Background: Although the GAB1 adaptor is the main partner of the MET receptor, its role under apoptotic stress is controversial.
Results: During apoptosis, GAB1 is caspase-cleaved, generating a p35-GAB1 fragment that plays a pro-survival role in HGF/SF-MET signaling by retaining some signal transduction properties of GAB1.
Conclusion: The caspase-cleaved GAB1 can maintain HGF/SF-MET survival signaling.
Significance: A caspase-generated protein fragment can be anti-apoptotic.
Keywords: AKT, Apoptosis, Caspase, Receptor-tyrosine Kinase, Signal Transduction, GAB1, SHP2, c-MET
Abstract
The GRB2-associated binder 1 (GAB1) docking/scaffold protein is a key mediator of the MET-tyrosine kinase receptor activated by hepatocyte growth factor/scatter factor (HGF/SF). Activated MET promotes recruitment and tyrosine phosphorylation of GAB1, which in turn recruits multiple proteins and mediates MET signaling leading to cell survival, motility, and morphogenesis. We previously reported that, without its ligand, MET is a functional caspase target during apoptosis, allowing the generation of a p40-MET fragment that amplifies apoptosis. In this study we established that GAB1 is also a functional caspase target by evidencing a caspase-cleaved p35-GAB1 fragment that contains the MET binding domain. GAB1 is cleaved by caspases before MET, and the resulting p35-GAB1 fragment is phosphorylated by MET upon HGF/SF binding and can interact with a subset of GAB1 partners, PI3K, and GRB2 but not with SHP2. This p35-GAB1 fragment favors cell survival by maintaining HGF/SF-induced MET activation of AKT and by hindering p40-MET pro-apoptotic function. These data demonstrate an anti-apoptotic role of caspase-cleaved GAB1 in HGF/SF-MET signaling.
Introduction
Scaffold/docking/adaptor proteins play an important role in receptor-tyrosine kinase-mediated signaling pathways (1) as well as in many other transduction pathways (2, 3). Among them, GAB15 was initially identified as a GRB2-associated binder (GAB) protein (4) and as an interaction partner of the MET-tyrosine kinase receptor (5). GAB1 belongs to the GAB/DOS multisubstrate docking/adaptor protein subfamily that includes the Drosophila daughter of sevenless (DOS), Caenorhabditis suppressor of Clr (SOC)-1, and mammalian GAB1, GAB2, and GAB3 (6–8). Although all GAB/DOS family members contain an N-terminal pleckstrin homology domain, two GRB2 binding proline-rich sequences, and multiple phosphorylation sites (4, 5), GAB1 is the only subfamily member to carry the MET binding site (GAB1 MBS) within a larger domain called the MET binding domain (GAB1 MBD) (3, 9–11).
Upon HGF/SF binding, the MET receptor autophosphorylates on tyrosine residues, in particular on two tyrosine residues of the MET multi-docking site located within its C-terminal part (12). This phosphorylated MET multi-docking site is responsible for the recruitment of most MET signaling mediators (13), including GAB1, through both a direct binding via the corresponding GAB1-MBS (5) and an indirect binding via GAB1 interaction with the GRB2 adaptor bound to the MET multi-docking site (9). Subsequent phosphorylation of GAB1 on tyrosine residues by the activated MET receptor leads to the recruitment and activation of multiple SH2 domain-containing proteins, including the GRB2 adaptor, the p85 subunit of the phosphatidylinositol 3-kinase (p85-PI3K), and the SH2-containing tyrosine phosphatase (SHP2) (7, 8, 14, 15). These proteins can in turn activate PI3K-AKT and RAS-ERK pathways, which are known to be responsible for the major phenotypes observed in response to MET activation (16, 17). Moreover, activated PI3K catalyzes the production of PIP3 within the plasma membrane and, therefore, facilitates additional GAB1 recruitment via its PH domain in the vicinity of MET. This leads to an increase in p85-PI3K recruitment on GAB1 and thus to a GAB1-PI3K positive feedback loop (18). GAB1 phosphorylation also leads to termination of HGF/SF-MET-induced signaling via its c-CBL-dependent ubiquitination and proteasomal degradation (19).
Genetic evidence for the critical role of GAB1 in MET signaling is based on the extensive similarities between the phenotypes of GAB1−/−-, MET−/−-, and HGF/SF−/−-deficient mice. GAB1-null mice die between embryonic days 13.5 and 18.5 with severe defects in liver, placenta, and muscle development (20, 21), reminiscent of what was also observed in mice knocked-out for either the HGF/SF (22, 23) or the MET gene (24). In addition, the ERK kinases were activated at much lower levels in cells from GAB1-deficient embryos in response to HGF/SF, demonstrating that ERKs are downstream kinase targets of GAB1 in HGF/SF signaling (20, 21). In agreement with cellular responses to HGF/SF, overexpression of GAB1 leads to constitutive scattering of MDCK epithelial cells (5), whereas overexpression of truncated versions of GAB1 impairs HGF/SF signaling, with defects in cell morphogenesis, as well as in cell-cell contacts (5, 25, 26). GAB1 is, therefore, a multisubstrate docking protein that amplifies, diversifies, and integrates the HGF/SF-MET-induced signaling pathways (7).
The HGF/SF-activated MET is known to stimulate proliferation, survival, scattering, invasion, and morphogenesis of epithelial cells as well as of other cell types, indicating that HGF/SF functions mainly as a trophic and/or as an invasive growth factor (12, 27, 28). Nonetheless, we have previously shown that under stress induction and in the absence of HGF/SF, MET operates as a pro-apoptotic protein, with two sequential caspase-dependent cleavages of MET generating an intracellular p40-MET fragment (29, 30) and an extracellular membrane-bound p100-MET decoy fragment (31). In contrast, under stress induction and in the presence of HGF/SF, the phosphorylated MET receptor itself hinders its caspase-dependent cleavage, pointing out a possible fine-tuning of MET activity by stress and/or HGF/SF (32). These data indicate that the MET receptor can be pro- or anti-apoptotic according to environmental conditions. Given the various links between MET and GAB1 in HGF/SF-induced signaling, we have investigated whether GAB1 is also a functional target of caspases and, accordingly, whether it plays a role in the anti- or pro-apoptotic functions of the HGF/SF-MET signaling complex.
EXPERIMENTAL PROCEDURES
Cell Lines
Canine kidney MDCK epithelial cells and human cell lines (hepatocellular carcinoma HepG2, osteosarcoma U2OS, and cervical adenocarcinoma HeLa cells) were cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum (FBS) (Invitrogen). Human mammary MCF10A epithelial cells were cultured as described previously (33). A p40-MET Tet-on inducible MDCK cell line was kindly provided by Dr. D. Tulasne (CNRS-UMR8161, Lille, France).
Reagents
Recombinant human HGF/SF, TNF-related apoptosis-inducing ligand (TRAIL), and TNFα were purchased from Peprotech, anisomycin and ALLN were from Biomol, cycloheximide was from ICN, staurosporine was from Sigma, and Z-Val-Ala-Asp (OMe)-fluoromethyl ketone (Z-VAD-FMK) was from Kamiya Biomedical Co. β-Nerve growth factor (β-NGF) and epidermal growth factor (EGF) were purchased from R&D Systems, and insulin was from Sigma. Purified active caspase-3, -6, -7, -8, and -9 were generously provided by Dr. G.S. Salvesen (The Burnham Institute, La Jolla, CA). This laboratory verified the titration of the active recombinant caspases using in vitro cleavages against synthetic substrates (caspase-3, 13.9 mm; caspase-6, 7.5 mm; caspase-7, 19.7 mm; caspase-8, 15 mm; caspase-9, 41.5 mm).
Antibodies
A rabbit monoclonal antibody directed against residues surrounding Tyr-472 within the GAB1-MBD was purchased from Epitomics (clone Y423), and rabbit polyclonal antibodies directed against the C-terminal tail of GAB1 (amino acids (aa) 664–694; GAB1-C) were purchased from Upstate Biotechnology. We purchased and/or generated six other anti-GAB1 antibodies from distinct sources and/or targeting various GAB1 regions that were not useful, indicating that workable GAB1 antibodies are not easily available. Mouse monoclonal antibodies directed against the kinase domain of MET (clone 3D4) or GAPDH (clone 6C5) were purchased from Invitrogen, anti-phospho-MET (Tyr-1234/1235), anti-total AKT, anti-phospho-AKT (Ser-473), anti-phospho-ERK1/2 (Thr-202/Tyr-204), and anti-active caspase-3 (Asp175) antibodies were from Cell Signaling, anti-ERK2 (C-14), anti-poly(ADP-ribose) polymerase (PARP; H-250), and anti-SHP2 (N-16) antibodies were from Santa Cruz, anti-Na+/K+-ATPase-α1 (clone 464.6) was from Novus Biologicals, anti-LaminB1 (ab16048) was from AbCam, anti-p85-PI3K was from Upstate Biotechnology, and anti-GRB2 antibody was from BD Transduction Laboratories. Horseradish Peroxidase (HRP)-conjugated antibodies directed against rabbit or mouse IgG were purchased from Jackson ImmunoResearch Laboratories. Mouse monoclonal antibody against Myc-tag and anti-GFP antibody were purchased from Santa Cruz Biotechnology. Rabbit polyclonal antibody against HA-tag was purchased from Berkeley Antibody Co., Inc./Covance Research. An antibody directed against the N-terminal tail of GAB1 was purchased from Santa Cruz Biotechnology (N-17).
Plasmids and Transfections
Experimentally manipulated DNA sequences of GAB1 are from human origin (GenBankTM sequence NM_002039 encoding GAB1 isoform b, 694 aa), except for the canine siRNA GAB1 (GenBankTM predicted sequence XM_540929.2). The C-terminal-Myc-tagged GAB1 (pcDNA3-GAB1-Myc) as well as the N-terminal-HA-tagged GAB1 (pcDNA3-HA-GAB1) vectors were kindly provided by Prof. P. Raynal (EA4568, Université Toulouse III, Toulouse, France). The size of the Myc-tag is 20 kDa, as the pcDNA3-GAB1-Myc plasmid contains multiple repeats of the EQKLISEEDL Myc-tag sequence. Single or multiple mutated (replacement of aspartate, Asp, by asparagine, Asn) versions of GAB1 were generated from the pcDNA3-GAB1-Myc plasmid using the QuikChange Lightning site-directed mutagenesis kit (Stratagene). Introduced mutations were verified by standard Sanger DNA sequencing.
The following are the siRNA sequences (Eurogentec) targeting: human GAB1 (5′-GAGAGUGGAUUAUGUUGUUTT-3′), canine GAB1 (5′- GAGAGUAGAUUAUGUUGUGTT-3′), or canine MET (5′-GUGAGAGCACAACAAAUAUTT-3′). The AllStars Negative Control siRNA (Qiagen) was used as a control.
Vector transient transfections were performed using PEI/ExGen 500 (Euromedex) for HeLa cells and Lipofectamine 2000 reagent (Invitrogen) for MDCK cells, according to the manufacturer's instructions. For siRNA, transfections were performed using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's reverse transfection protocol.
