Summary
The flk locus of Salmonella typhimurium was identified as a regulator of flagellar gene expression in strains defective in P- and l-ring formation. Flk acts as a regulator of flagellar gene expression by modulating the protein levels of the anti-σ28 factor FlgM. Evidence is presented which suggests that Flk is a cytoplasmic-facing protein anchored to the inner membrane by a single, C-terminal transmembrane-spanning domain (TMS). The specific amino acid sequence of the TMS is not essential for Flk activity, but membrane anchoring is essential. Membrane fractionation and visualization of protein fusions of green fluorescent protein derivatives to Flk suggested that the Flk protein is present in the membrane as punctate spots in number that are much greater than the number of flagellar basal structures. The turnover of the anti-σ28 factor FlgM was increased in flk mutant strains. Using FlgM–β-lactamase fusions we show the increased turnover of FlgM in flk null mutations is due to FlgM secretion into the periplasm where it is degraded. Our data suggest that Flk inhibits FlgM secretion by acting as a braking system for the flagellar-associated type III secretion system. A model is presented to explain a role for Flk in flagellar assembly and gene regulatory processes.
Introduction
The bacterial flagellum of Salmonella typhimurium is composed of three main substructures: the basal body (which functions as a transmembrane motor), the hook (which serves as universal joint permitting articulation between the motor and the filament) and the filament (the propeller) (Macnab, 1996). A flagellar-specific type III secretion (T3S) system transports precursor proteins through the hollow centre of the structure during assembly (Macnab, 2004). After secretion, protein subunits travel to the tip of the elongating structure where they assemble into place (Iino, 1969; Emerson et al., 1970). Flagellar assembly follows an ordered pathway of subunit addition (Fig. 1; reviewed in Aizawa, 1996). Insertion of the MS-ring (FliF) into the inner membrane is the initial step of basal body assembly. This is followed by the addition of the switch complex and T3S apparatus (Macnab, 1999). A driveshaft (rod) structure extends from the MS-ring through the peptidoglycan layer. At this point hook initiation can begin, but elongation outside the cell requires that the P- and l-rings be assembled first (Kubori et al., 1992). The P- and l-rings act as bushings around the rod in the peptidoglycan and lipopolysaccharide layers respectively. Upon hook completion, the flagellar T3S apparatus changes in specificity from hook-basal body (HBB) substrates to late assembly substrates (Minamino et al., 1999; Makishima et al., 2001; Hirano et al., 2003). The hook–filament junction proteins and cap (collectively known as the hook-associated proteins) are assembled next followed by the long external filament. The filament is assembled beneath the cap, which acts as a nucleation site for self-polymerization of flagellin (Yonekura et al., 2000).
Fig. 1.
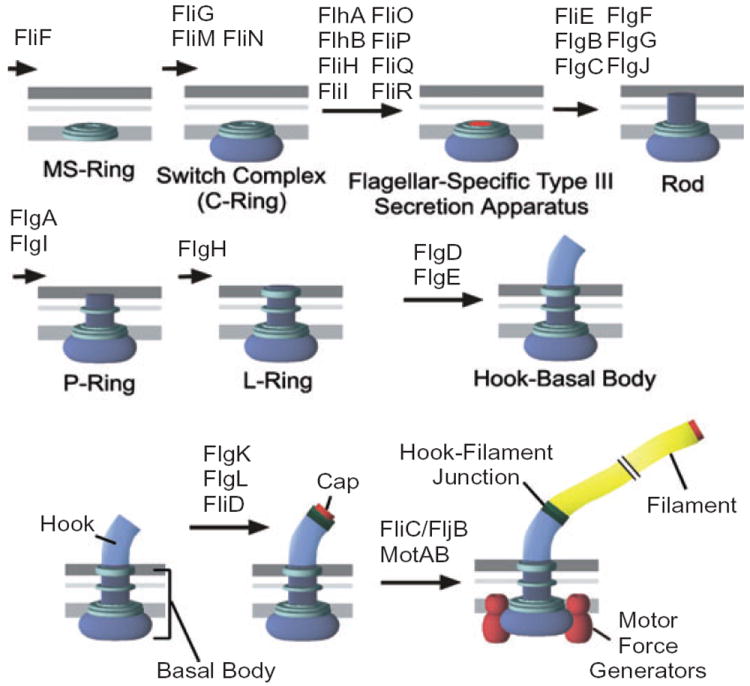
The flagellar assembly pathway of Salmonella typhimurium.
The regulation of flagellar gene expression is coupled to the assembly of the flagellar organelle. In S. typhimurium, the flagellar regulon comprises over 50 genes and is organized into a transcriptional hierarchy of three promoter classes (reviewed in Chilcott and Hughes, 2000). The class 1 promoter transcribes a single operon, the flhDC operon. The FlhC and FlhD proteins form a heterotetrameric complex, FlhC2FlhD2, that direct σ70-RNA polymerase complex to activate transcription from class 2 promoters. The products of class 2 transcripts are primarily required for the structure and assembly of the HBB structure. Among class 2-transcribed genes is the fliA gene that encodes the flagellar-specific transcription factor σ28. The σ28 holoenzyme of RNA polymerase transcribes class 3 promoters. In general, class 3 transcripts code for proteins required late in the flagellar assembly process and genes that code for the chemosensory system.
A negative regulatory protein, FlgM, co-ordinates the transition from HBB completion to initiation of class 3 transcription (Karlinsey et al., 2000). FlgM is an anti-σ28 factor that inhibits σ28-dependent transcription from class 3 promoters prior to HBB completion (Ohnishi et al., 1992). Upon HBB completion, FlgM is secreted from the cell and σ28 is free to transcribe the late assembly genes now needed (Hughes et al., 1993; Kutsukake, 1994). In this way, genes whose products are assembled outside the cytoplasm (in particular the large external filament) are not transcribed until the HBB structure onto which these late subunits will be added is completed.
In strains that are defective in the formation of the HBB (HBB−), FlgM is not secreted and accumulates to inhibit σ28-dependent transcription from class 3 promoters. Genetic screening of transposon mutants in a HBB strain defective in P-ring formation (Gillen and Hughes, 1991a), identified two novel loci that allowed class 3 transcription and were later named the flgM and flk loci (Gillen and Hughes, 1991b; Karlinsey et al., 1997). Loss of FlgM allows class 3 transcription in all HBB mutant strains. In contrast, loss of Flk allowed class 3 transcription only in HBB mutant strains defective in the assembly of the P- and l-rings (Karlinsey et al., 1997). It was later shown that loss of Flk resulted in a reduction of intracellular FlgM in P- or l-ring mutant strains (Karlinsey et al., 1998). Although how FlgM regulates flagellar gene expression has been identified, the role of Flk during flagellar assembly still remains unclear.
The flk locus was also identified (as the rflH locus) by an independent genetic selection (Kutsukake, 1997). Upon HBB completion, a component of the flagellar secretion system (FlhB) is altered to switch from hook-type secretion substrates to late secretion substrates such as FlgM and flagellin. The FlhB protein can be altered to allow late secretion in the absence of the hook completion signal. However, FlgM secretion in the altered flhB mutant strain did not occur in the absence of the hook unless the Flk protein was also defective (Kutsukake, 1997). Thus, Flk appears to play a role in preventing FlgM secretion at the stage of hook elongation. How this affects flagellar assembly is still not understood because loss of Flk has no discernable effect on flagellar assembly in wild-type strains.
To understand a role for Flk in normal flagellar assembly, we characterized the Flk protein. The translated flk sequence suggested that the presumed Flk protein is membrane-anchored by a C-terminal hydrophobic transmembrane segment (Karlinsey et al., 1997). To our knowledge, there are very few characterized membrane proteins with such a structure. In this paper, we show that Flk is anchored to the cytoplasmic membrane by the C-terminus with the bulk of the protein facing the cytoplasm. Membrane anchoring is essential for Flk function in flagellar gene regulation. We also show that loss of Flk in a P- and l-ring double mutant strain leads to a switch in the flagellar secretion apparatus to late secretion specificity resulting in the premature secretion of FlgM into the periplasm where it is degraded.
Results
Flk: a flagellar regulatory protein with a rare C-terminal membrane anchor
The flk gene was identified as a lock on the flagellar secretion gate to control flagellar hook-length (Kutsukake, 1997) and as a regulator of the flagellar FlgM anti-σ28 factor (Karlinsey et al., 1997). DNA sequence analysis revealed that the C-terminal 22 amino acid codons of the predicted 333 amino acid Flk protein from S. typhimurium includes a stretch of 20 hydrophobic amino acids at the C-terminus (Karlinsey et al., 1997). If this hydrophobic stretch represented a transmembrane segment then Flk would be predicted to be associated with the membrane fraction.
