Background: The mechanism by which antibody constant region alters fine specificity is unknown.
Results: Different constant regions were found to change electronic and chemical properties of the antigen-binding site.
Conclusion: Constant regions can affect the energy landscape of the variable region.
Significance: These results are potentially critical for understanding fast, correct immune responses at the systems level and for future immunotherapy development.
Keywords: Antibodies, Antigen, Immunology, NMR, Peptides, Isotype, Paratope
Abstract
The finding that the antibody (Ab) constant (C) region can influence fine specificity suggests that isotype switching contributes to the generation of Ab diversity and idiotype restriction. Despite the centrality of this observation for diverse immunological effects such as vaccine responses, isotype-restricted antibody responses, and the origin of primary and secondary responses, the molecular mechanism(s) responsible for this phenomenon are not understood. In this study, we have taken a novel approach to the problem by probing the paratope with 15N label peptide mimetics followed by NMR spectroscopy and fluorescence emission spectroscopy. Specifically, we have explored the hypothesis that the C region imposes conformational constraints on the variable (V) region to affect paratope structure in a V region identical IgG1, IgG2a, IgG2b, and IgG3 mAbs. The results reveal isotype-related differences in fluorescence emission spectroscopy and temperature-related differences in binding and cleavage of a peptide mimetic. We conclude that the C region can modify the V region structure to alter the Ab paratope, thus providing an explanation for how isotype can affect Ab specificity.
Introduction
Since the completion of detailed Ig structure-function studies in the 1960s, Ab4 molecules have been viewed as multifunctional molecules with two major domains defined by the variable and constant regions. The V region binds antigen (Ag) and is capable of enormous combinatorial diversity that can recognize a myriad of molecular conformations, whereas the C region provides such functional capacities as the ability to interact with host receptors and activate the complement system (1). In this conception, the V and C regions functioned as two virtually independent domains, with the V region being responsible for binding Ag and the C region providing other biological functions. This tidy view of separate structural and functional domains comprising an immunoglobulin G molecule has unraveled in recent years with various observations that C regions can affect the interaction of certain V regions with their Ag (2–7). At least six independent groups have reported findings that isotype switching is associated with altered specificity despite conservation of V region sequences (7–13). However, the molecular mechanisms for these phenomena are not understood.
The notion that B cell class switching can result in new Ab specificity without somatic mutation raises new possibilities for the ontogeny of humoral responses because B cells expressing different V region-identical isotypes with different specificities could presumably respond to different Ags. The observation that class switching of certain Abs can result in the acquisition of reactivity for self Ags despite identical V regions suggests that this phenomenon could contribute to certain pathological autoimmune responses (5, 14). Finally, an understanding of how C regions affect specificity is important for the development of therapeutic mAbs for immunotherapy, because the choice of isotype and/or exchanging rodent and human C domains to generate chimeric Abs could affect the binding characteristics of engineered mAbs (4, 6). In this regard human-mouse chimeric Abs have been shown to differ in specificity from their parent murine Abs (6).
Previous studies done in our lab using four murine mAb isotypes, IgG1, IgG2a, IgG2b, and IgG3 suggest that the C region imposes structural constraints on the V region that alter its structure and/or ability to undergo a conformational change upon Ag binding (7, 15–18). Thus, although they are identical in sequence, the V regions of this group of mAbs may have secondary structures capable of different interactions that manifest themselves as changed fine specificity. This view is supported by several lines of evidence. First, isotype switching was accompanied by altered reactivity with anti-idiotypic mAbs, implying a changed binding surface (19). Other groups have also shown that both idiotype reactivity (20) and immunogenicity (21) can be lost when the constant region is changed. Second, recent spectroscopic evidence on the 3E5 family of mAbs suggests that V and C domains are tightly coupled such that Ag binding can result in secondary structure changes that propagate into the C domain (17). Third, surface plasmon resonance and isothermal titration calorimetry, performed on the 3E5 family of mAbs using the monovalent peptide mimetic P1, revealed different activation energies and association constant (Ka) values for the different isotypes (3, 5, 17). A corollary of this mechanism is that isotype switching would result in an altered paratope, or Ag binding surface, but this inference has previously lacked direct experimental evidence.