Immunoblotting
Unattached floating cells were collected by gentle centrifugation and pooled with adherent cells in a lysis buffer (20 mm HEPES, pH 7.4, 142 mm KCl, 5 mm MgCl2, 1 mm EDTA, 5% glycerol, 1% Nonidet P-40, and 0.1% SDS) supplemented with freshly added protease and phosphatase inhibitors (#P8340 and #P5726, respectively, Sigma). Cells were lysed with this buffer on ice. Lysates were clarified by centrifugation (20,000 g × 15 min), and protein concentration was determined (BCA protein assay method, Pierce). The same protein amount of cell extracts was separated by either classic SDS-PAGE or NuPAGE (4–12% or 10% Bis-Tris precast gels) (Invitrogen) and electro-transferred to polyvinylidene difluoride membranes (Millipore). Membranes were probed with indicated primary antibodies followed by incubation with appropriate HRP-conjugated secondary antibodies. Protein-antibody complexes were visualized by chemiluminescence with the SuperSignal West Dura Extended Duration Substrate (Pierce) using a LAS-3000 camera acquisition system, and quantification of the acquired bioluminescent images was performed using the MultiGauge v3.0 image analysis program (FujiFilm). For image processing, contrast and brightness adjustment was applied to the whole images using Photoshop graphic software (Adobe).
Caspase Cleavage Assay
Caspase cleavages using cell lysates were performed as described previously (29). Samples were then analyzed by SDS-PAGE and immunoblotting. For GST-GAB1558–694, analyses were performed using mass spectrometry. The total mass of a cleavage fragment was determined in linear, positive ion linear mode. External calibration was performed over yeast enolase protein.
Immunoprecipitation Assays
Cells were lysed on ice in PY lysis buffer (20 mm Tris-HCl, pH 7.4, 50 mm NaCl, 5 mm EDTA, and 1% Triton X-100) containing freshly added protease and phosphatase inhibitor cocktails (protease inhibitor mixture (catalog no. P8340) and phosphatase inhibitor mixture 2 (catalog no. P5726) from Sigma). Lysates were clarified by centrifugation (20,000 g × 15 min), and protein concentration was determined using a BCA protein assay method (Pierce). Cell lysates (700 μg) were incubated with the GAB1 MBD antibody (3.5 μl) for 2 h at room temperature on an end-over-end rotator. The immune complexes were captured using protein A/G-agarose resin (Pierce) for 20 min at room temperature under gentle rotation. Beads were washed 3 times with PY lysis buffer, and immunoprecipitates were eluted with 0.2 m glycine pH 2 and subsequently analyzed by immunoblotting.
GST Fusion Protein Constructs
The cDNA sequence of GAB1 was inserted into pGEX4T-1 using EcoRI and SalI cloning site. GAB1 DNA sequence contains a single XhoI site located at the codon corresponding to amino acid 558. Accordingly, the pGEX4T-1-GAB1 plasmid was digested with EcoRI and XhoI. The pGEX4T-1-GAB1558–694 vector was obtained by self-ligation of EcoRI and XhoI sites after blunting with Klenow DNA polymerase fragment. The resultant glutathione S-transferase (GST) fusion protein (GST-GAB1558–694) was then expressed in Escherichia coli BL21 strain using 0.5 mm isopropyl 1-thio-β-d-galactopyranoside as an inducer and purified by glutathione affinity chromatography according to the manufacturer's instructions (Amersham Biosciences). The obtained recombinant protein was quantified using the Bio-Rad protein assay.
His-tagged Recombinant Proteins
Purified recombinant polyhistidine-tagged full-length GAB1 (His-Full GAB1) or p35-GAB1 (His-p35-GAB1) proteins were functionally expressed in bacteria using the ChampionTM pET200 Directional TOPO® expression kit with BL21 StarTM (DE3) One Shot® (Invitrogen). cDNA fragments corresponding to the full-length GAB1 gene (aa 1–694) and p35-GAB1 sequence (aa 371–610) were amplified by PCR using the primers pairs 5′-CACCAGTAGTTACTGTATCCCTACAGCAGGG-3′ and 5′-TTAATCAAGACTGTTACTGCCAAAGAGATT-3′ for full-length GAB1 and 5′-CACCAGCGGTGGTGAAGTGGTCTGCTCCGGA-3′ and 5′-TTATCTAAATTCAAGTCCTCTTCAGAAATG-3′ for p35-GAB1. Plasmid sequences were verified by standard Sanger DNA sequencing. The His-Full-GAB1 and His-p35-GAB1 vectors were then expressed in E. coli BL21 strain using isopropyl 1-thio-β-d-galactopyranoside as an inducer. After an overnight incubation, cells were harvested by centrifugation, and bacterial pellets were suspended in lysis buffer (25 mm HEPES, pH 7.4, 100 mm KCl, 10% (w/v) glycerol, 2 mm β-mercaptoethanol, and 4% Triton X-100) containing freshly added protease inhibitors (#P8340, Sigma). Lysates were sonicated on ice and centrifuged (15,000 g × 5 min); this process was repeated 4 times. Supernatants were subjected to ultracentrifugation in a SW41 rotor at 4 °C. The resultant recombinant His-Full-GAB1 and His-p35-GAB1 fusion proteins were purified by immobilized metal affinity chromatography procedure using HisPur Cobalt resin (Pierce). The clarified supernatants were adsorbed onto Cobalt-agarose beads. Beads were then washed with lysis buffer containing 10 mm imidazole. Elution of recombinant proteins was performed with an imidazole gradient (50–400 mm) using an appropriate buffer (25 mm HEPES, pH 7.4, 100 mm KCl, 10% (w/v) glycerol, 2 mm β-mercaptoethanol, and 0.5% Triton X-100). Eluted fractions were analyzed using NuPAGE gels, and protein purity was assessed and quantified by Coomassie Blue staining using a BSA standard.
Pulldown Protein-Protein Interaction Assays
Whole-cell lysates were prepared as described under “Immunoblotting” above, and a minimum of 600 μg of protein extract was incubated with 5–10 μg of the purified recombinant His-Full-GAB1 or His-p35-GAB1 proteins for 1 h 30 at 4 °C on an end-over-end rotator. Control samples without recombinant protein were prepared in parallel. After saving an aliquot of each sample (inputs), HisPur Cobalt resin (Pierce) was added to each sample for an additional 1 h incubation at 4 °C under gentle rotation to allow binding of His-tagged proteins complexes to the metal ion. Samples were then washed 3 times with lysis buffer (see “Immunoblotting”) containing 1 mm imidazole. Bound proteins were eluted with the lysis buffer containing 500 mm imidazole and analyzed by SDS-PAGE and immunoblotting.
Generation of Stable MDCK Cell Lines
A DNA fragment corresponding to the p35-GAB1 sequence (aa 371–610) was amplified by PCR using pcDNA3-GAB1-Myc expression vector as a template and the primers 5′-GAGAGAAAGCTTAGTAGTTACTGTATCCC-3′ and 5′-GAGAGAGGATCCGGATCAAGACTGTTACTGCC-3′. The amplified cDNA was cloned into the modified pL(FLAG-X-GFP)SH expression vector (34) using the HindIII/BamHI restriction sites included in the primers, respectively. The pL(FLAG-p35-GAB1-GFP)SH plasmid was verified by sequencing. MDCK or MDCK-p40-MET-Tet-On inducible cells were then transfected using pL(FLAG-p35-GAB1-GFP)SH or the empty pL(FLAG-X-GFP)SH as a control and were selected for 1 week in the presence of hygromycin (0.2 mg/ml). Cells stably expressing either FLAG-p35-GAB1-GFP or FLAG-GFP were analyzed by flow cytometry for their GFP expression and were subsequently isolated by FACS.
Subcellular Fractionation
Stably expressing FLAG-GFP and FLAG-p35-GAB1-GFP MCDK cells were seeded in 100-mm plates (80% confluence). Endogenous canine GAB1-targeted siRNA or control siRNA were transfected using Lipofectamine RNAiMax. After overnight starving (0.1% FBS), cells were treated with HGF/SF (100 ng/ml) for 5 min and immediately washed twice with ice-cold PBS. Differential subcellular cell lysis was performed according to manufacturer's instructions (Subcellular Proteome Extraction kit, Calbiochem). Nuclear, membrane, and cytoplasmic fractions were next analyzed by Western blotting. Relative fractions loads were normalized based on cytoplasmic protein content.
3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyltetrazolium Bromide (MTT) Assay
Cells were washed with PBS to eliminate dead cells and were incubated in medium containing 0.5 mg/ml MTT; Invitrogen) for 1 h. After a washing step with PBS, the formazan crystals were solubilized and mixed thoroughly with 0.04 n HCl in isopropyl alcohol. For each condition, 60 μl of formazan solution was loaded in triplicate onto a 96-well plate. Absorbance was then measured with a microplate spectrophotometer at 550 nm and 620 nm as test and reference wavelengths, respectively. The absorbance correlates with cell number.
Scratch Wound-healing Assay
Cells were grown to 80–90% confluence in a 6-well plate, and three wounds were then performed in each plate using a 1-ml pipette tip. The cell monolayers were then washed with PBS to remove cell debris and subsequently incubated under the indicated conditions in culture medium containing 0.1% FBS. Representative images were snap-captured using a phase contrast microscopy with ×40 magnification (Nikon Eclipse TS100) at 0 and 24 h post-wounding.
Caspase-3 Activity Assay
Caspase-3 enzymatic activity for caspase-3 was assayed using the EnzChek caspase-3 assay kit #1 (Invitrogen) following the manufacturer's instructions. Briefly, floating and adherent cells from a well of a 6-well plate were washed once in PBS and lysed in 40 μl of lysis buffer, and samples were cleared by centrifugation. Protein concentration was measured using the Pierce BCA Protein Assay, and 50 μg of cell lysate was diluted in lysis buffer and then incubated with substrate solution at 37 °C for 2 h in a Fluostar Optima (BMG Labtech) fluorescence reader which measures (excitation = 355 nm, emission = 430 nm) every 15 min. Experiments were performed in triplicate, and data are expressed as -fold change in fluorescence (% treatment/control).