In order to determine the effect of the C-terminal hydrophobic tail on the membrane localization Flk, two plasmid-encoded flk constructs were used, one with full-length Flk fused to a FLAG epitope at its N-terminus (FLAG–Flk) and an identical construct deleted for the C-terminal hydrophobic tail (FLAG–Δ307-333Flk). The vector used placed the FLAG fusion under control of the T7 promoter. To determine the localization of FLAG–Flk and FLAG–Δ307-333Flk in the cell, cell extracts were separated into cytoplasmic, periplasmic and membrane fractions following induction of T7 RNA polymerase. Both FLAG–Flk and FLAG–Δ307-333Flk were detected with either anti-FLAG or anti-Flk antibodies (Fig. 2). FLAG–Flk was detected primarily in the cytoplasmic fraction, but also in the membrane fraction and a small amount could be detected in the periplasmic fraction. The high concentration of cytoplasmic FLAG–FLK is likely an artefact of the high levels of overexpression that can be obtained using T7 RNA polymerase. By comparison, deletion of the C-terminal hydrophobic tail (FLAG–Δ307-333Flk) resulted in a reduced but detectable membrane association and an increase in fractionation to the periplasm. However, the cytoplasmic control protein chloramphenicol acetyl transferase (Cat) was found primarily in the cytoplasmic fraction with a small amount detected in the periplasmic fraction and none in the membrane fraction. This suggests that the presence of the FLAG–Flk fusions in the periplasmic fraction is not significant and is a limitation of the assay. The periplasmic control protein, MalE (maltose binding protein) was detected primarily in the periplasmic fraction although a significant level was found in the cytoplasmic fraction. In contrast, the membrane control protein OmpA was detected only in the membrane fraction. These results suggest that FLAG–Flk is localized to the membrane fraction although a significant amount was found in the cytoplasmic fraction. Thus, loss of the C-terminal hydrophobic tail did not completely eliminate the association of Flk with the membrane fraction.
Fig. 2.
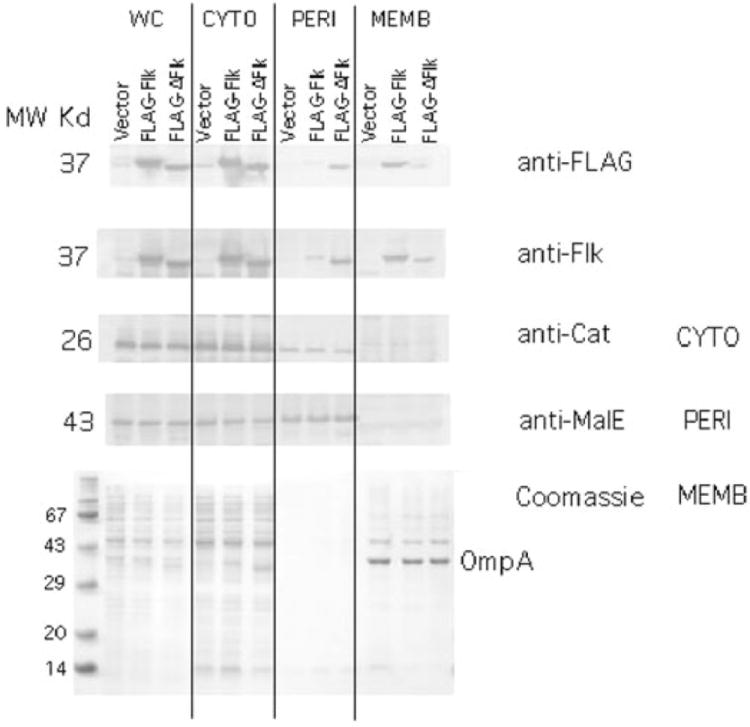
Cellular fractionation of the Flk protein. Plasmids pJK462 and pJK463 express Flk or FLAG–flk from the T7 promoter respectively. These plasmids and a vector control, pJK447 were placed in strain TH4657 that harbours T7 RNA polymerase under control of the lac promoter (Table 2). Following induction of T7 RNA polymerase, the whole cells (WC) were separated into cytoplasmic (CYTO), periplasmic (PERI) and membrane (MEMB) fractions (Experimental procedure). The proteins chloramphenicol acetyl transferase (Cat), maltose binding protein (MalE) and outer membrane protein A (OmpA) were used as controls for proteins known to be cytoplasmic, periplasmic and membrane-associated respectively. The fractions were run on SDS-PAGE and analysed by Western blot with anti-FLAG, anti-Flk, anti-Cat, anti-MalE antibodies. MW, molecular weight.
Flk is a cytoplasmic protein anchored to the inner membrane
Membrane fractionation assays determined that Flk was distributed between the cytoplasm and the membrane fractions. The presence of a single transmembrane-spanning domain (TMS) at the C-terminus of Flk, would suggest that it was inserted into the inner membrane by the TMS leaving the bulk of the protein in the cytoplasm. We used a genetic approach for membrane localization to determine the orientation of Flk in the inner membrane. Flk N-terminal fusions to either β-galactosidase (LacZ) or alkaline phosphatase (PhoA) were made that resulted in a fusion protein lacking the C-terminal hydrophobic sequence. Fusion proteins were then assayed for LacZ or PhoA activity on indicator media. Fusions of LacZ and PhoA have been successfully used previously to determine the topology of inner membrane proteins (Manoil et al., 1988). Fusions to PhoA and LacZ are PhoA+ and LacZ− when the PhoA or LacZ portion of the fusion is targeted to the periplasm and PhoA− and LacZ+ when the PhoA or LacZ portion of the fusion is targeted to the cytoplasm.
Insertions of TnlacZ/in and TnphoA/in transposons were isolated yielding fusions of N-terminal Flk to either ′LacZ of ′PhoA at positions 10, 14, 45, 103, 216, 283 and 295 (see Experimental procedures). All of the constructs harbouring LacZ fusions expressed β-galactosidase at high levels indicated by the Lac+ (Red) phenotype on MacConkey-lactose indicator plates (Fig. 3). All of the constructs harbouring PhoA fusions expressed PhoA at low levels indicated by the PhoA− (light blue) phenotype on plates containing the chromogenic substrate X-PhoA (Fig. 3). These results suggest that the N-terminal portion of Flk up to amino acid 295 is cytoplasmic and that the C-terminal hydrophobic tail would be inserted into the inner membrane in the orientation such that the very C-terminus of Flk is in the periplasm. Fusions of full-length Flk to ′LacZ and ′PhoA were generated by in vitro cloning techniques (Experimental procedures). In these cases, the Flk–′LacZ fusion was Lac− (Mac-lac White) while the Flk–′PhoA fusion was PhoA+ (Xpho blue) (Fig. 3). These data support a membrane topology where the bulk of the Flk protein is cytoplasmic and anchored to the inner membrane by a C-terminal hydrophobic stretch of amino acids.
Fig. 3.
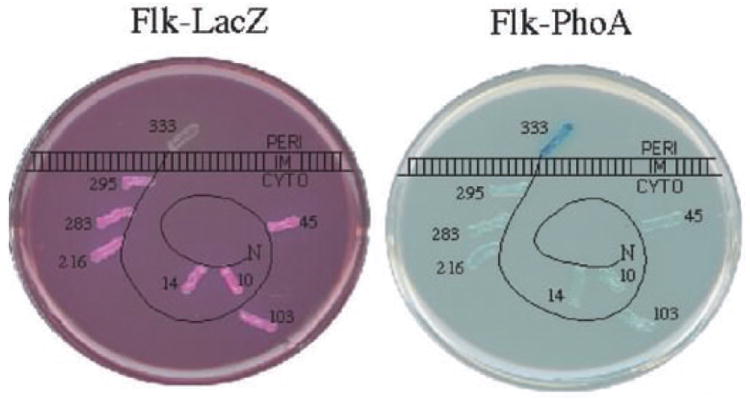
Flk is inserted into the membrane by a C-terminal hydrophobic tail. A diagram of the membrane topology of the Flk protein in the inner (cytoplasmic) membrane based on β-galactosidase (LacZ) and alkaline phosphatase (PhoA) fusion activities. The protein is represented as being primarily cytoplasmic (CYTO) anchored in the inner membrane with a C-terminal hydrophobic transmembrane segment that extends to the periplasm (PERI). Translational fusions of various proteins of the N-terminus of Flk were fused to LacZ (left) and PhoA (right) and assayed on indicator media. The numbers indicate the amino acid fusion junction of the LacZ or PhoA reporter to the specified amino acid position in Flk. On MacConkey-lactose medium (left) Lac+ cells are red and Lac− cells are white. On medium containing the chromogenic substrate for PhoA, Xpho (right) PhoA+ cells are blue and PhoA− cells are white. All LacZ fusions were Lac+ except the fusion at position 333. Conversely, all PhoA fusions were PhoA−, except the fusion at position 333.