In this study we explored isotype-related differences in V region identical antibodies by tryptophan fluorescence and 15N-labeled peptide NMR. There are a variety of extensively used NMR techniques and approaches that can be used to stably label either the Ab or the Ag with isotopes (11, 22–24) for mapping residue-specific protein-ligand interactions (25, 26). By monitoring the chemical shift perturbations of 15N-labeled methionine ([15N]Met-10) and leucine ([15N]Leu-11) residues, we demonstrate that Ag P1 binds to all IgG isotypes. Furthermore, when P1 is bound to IgG2b, its behavior is significantly different from that of the other 3E5 IgGs, although all except IgG3 are capable of cleaving P1. Our results provide direct experimental support for the notion that the C domain can affect antibody fine specificity by influencing the chemical and electronic environment of the Ab paratope.
EXPERIMENTAL PROCEDURES
mAb Preparation
The IgG1, IgG2a, and IgG2b switch variants of 3E5-IgG3 have been described previously (3, 6). mAb 18B7, a Cryptococcus neoformans capsule-specific IgG1 was obtained as previously described (27). The murine mAbs were purified by protein A or G affinity chromatography (Pierce) from hybridoma cell culture supernatants in the presence of protease inhibitors (Roche) and concentrated, and buffer was exchanged against 0.1 m Tris-HCl, pH 7.4. mAb concentration was determined by A280 measurement.
GXM Preparation
GXM was isolated from C. neoformans strain 24067 (serotype D) and purified with minor modifications by the filtration method (28). An amount of 400 μg of proteinase K (Sigma) was then added to the suspension and incubated overnight in a 37 °C water bath. Two successive one-fifth volume butane:chloroform (1:5) extractions were then done by mixing well and allowing a 1-h incubation at −20 °C. For better separation of the layers, after extraction, the samples were centrifuged at 10,000 × g. The sample was then lyophilized again.
Peptides
The unlabeled P1 peptide and its derivatives were synthesized using Fmoc (N-(9-fluorenyl)methoxycarbonyl) chemistry on a microwave-assisted peptide synthesizer (Liberty; CEM Corp.) at the Proteomics Resource Center (Rockefeller University, NY). For the NMR studies, the P1 peptide was synthesized by Chem Pep to include 15N-labeled methionine and 15N-labeled leucine (SPNQHTPPW-[15N]M-[15N]L-K) or a single 15N-labeled leucine (SPNQHTPPWM-[15N]L-K).
Antibody Binding to Peptide ELISA
Polystyrene plates were first coated with 1 μg/ml of streptavidin in PBS followed by blocking with 1% BSA in PBS. Biotinylated peptide P1 and its mutated variants were then added at a concentration of 2 μg/ml followed by the addition of the mAb 3E5 variants (5 μg/ml). Ab binding to peptide was detected by the addition of alkaline phosphatase-conjugated goat anti-mouse κ (10 μg/ml) followed by color development with p-nitrophenyl phosphate substrate (1 mg/ml). All incubations were performed for 1.5 h at 37 °C, and absorbance was measured at 405 nm.
Fluorescence Analysis of Antibody-Polysaccharide Complex
13 pmol of mAb was added to 83 pmol of GXM for each sample in 0.1 m Tris-HCl, pH 7.4, for a total volume of 185 μl. The molar concentration of GXM was calculated assuming the molecular mass of 1,200 kDa derived from light scattering measurements (29). The mAb-GXM solution was then allowed to equilibrate at room temperature for 1 h before spectral measurements were done. Fluorescence measurements were done on a Jobin Yvon (Edison, NJ) Fluoromax-3 spectrofluorometer using 285-nm excitation and 354-nm emission wavelengths. The spectra are averages of five or six independent experiments in which two successive 120-s scans were averaged. The base-line buffer spectrum was subtracted from all the mAb spectra without GXM addition, and the spectrum of GXM alone was subtracted from all measurements using GXM.
NMR Spectroscopy
Antibodies were concentrated to 27 μm (IgG2a, IgG3) or 54 μm (IgG1, IgG2b) in 0.1 m Bis-Tris and 0.15 m NaCl pH 6.5 buffer. 100 μm (IgG2b, IgG1) or 50 μm (IgG2a, IgG3) P1 (Chem Pep) was added just before NMR analysis for the 25 °C experiments. The antibodies were concentrated to 27 (IgG2a, IgG3), 47 (IgG1), and 77 (IgG2b) μm in the above buffer, and 100 (IgG2a, IgG1, IgG3) or 140 (IgG2b) μm P1 was added just before NMR analysis for the 37 °C experiments. 15N-1HN heteronuclear single quantum coherence (HSQC) spectra (30, 31) were recorded using 15N-labeled P1, P1 alone, and IgG/P1 complexes on a Bruker Avance spectrometer at 600 MHz capable of applying pulse field gradients along the z axis. Experiments done at 25 °C were run as 2-h blocks, after incubating P1 with the mAbs at 4 °C for 2 h, 4 h, 24 h, and 7 days. Studies done at 37 °C were done immediately upon incubation of P1 with the mAb, as a series of 8–12 HSQC runs, spanning 17–23 h. Experiments were processed using NMRPipe. Analysis was done using either NMRPipe (32) or NMRViewJ (33).