RESULTS
The Down-expression of GAB1 in Stress Conditions Is Associated with a Reduced Activation of Both AKT and ERK in Response to HGF/SF
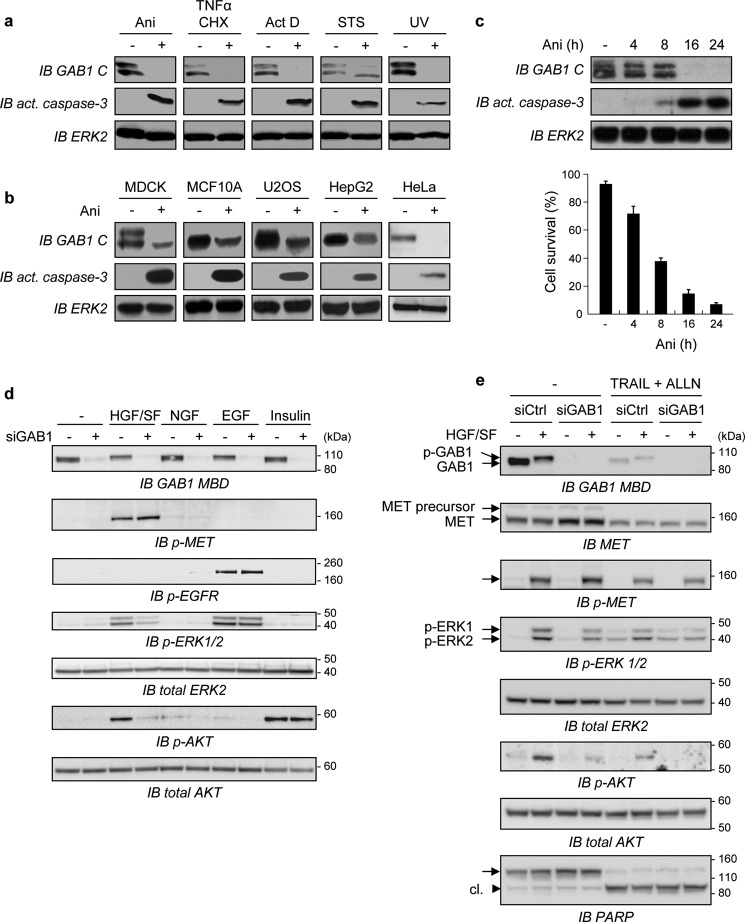
MDCK epithelial cells were treated with agents known to promote apoptosis through different mechanisms. Every treatment was found to induce GAB1 degradation together with apoptosis, as evidenced by detection of active caspase-3 (Fig. 1a). A decrease of GAB1 expression was also observed in various cell lines treated by anisomycin, making a cell line-specific effect unlikely (Fig. 1b). During time-course experiments, GAB1 degradation occurred within few hours, and this result was correlated with increased level of active caspase-3 and inhibition of cell survival (Fig. 1c).
FIGURE 1.
GAB1 down-expression by either stress conditions or by siGAB1 knockdown contributes to inhibition of HGF/SF-MET downstream signaling. a, MDCK cells were cultured in medium, 0% FBS for 8 h in the absence (−) or presence (+) of different apoptotic inducers: anisomycin (Ani, 50 μm), a combination of TNFα (30 ng/ml) and cycloheximide (CHX, 10 μg/ml), actinomycin D (ActD, 5 μg/ml), or staurosporine (STS, 1 μm). For UV treatment, cells were exposed to UV-B light 400 J/m2 for 2.5 min and then cultured in medium-0% FBS for an additional 8 h. Whole-cell lysates were immunoblotted (IB) using the anti-C terminus GAB1 antibody (GAB1 C) and an anti-active caspase-3 antibody. The filter was stripped and reprobed using an anti-ERK2 antibody to verify comparable loading. GAB1 was detected as a doublet that may correspond to the known A and B isoforms of GAB1 produced by alternative splicing. b, MDCK, MCF10A, U2OS, HepG2, or HeLa cells were treated for 8 h without (−) or with (+) anisomycin (Ani, 50 μm) in medium-0% FBS. Whole-cell lysates were immunoblotted using antibodies, as described for panel a. c, upper panel, MDCK cells were treated with anisomycin (Ani) or left untreated as described above for the indicated times in hours. Cell lysates were collected and then analyzed by immunoblotting using the same antibodies as described for panel a (IB act. caspase-3). Lower panel, the percentage of cell survival was determined by trypan blue staining. d, HeLa cells were transiently transfected with either siRNA negative control (−) or siRNA targeting the endogenous Gab1 transcript (siGab1, +). The next day cells were cultured in medium, 0.1% FBS for 6 h and subsequently treated with either HGF (50 ng/ml, 10 min), NGF (100 ng/ml, 10 min), EGF (10 ng/ml, 5 min), or insulin (0.1 μm, 3 min) or left untreated for 10 min. Cell lysates were then subjected to immunoblot analysis using an anti-GAB1 middle antibody (GAB1 M) directed against the residues surrounding Tyr-472 of GAB1. The same blot was stripped and reprobed several times using the indicated antibodies. e, HeLa cells were transiently transfected with either siRNA negative control (siCtrl) or siRNA targeting the endogenous GAB1 transcript (siGAB1). The next day cells were cultured overnight in medium, 0.1% FBS and subsequently treated with TRAIL (30 ng/ml) in combination with the proteasome inhibitor ALLN (50 μm) or left untreated (−) for 4 h. Cells were then treated (+) or not (−) with HGF/SF (50 ng/ml) for 10 min. Cell lysates were subjected to immunoblot analysis using an antibody directed against the GAB1-MBD (IB GAB1-MBD)). The same blot was stripped and reprobed several times using the indicated antibodies. Detection of PARP cleavage (cl.) was assessed to monitor caspases activation.
To evaluate the functions of GAB1 in mediating downstream signals of diverse growth factors in HeLa cells, GAB1 expression was knocked down using transient transfection of siGAB1, and cells were then treated or not by HGF/SF, NGF, EGF, or insulin (Fig. 1d). GAB1 was found to be crucial for activation of AKT and ERK only in response to HGF/SF, as demonstrated using antibodies recognizing phosphorylation sites of AKT (Ser-473) or ERK (Thr-202/Tyr-204). Such a GAB1 critical role in MET signal transduction is likely due to its specific and unique GAB1 MBS that confers its direct interaction with MET (9–11).
We then investigated the consequences of GAB1 down-expression on HGF/SF-MET signaling under stress conditions. Upon HGF/SF treatment, phosphorylation of GAB1 was induced, as evidenced by an up-shifted migration band, and in parallel both AKT and ERK were activated (Fig. 1e). In agreement with Fig. 1d, transient transfection of siGAB1, but not of the negative control siRNA (siCtrl), led to a decrease of both HGF/SF-induced AKT and ERK activation, whereas MET phosphorylation was unmodified. The combination of TRAIL with the proteasome inhibitor ALLN caused sustained down-expression of GAB1 and, to a lesser extent, down-expression of MET, which correlated with the increased cleavage of PARP that reflects caspases activation. Under these apoptotic conditions, HGF/SF-mediated activation of AKT and ERK was consistently decreased, and this effect was further amplified using siGAB1 (Fig. 1e). These data demonstrated down-expression of GAB1 in cells undergoing apoptosis, leading to a deleterious impact on downstream activation of both AKT and ERK, similarly to the forced siRNA-mediated GAB1 knockdown.
Stress Conditions Cause the Generation of a Stable GAB1 Fragment, Which Is Hindered by HGF/SF
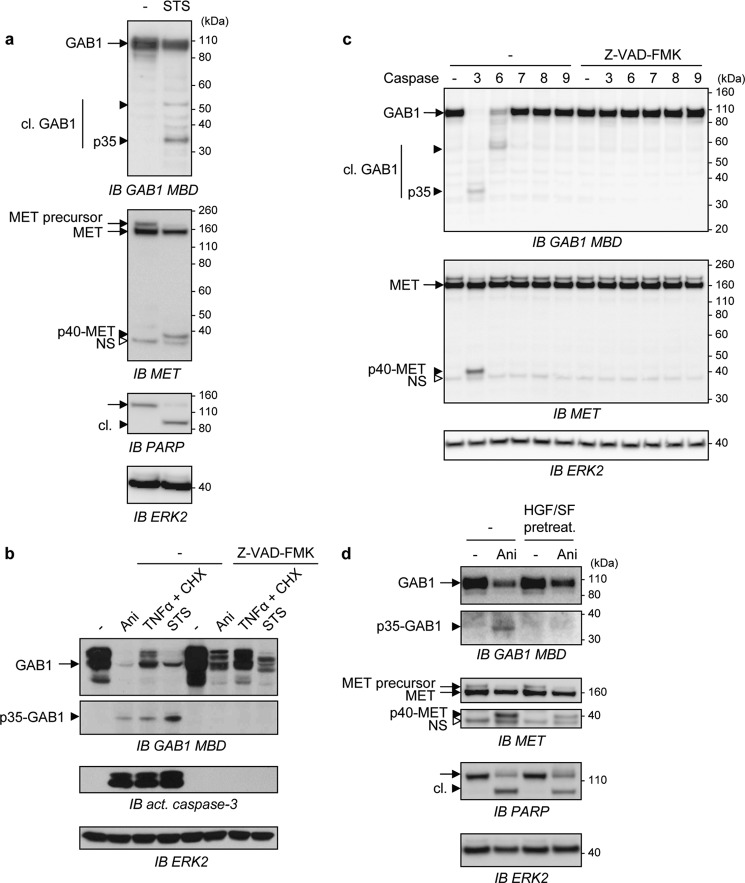
We further examined the mechanism leading to GAB1 down-expression during apoptosis. Upon staurosporine-induced apoptosis, GAB1 was found to be both degraded and cleaved, with the generation of fragments detectable with an antibody specifically recognizing the central part of GAB1 (anti-GAB1 MBD) (Fig. 2a). Despite the detection of many fragments, only a 35-kDa GAB1 fragment was consistently found and generated in parallel of the previously described intracellular p40-MET fragment (29–31). GAB1 was also found to be down-expressed by various stress inducers, and its expression was partially restored by treatment with Z-VAD-FMK, a general caspase inhibitor (Fig. 2b), indicating that GAB1 is a target of caspases and possibly of other stress-induced proteases. In parallel, the p35-GAB1 fragment was detected using the same stress inducers, and its generation was prevented by Z-VAD-FMK (Fig. 2b).
FIGURE 2.
Stress conditions cause the generation of a stable fragment of GAB1 and this caspase-dependent mechanism is hindered by HGF/SF. a, MDCK cells were treated with staurosporine (STS, 1 μm) or left untreated (−) for 8 h and then lysed. Immunoblot analysis (IB) was performed on cell lysates, and the membrane was probed using anti-GAB1 MBD or anti-MET antibodies. The filter was stripped and reprobed with anti-PARP antibody and anti-ERK2 antibody. b, MDCK cells were cultured in medium, 0% FBS and pretreated without (−) or with the general caspase inhibitor Z-VAD-FMK (20 μm) for 30 min followed by a treatment with either anisomycin (Ani, 50 μm), a combination of TNFα (30 ng/ml) and cycloheximide (CHX, 10 μg/ml), or staurosporine (STS, 2 μm) for 16 h. Cell lysates were analyzed by immunoblotting using the indicated antibodies. c, MDCK cells were lysed, and the same amount of protein for each sample was incubated without (−) or with active purified caspase-3, -6, -7, -8, or -9 at 37 °C for 4 h in the absence (−) or presence of the general caspase inhibitor Z-VAD-FMK (40 μm). Samples were then analyzed by immunoblotting using the anti-GAB1 MBD or the anti-MET antibodies. The filter was stripped and reprobed using an anti-ERK2 antibody to verify comparable loading. d, MDCK cells were pretreated with HGF/SF (HGF/SF pretreat., 10 ng/ml) or left untreated (−) for 1 h and incubated for an additional 10 h with or without (−) anisomycin (Ani, 50 μm). Cell lysates were analyzed by immunoblotting using the indicated antibodies. a-c, arrows point to full-length (black arrow) and cleaved (black arrowhead) proteins. The small white arrowhead corresponds to a nonspecific immunoreactive band (NS). Cleaved proteins are abbreviated as cl.