The method by which fusions of LacZ to plasmid-expressed Flk was made at positions 10, 14, 45, 103, 216, 283 utilizes transposon Tn5lacZ/IN (Manoil and Bailey, 1997). Tn5lacZ/IN is designed so that isolation on BamHI restriction and religation removes all of the transposon leaving 93 base pairs in frame at the insertion site provided the original insertion was LacZ+. Removal of the Tn5lacZ/IN sequences in flk by BamH1 digestion and religation resulted in in-frame insertion of 31 amino acids after positions 10, 14, 45, 103, 216 and 283 of Flk. All six insertion mutants complemented a chromosomal flk null allele for activity [they inhibited expression of a fliC–lac fusion in the ring mutant background (Karlinsey et al., 1997)] suggesting that the three-dimensional structure Flk is not constrained to a limited conformation.
Membrane anchoring, but not membrane sequence is essential for Flk activity
The only Salmonella or Escherichia coli proteins shown (to our knowledge) to possess a hydrophobic C-terminal membrane-anchoring tail are the beta subunit isoenzymes FdoH and FdnH of the minor formate dehydrogenase complex (FDH-O) from E. coli (Benoit et al., 1998; Boyd et al., 1998). Both proteins possess single C-terminal TMS followed by a short periplasmic localized tail. Other than FdoH and FdnH penicillin binding proteins have also been suggested to anchor to the membrane via C-terminal amphipathic α-helices (Harris et al., 2002).
A flk mutant deleted for the last 19 amino acids failed to complement a flk null allele for the ability to prevent flagellar class 3 promoter transcription in the P- or l-ring mutant background (data not shown). This suggests that the membrane anchor is essential for Flk activity. We further tested whether the sequence of amino acids in the TMS was essential for Flk activity by replacing the Flk TMS with TMS-5 from lactose permease (LacY). The Flk-TMS-5 was able to complement a flk null mutant suggesting that only membrane anchoring and not specific amino acid sequences of the TMS are required for Flk function. Finally, addition of TMS-6 of LacY to the very C-terminus of Flk or replacement of the Flk TMS with both TMS-5 through TMS-6 of LacY did not inhibit Flk activity. This suggested that a strengthening of the membrane anchor by adding a second TMS did not significantly affect Flk function.
Visualization of Flk cellular localization
Hook-basal body structures have been visualized as discrete spots on the E. coli cell using yellow fluorescent protein (YFP) fusions to the switch complex protein FliM (Sourjik and Berg, 2000). We created Flk–YFP N- and C-terminal protein fusions to visualize Flk in Salmonella cells. The fusion of YFP to Flk at the N-terminus, YFP–Flk, was generated such that it was expressed from the chromosomal flk promoter (see Experimental procedures). YFP–Flk was active for yellow fluorescence (Fig. 4) and Flk regulatory activity. Similarly, a YFP fusion to the C-terminus of Flk was also active for both Flk and yellow fluorescence provided that the LacY TM5 and TM6 was included between the end of Flk and the start of YFP. The Flk-TM5/6-YFP derivative also included the linker (RRGAGAGA) between LacY TM6 and YFP to ensure that the YFP portion remained in the cytoplasm.
Fig. 4.
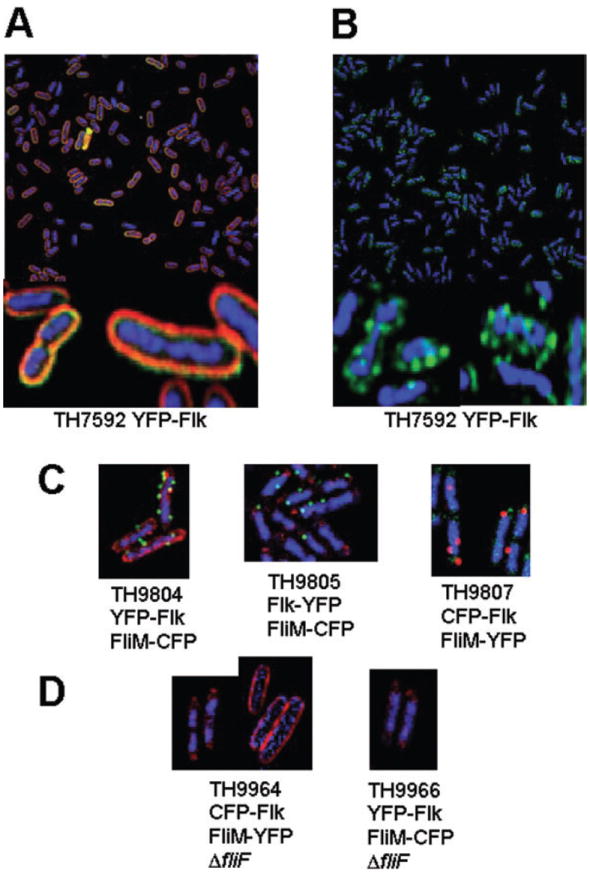
Flk appears as discrete spots in the cell membrane.
A. Expression of YFP–Flk is associated with the membrane fraction as indicated by the membrane stain FM 4-64.
B. Visualization of YFP–Flk in the absence of membrane stain indicates that it forms discrete spots on the cell membrane.
C. Strains TH9804 expressing an N-terminal YFP fusion to Flk (YFP–Flk) and a CFP-labelled HBB (FliM–CFP), TH9805 expressing a C-terminal YFP fusion to Flk (Flk–YFP) and TH9807 expressing an N-terminal CFP fusion to Flk (CFP–Flk) and a YFP-labelled HBB (FliM–YFP).
D. Strains TH9964 and TH9966 express CFP–Flk and FliM–YFP or YFP–Flk and FliM–CFP respectively, but are also deleted for the fliF gene, which is required for FliM assembly into HBB structures. Strain TH9964 was stained either with DAPI and the CFP–Flk visualized in red (TH9964, left photo) or with FM 4-64 and CFP–Flk visualized in blue (TH9964 right photo). TH9966 was stained with DAPI and YFP–FLK visualized in red. For TH9964 or TH9966, FliM–YFP or FliM–CFP, respectively, was not detected.
The Flk YFP derivatives were examined by fluorescence microscopy (Fig. 4). Deconvolution of acquired images allowed the examination of a 0.2 micron thick section of the cells for YFP–Flk localization in the membrane (Fig. 4A). Using a live membrane stain, FM 4-64 (red), we observed the fluorescence to be primarily in the membrane fraction. Also, the YFP–Flk fluorescence appeared punctate in the membrane. The YFP–Flk derivative was also examined in the absence of membrane stain on whole cell stacks (Fig. 4B). Here we see that Flk appears to form multiple foci throughout the cell.
To determine if Flk colocalized with flagellar HBB structures, strains were constructed that had YFP–Flk or Flk–YFP expressed in the same strain as a FliM–CFP fusion (Fig. 4C, TH9804 and TH9805 respectively). We also constructed a strain, TH9807 that expresses CFP–Flk and FliM–YFP derivatives (Fig. 4C). All strains have YFP or CFP fusion derivatives inserted at the normal chromosomal location for either flk or fliM. The strains are motile indicating that the FliM–CFP and FliM–YFP constructs are functional and do not adversely affect expression of the downstream fliNOPQR genes. The FliM–CFP and FliM–YFP fusions are fewer in number and much brighter than the YFP–Flk, Flk–YFP and CFP–Flk fusions. The number of FliM subunits in a basal body is estimated at 34, which would account for the brightness of the FliM fusion constructs (Zhao et al., 1996; Lowder et al., 2005). As expected, deletion of the MS-ring structural gene, fliF, resulted in the disappearance of FliM–CFP and FliM–YFP spots because FliM assembles onto the FliF MS-ring structure (Fig. 4D). However, deletion of fliF had no effect on the punctate appearance of CFP–Flk (TH9964) and YFP–Flk (TH9966) in the cell (Fig. 4D). It is evident that the majority of the Flk protein is not associated with HBB structures. However, it cannot be determined whether or not HBB structures are associated with Flk.
