1021/tx300044t. [DOI] [PubMed] [Google Scholar]
- 14.Kang HS, Krunic A, Shen Q, Swanson SM, Orjala J. J Nat Prod. 2011;74:1597–1605. doi: 10.1021/np2002226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Falch BS, Konig GM, Wright AD, Sticher O, Angerhofer CK, Pezzuto JM, Bachmann H. Planta Med. 1995;61:321–328. doi: 10.1055/s-2006-958092. [DOI] [PubMed] [Google Scholar]
- 16.Sabareesh V, Balaram P. Rapid Commun Mass Spectrom. 2006;20:618–628. doi: 10.1002/rcm.2349. [DOI] [PubMed] [Google Scholar]
- 17.Fujii K, Ikai Y, Oka H, Suzuki M, Harada K. Anal Chem. 1997;69:5146–5151. [Google Scholar]
- 18.Harada K, Fujii K, Hayashi K, Suzuki M. Tetrahedron Lett. 1996;37:3001–3004. [Google Scholar]
- 19.Fujii K, Shimoya T, Ikai Y, Oka H, Harada K. Tetrahedron Lett. 1998;39:2579–2582. [Google Scholar]
- 20.Christiansen G, Philmus B, Hemscheidt T, Kurmayer R. J Bacteriol. 2011;193:3822–3831. doi: 10.1128/JB.00360-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Komárek J, Komárková J, Kling H. Filamentous Cyanobacteria. In: Wehr JD, Sheath RG, editors. Freshwater Algae of North America. Vol. 2003. Academic Press; San Diego: 2003. pp. 117–196. [Google Scholar]
- 22.Chlipala GE, Sturdy M, Krunic A, Lantvit DD, Shen Q, Porter K, Swanson SM, Orjala J. J Nat Prod. 2010;73:1529–1537. doi: 10.1021/np100352e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nübel U, Garcia-Pichel F, Muyzer G. Appl Environ Microbiol. 1997;63:3327–3332. doi: 10.1128/aem.63.8.3327-3332.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shendage DM, Fröhlich R, Haufe G. Org Lett. 2004;6:3675–3678. doi: 10.1021/ol048771l. [DOI] [PubMed] [Google Scholar]
- 26.Rae JM, Creighton CJ, Merck JM, Haddad BR, Johnson MD. Breast Cancer Res Treat. 2007;104:13–19. doi: 10.1007/s10549-006-9392-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.