To establish which caspases cleave GAB1, cell lysates were subjected to an in vitro caspase cleavage assay using purified active recombinant caspases (Fig. 2c). Among them, two effector caspases (caspases-3 and -6) were capable to cleave GAB1 even though only caspase-3 led to p35-GAB1 generation. In contrast caspase-7, -8, and -9 have no significant effects on GAB1 cleavages, and no cleavages were evidenced in the presence of Z-VAD-FMK (Fig. 2c). In this experiment we have verified the correct processing of caspase-7, -8, and -9 in their active form (supplemental Fig. S1a). These results show the preferential cleavage of GAB1 by caspase-3, similarly to MET (29, 30). To investigate the possible existence of other GAB1 fragments, cells were transfected with either a C-terminal Myc-tagged human GAB1 (GAB1-Myc, ∼120 kDa) or an N-terminal hemagglutinin-tagged human GAB1 (HA-GAB1, ∼100 kDa)-encoding plasmids, and cell lysates were then subjected to an in vitro caspase-3 cleavage assay (supplemental Fig. S1b). A 35-kDa fragment was observed in both samples using the GAB1 MBD antibody, whereas no detectable fragment was found using either an anti-HA antibody or an anti-GAB1 antibody directed against the C terminus of human GAB1 (anti-GAB-C). A fragment of about 24 kDa was recognized by the anti-Myc antibody but not by the C-terminal antibody, indicating the presence of a cleavage site in the extreme C-terminal region of GAB1, as the size of the Myc tag is of 20-kDa (constituted of Myc-tag repeats). Nonetheless, the absence of detectable N- and C-terminal fragments using anti-HA or anti-GAB1-C antibodies, respectively, confirmed that p35-GAB1 is the only stable GAB1 fragment generated by caspase cleavages (supplemental Fig. S1b).
Finally, pretreatment with HGF/SF before stress induction was found to partially protect GAB1 from degradation and to impair generation of p35-GAB1 fragment. In parallel, HGF/SF also prevented the caspase-dependent cleavage of MET and PARP (Fig. 2d). These data are in agreement with the known anti-apoptotic effects of HGF/SF (16, 17) and with protection of MET-dependent caspase cleavages by HGF/SF (30, 32). These results demonstrated that the GAB1 adaptor is cleaved by caspases under stress induction, leading to the generation of a unique and stable 35-kDa GAB1 fragment containing its central region.
The Caspase-cleaved p35-GAB1 Fragment Contains the Previously Described GAB1 MBD
According to the detection of the p35-GAB1 fragment with an antibody recognizing the GAB1 MBD domain (aa 450–532), we examined whether GAB1 is cleaved at multiple sites on both sides of the GAB1 MBD. It is generally accepted that caspases cleave substrates after an aspartate residue located at the end of a tetrapeptide motif followed preferentially by a small residue (such as glycine, serine or alanine) even though phenylalanine and tyrosine aromatic residues are also well tolerated (35). We analyzed putative caspase cleavage sites in GAB1 using the Server for SVM Prediction of Caspase Substrates Cleavage Sites (CASVM) (36). This analysis revealed 17 predicted motifs within the GAB1 sequence (694 aa) (supplemental Table 1).
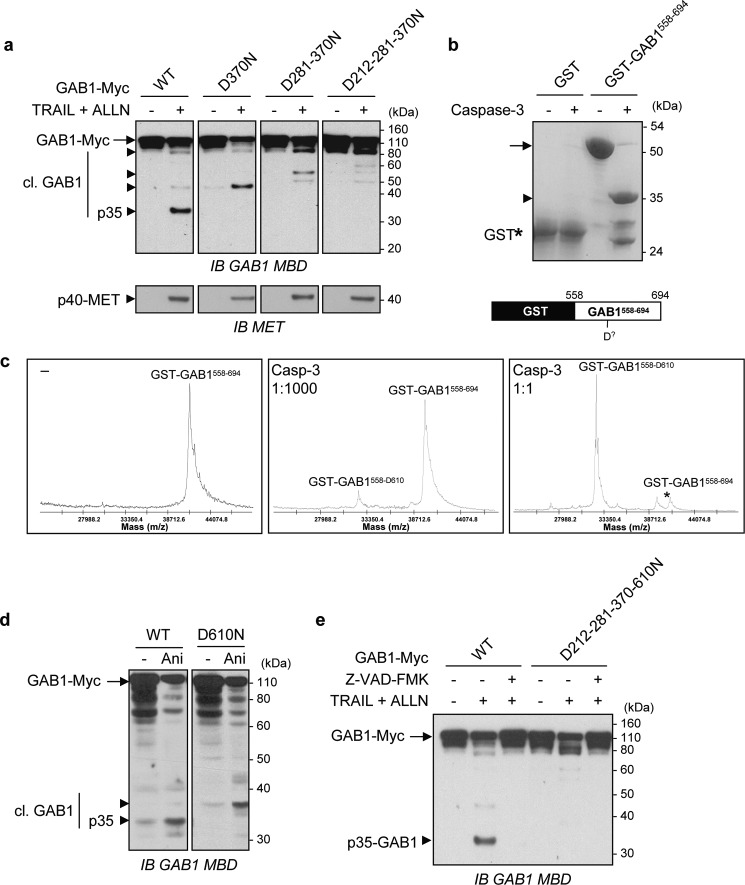
Among these sites, the SDTD370 motif is a caspase cleavage site for two distinct proteins, Rad9 (37) and JNK (38), and this motif is followed by a favorable residue (serine) only for GAB1. We proceed to a D370N substitution in the sequence of the GAB1-Myc plasmid to examine its functionality as a caspase cleavage site (Fig. 3a). Transient cellular expression of the GAB1 D370N mutant caused an upward shift in the size of the GAB1 fragment from 35 to ∼45 kDa, showing that Asp-370 is a caspase cleavage site. The modest upward shift of the fragment also revealed the existence of another upstream cleavage site. By further mutating upstream putative sites, individually or in combination with D370N, we progressively found that the GAB1 D281N/D370N and D212N/D281N/D270N multiple mutants increased the size of p35-GAB1 to ∼55 and ∼90 kDa, respectively (Fig. 3a). Three caspase-cleavage sites Asp-212, -281, and 370 were therefore identified, located upstream of the GAB1 MBD.
FIGURE 3.
The p35-GAB1 fragment is surrounded by four caspase cleavage sites and contains the MET binding domain. a, after transient transfection for 24 h using Myc-tagged wild type (WT) GAB1 or its mutated versions D370N, D281N/D370N, or D212–281-370N, HeLa cells were cultured overnight in medium, 0.1% FBS. Cells were then treated (+) with TRAIL (30 ng/ml) in combination with the proteasome inhibitor ALLN (50 μm) or left untreated (−) for 5 h. For each condition, the same amount of whole-cell extracts was analyzed by immunoblotting (IB) using anti-GAB1 MBD or anti-MET antibodies. b, purified recombinant GST and GST-GAB1558–694 proteins were incubated with (+) or without (−) recombinant active caspase-3 at 37 °C for 1 h. Products were then separated by SDS-PAGE and stained using Coomassie Blue (upper panel). Schematic representation of the GST-GAB1558–694 structure is also shown on the lower panel. c, purified recombinant GST-GAB1558–694 protein was analyzed on a MALDI-TOF spectrometer before (left panel) and after cleavage with caspase-3 at a low (1:100 dilution, middle panel) or high concentration (1:1 dilution, right panel) at 37 °C for 1 h. Experimentally determined masses of GST-GAB1 and the main cleavage product are indicated. d, MDCK cells were transfected for 24 h with Myc-tagged wild type (WT) GAB1 or its mutant D610N. Cells were then treated with anisomycin (Ani, 50 μm) or left untreated (−) for 8 h. Cell lysates were collected and analyzed by immunoblotting using an anti-GAB1 MBD antibody. e, after transient transfection for 24 h with Myc-tagged wild type (WT) GAB1 or its mutated version D212N/D281N/D370N/D610N, HeLa cells were cultured overnight in medium, 0.1% FBS. Cells were then left untreated (−) or pretreated (+) with Z-VAD-FMK (20 μm) for 30 min immediately followed by an additional 5-h incubation with (+) or without (−) TRAIL and ALLN (as described in panel a). Immunoblot analysis was then performed on whole-cell lysates using an anti-GAB1 MBD antibody. a–e, arrows point to full-length (black arrow) and cleaved (black arrowhead) proteins. Cleaved GAB1 is abbreviated as cl. GAB1.
Attempts to identify C-terminal cleavage sites by site-directed mutagenesis strategies were unsatisfactory. We, therefore, generated a GST-GAB1 recombinant fusion protein containing the last aa of GAB1 (GST-GAB1558–694, ∼50 kDa). The purified GST- GAB1558–694 fusion protein was subjected to a caspase-3 cleavage assay, which led to the generation of a main GAB1 ∼35-kDa fragment (Fig. 3b). Using mass spectrometry analysis, a major peak was observed and, according to its molecular weight of 32,382, was attributed to a caspase cleavage at Asp-610 (Fig. 3c). A GAB1 D610N mutant was then generated and introduced into cells by transfection before treatment by anisomycin. After immunoblotting of cell lysates, an upward shift in the size of the GAB1 fragment from 35 to ∼38 kDa (Fig. 3d), confirmed that Asp-610 was a caspase cleavage site. Finally, the four putative caspase cleavage sites were introduced in GAB1 (D212N/D281N/D370N/D610N), and no more detectable GAB1 fragments were observed by immunoblotting using the anti-GAB1 MBD antibody (Fig. 3e), showing that the four sites are the main caspase cleavage sites in GAB1. Overall, the p35-GAB1 fragment contains the previously described GAB1 MBD domain, and we have mapped its upstream (Asp-212, Asp-281, and Asp-370) and downstream (D610) caspase cleavage sites (see Fig. 8b).
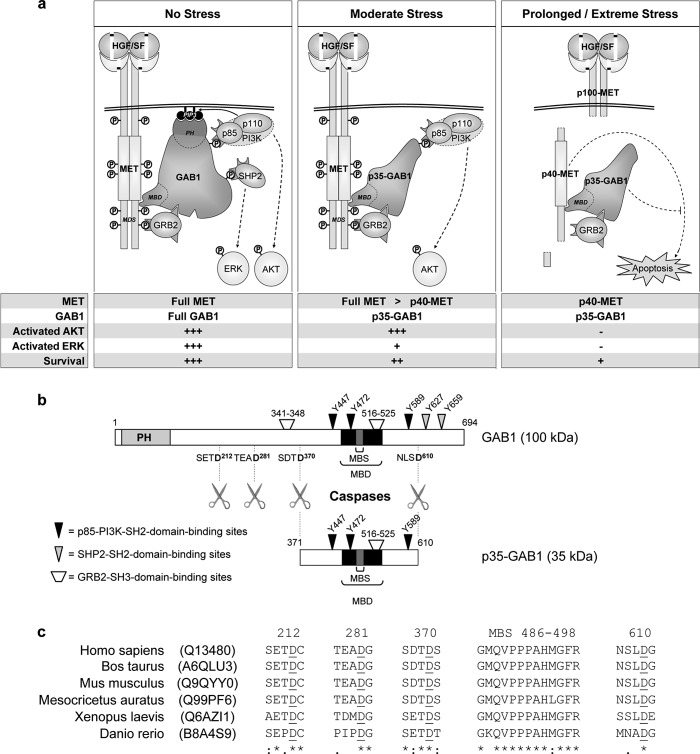
FIGURE 8.