FlgM turnover is increased in flk mutant strains
Previously, we had reported that the reduction in FlgM levels in ring mutant strains by loss of Flk was not due to an increase in FlgM turnover (Karlinsey et al., 1998). Our previous assays only monitored FlgM turnover for a period of 7.5 min by pulse-chase assays followed by immunoprecipitation with anti-FlgM antibodies (Karlinsey et al., 1998). For this short period of time, no apparent turnover of newly synthesized FlgM was found in the flk background. Using a different assay, FlgM levels were assayed over 60 min following addition of the translation inhibitor spectinomycin (see Experimental procedures) (Aldridge et al., 2003). Consistent with our previous data the expected accumulation of FlgM in the HBB mutant background was observed (Fig. 5). Significantly, this assay showed that the flk null allele had a strong effect on FlgM stability reducing FlgM half-life from 98 min in the ΔflgHI (ring mutant) strain to 6 min in the ΔflgHI flk strain (Fig. 5). Thus, the reduction in FlgM levels observed in ring mutant strains in the absence of flk is due to increased FlgM turnover.
Fig. 5.
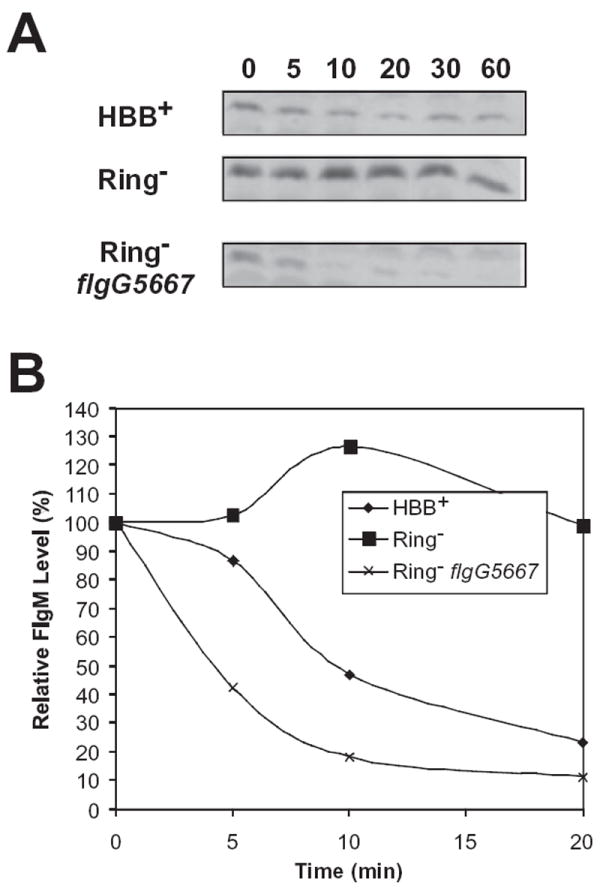
FlgM is unstable in the absence of Flk.
A. Immunoblot analysis of FlgM protein at different time points following the addition of the translation inhibitor spectinomycin. Cellular FlgM turns over more rapidly in a strain with a functional hook-basal body (TH4803, HBB+) because of FlgM secretion. In a strain deleted for the P- and l-rings (TH7025, Ring−), FlgM accumulates in the cell and is relatively stable. A null mutation in flk (TH7026) results in a sharp decrease in FlgM stability in the ring mutant strain.
B. A graph representation of the data in (A).
Secretion of FlgM–β-lactamase fusions into the periplasm in the absence of Flk
These data presented above are consistent with a model proposed by Kutsukake for Flk acting in a double-locked gate mechanism to control the flagellar substrate specificity switch (Kutsukake, 1997). If FlgM instability was due to premature secretion in the absence of Flk, we would have expected to find FlgM in the periplasm of a flk ΔflgHI strain, but not in the ΔflgHI background. A standard method to determine the presence of proteins in the periplasm is by osmotic shock as we did to determine cellular localization of Flk (Fig. 2). However, osmotic shock results in the ‘leakage’ of small proteins from the cytoplasm (Vazquez-Laslop et al., 2001). As FlgM is only 97 amino acids in length (Gillen and Hughes, 1991b), we decided to use an alternative genetic approach to study our prediction by fusing FlgM to the periplasmic reporter protein, β-lactamase.
For β-lactamase to confer resistance to ampicillin in Gram-negative bacteria, it must be exported to the periplasm (ApR) (Chervaux et al., 1995; Sauvonnet and Pugsley, 1996). The N-terminus of FlgM includes its flagellar type III secretion signal (Chilcott and Hughes, 1998) so Bla was fused to the very C-terminus of FlgM. It was hoped that if the FlgM–Bla fusion were secreted into the periplasm, the Bla portion of the fusion would be resistant to proteolysis and confer ApR.
Expression of the FlgM–Bla fusion was monitored in HBB+ and ΔflgHI (ring mutant) strains that were either flk+ or Δflk. We used a combination of osmotic shock to detect the presence of FlgM–Bla in the periplasm (Fig. 6) and resistance to ampicillin on solid media with or without 30 or 100 μg ml−1 ampicillin and in liquid media, in which a minimal inhibitory concentration (MIC, see Experimental procedures) of ampicillin was determined (Table 1). As expected, the control strain TH4803 (HBB+flk+) expressing no FlgM–Bla fusion was highly sensitive to ampicillin and did not export detectable FlgM into the periplasm (Fig. 6, lane 1) nor did an isogenic ΔflgHI flk+ strain (TH7025; Fig. 6, lane 2). Expression of the FlgM–Bla fusion in isogenic HBB+flk+ (TH9480; Fig. 6, lane 3), ΔflgHI flk+ (TH9481; Fig. 6, lane 4) and HBB+ Δflk (TH9486; Fig. 6, lane 5) strains showed some FlgM–Bla protein in the periplasmic fractions. The HBB+flk+ (TH9480) and ΔflgHI flk+ (TH9481) strains showed comparable levels of sensitivity to ampicillin both in liquid and on plates (Table 1), while the HBB+ Δflk (TH9486) strain showed significant levels of ampicillin resistance in liquid and on plates. However, the ΔflgHI Δflk double mutant strain expressing the FlgM–Bla fusion (TH9488) had significantly more FlgM–Bla present in the periplasmic fraction (Fig. 6, lane 6) and the highest levels of resistance to ampicillin on solid and in liquid media (Table 1). These data suggest that FlgM is exported into the periplasm in a Δflk strain in the presence of a functional HBB, and significantly more in a Δflk strain missing the P- and l-ring structures (ΔflgHI). Thus, loss of Flk has the effect of allowing FlgM export into the periplasm.
Fig. 6.

FlgM–Bla is secreted into the periplasm in flk ΔflgHI strains. Immunoblot analysis of FlgM and a FlgM–Bla fusion in cellular and periplasmic fractions (see Experimental procedures). Strains were assayed for the ability to secrete either FlgM or a chromosomal FlgM–Bla fusion into the periplasm. Lanes 1 and 2, TH4803 (HBB+flgM+) and TH7025 (Ring−flgM+), respectively, has FlgM in the cytoplasm, but not in the periplasm. Lanes 3–5, TH9480 (HBB+flgM–bla), TH9481 (Ring−flgM–bla) and TH9486 (HBB+ Δflk flgM–bla), respectively, has FlgM–Bla in the cytoplasm, but not in the periplasm. Lane 6, TH9488 (Ring− Δflk flgM–bla) and has FlgM–Bla secreted into the periplasm. Maltose binding protein (MBP) was used as a periplasmic protein control.
Table 1.
Growth of flgM–bla strains without or with ampicillin (100 μg ml−1; 30 μg ml−1) on plates.
| Strain | Genotype | LB plate | Amp plate 100 μg ml−1 | Amp plate 30 μg ml−1 | MICa |
|---|---|---|---|---|---|
| TH4803 | Wild type | +++ | − − − | − − − | < 3 |
| TH9480 | HBB+flgM–bla | +++ | − − − | − − −/+ | 12.5 |
| TH9481 | Ring−flgM–bla | +++ | − − − | − − − | 6.25 |
| TH9486 | Δflk HBB+flgM–bla | +++ | − −/+ | −/+ | 50 |
| TH9488 | Δflk Ring−flgM–bla | +++ | ++ | +++ | 100 |
The minimal inhibitory concentration (MIC) for sensitivity to ampicillin (μg ml−1) in liquid culture was determined for each strain as described in Experimental procedures.
Five levels of growth on the solid medium in the presence or absence of added ampicillin was reproducibly observed ranging from no growth at all (− − −) to a robust, wild-type growth phenotype (+++).