Mass Spectrometry Analysis of P1 before and after IgG2b and IgG3 Binding
[15N]M-[15N]L-labeled P1 was sent for MALDI-TOF mass analysis at the Protein Core Facility of Columbia University, before and after NMR analysis with 3E5-IgG2b, in the NMR buffer (above).
Full Atom Molecular Dynamics Simulations
The anti-GXM IgG1, 2H1, differs from 3E5-IgG1 by 12 amino acids in the VH (8 amino acids) and VL (4 amino acids) chains. Its cognate peptide PA1 has a sequence (GLQYTPSWMLVG) similar to that of P1 (SPNQHTPPWMLK). The crystal structure (Protein Data Bank code 2H1P) is found for 2H1 mAb in complex with PA1 (34). Using this crystal structure and performing in silico mutations on both 2H1 and PA1, we have generated a model for 3E5-IgG1+P1 complex. On this complex, we have performed constant temperature and pressure (300 K, 1 atm) 10-ns all atom MD simulation with AMBER11 (35). An AMBER99SB (36) force-field was used with explicit solvent model TIP3P (37) in a rectilinear box of dimensions 93, 83, and 94 Å. Prior to the 10-ns production run, a short minimization was employed followed by a 20-ps heating (0–300 K), a 20-ps density equilibration, and a 100-ps constant pressure/temperature (1 atm/300 K) equilibration steps.
Statistical Analysis
A one-way analysis of variance for the mAb ELISA binding studies was done with a Tukey multiple comparison test revealed statistical significance (*, p < 0.0005; **, p < 0.0004; ***, p < 0.0001) in the comparison between some pairs of isotypes for the alanine mutations shown. t tests were used in comparing the peak fluorescence emissions. The errors in rates of intensity changes over time in the NMR rate analysis were calculated as the S.E.
RESULTS
Reactivity of V Region Identical IgG Subclasses with Mutated Peptide Mimetics
The mAb 3E5 family reacts with the GXM of the capsular polysaccharide of C. neoformans. These mAbs have identical heavy and light chain V region sequences and bind to the 12-amino acid peptide mimetic SPNQHTPPWMLK known as P1 (38). To explore the contribution of the various amino acid residues and in an attempt to generate peptides that would discriminate between the four subclasses, we tested two sets of mutated peptides. One set involved the substitution of each residue with alanine. Binding of the four IgG subclasses to the alanine-substituted peptides in this set was very similar. IgG2a responses decreased most upon alanine substitution for each of the residues evaluated (see supplemental Fig. S1A). Replacement of residues His-5, Thr-6, Pro-7, Trp-9, Met-10, and Leu-11 decreased IgG binding to 10–25%. One-way analysis of variance analysis reveals statistical significance between the binding of the 3E5 mAb pairs to the mutations and P1 (see supplemental Fig. S1B). We note that the binding of each of these peptides to the plate is through an avidin-biotin interaction, and thus differences in the reactivity by ELISA were not a result of differences in peptide binding to polystyrene.
The second peptide analog set consisted of peptides with conserved substitutions. When the four subclasses were tested for binding on the peptide set with conserved substitutions, binding decreased for all four isotypes to 10% with the exception of the T6S substitution, which resulted in binding of up to 35% (see supplemental Fig. S1C). There was no significant difference between the binding curves for the conserved replacement peptides among all four isotypes. In addition, previous studies reveal that N-linked glycans do not play a significant role in 3E5 mAb-P1 binding (6). These data suggest that some constant regions change the interaction site of the V region with peptide and provide critical information for the selection of residues Met-10 and Leu-11 to isotope label for NMR studies (see below).