The GAB1 adaptor is a checkpoint regulator of HGF/SF-MET signaling under stress conditions. a, shown is a model for the regulatory role of the GAB1 adaptor and its caspase-cleaved p35-GAB1 fragment in HGF/SF-MET signaling under distinct stress conditions (see “Discussion”). Full MET or Full GAB1 stands for full-length MET or full-length GAB1, respectively; p40-MET and p35-GAB1 stands for the caspase-dependent fragment of MET and GAB1, respectively; +++, strong response to HGF/SF; +, decreased response to HGF/SF; −, no response. b, shown is a schematic representation of the presently identified four caspase cleavage sites in the GAB1 adaptor and main signaling partners binding sites for either full-length GAB1 protein or the p35-GAB1 fragment. Specific aa in GAB1 are indicated corresponding in particular to phosphorylated tyrosine residues known to be involved in binding of SH2 domain-containing proteins. The apparent molecular weights of GAB1 and p35-GAB1 in SDS gels are indicated. c, sequence alignment shows conservation of the GAB1-MBS and the presently identified caspase cleavage sites in GAB1 from various species. All sequences were retrieved from the UniProt database, and UniProt entries are indicated. Numbering is indicated according to the human sequence. Aspartate sites are underlined. See also the full sequence alignment of various species in supplemental Fig. S5.
The GAB1 Adaptor Is Both Stress-degraded and Caspase-cleaved Faster and More Progressively Compared with the MET Receptor
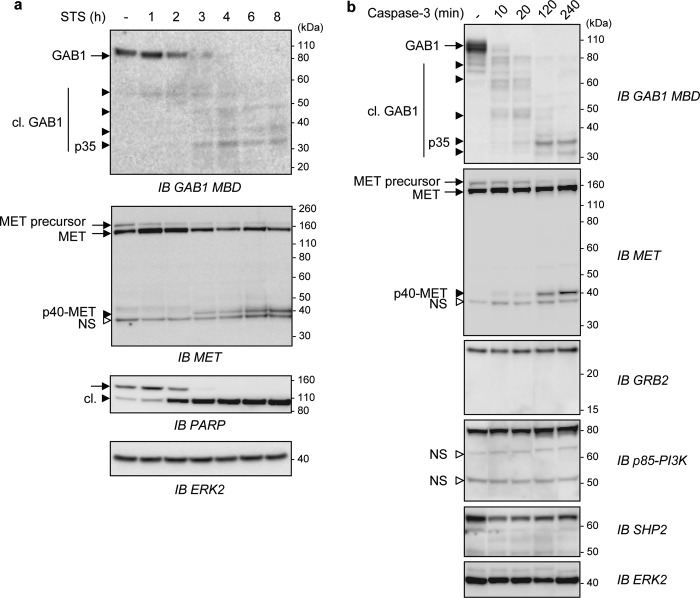
We next investigated the kinetics of degradation and caspase cleavage of MET and GAB1. Time-course experiments were performed from either cells treated by stress inducers (Fig. 4a) or from cell lysates subjected to a caspase-3 cleavage assay (Fig. 4b) to mobilize endogenous stress proteases or to analyze the effect of caspase-3, respectively. The two approaches gave similar results; GAB1 was found to be cleaved faster than MET, leading to the generation of unstable truncated forms of GAB1 and finally to the stable p35-GAB1 fragment, whereas p40-MET was detected without any detectable intermediate processing. Nevertheless, both p35-GAB1 and p40-MET were detected at the same time point. Interestingly, other binding partners of the activated MET signaling complex, e.g. GRB2, p85-PI3K, and SHP2, were neither degraded nor caspase-3-cleaved. Accordingly, it is possible that in response to stress induction, the full MET receptor is rapidly unable to interact with intact full GAB1 but rather may interact with truncated GAB1, which in turn may impair normal MET receptor signaling.
FIGURE 4.
Both MET and GAB1 are degraded and fragmented by stress or by caspase 3, with the GAB1 adaptor being caspase-cleaved faster and more progressively than the MET receptor. a, HeLa cells were left untreated (−) or treated with staurosporine (STS, 1 μm) for the indicated times in hours. Cell lysates were then immunoblotted (IB) using the indicated antibodies. b, lysates from MDCK cells were incubated without (−) or with active purified caspase-3 at 37 °C for the indicated times in minutes. Samples were then analyzed by immunoblotting using the indicated antibodies. a and b, arrows point to full-length (black arrow) and cleaved (black arrowhead) proteins. The small white arrowhead corresponds to a nonspecific immunoreactive band (NS). Cleaved proteins are abbreviated as cl.
The p35-GAB1 Fragment Maintains Parts of Full-length GAB1 Recruitment Capacities
The p35-GAB1 fragment is potentially able to interact with both MET and p85-PI3K. It contains the GAB1-MBD and has conserved one of the two GRB2 binding motifs (aa 516–525) and all three SH2 binding sites for p85-PI3K but has lost the C-terminal SHP2-interacting domains containing crucial Tyr phosphorylation sites (Tyr-627 and -659) (7, 8, 14, 15) (Fig. 8b).
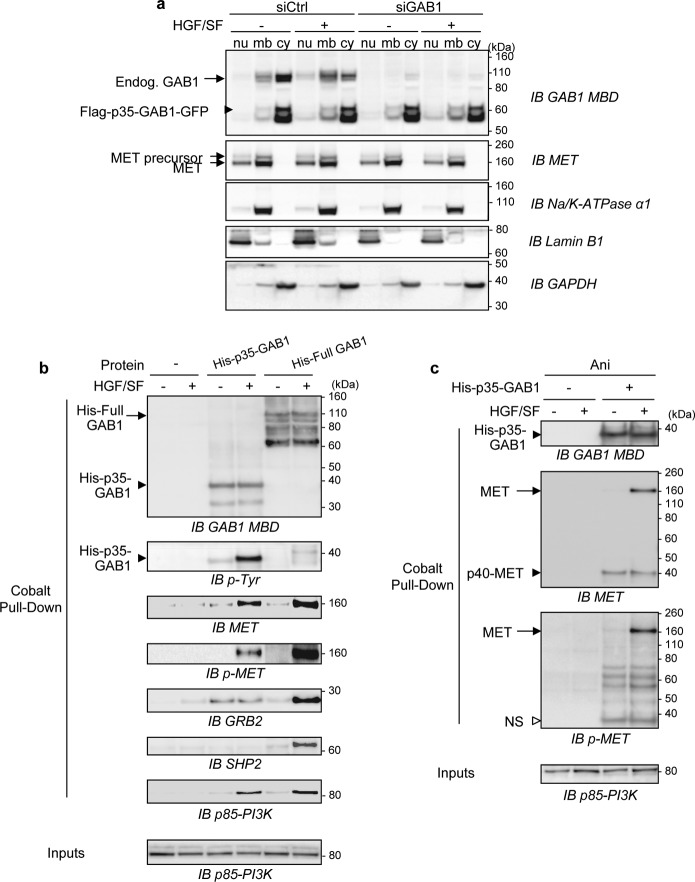
To examine the subcellular distribution of p35-GAB1, we generated MDCK cells stably expressing the p35-GAB1 fragment with an N-terminal FLAG tag and a C-terminal GFP (FLAG-p35-GAB1-GFP) or the control vector (FLAG-GFP) (supplemental Fig. S2). The FLAG-p35-GAB1-GFP fusion proteins were detected at the expected size of 60 kDa using anti-GAB1 MBD or anti-GFP antibodies and were largely overexpressed compared with endogenous GAB1. The FLAG-p35-GAB1-GFP protein was tyrosine-phosphorylated in response to HGF/SF, indicating that this protein can interact with the activated MET receptor (supplemental Fig. S2b). Cells were transiently transfected with siGAB1 directed against the canine sequence to knock down endogenous GAB1 expression and were then treated or not by HGF/SF and subjected to cell fractionation. The p35-GAB1 fragment was found to be mainly cytosolic and to relocalize to the membrane compartment upon HGF/SF treatment, similarly to endogenous full GAB1 (Fig. 5a). In agreement with these results, FLAG-p35-GAB1-GFP was detected at the plasma membrane in response to HGF/SF by confocal microscopy analysis (supplemental Fig. S3).
FIGURE 5.
Subcellular localization and association capacities of the p35-GAB1 fragment. a, MDCK cells stably expressing FLAG-p35-GAB1-GFP proteins were transiently transfected with siRNA control (siCtrl) or targeting endogenous GAB1 transcripts (siGAB1). The next day cells were grown overnight in medium, 0.1% FBS and subsequently treated with HGF/SF (50 ng/ml) for 10 min (+) or left untreated (−). Cell lysates were sequentially fractionated in subcellular compartments (nu, nucleus; mb, membrane; cy, cytosol) and subjected to immunoblot (IB) analysis using GAB1 or MET antibodies as well as antibodies directed against classical compartment markers (Na+/K+-ATPase, Lamin B1, and GAPDH). b and c, MDCK cells were cultured overnight in medium, 0.1% FBS in the absence (b) or presence (c) of anisomycin (Ani, 50 μm) and were then left untreated (−) or treated (+) with HGF/SF (100 ng/ml) for 5 min. Cell lysates were prepared and incubated without (−) or with purified recombinant His-tagged p35-GAB1 (His-p35-GAB1) or full-length GAB1 (His-Full GAB1) proteins. Pulldown assays were performed, and eluates were analyzed by immunoblotting using the indicated antibodies. Total cell lysates before pull down (Inputs) were also loaded and analyzed (supplemental Fig. S4, b and c). In c, the white arrowhead corresponds to a nonspecific immunoreactive band (NS).
To investigate the p35-GAB1 binding properties, we generated and expressed recombinant proteins, corresponding to p35-GAB1 (His-p35-GAB1) and full-length GAB1 (His-Full GAB1). The purified recombinant proteins were detectable by Coomassie Blue staining at the expected sizes (∼38 kDa for His-p35-GAB1; ∼110 kDa for His-Full GAB1) (supplemental Fig. S4). Pulldown assays were performed by incubating these purified recombinant proteins with lysates from cells treated or not by HGF/SF and pretreated (Fig. 5c) or not (Fig. 5b) with stress. Samples were processed for immunoblotting.
In response to HGF/SF, p35-GAB1 was found tyrosine-phosphorylated and able to interact with the phosphorylated MET receptor, showing that p35-GAB1 can be recruited and phosphorylated by the activated MET receptor. Similar data were obtained using full GAB1 as expected. We also found that p35-GAB1 interacts with GRB2 independently of HGF/SF action, whereas full GAB1 interacts more efficiently with GRB2 in response to HGF/SF. Both p35-GAB1 and full GAB1 were found to bind p85-PI3K in the presence of HGF/SF. In contrast to full GAB1, p35-GAB1 was found unable to bind SHP2, as expected (see Fig. 5b and supplemental Fig. S4b for input details). These data demonstrated that in response to HGF/SF, the p35-GAB1 fragment is still able to interact with MET and p85-PI3K, but not with SHP2, and interacts constitutively with GRB2.