Discussion
Flagellar assembly is a temporal process that requires the correct structural subunit to be secreted and assembled at the correct time (Macnab, 2003). To facilitate the co-ordinate secretion of flagellar subunits, bacteria have evolved a complex regulatory circuits that couple flagellar gene expression to assembly (Soutourina and Bertin, 2003). This allows for only a specific subset of protein subunits to be produced and assembled at any given time point of assembly. Coupling gene expression to assembly also reduces substrate competition for the secretion apparatus. Two major components of the regulation of flagellar assembly in many bacterial species are σ28 and its anti-σ28 factor FlgM. σ28 is a flagellar-specific alternative sigma factor that controls the expression of flagellar subunits required after the HBB is assembled (Ohnishi et al., 1990). Early in flagellar assembly, while the HBB is being constructed, σ28 is inhibited via direct interaction with FlgM (Chadsey and Hughes, 2000). On HBB completion there is a substrate specificity switch of the flagellar-associated T3S apparatus that allows for FlgM secretion and thus σ28 becomes active as a transcription factor (Minamino and Namba, 2004). The number of mechanisms that sense HBB completion supports a central role of this critical checkpoint in flagellar assembly. Coincidental to HBB completion two independent events occur that lead to late flagellar gene expression. First, the regulatory protein FliK interacts with the secretion apparatus subunit FlhB causing a substrate specificity switch from hook-rod type subunit secretion to filament-type subunit secretion (Minamino and Pugsley, 2005). Second, there is the physical removal of FlgM from the cell via secretion by the flagellar T3S apparatus (Karlinsey et al., 2000). In this paper we have characterized a third regulatory mechanism, involving a membrane-bound protein Flk that monitors FlgM secretion by flagellar T3S systems associated with unfinished HBB structures.
Null mutants in flk were first identified by the phenotype of allowing σ28-dependent transcription in a HBB− background (Karlinsey et al., 1997). Unlike null mutants in flgM that exhibit σ28 activity in all HBB− backgrounds, the flk mutant phenotype required a specific background defective in the P- and/or l- ring assembly. Flk is an interesting protein because it possesses a single C-terminal TMS. This configuration is very rare and to our knowledge has only been demonstrated for two other bacterial proteins FdoH and FdnH, two isoenzymes that are beta subunits of a formate dehydrogenase complex (Benoit et al., 1998; Boyd et al., 1998). It has also been suggested that penicillin binding proteins may associate with the inner membrane via a C-terminal amphipathic α-helix (Harris et al., 2002). During this study we have shown that the C-terminal TMS is essential for the activity of Flk. Even though membrane association is essential for Flk activity there is no sequence specificity associated with the C-terminal TMS as it could be replaced with either one or two TMS domains from the integral membrane protein LacY.
We have previously utilized pulse chase assays to monitor FlgM turnover in flk mutants over a 7 min time-course and concluded that Flk did not play a role in FlgM stability (Karlinsey et al., 1998). During this study we repeated these assays by a different method over a 60 min time period. To our surprise, we found that in a flk null mutant deleted for the P- and l-ring structural genes, FlgM was rapidly degraded compared with the isogenic flk+ ring mutant where FlgM was found to be highly stable. We conclude that the extended time period in our current FlgM stability assays reflects the true effect of Flk on FlgM stability. This result is consistent with the model proposed by Kutsukake that Flk is involved in a double-lock gating mechanism for the flagellar-associated T3S apparatus (Kutsukake, 1997). This model predicts that when Flk is absent substrate selectivity of the secretion apparatus is reduced thus leading to premature secretion of late secretion subunits such as FlgM. FlgM–Bla fusions confirmed this prediction as ampicillin resistance was fully achieved only in the flk ring double mutant. As Bla must be in the periplasm to confer ampicillin resistance on solid media we can conclude that in the ring mutant background FlgM is secreted into the periplasm through the flagellar basal structure when Flk is absent. The fact that a lower, but significant level of ampicillin resistance was observed in an isogenic HBB+ strain lacking Flk suggests that FlgM–Bla is also being exported into the periplasm even in the presence of a complete HBB. The fact that more is exported in ring mutant strain probably reflects that there is more time for FlgM export. The HBB+ strains would presumably have only a narrow window of time after initiation of rod growth and prior to P- and l-ring completion where FlgM could be exported into the periplasm. However, the significance of the result that FlgM is exported into the periplasm in HBB+ strains is the suggestion that the secretion substrate specificity switch from rod-hook subunits to late secretion substrates occurs prior to HBB completion.
What can we conclude about the role of Flk in regulating protein secretion via the flagellar T3S apparatus? The transcription of the flk gene is not dependent on flagellar regulatory proteins, FlhDC or σ28 (Karlinsey et al., 1997). The cellular distribution of Flk was also unaffected by the absence of flagellar structures. Therefore, we conclude that Flk has been sequestered to regulate flagellar secretion in addition to its ‘normal’ cellular function, which is not known.
We envision at least two roles for Flk in flagellar assembly. Either (i) Flk is present to prevent FlgM secretion prior to HBB completion in normal flagellar synthesis or (ii) its role is to prevent FlgM secretion in basal body structures that for whatever reason are stalled between rod initiation and hook completion. If the first case is true, the flagellar T3S apparatus can switch to late substrate secretion prior to HBB completion. This seems unlikely as flgE mutants lacking hook structures never switch to late substrate secretion (Kutsukake, 1997). Furthermore, mutants in the secretion-specificity switch protein, FlhB, which can secrete late substrates in the absence of flgE, will not do so unless flk is also mutated (Kutsukake, 1997). If the second case were true (Flk preventing secretion through basal structures stalled in the assembly process), it would suggest that there is some imperfection in flagellar assembly. FliK is designed to interact with FlhB upon hook completion and to flip the secretion-specificity switch (Minamino et al., 2004). It is possible that this mechanism is not 100% perfect and sometimes FliK interacts with FlhB during rod assembly to flip the secretion-specificity switch too soon (Hirano et al., 2005). This would result in a small fraction of basal structures that undergo the secretion-specificity switch to the late secretion-specificity mode prior to HBB completion. This switch would allow the premature secretion of FlgM and other late secretion substrates into the periplasm and a huge energy drain on the cell. The work presented here shows that Flk would prevent this.
Another possibility in line with the first role (prevention of FlgM secretion prior to HBB completion in normal flagellar synthesis) is that the flagellar substrate specificity switch can occur during hook elongation. Mutants in the flagellar C-ring (fliG, fliM and fliN) have been reported that result in hooks that are 1/2 or 3/4 the normal hook length (Makishima et al., 2001). This led to a model that hook length is controlled by the C-ring acting as a measuring cup of hook subunits. It is possible that the wild-type Cring measuring cup is not always full of FlgE subunits when it is emptied, leading to a short hook phenotype. Our data suggest that Flk could be designed to prevent premature late gene secretion through such structures. As a result, HBB structures with short hooks are allowed to finish the growth of hooks to normal length.
It is clear that to achieve efficient substrate recognition during the transition from HBB assembly to filament assembly, multiple regulatory circuits are sensing the status of hook elongation. Flk appears to monitor the progress of hook elongation, FliK regulates the switching of substrate specificity via interactions with FlhB, and FlgM secretion is timing late subunit gene expression to coincide with HBB completion ultimately leading to the completion of the flagellar structure. We are now in an ideal position to begin to study exactly how these regulatory components are interacting with each other and/or the T3S apparatus and deduce exactly how they communicate their findings to the flagellar T3S apparatus.
Experimental procedures
Bacterial strains and plasmids
Bacterial strains used in this study and their origin are listed in Table 2.
Table 2.
List of bacterial strains.
| Strain | Genotype | Sourcea |
|---|---|---|
| Salmonella typhimurium | ||
| LT2 (wild-type) | John Roth | |
| TH4657 ΔflgHI958 fliC5050∷MudA flk-5206∷Tn10dCm trp∷[SpR/T7-RNAP lacOPI] | ||
| TH4803 fliC5504∷MudJ fljBenx vh2 | ||
| TH4987 ΔflgHI958 motA5461∷MudJ flhA(ts) | ||
| TH4988 ΔflgHI958 motA5461∷MudJ flk-5212∷Tn10dTc | ||
| TH4989 ΔflgHI958 motA5461∷MudJ flk-5212∷Tn10dTc flhA(ts) | ||
| TH7025 ΔflgHI958 fliC5504∷MudJ fljBenx vh2 | ||
| TH7026 ΔflgHI958 flk-5206∷Tn10dCm fliC5504∷MudJ fljBenx vh2 | ||
| TH7592 flk-6033(YFP-GAGAGA-Flk) | ||
| TH7688 flk-6037(Flk-LacY-TMD6-IR-GAGAGA-YFP) | ||
| TH9476 flgM6427(FlgM–Bla) | ||
| TH9480 flgM6427(FlgM–Bla) fliC5504∷MudJ fljBenx vh2 | ||
| TH9486 flgM6427(FlgM–Bla) Δflk-5867∷tetRA fliC5504∷MudJ fljBenx vh2 | ||
| TH9488 flgM6427(FlgM–Bla) ΔflgHI958 Δflk-5867∷tetRA fliC5504∷MudJ fljBenx vh2 | ||
| TH9804 Δhin-5717∷FRT flk-6033(YFP–Flk) fliM5980(FliM–CFP) fliC5569∷tetRA | ||
| TH9805 Δhin-5717∷FRT flk-6037(Flk–YFP) fliM5980(FliM–CFP) fliC5569∷tetRA | ||
| TH9807 Δhin-5717∷FCF flk-6031(CFP–Flk) fliM5982(FliM–YFP) fliC5569∷tetRA | ||
| TH9964 Δhin-5717∷FCF flk-6031 fliM5982 ΔfliFG6021∷FKF fliC5569∷tetRA | ||
| TH9966 Δhin-5717∷FRT flk-6033 fliM5980 ΔfliF5629∷FKF fliC5569∷tetRA | ||
| Escherichia coli | ||
| pCM400/DH5α | Manoil and Bailey (1997) | |
| pCM1000/DH5α | Manoil and Bailey (1997) | |
| CC160 Δ(ara-leu)7697 araD139 ΔlacX74 galE galK thi rpsL dam | Manoil and Bailey (1997) | |
| CC245 F−supF supE hsdR galK trpR mctB lacY tonA dam∷Km | Manoil and Bailey (1997) | |
| Plasmids | ||
| pJK447 (pT7 vector control) | ||
| pJK462 (pT7 FLAG–flk) | ||
| pJK463 (pT7FLAG–flkΔ307-333) |
Unless indicated otherwise, all strains were constructed during the course of this work.