Fluorescence Emission Spectra of V Region Identical IgG Subclasses with GXM
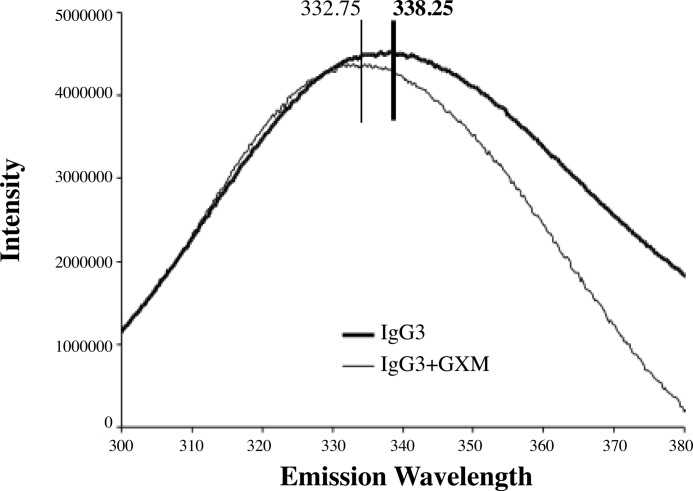
Each of the Abs in the mAb 3E5 family has four Trp residues in their V region: two in the heavy chain and two in the light chain. One of these residues is implicated in being directly involved in Ag interactions (34). Consequently, we measured the Trp emission spectra of each IgG subclass after saturation with GXM (Fig. 1). Because each mAb has identical V region sequences, the expectation is that Abs with identical structures will have similar fluorescence emission spectral changes upon binding antigen. The peak emission wavelength of Trp is 354 nm, so emission spectra were recorded in the 300–400-nm range after excitation with light of 285 nm. For all mAb 3E5 IgGs, GXM binding resulted in a blue shift in the Trp emission maxima. The magnitude of the change in emission wavelength differed among the various isotypes (Table 1), providing supporting evidence for the notion that these isotypes differ in their paratope.
FIGURE 1.
Tryptophan fluorescence emission spectra. Fluorescence emission spectra following the binding to 3E5-IgG3 after binding to GXM are shown. Binding is accompanied by blue shifting of the fluorescence emission peak wavelength of the Trp residues. The IgG3 spectrum is an average of seven separate experiments, whereas the IgG3+GXM spectrum is an average of two separate experiments.
TABLE 1.
Changes in fluorescence maxima of Ab-GXM complexes and corresponding energy calculation
| Isotype | Wavelength changea | Change in energyb |
|---|---|---|
| nm | kJ/mol | |
| IgG1 | 1.7 ± 1.2 | −1.9 |
| IgG2a | 2.4 ± 1.4 | −2.6 |
| IgG2b | 2.7 ± 1.5 | −2.9 |
| IgG3 | 4.8 ± 1.0 | −5.1 |
a Wavelength change values and standard deviations are averages of three to six independent measurements taken in triplicate each time. Comparison of native fluorescence spectra reveals statistical significance (p < 0.05) in the comparison between IgG2b and IgG1, as well as IgG3 and IgG2a. Upon addition of GXM, t tests reveal statistical significance (p < 0.005) when IgG3 is compared with the rest of the isotypes.
b The changes in energy were calculated using the de Broglie equations, E = hc/λ, where E is energy in Joules (J), h is the Planck constant (6.62606896 × 10−34 J*s), c is the speed of light (2.997924 × 108 m/s), and λ is either the initial or final emission wavelength measured.