Cells were also exposed overnight to stress induction by anisomycin and then challenged for 10 min by HGF/SF. The p35-GAB1 fragment was still found to interact with the MET receptor activated by HGF/SF, whereas it was found to interact with p40-MET or GRB2 independently of HGF/SF treatment. Endogenous p40-MET was not found to be phosphorylated in response to HGF/SF (see Fig. 5c and supplemental Fig. S4c for inputs details), in agreement with the fact that this caspase-cleaved fragment corresponds to the intracellular kinase domain, not bound to the p100-MET membrane fragment (30, 31). These data demonstrated that under prolonged stress conditions, the p35-GAB1 fragment has conserved the capacities to interact with MET activated by HGF/SF, whereas it interacts with p40 MET independently of HGF/SF action.
In Response to HGF/SF, the p35-GAB1 Fragment Allows Resistance to Apoptotic Stress
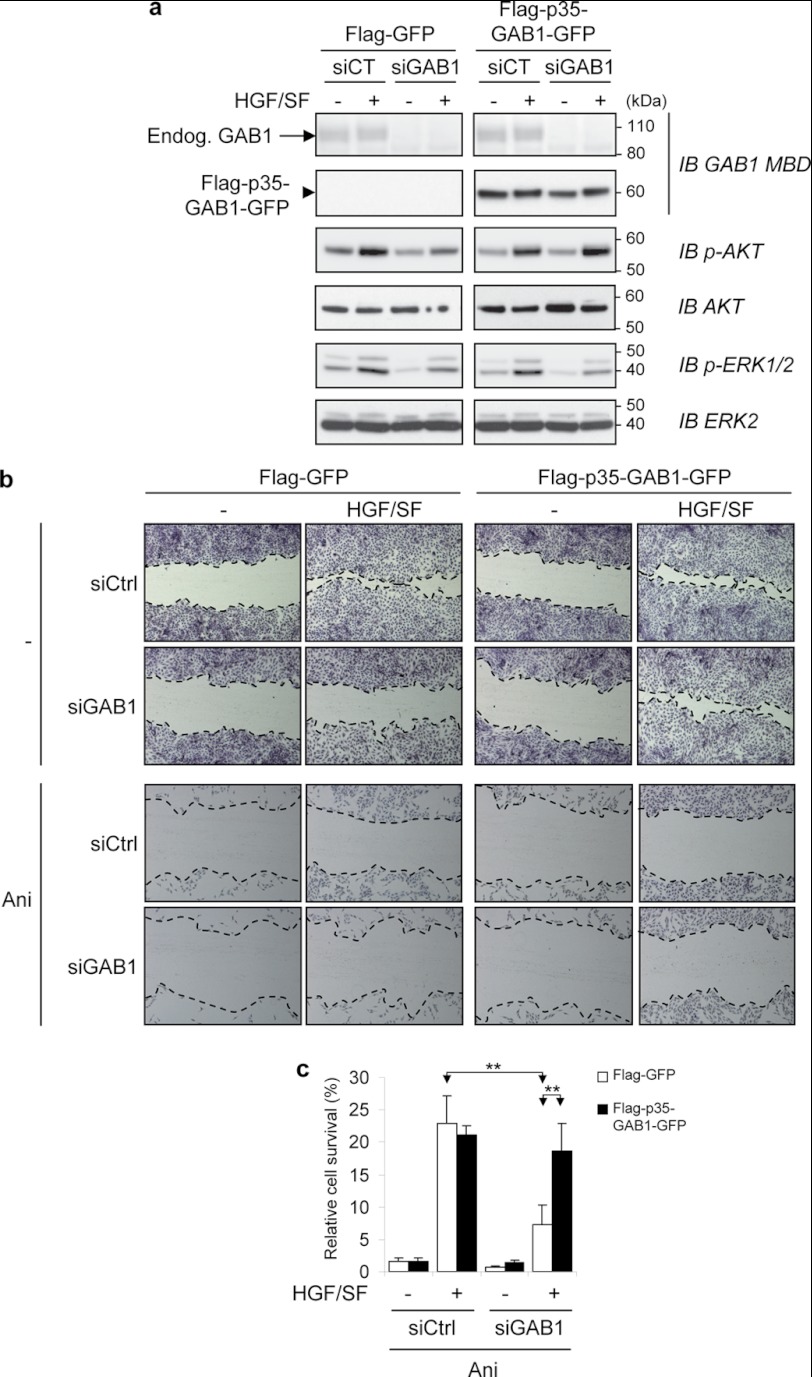
We evaluated HGF/SF-MET signaling in MDCK cells stably expressing the FLAG-p35-GAB1-GFP construct. To optimize examination of the functional consequences of overexpression of FLAG-p35-GAB1-GFP, we knocked-down endogenous expression by siGAB1 before examining HGF/SF downstream targets (Fig. 6a). In response to HGF/SF, we found that AKT activation was maintained in FLAG-p35-GAB1-GFP compared with FLAG-GFP-expressing cell lines, whereas ERK activation was decreased in both cell lines (Fig. 6a). It is worth mentioning that the ability of p35-GAB1 to sustain AKT activation was evidenced in these conditions of p35-GAB1 overexpression and not in endogenous conditions (see Fig. 1e). Indeed, in endogenous conditions and in response to HGF/SF and apoptotic stimuli, the downstream activation of AKT or ERK is a combination of the complex contributions of MET, GAB1, and their corresponding fragments (see Fig. 1e).
FIGURE 6.
In response to HGF/SF, the p35-GAB1 fragment maintains AKT activation and cell migration and allows resistance to apoptotic stress. a, MDCK cells stably expressing FLAG-GFP or FLAG-p35-GAB1-GFP proteins were transiently transfected with either siRNA negative control (siCT) or siRNA targeting the endogenous GAB1 transcript (siGAB1). The next day cells were grown overnight in medium, 0.1% FBS and subsequently treated with HGF/SF (50 ng/ml) for 10 min (+) or left untreated (−). Cell lysates were subjected to immunoblot (IB) analysis using the indicated antibodies. Note that expression of endogenous GAB1 (Endog. GAB1) was faint compared with FLAG-p35-GAB1-GFP. b, MDCK cells stably expressing FLAG-GFP or FLAG-p35-GAB1-GFP protein were transiently transfected with either siRNA negative control (siCtrl) or siRNA targeting the endogenous GAB1 transcript (siGAB1). The next day a scratch wound-healing assay was performed. After 24 h of incubation without (−) or with HGF/SF (30 ng/ml) in medium, 0.1% FBS, cells were stained using Hemacolor kit, and the wound closure areas were visualized under an inverted microscope (×40). Dotted lines were added to delineate the migrating cell front. Note that the width of scratched wounds was comparable between samples just before incubation of HGF/SF. c, MDCK cells stably expressing FLAG-GFP or FLAG-p35-GAB1-GFP proteins were transiently transfected with siRNA as described above. The next day cells were treated for 16 h with or without anisomycin (Ani, 1 μm) in medium-0.1% FBS in the absence (−) or presence (+) of HGF/SF (30 ng/ml). An MTT assay was performed to evaluate cell survival. Results are expressed as a percentage of untreated control cells. All values are shown as the mean ± S.D. of three independent MTT assays, each condition measured in triplicate (**, p < 0.01 using Student's t test with Bonferroni correction).
These results show that p35-GAB1 maintains part of the intact GAB1 protein functional capacities in response to HGF/SF. Indeed, p35-GAB1 is still recruited to the activated MET receptor along with GRB2 and subsequently tyrosine-phosphorylated, which allows p85-PI3K recruitment (Fig. 5b) and downstream activation of the PI3K-AKT pathway (Fig. 6a). In contrast, p35-GAB1 is unable to bind SHP2 (Fig. 5b), which alters downstream activation of the RAS-ERK pathway (Fig. 6a).
The effect of the p35-GAB1 fragment was examined on cell motility using a scratch wound-healing assay. After siRNA-mediated extinction of endogenous GAB1, stably transfected cells were incubated or not with HGF/SF for 24 h (Fig. 6b). HGF/SF-induced migration was conserved in FLAG-p35-GAB1-GFP cells but not in control FLAG-GFP cells, demonstrating the efficiency of the p35-GAB1 fragment in favoring cell motility by reversing the knockdown of GAB1.
A similar experiment was performed in the presence of both HGF/SF and anisomycin. Under these conditions, the survival rather than the scattering effect of HGF/SF was evidenced, and this survival effect was more pronounced in FLAG-p35-GAB1-GFP cells than in control FLAG-GFP cells (Fig. 6b). To more directly address this question, we examined cell viability using a MTT assay (Fig. 6c). The results demonstrate that in response to HGF/SF, the p35-GAB1 fragment allows resistance to apoptotic stress.
In the Absence of HGF/SF, the p35-GAB1 Fragment Impedes Apoptosis Amplified by the p40-MET Fragment
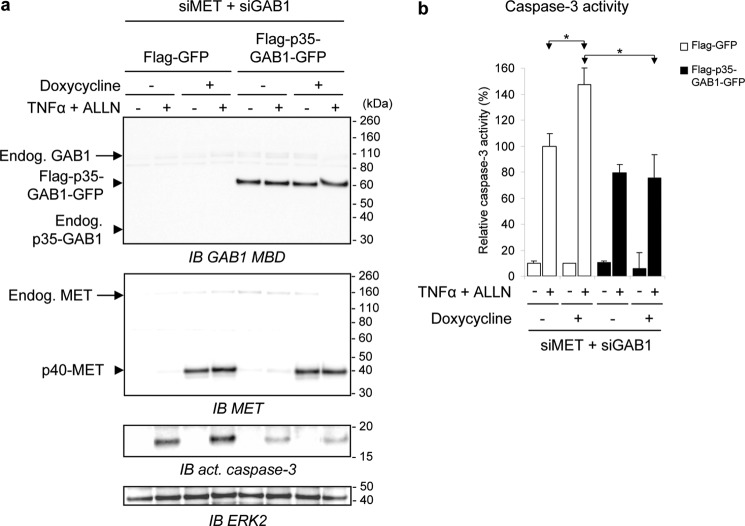
Because p35-GAB1 can interact with p40-MET even in the absence of HGF/SF (Fig. 5c), we investigated whether p35-GAB1 can modulate the pro-apoptotic p40-MET function. To this end we stably expressed the FLAG-p35-GAB1-GFP construct or the corresponding control (FLAG-GFP) in a MDCK cell line stably carrying a doxycycline-inducible p40-MET fragment (MDCK-p40-MET-Tet-On6). To avoid interference with endogenous GAB1 and MET and their corresponding fragments, we knocked down the canine endogenous transcripts by double-silencing MET and GAB1. In these conditions induction of p40-MET effectively amplified TNFα/ALLN-induced apoptosis, as revealed by the augmentation of activated caspase-3 in FLAG-GFP-expressing cells (Fig. 7a). In FLAG-p35-GAB1-GFP-expressing cells, production of the activated caspase-3 was lowered and was no longer increased upon induction of p40-MET expression (Fig. 7a). These results were confirmed by quantification of caspase-3 activity (Fig. 7b). These results show that the caspase-generated p35-GAB1 fragment can also counteract amplification of apoptosis by p40-MET.
FIGURE 7.