Media and standard genetic manipulations
Media, growth conditions, transductional methods and motility assays were as described previously (Gillen and Hughes, 1991a, b).
Determination of β-lactamase minimal inhibitory concentrations
One millilitre culture (strains TH9480, TH9481, TH9486 and TH9488 and LT2 as a negative control) was grown overnight at 37°C in Luria–Bertani (LB) medium. The cells were diluted 100-fold into 5 ml of LB medium and grown at 37°C with aeration to mid-log (OD600 = 0.6). The cells were diluted 50-fold in LB medium containing variable concentrations of ampicillin (Ap) (serial diluted twofold from 800 μg ml−1 to 3.125 μg ml−1). After 4 h of growth at 37°C, the OD600 of each sample was measured. If OD600 was lower than 0.05, the Ap concentration in LB was considered as MIC.
Cell fractionation
Isolation of soluble, periplasmic and membrane fractions were made as follows. Cells were grown to an OD~600 nm in LB broth + Km (50 μg ml−1). Three millilitres of cells was pelleted by microcentrifugation and the cells were resuspended in 300 μl of cold periplast buffer (20% Sucrose/1 mM EDTA/20 μg ml−1 PMSF). A 100 μl portion was spun down and the cells were resuspended in 50 μl of 1× SDS-PAGE sample buffer (whole cell fraction WC). Lysozyme was added to the rest of the cells at 0.1 mg ml−1 and incubated on ice for 5 min. A 200 μl portion of cold water was added followed by a 5 min incubation at 0°C. The cells were spun at 4°C for 3 min at 12 000 g. The supernatant was saved (periplasmic faction P). The cells were then resuspended in 600 μl of spheroplast buffer (10 mM Tris-HCl, pH 7.5/50 mM KCL/1 mM EDTA/0.1% deoxycholate) and incubated at room temperature for 5 min. The cells were sonicated at 2×/2 second bursts, spun at 4°C for 3 min at 12 000 g to remove any unlysed cells. The supernatant was transferred to a new tube and spun at 4°C for 30 min at 16 000 g. The supernatant was saved (cytoplasmic fraction C). The membrane pellet was rinsed with 10 mM Tris-HCl, pH 8.0/20 μg ml−1 PMSF) and spun at 4°C for 30 min at 16 000 g. The supernatant was discarded and the membrane pellet was resuspended in 60 μl 2× SDS-PAGE sample buffer and heated at 65°C for 10 min (membrane fraction M). Samples were run on 10% Tricine-SDS-PAGE (Schägger and Jagow, 1987) and electrotransferred to Hybond-P membranes (Amersham Biosciences, Piscataway, NJ) in CAPS buffer (Matsudaira, 1987). The gels were hybridized with various antibodies and developed using ECL-Plus detection system (Amersham Biosciences, Piscataway, NJ).
Antibodies for cell fractionation
With the exception of anti-Flk antibody, all others were available commercially. These included Mouse Anti-FLAG M2 (Sigma, St Louis, MO), rabbit anti-CAT (5′PRIME3′ Boulder, CO), rabbit anti MBP (New England Biolabs, Beverly, MA), goat anti-rabbit IgG-HRP and goat anti-mouse IgG (Bio-Rad, Hercules, CA).
For purification of His-tagged Flk protein for antibody production, a 1042 bp NsiI-SacII, blunt-ended Mung Bean nuclease fragment from pJK232 containing the coding region of flk, was ligated into pET15b (Novagen, Madison, WI) cut with XhoI and blunt-ended with Klenow. The construct pJK264 was electroporated into strain BL21(DE3) (Novagen, Madison, WI) for induction and purification of His-tagged Flk fusion protein as described by Novagen. Anti-Flk antibody was made by BioWorld (Dublin, OH) using purified His-tagged Flk protein and the serum further purified as previously described (Hughes et al., 1993).
Isolation of LacZ and PhoA fusions to Flk
Two methods were used to isolate C-terminal fusions of β-galactosidase or alkaline phosphatase to different lengths of the Flk protein. One method utilized in vivo transposon mutagenesis of TnlacZ/in and TnphoA/in as described by Manoil and Bailey (Manoil and Bailey, 1997).
Plasmid pJK232 is a pBR322-based plasmid that carries the flk and adjacent pdxB genes expressed from their own promoters and will complement both flk and pdxB null mutants (Karlinsey et al., 1997). Strain pJK232/CC160 was mutagenized with either TnlacZ/in or TnphoA/in as described (Manoil and Bailey, 1997). For TnlacZ/in, about 1000 CmR insertion mutants were pooled together; plasmid DNA was prepared on 27 independent pools, electroporated into strain CC191 (Δlac ΔphoA) and screened for plasmids that carried insertions of the TnlacZ/in transposon (ApR CmR) such that a fusion of β-galactosidase to a gene encoded on pJK232 was obtained [Xgal blue phenotype (LacZ+)]. Twenty-five LacZ+ insertions in the flk gene were isolated from nine independent pools, 31 LacZ+ insertions in the pdxB gene were isolated from 11 independent pools, and 44 LacZ+ insertions in plasmid vector sequences were isolated from 19 independent pools. The 25 LacZ+ insertions in Flk were analysed by DNA sequencing and resulted in fusions of ′LacZ to Flk after amino acids 10, 14, 45, 103, 216, 283 and 295 of Flk. Several LacZ+ insertion in Flk turned out to be fused out-of-frame and were not characterized further. For TnphoA/in, about 1000 CmR insertion mutants were pooled together; plasmid DNA was prepared on 10 independent pools, electroporated into strain CC191 (Δlac ΔphoA) and screened for plasmids that carried insertions of the TnphoA/in transposon (ApR CmR) such that a fusion of ′phoA to a gene encoded on pJK232 was obtained [Xpho blue phenotype (PhoA+)]. Eighteen PhoA+ insertions in plasmid vector sequences were isolated from the 10 independent pools; no PhoA+ insertions were isolated in either the flk or pdxB genes.
The TnlacZ/in mutants that were isolated in frame at seven positions within the Flk were converted to ′phoA fusions as described (Manoil and Bailey, 1997). The TnlacZ/in and TnphoA/in transposons were designed to be interconvertable. Basically, the BamHI fragment of TnlacZ/in containing the ′lacZ cat sequences was swapped with the ′phoA cat sequences within the BamHI fragment from TnphoA/in. Fusions of ′lacZ and ′phoA to the very C-terminus of flk were constructed in vitro.
Protein stability assays, immunoblots and quantification
For protein stability assays of intracellular FlgM plus surface-associated FlgM, bacteria from overnight cultures were diluted into 30 ml of LB and grown at 37°C to an OD600 = 0.6 and 0.5 mg ml−1 spectinomycin (20 mg ml−1) was added to inhibit further protein synthesis. Total protein extracts were collected at 0, 5, 10, 20, 30 and 60 min after addition of spectinomycin from 1 ml of samples by centrifugation and resuspension of the cell pellet in SDS sample buffer (Aldridge and Jenal, 1999). The protein samples were normalized to cell density and subjected to Tricine-SDS-PAGE (Karlinsey et al., 1998) prior to immunoblot analysis. All other protein samples used throughout this study were taken from late log phase cultures of an OD 600 between 0.6 and 0.8.