NMR Spectroscopy with [15N]Met-10–[15N]Leu-11-labeled P1
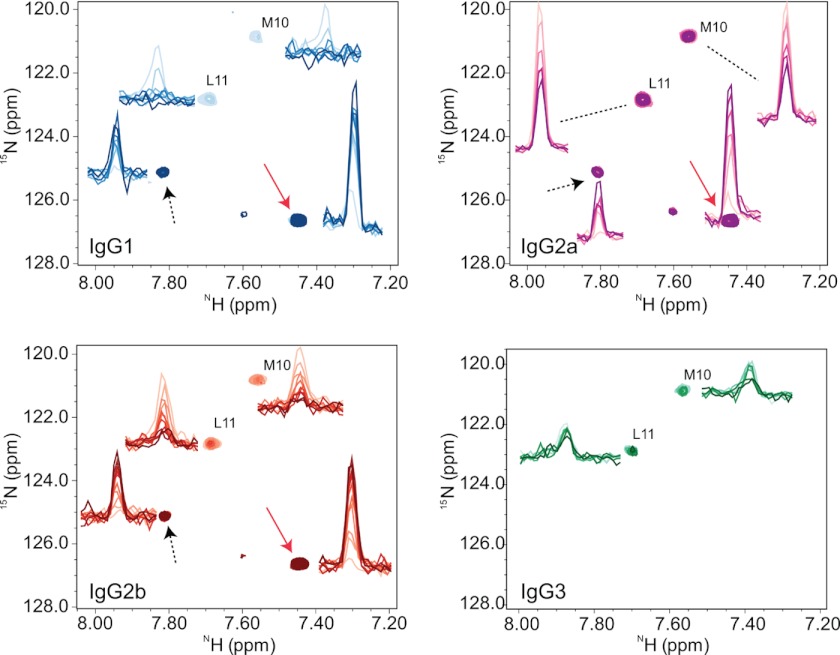
To explore the paratopes of the 3E5 mAbs, we studied the in-solution binding of each isotype to a P1 peptide with two 15N-labeled amino acids by HSQC and mapped chemical shift perturbations. We measured the 15N and 1HN HSQC correlations of P1 when bound to individual mAbs at 25 °C and 37 °C and compared them to the spectra of P1 without mAb, as well as to the spectra of a control mAb, MOPC195 (murine IgG2b), incubated with P1. As with the fluorescence experiments, identical NMR signals are expected for Abs with identical structures from the isotope-labeled peptide. We found that all of the isotypes bound P1 at both temperatures, as is evident by a significant decrease in the intensities of the resonance peaks, where each peak represents one of the 15N-labeled amide bond on either Met-10 or Leu-11.
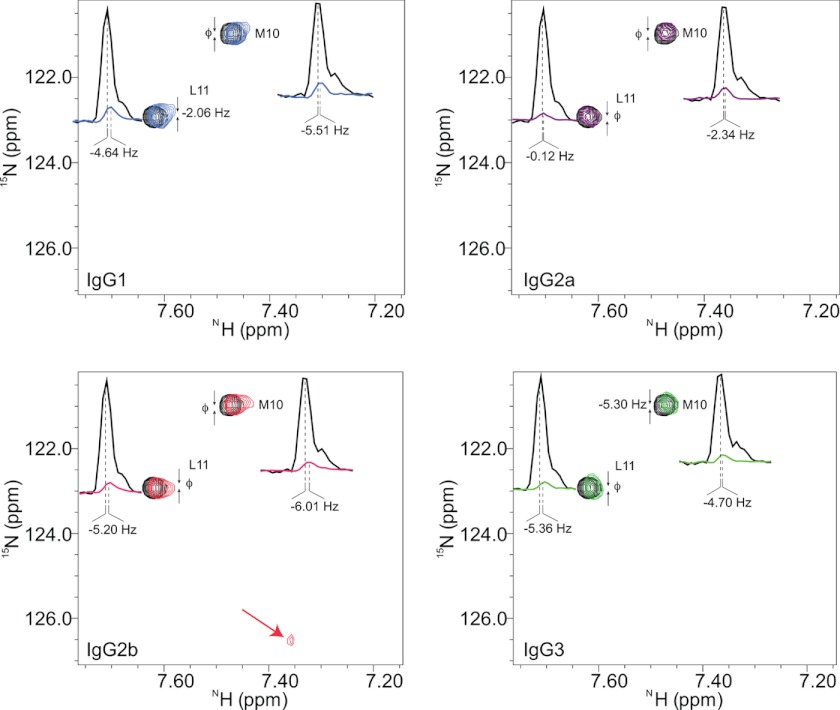
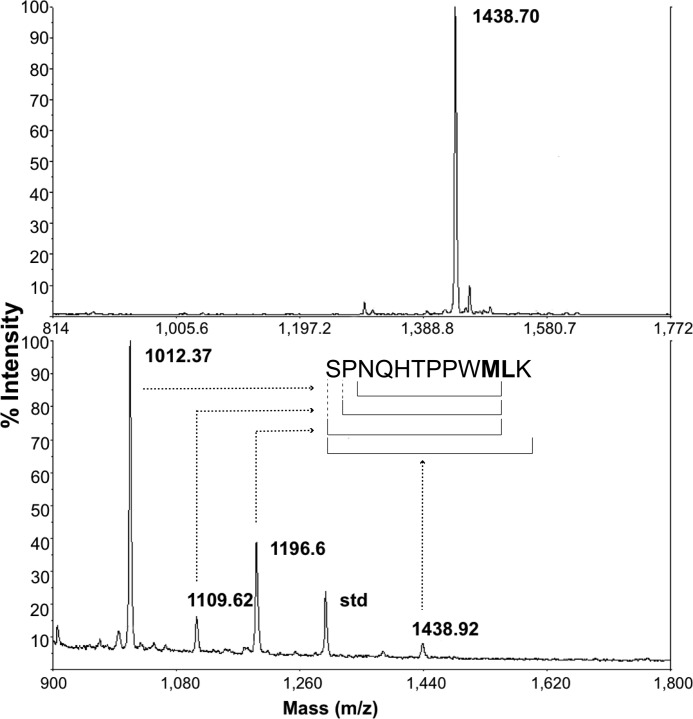
The differences in chemical shift positions in the P1-bound complexes were marginal, which is expected because the V regions are identical in sequence and the environments experienced by 15N-labeled residues of P1 are expected to be similar. However, at 37 °C, prolonged incubation of each isotype with P1 resulted in the appearance of new resonance peaks for all isotypes, except for IgG3 (Fig. 2). The rate of decrease in P1 resonances and the rate of increase in new resonances are identical, suggesting that the peptide was being modified and possibly cleaved. When the experiment was repeated at 25 °C, we observed this pattern only for IgG2b; the rest of the isotypes showed P1 binding but no new peaks (Fig. 3). The HSQC spectra were also collected at 37 °C for P1 alone; resonance peaks did not change, and there were no visible new peaks, suggesting that P1 is stable in the buffer used (data not shown). Mass spectrometric analysis of the IgG2b+P1 solution after NMR analysis at 37 °C revealed the appearance of three fragments with masses (m/z) of ∼1012, ∼1109, and ∼1196, confirming hydrolysis of the peptide (Fig. 4).
FIGURE 2.