The p35-GAB1 fragment hinders amplification of caspase 3 activity by the p40-MET fragment. MDCK-p40-MET-Tet-On cells stably expressing FLAG-GFP or FLAG-p35-GAB1-GFP proteins were transiently transfected with siRNAs targeting both endogenous GAB1 and MET transcripts (siMET + siGAB1). After 48 h of cell culture in a complete medium, cells were starved overnight in medium, 0.1% FBS with (+) or without (−) doxycycline (0.2 μg/ml) and subsequently treated with TNFα (30 ng/ml) and ALLN (50 μm) for 4 h (+) or left untreated (−). Cell lysates were subjected to immunoblot analysis (IB) using indicated antibodies (a), or caspase-3 activity assay was performed from an independent experiment (b) (*, p < 0.05 using Student's t test with Bonferroni correction). Similar results were obtained in distinct experiments.
DISCUSSION
Although the multisubstrate docking protein GAB1 is known to amplify, diversify, and integrate the HGF/SF-MET-induced signaling pathways (7), its role in mediating cell survival has led to controversial results. Indeed, overexpression of GAB1 was found to inhibit the ability of HGF/SF to protect cells against adriamycin, a DNA-damaging agent (39), whereas cells with a homozygous deletion of the GAB1 gene were more sensitive to the deleterious effects of H2O2 (40) or of UV irradiation (41). Our present results reveal the importance of examining the fate of GAB1 in stress conditions to understand its role during apoptosis. We found that under apoptotic stresses, GAB1 undergoes proteolytic processing by caspases, leading to the generation of a unique 35-kDa cleavage fragment, p35-GAB1, that accompanies the caspase-dependent cleavage of MET in a pro-apoptotic p40-MET fragment that we previously reported (29–31). Although it could be expected that the cleavage of GAB1 will simply reinforce the pro-apoptotic function of MET, we found that the p35-GAB1 fragment can maintain the survival function of HGF/SF-MET under moderate stress.
We mapped four main caspases sites in human GAB1 sequence located on both sides of the GAB1 MET MBD, i.e. Asp-212, Asp-281, Asp-370, and Asp-610 (Fig. 8b). Caspase sites are usually defined as sequences of four amino acids, with a terminal aspartate, after which caspases cleave. None of the identified caspase sites, i.e. SETD212, TEAD281, SDTD370, and NSLD610, resemble identified caspase motifs, except SDTD370 (42), which is a caspase cleavage site for at least two other proteins, Rad9 (37) and JNK (38). However, for all these sites, the aspartate position is immediately followed by a small amino acid residue (either glycine, serine, or cysteine), which is often a common signature for caspase cleavage sites (35). Interestingly, by aligning known GAB1 amino acid sequences from different species, we found that these caspase cleavage motifs are extremely conserved along evolution, with the four aspartate sites being found in all vertebrates (Fig. 8c and supplemental Fig. S5). These cleavage sites allow the generation of a p35-GAB1 fragment that contains the GAB1 MET MBS, which also align almost perfectly in vertebrates. Therefore, preservation of both caspase sites and MBS bring us to propose that generation of p35-GAB1 in apoptotic cells is a highly conserved process throughout evolution that might not be limited to human and canine GAB1, as presently demonstrated.
We demonstrated that full-length GAB1 is severely and rapidly cleaved by caspases, with the p35-GAB1 fragment being progressively generated. In contrast, the full membrane-bound MET receptor remained present for a longer period of time than GAB1 and can coexist with the generated cytosolic p40-MET fragment. In agreement with these data, we demonstrated that p35-GAB1 is responsive to HGF/SF through activation of MET. Indeed, p35-GAB1 is recruited and phosphorylated by the activated MET receptor and can recruit p85-PI3K. We also found a constitutive interaction of GAB1 with the GRB2 adaptor, in agreement with its original identification (4). Such GRB2 interaction with p35-GAB1 could reinforce the complex between MET and the caspase-cleaved fragment of GAB1. Overexpression of p35-GAB1, itself activated by HGF/SF, was capable of causing the subsequent downstream activation of the PI3K-AKT pathway as well as cell migration and resistance to apoptotic stress. On the other hand, p35-GAB1, in agreement with the loss of the C-terminal part of GAB1, was not capable to recruit SHP2 any more, and its overexpression in cells was associated with a reduced activation of the RAS-ERK pathway, as expected for the role of SHP2 in GAB1 signaling (7, 8, 14, 15). Taken together, these data indicate that the caspase cleavage of the GAB1 docking adaptor protein can maintain HGF/SF-induced cell survival/resistance to apoptosis (Fig. 8a).
In the time-course of the apoptotic response, we evidenced the simultaneous generation of both p40-MET and p35-GAB1, which can interact independently of HGF/SF action. Under prolonged or extreme stress conditions, we also observed that both MET and GAB1 are fully degraded and cleaved by caspases, indicating that cells will not be responsive to HGF/SF anymore. To address the relative importance of both fragments in regulating apoptosis, we coexpressed the p40-MET and p35-GAB1 fragments in the context of double silencing of endogenous MET and GAB1. In response to stress, we found that the p35-GAB1 fragment can hinder apoptosis by the p40-MET fragment. These results indicate that the p35-GAB1 fragment can play an additional anti-apoptotic role by modulating the pro-apoptotic function of the cleaved MET receptor (Fig. 8a). Although we previously demonstrated that p40-MET exhibits pro-apoptotic functions (28, 29), the mechanism by which p40-MET induces apoptosis is still undefined.
The N-terminal PH domain mediates localization of GAB1 to the plasma membrane where receptor-tyrosine kinases are activated. Nonetheless, due to the presence of the MBD, which confers the specificity to the MET/GAB1 interaction (5, 9, 10), the PH domain would be likely dispensable for part of the MET receptor biological activities. This explains why the p35-GAB1 fragment still functions as a cell survival and migration mediator despite a reduced ability to concentrate to the cell membrane.
Several component members of ERK and AKT signaling pathways, including RasGAP, RAF-1, and AKT-1, are cleaved by caspases in apoptotic cells, which results in the inhibition of survival and cell growth, allowing the death signals to predominate (43–46). However, AKT cleavage occurs relatively late during the apoptotic response (45), which is consistent with our observations that PI3K/AKT survival signals persist when p35-GAB1 is generated. More generally, only a limited set of proteins with signaling functions could be cleaved in cells undergoing cell death, thus inducing the specific turn-off of survival pathways, which could otherwise interfere with the apoptotic response.
GAB1 is not the unique example of an adaptor protein that is a substrate for caspase cleavage. For instance, insulin receptor substrate-1 is cleaved by caspase-10, which leads to the suppression of insulin-like growth factor-I-mediated signals (47). Gads cleavage results in the uncoupling of signaling pathways downstream of the T cell receptor (48). Proteolytic processing of kinase suppressor of RAS 1 (KSR1) alters ERK activity in apoptotic cells (49). The functional consequence of caspase cleavage of these proteins is the loss/destruction of their full-length scaffold/adaptor role as well as the production of cleavage product, which can act in a dominant negative/inhibitory manner, resulting in the down-regulation of survival signals to promote cellular apoptosis. GAB1 distinguishes itself from other adaptor proteins by the fact that caspase-mediated cleavage of GAB1 does not lead to the simple disruption of HGF/SF-MET signaling and/or to amplification of apoptosis but rather can maintain MET receptor survival signals. These data originally document the fine-tuning provided by a docking protein to its specific receptor.
Supplementary Material
Acknowledgments
We thank H. Drobeck for mass spectrometry assistance, C. de Witte for technical help, W. Towler and E. Martini for critical reading of the manuscript, and J. Deheuninck for fruitful discussions. We are grateful to Prof. P. Raynal (EA4568, Université Toulouse III, Toulouse, France) for providing GAB1 constructs, Dr. G.S. Salvesen (The Burnham Institute, La Jolla, CA) for providing active caspases, and Dr. D. Tulasne (CNRS-UMR8161, Lille, France) for providing the MDCK-p40-MET-Tet-On cell line and for fruitful discussions.
This work was supported by CNRS, Pasteur Institute of Lille, Université Lille-Nord de France and INSERM and by grants from Fondation de France, FEDER-Region Nord Pas de Calais, and the Ligue Regionale Contre le Cancer-Comité de l'Aisne.

This article contains supplemental Table 1 and Figs. 1–5.
D. Tulasne, unpublished results.
- GAB1
- GRB2-associated binder 1
- HGF/SF
- hepatocyte growth factor/scatter factor
- AKT
- v-akt murine thymoma viral oncogene homolog 1 or protein kinase B
- GRB2
- growth factor receptor-bound protein 2: SH2, Src homology
- SHP2
- SH2 domain-containing protein tyrosine phosphatase-2
- MBS
- MET binding site
- MBD
- MET binding domain
- MDCK
- Madin-Darby canine kidney cell
- TRAIL
- TNF-related apoptosis-inducing ligand
- Z-VAD-FMK
- Z-Val-Ala-Asp (OMe)-fluoromethyl ketone
- PARP
- poly(ADP-ribose) polymerase
- MTT
- 3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyltetrazolium bromide
- aa
- amino acids
- PH
- pleckstrin homology
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- ALLN
- N-acetyl-l-leucinyl-l-leucinyl-l-norleucinal.