Immunoblot analysis was performed as described (Aldridge and Jenal, 1999). Immunoblots were developed using the ECL plus kit (Amersham/Pharmacia) and gels were visualized using a Strom840 phosphoimager according to the manufacturer’s instructions. Quantification of immunoblots was performed with Imagequant for Macintosh, IQMac v1.2 (Molecular Dynamics). All experiments were performed in triplicate. Half-lives of FlgM were calculated using the ‘Trendline’ function from Microsoft Excel. Trendlines were assessed by the accompanying R2 value. Half-lives were calculated from trendlines showing R2 values of greater than 0.6.
Preparation of spheroplast cells for periplasmic fraction assays
The standard osmotic shock protocol for E. coli cells (Kaback, 1971) was modified for S. typhimurium strains. For the periplasmic fraction: cells were grown at 37°C in minimal E salts medium with 0.2% glycerol and auxotrophic supplement pools 6 through 11 (Davis et al., 1980) until they reached an OD600 of ~0.4 (Net Klett of ~77). A 5 ml portion of cells was pelleted (8000 g for 10′ at 4°C) and resuspended in 500 μl 100 mM Tris-HCl pH 8.0/20% Sucrose/10 mM EDTA. To this solution was added 12.5 μl of a 100 mg ml−1 stock lysozyme solution (2.5 mg ml−1 lysozyme in 10 mM Tris-HCl pH 8.0 and incubated at 0°C for 1.5 h. We found ~72% of the cells to be spheroplasts under these conditions. The cells were spun for 4′ at 3000 g 4°C. (5.7 rpm/2 min/4°C). A 53 μl portion of saturated trichloroacetic acid was added to 480 μl of supernatant, incubated at 0°C for 15 min, spun for 10 min in a microfuge and resuspended in sample buffer. For the cellular fraction: cells were resuspended to 20 OD600 μl−1 calculated as follows: (ODcells)·(Amount of volume in μl, i.e. 5000)/20; the cell fraction was resuspend cell fraction to 60 OD μl μl−1 calculated as follows: (ODcells)·(Amount of volume in μl, i.e. 4800)/60. The calculations were made so that the same OD units per sample was loaded onto the gel: a 10 μl of 200 OD amount of the cellular fraction and a 10 μl of 600 OD amount of periplasmic fraction were loaded onto the Tricine gels.
Chromosomally expressed YFP–Flk and Flk–YFP construction
A chromosomal YFP–Flk fusion was constructed by inserting a TcR-element (tetRA) from transposon Tn10 at the 5′ end of the flk gene just after the initiator methionine codon by polymerase chain reaction (PCR) amplification of YFP with primers that had 5′- and 3′-flanking sequences with homology to either the 5′-40 bases of flk upstream of amino acid codon 2 or the 3′-40 bases of flk downstream of amino acid codon 1 followed by l-Red-mediated recombination (Datsenko and Wanner, 2000) to replace the tetRA cassette with YFP to create a yfp–flk gene fusion expressed from the chromosomal flk promoter. Similarly, a chromosomally expressed Flk–YFP fusion was constructed by inserting a TcR-element (tetRA) from transposon Tn10 at the 3′ end of the flk gene just before the translation termination codon by PCR amplification of YFP fused at its N-terminus to transmembrane segment from N-terminal LacY (TMS6) with primers that had 5′- and 3′-flanking sequences with homology to either the 5′-40 bases of flk upstream of the stop codon or the 3′-40 bases of flk downstream of the last amino acid (333) using λ-Red-mediated recombination (Datsenko and Wanner, 2000) to replace the tetRA cassette with YFP to create a yfp–flk gene fusion expressed from the chromosomal flk promoter. All constructs were verified by DNA sequence analysis.
Microscopy and image analysis
For visualization of cells expressing the chromosomal YFP–Flk fusion cells were grown to mid-logarithmic stage and harvested by centrifugation. Cells were concentrated 10-fold by resuspension in supernatant and 2 μl of cells was applied to a slide. After addition of 1 μl of stain solution (0.6 μg ml−1 DAPI and 1 μg ml−1 Fm 4-64) the cells were immobilized with poly l-lysine-treated coverslips and visualized with an Applied Precision optical sectioning microscope (Sharp and Pogliano, 1999). Images were acquired and deconvolved using Delta Vision v3.3.6 software (Applied Precision) (Perez et al., 2000); typically five optical sections were taken for each sample. Figures were assembled using Photoshop v7.0 software (Adobe) or Canvas v9 (Deneba).
Acknowledgments
This work was supported by Public Health Service grant GM56141 from the National Institutes of Health. Dr Chevance received salary support from the University of Utah
References
- Aizawa S-I. Flagellar assembly in Salmonella typhimurium. Mol Microbiol. 1996;20:1–4. doi: 10.1046/j.1365-2958.1996.344874.x. [DOI] [PubMed] [Google Scholar]
- Aldridge P, Jenal U. Cell cycle-dependent degradation of a flagellar motor component requires a novel-type response regulator. Mol Microbiol. 1999;32:379–391. doi: 10.1046/j.1365-2958.1999.01358.x. [DOI] [PubMed] [Google Scholar]
- Aldridge P, Karlinsey J, Hughes KT. The type III secretion chaperone FlgN regulates flagellar assembly via a negative feedback loop containing its chaperone substrates FlgK and FlgL. Mol Microbiol. 2003;49:1333–1345. doi: 10.1046/j.1365-2958.2003.03637.x. [DOI] [PubMed] [Google Scholar]
- Benoit S, Abaibou H, Mandrand-Berthelot MA. Topological analysis of the aerobic membrane-bound formate dehydrogenase of Escherichia coli. J Bacteriol. 1998;180:6625–6634. doi: 10.1128/jb.180.24.6625-6634.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd D, Schierle C, Beckwith J. How many membrane proteins are there? Protein Sci. 1998;7:201–205. doi: 10.1002/pro.5560070121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadsey MS, Hughes KT. A multipartite interaction between Salmonella transcription factor, σ28, and its anti-sigma factor FlgM: implications for σ28 holoenzyme destabilization through stepwise binding. J Mol Biol. 2000;306:915–929. doi: 10.1006/jmbi.2001.4438. [DOI] [PubMed] [Google Scholar]
- Chervaux C, Sauvonnet N, Le Clainche A, Kenny B, Hung AL, Broome-Smith JK, Holland IB. Secretion of active beta-lactamase to the medium mediated by the Escherichia coli haemolysin transport pathway. Mol Gen Genet. 1995;249:237–245. doi: 10.1007/BF00290371. [DOI] [PubMed] [Google Scholar]
- Chilcott GS, Hughes KT. The type III secretion determinants of the flagellar anti-transcription factor, FlgM, extend from the amino-terminus into the anti-σ28 domain. Mol Microbiol. 1998;30:1029–1040. doi: 10.1046/j.1365-2958.1998.01131.x. [DOI] [PubMed] [Google Scholar]
- Chilcott GS, Hughes KT. The coupling of flagellar gene expression to flagellar assembly in Salmonella typhimurium and Escherichia coli. Microbiol Mol Biol Rev. 2000;64:694–708. doi: 10.1128/mmbr.64.4.694-708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RW, Botstein D, Roth JR. Advanced Bacterial Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1980. [Google Scholar]
- Emerson SU, Tokuyasu K, Simon MI. Bacterial flagella: polarity of elongation. Science. 1970;169:190–192. doi: 10.1126/science.169.3941.190. [DOI] [PubMed] [Google Scholar]
- Gillen KL, Hughes KT. Negative regulatory loci coupling flagellin synthesis to flagellar assembly in Salmonella typhimurium. J Bacteriol. 1991a;173:2301–2310. doi: 10.1128/jb.173.7.2301-2310.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillen KL, Hughes KT. Molecular characterization of flgM, a gene encoding a negative regulator of flagellin synthesis in Salmonella typhimurium. J Bacteriol. 1991b;173:6453–6459. doi: 10.1128/jb.173.20.6453-6459.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris F, Brandenburg K, Seydel U, Phoenix D. Investigations into the mechanisms used by the C-terminal anchors of Escherichia coli penicillin-binding proteins 4, 5, 6 and 6b for membrane interaction. Eur J Biochem. 2002;269:5821–5829. doi: 10.1046/j.1432-1033.2002.03295.x. [DOI] [PubMed] [Google Scholar]