REFERENCES
- 1. Schlessinger J. (2000) Cell signaling by receptor-tyrosine kinases. Cell 103, 211–225 [DOI] [PubMed] [Google Scholar]
- 2. Pawson T., Scott J. D. (1997) Signaling through scaffold, anchoring, and adaptor proteins. Science 278, 2075–2080 [DOI] [PubMed] [Google Scholar]
- 3. Buday L., Tompa P. (2010) Functional classification of scaffold proteins and related molecules. FEBS J. 277, 4348–4355 [DOI] [PubMed] [Google Scholar]
- 4. Holgado-Madruga M., Emlet D. R., Moscatello D. K., Godwin A. K., Wong A. J. (1996) A Grb2-associated docking protein in EGF and insulin receptor signaling. Nature 379, 560–564 [DOI] [PubMed] [Google Scholar]
- 5. Weidner K. M., Di Cesare S., Sachs M., Brinkmann V., Behrens J., Birchmeier W. (1996) Interaction between Gab1 and the c-Met receptor-tyrosine kinase is responsible for epithelial morphogenesis. Nature 384, 173–176 [DOI] [PubMed] [Google Scholar]
- 6. Gu H., Neel B. G. (2003) The “Gab” in signal transduction. Trends Cell Biol. 13, 122–130 [DOI] [PubMed] [Google Scholar]
- 7. Nishida K., Hirano T. (2003) The role of Gab family scaffolding adapter proteins in the signal transduction of cytokine and growth factor receptors. Cancer Sci. 94, 1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wöhrle F. U., Daly R. J., Brummer T. (2009) Function, regulation, and pathological roles of the Gab/DOS docking proteins. Cell Commun. Signal 7, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lock L. S., Frigault M. M., Saucier C., Park M. (2003) Grb2-independent recruitment of Gab1 requires the C-terminal lobe and structural integrity of the Met-receptor kinase domain. J. Biol. Chem. 278, 30083–30090 [DOI] [PubMed] [Google Scholar]
- 10. Lock L. S., Maroun C. R., Naujokas M. A., Park M. (2002) Distinct recruitment and function of Gab1 and Gab2 in Met receptor-mediated epithelial morphogenesis. Mol. Biol. Cell 13, 2132–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schaeper U., Gehring N. H., Fuchs K. P., Sachs M., Kempkes B., Birchmeier W. (2000) Coupling of Gab1 to c-Met, Grb2, and Shp2 mediates biological responses. J. Cell Biol. 149, 1419–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Birchmeier C., Birchmeier W., Gherardi E., Vande Woude G. F. (2003) Met, metastasis, motility, and more. Nat. Rev. Mol. Cell Biol. 4, 915–925 [DOI] [PubMed] [Google Scholar]
- 13. Ponzetto C., Bardelli A., Zhen Z., Maina F., dalla Zonca P., Giordano S., Graziani A., Panayotou G., Comoglio P. M. (1994) A multifunctional docking site mediates signaling and transformation by the hepatocyte growth factor/scatter factor receptor family. Cell 77, 261–271 [DOI] [PubMed] [Google Scholar]
- 14. Liu Y., Rohrschneider L. R. (2002) The gift of Gab. FEBS Lett. 515, 1–7 [DOI] [PubMed] [Google Scholar]
- 15. Sármay G., Angyal A., Kertész A., Maus M., Medgyesi D. (2006) The multiple function of Grb2 associated binder (Gab) adaptor/scaffolding protein in immune cell signaling. Immunol. Lett. 104, 76–82 [DOI] [PubMed] [Google Scholar]
- 16. Bertotti A., Burbridge M. F., Gastaldi S., Galimi F., Torti D., Medico E., Giordano S., Corso S., Rolland-Valognes G., Lockhart B. P., Hickman J. A., Comoglio P. M., Trusolino L. (2009) Only a subset of Met-activated pathways are required to sustain oncogene addiction. Sci. Signal. 2, er11. [DOI] [PubMed] [Google Scholar]
- 17. Xiao G. H., Jeffers M., Bellacosa A., Mitsuuchi Y., Vande Woude G. F., Testa J. R. (2001) Anti-apoptotic signaling by hepatocyte growth factor/Met via the phosphatidylinositol 3-kinase/Akt and mitogen-activated protein kinase pathways. Proc. Natl. Acad. Sci. U.S.A. 98, 247–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rodrigues G. A., Falasca M., Zhang Z., Ong S. H., Schlessinger J. (2000) A novel positive feedback loop mediated by the docking protein Gab1 and phosphatidylinositol 3-kinase in epidermal growth factor receptor signaling. Mol. Cell. Biol. 20, 1448–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Goormachtigh G., Ji Z., Le Goff A., Fafeur V. (2011) Degradation of the GAB1 adaptor by the ubiquitin-proteasome pathway hampers HGF/SF-MET signaling. Biochem. Biophys. Res. Commun. 411, 780–785 [DOI] [PubMed] [Google Scholar]
- 20. Itoh M., Yoshida Y., Nishida K., Narimatsu M., Hibi M., Hirano T. (2000) Role of Gab1 in heart, placenta, and skin development and growth factor- and cytokine-induced extracellular signal-regulated kinase mitogen-activated protein kinase activation. Mol. Cell. Biol. 20, 3695–3704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sachs M., Brohmann H., Zechner D., Müller T., Hülsken J., Walther I., Schaeper U., Birchmeier C., Birchmeier W. (2000) Essential role of Gab1 for signaling by the c-Met receptor in vivo. J. Cell Biol. 150, 1375–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schmidt C., Bladt F., Goedecke S., Brinkmann V., Zschiesche W., Sharpe M., Gherardi E., Birchmeier C. (1995) Scatter factor/hepatocyte growth factor is essential for liver development. Nature 373, 699–702 [DOI] [PubMed] [Google Scholar]
- 23. Uehara Y., Minowa O., Mori C., Shiota K., Kuno J., Noda T., Kitamura N. (1995) Placental defect and embryonic lethality in mice lacking hepatocyte growth factor/scatter factor. Nature 373, 702–705 [DOI] [PubMed] [Google Scholar]
- 24. Bladt F., Riethmacher D., Isenmann S., Aguzzi A., Birchmeier C. (1995) Essential role for the c-met receptor in the migration of myogenic precursor cells into the limb bud. Nature 376, 768–771 [DOI] [PubMed] [Google Scholar]
- 25. Maroun C. R., Holgado-Madruga M., Royal I., Naujokas M. A., Fournier T. M., Wong A. J., Park M. (1999) The Gab1 PH domain is required for localization of Gab1 at sites of cell-cell contact and epithelial morphogenesis downstream from the met receptor-tyrosine kinase. Mol. Cell. Biol. 19, 1784–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Maroun C. R., Moscatello D. K., Naujokas M. A., Holgado-Madruga M., Wong A. J., Park M. (1999) A conserved inositol phospholipid binding site within the pleckstrin homology domain of the Gab1 docking protein is required for epithelial morphogenesis. J. Biol. Chem. 274, 31719–31726 [DOI] [PubMed] [Google Scholar]
- 27. Gentile A., Trusolino L., Comoglio P. M. (2008) The MET-tyrosine kinase receptor in development and cancer. Cancer Metastasis Rev. 27, 85–94 [DOI] [PubMed] [Google Scholar]
- 28. Matsumoto K., Nakamura T. (1996) Emerging multipotent aspects of hepatocyte growth factor. J. Biochem. 119, 591–600 [DOI] [PubMed] [Google Scholar]
- 29. Foveau B., Leroy C., Ancot F., Deheuninck J., Ji Z., Fafeur V., Tulasne D. (2007) Amplification of apoptosis through sequential caspase cleavage of the MET-tyrosine kinase receptor. Cell Death Differ. 14, 752–764 [DOI] [PubMed] [Google Scholar]
- 30. Tulasne D., Deheuninck J., Lourenco F. C., Lamballe F., Ji Z., Leroy C., Puchois E., Moumen A., Maina F., Mehlen P., Fafeur V. (2004) Proapoptotic function of the MET-tyrosine kinase receptor through caspase cleavage. Mol. Cell. Biol. 24, 10328–10339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Deheuninck J., Foveau B., Goormachtigh G., Leroy C., Ji Z., Tulasne D., Fafeur V. (2008) Caspase cleavage of the MET receptor generates an HGF interfering fragment. Biochem. Biophys. Res. Commun. 367, 573–577 [DOI] [PubMed] [Google Scholar]
- 32. Deheuninck J., Goormachtigh G., Foveau B., Ji Z., Leroy C., Ancot F., Villeret V., Tulasne D., Fafeur V. (2009) Phosphorylation of the MET receptor on juxtamembrane tyrosine residue 1001 inhibits its caspase-dependent cleavage. Cell. Signal. 21, 1455–1463 [DOI] [PubMed] [Google Scholar]
- 33. Leroy C., Deheuninck J., Reveneau S., Foveau B., Ji Z., Villenet C., Quief S., Tulasne D., Kerckaert J. P., Fafeur V. (2006) HGF/SF regulates expression of apoptotic genes in MCF-10A human mammary epithelial cells. Ann. N.Y. Acad. Sci. 1090, 188–202 [DOI] [PubMed] [Google Scholar]
- 34. Grasset M. F., Gobert-Gosse S., Mouchiroud G., Bourette R. P. (2010) Macrophage differentiation of myeloid progenitor cells in response to M-CSF is regulated by the dual-specificity phosphatase DUSP5. J. Leukoc. Biol. 87, 127–135 [DOI] [PubMed] [Google Scholar]
- 35. Stennicke H. R., Renatus M., Meldal M., Salvesen G. S. (2000) Internally quenched fluorescent peptide substrates disclose the subsite preferences of human caspases 1, 3, 6, 7, and 8. Biochem. J. 350, 563–568 [PMC free article] [PubMed] [Google Scholar]
- 36. Wee L. J., Tan T. W., Ranganathan S. (2006) BMC Bioinformatics 7, S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lee M. W., Hirai I., Wang H. G. (2003) caspase-3-mediated cleavage of Rad9 during apoptosis. Oncogene 22, 6340–6346 [DOI] [PubMed] [Google Scholar]
- 38. Enomoto A., Suzuki N., Morita A., Ito M., Liu C. Q., Matsumoto Y., Yoshioka K., Shiba T., Hosoi Y. (2003) caspase-mediated cleavage of JNK during stress-induced apoptosis. Biochem. Biophys. Res. Commun. 306, 837–842 [DOI] [PubMed] [Google Scholar]
- 39. Fan S., Ma Y. X., Gao M., Yuan R. Q., Meng Q., Goldberg I. D., Rosen E. M. (2001) The multisubstrate adapter Gab1 regulates hepatocyte growth factor (scatter factor)-c-Met signaling for cell survival and DNA repair. Mol. Cell. Biol. 21, 4968–4984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Holgado-Madruga M., Wong A. J. (2003) Gab1 is an integrator of cell death versus cell survival signals in oxidative stress. Mol. Cell. Biol. 23, 4471–4484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sun Y., Yuan J., Liu H., Shi Z., Baker K., Vuori K., Wu J., Feng G. S. (2004) Role of Gab1 in UV-induced c-Jun NH2-terminal kinase activation and cell apoptosis. Mol. Cell. Biol. 24, 1531–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fischer U., Jänicke R. U., Schulze-Osthoff K. (2003) Many cuts to ruin. A comprehensive update of caspase substrates. Cell Death Differ. 10, 76–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bachelder R. E., Ribick M. J., Marchetti A., Falcioni R., Soddu S., Davis K. R., Mercurio A. M. (1999) p53 inhibits α6β4 integrin survival signaling by promoting the caspase 3-dependent cleavage of AKT/PKB. J. Cell Biol. 147, 1063–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bachelder R. E., Wendt M. A., Fujita N., Tsuruo T., Mercurio A. M. (2001) The cleavage of Akt/protein kinase B by death receptor signaling is an important event in detachment-induced apoptosis. J. Biol. Chem. 276, 34702–34707 [DOI] [PubMed] [Google Scholar]
- 45. Widmann C., Gibson S., Johnson G. L. (1998) caspase-dependent cleavage of signaling proteins during apoptosis. A turn-off mechanism for anti-apoptotic signals. J. Biol. Chem. 273, 7141–7147 [DOI] [PubMed] [Google Scholar]
- 46. Xu J., Liu D., Songyang Z. (2002) The role of Asp-462 in regulating Akt activity. J. Biol. Chem. 277, 35561–35566 [DOI] [PubMed] [Google Scholar]
- 47. Green K. A., Naylor M. J., Lowe E. T., Wang P., Marshman E., Streuli C. H. (2004) caspase-mediated cleavage of insulin receptor substrate. J. Biol. Chem. 279, 25149–25156 [DOI] [PubMed] [Google Scholar]
- 48. Berry D. M., Benn S. J., Cheng A. M., McGlade C. J. (2001) caspase-dependent cleavage of the hematopoietic-specific adaptor protein Gads alters signaling from the T cell receptor. Oncogene 20, 1203–1211 [DOI] [PubMed] [Google Scholar]
- 49. McKay M. M., Morrison D. K. (2007) caspase-dependent cleavage disrupts the ERK cascade scaffolding function of KSR1. J. Biol. Chem. 282, 26225–26234 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.