- Hirano T, Minamino T, Namba K, Macnab RM. Substrate specificity classes and the recognition signal for Salmonella type III flagellar export. J Bacteriol. 2003;185:2485–2492. doi: 10.1128/JB.185.8.2485-2492.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Shibata S, Ohnishi K, Tani T, Aizawa S. N-terminal signal region of FliK is dispensable for length control of the flagellar hook. Mol Microbiol. 2005;56:346–360. doi: 10.1111/j.1365-2958.2005.04615.x. [DOI] [PubMed] [Google Scholar]
- Hughes KT, Gillen KL, Semon MJ, Karlinsey JE. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science. 1993;262:1277–1280. doi: 10.1126/science.8235660. [DOI] [PubMed] [Google Scholar]
- Iino T. Genetics and chemistry of bacterial flagella. Bacteriol Rev. 1969;33:454–475. doi: 10.1128/br.33.4.454-475.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaback HR. Bacterial membranes. Methods Enzymol. 1971;22:99–120. [Google Scholar]
- Karlinsey JE, Pease AJ, Winkler ME, Bailey JL, Hughes KT. The flk gene of Salmonella typhimurium couples flagellar P- and 1-ring assembly to flagellar morphogenesis. J Bacteriol. 1997;179:2389–2400. doi: 10.1128/jb.179.7.2389-2400.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlinsey JE, Tsui H-CT, Winkler ME, Hughes KT. Flk couples flgM translation to flagellar ring assembly in Salmonella typhimurium. J Bacteriol. 1998;180:5384–5397. doi: 10.1128/jb.180.20.5384-5397.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlinsey JE, Shugo Tanaka S, Bettenworth V, Yamaguchi S, Boos W, Aizawa SI, Hughes KT. Completion of the hook-basal body of the Salmonella typhimurium flagellum is coupled to FlgM secretion and fliC transcription. Mol Microbiol. 2000;37:1220–1231. doi: 10.1046/j.1365-2958.2000.02081.x. [DOI] [PubMed] [Google Scholar]
- Kubori T, Shimamoto N, Yamaguchi S, Namba K, Aizawa S. Morphological pathway of flagellar assembly in Salmonella typhimurium. J Mol Biol. 1992;226:433–446. doi: 10.1016/0022-2836(92)90958-m. [DOI] [PubMed] [Google Scholar]
- Kutsukake K. Excretion of the anti-sigma factor through a flagellar substructure couples flagellar gene expression with flagellar assembly in Salmonella typhimurium. Mol Gen Genet. 1994;243:605–612. doi: 10.1007/BF00279569. [DOI] [PubMed] [Google Scholar]
- Kutsukake K. Hook-length control of the export-switching machinery involves a double-locked gate in Salmonella typhimurium flagellar morphogenesis. J Bacteriol. 1997;179:1268–1273. doi: 10.1128/jb.179.4.1268-1273.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowder BJ, Duyvesteyn MD, Blair DF. FliG subunit arrangement in the flagellar rotor probed by targeted cross-linking. J Bacteriol. 2005;187:5640–5647. doi: 10.1128/JB.187.16.5640-5647.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnab RM. Flagella and motility. In: Neidhart FC, Curtiss R III, Ingraham LJ, Lin ECC, et al., editors. Escherichia Coli and Salmonella Typhimurium: Cellular and Molecular Biology. Washington, DC: American Society for Microbiology; 1996. pp. 123–145. [Google Scholar]
- Macnab RM. The bacterial flagellum: reversible rotary propellor and type III export apparatus. J Bacteriol. 1999;181:149–153. doi: 10.1128/jb.181.23.7149-7153.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnab RM. How bacteria assemble flagella. Annu Rev Microbiol. 2003;57:77–100. doi: 10.1146/annurev.micro.57.030502.090832. [DOI] [PubMed] [Google Scholar]
- Macnab RM. Type III flagellar protein export and flagellar assembly. Biochim Biophys Acta. 2004;1694:207–217. doi: 10.1016/j.bbamcr.2004.04.005. [DOI] [PubMed] [Google Scholar]
- Makishima S, Komoriya K, Yamaguchi S, Aizawa SI. Length of the flagellar hook and the capacity of the type III export apparatus. Science. 2001;291:2411–2413. doi: 10.1126/science.1058366. [DOI] [PubMed] [Google Scholar]
- Manoil C, Bailey J. A simple screen for permissive sites in proteins: analysis of Escherichia coli lac permease. J Mol Biol. 1997;267:250–263. doi: 10.1006/jmbi.1996.0881. [DOI] [PubMed] [Google Scholar]
- Manoil C, Boyd D, Beckwith J. Molecular genetic analysis of membrane protein topology. Trends Genet. 1988;4:223–226. doi: 10.1016/0168-9525(88)90154-0. [DOI] [PubMed] [Google Scholar]
- Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987;262:10035–10038. [PubMed] [Google Scholar]
- Minamino T, Namba K. Self- assembly and type III protein export of the bacterial flagellum. J Mol Microbiol Biotechnol. 2004;7:5–17. doi: 10.1159/000077865. [DOI] [PubMed] [Google Scholar]
- Minamino T, Pugsley AP. Measure for measure in the control of type III secretion hook and needle length. Mol Microbiol. 2005;56:303–308. doi: 10.1111/j.1365-2958.2005.04611.x. [DOI] [PubMed] [Google Scholar]
- Minamino T, Doi H, Kutsukake K. Substrate specificity switching of the flagellum-specific export apparatus during flagellar morphogenesis in Salmonella typhimurium. Biosci Biotechnol Biochem. 1999;63:1301–1303. doi: 10.1271/bbb.63.1301. [DOI] [PubMed] [Google Scholar]
- Minamino T, Saijo-Hamano Y, Furukawa Y, Gonzalez-Pedrajo B, Macnab RM, Namba K. Domain organization and function of Salmonella FliK, a flagellar hook-length control protein. J Mol Biol. 2004;341:491–502. doi: 10.1016/j.jmb.2004.06.012. [DOI] [PubMed] [Google Scholar]
- Ohnishi K, Kutsukake K, Suzuki H, Iino T. Gene fliA encodes an alternative sigma factor specific for flagellar operons in Salmonella typhimurium. Mol Gen Genet. 1990;221:139–147. doi: 10.1007/BF00261713. [DOI] [PubMed] [Google Scholar]
- Ohnishi K, Kutsukake K, Suzuki H, Iino T. A novel transcriptional regulatory mechanism in the flagellar regulon of Salmonella typhimurium: an anti sigma factor inhibits the activity of the flagellum-specific sigma factor, σF. Mol Microbiol. 1992;6:3149–3157. doi: 10.1111/j.1365-2958.1992.tb01771.x. [DOI] [PubMed] [Google Scholar]
- Perez AR, Abanes-De Mello A, Pogliano K. SpoIIB localizes to active sites of septal biogenesis and spatially regulates septal thinning during engulfment in Bacillus subtilis. J Bacteriol. 2000;182:1096–1108. doi: 10.1128/jb.182.4.1096-1108.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvonnet N, Pugsley AP. Identification of two regions of Klebsiella oxytoca pullulanase that together are capable of promoting beta-lactamase secretion by the general secretory pathway. Mol Microbiol. 1996;22:1–7. doi: 10.1111/j.1365-2958.1996.tb02650.x. [DOI] [PubMed] [Google Scholar]
- Schägger H, Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Sharp MD, Pogliano K. An in vivo membrane fusion assay implicates SpoIIIE in the final stages of engulfment during Bacillus subtilis sporulation. Proc Natl Acad Sci USA. 1999;96:14553–14558. doi: 10.1073/pnas.96.25.14553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourjik V, Berg HC. Localization of components of the chemotaxis machinery of Escherichia coli using fluorescent protein fusions. Mol Microbiol. 2000;37:740–751. doi: 10.1046/j.1365-2958.2000.02044.x. [DOI] [PubMed] [Google Scholar]
- Soutourina OA, Bertin PN. Regulation cascade of flagellar expression in Gram-negative bacteria. FEMS Microbiol Rev. 2003;27:505–523. doi: 10.1016/S0168-6445(03)00064-0. [DOI] [PubMed] [Google Scholar]
- Vazquez-Laslop N, Lee H, Hu R, Neyfakh AA. Molecular sieve mechanism of selective release of cytoplasmic proteins by osmotically shocked Escherichia coli. J Bacteriol. 2001;183:2399–2404. doi: 10.1128/JB.183.8.2399-2404.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura K, Maki S, Morgan DG, DeRosier DJ, Vonderviszt F, Imada K, Namba K. The bacterial flagellar cap as the rotary promoter of flagellin self-assembly. Science. 2000;290:2148–2152. doi: 10.1126/science.290.5499.2148. [DOI] [PubMed] [Google Scholar]
- Zhao R, Pathak N, Jaffe H, Reese TS, Khan S. FliN is a major structural protein of the C-ring in the Salmonella typhimurium flagellar basal body. J Mol Biol. 1996;261:195–208. doi: 10.1006/jmbi.1996.0452. [DOI] [PubMed] [Google Scholar]