NMR chemical shift perturbations at 37 °C. Binding of mAbs to [15N]M and [15N]L-labeled P1 at 37 °C is shown. The reference peaks are the [15N]Met-10 and [15N]Leu-11 residues from the P1 alone (51). All mAbs bind P1, which is evident by the attenuations seen in resonance peak intensities (color-coded: IgG1, blue, IgG2a, magenta, IgG2b, red; IgG3, green). The contour levels of the two-dimensional spectra are adjusted to be able to display shifts in positions efficiently; in all graphs the reference spectra are drawn with the same contour level, and the spectra of mAbs are all at the same contour level. The one-dimensional intensity graphs are at their actual contour levels depicting the level of attenuations. At 37 °C, all 3E5 mAbs except IgG3 show the appearance of new peaks and the original peaks disappear during a time course from 17 to 24 h (sequential dark color scheme). The new peaks correspond to the fragments of chopped P1 generated by the mAb in solution. The rates of increase in intensities of new peaks are equal to the rates of decrease in intensities of original peaks, indicating a turnover from full P1 to fragments of P1 catalyzed by mAbs. The peaks designated by arrows are proteolysis products that represent [15N]Met-10-containing cleaved fragments of P1.
FIGURE 3.
NMR chemical shift perturbations at 25 °C. Binding of mAbs to [15N]M and [15N]L-labeled P1 at 25 °C is shown. The mAbs isotypes were incubated with labeled P1 (51) at 4 °C for 2 h, 6 h, 24 h (data not shown), and 7 days before NMR experiments were performed. The HSQC spectra were recorded at 25 °C. The reference peaks are the [15N]Met-10 and [15N]Leu-11 residues from the P1 alone. All mAbs bind P1, which is evident by the attenuations seen in resonance peak intensities (color-coded: IgG1, blue; IgG2a, magenta; IgG2b, red; and IgG3, green). All mAbs cause marginal perturbations at resonance positions, but unlike other mAbs, in IgG2b spectra an extra weak but narrow peak is visible in the same position as in the 37 °C data, and an additional peak at noise level is also present (not shown in this figure); both of which represent [15N]Met-10-containing cleaved fragments of P1. The contour levels of the two-dimensional spectra are adjusted to be able to display shifts in positions efficiently; in all graphs the reference spectra are drawn with the same contour level, the spectra of mAbs are all at the same contour level. The one-dimensional intensity graphs are at their actual contour levels depicting the level of attenuations.
FIGURE 4.
Mass spectrometry post IgG2b NMR analysis. Top panel, the purified P1 sample; bottom panel, the P1 sample after overnight NMR analysis in the presence of IgG2b. Three new P1 fragments are visible after overnight incubation of P1 with IgG2b at 37 °C. MS was performed by MALDI-TOF experiments.
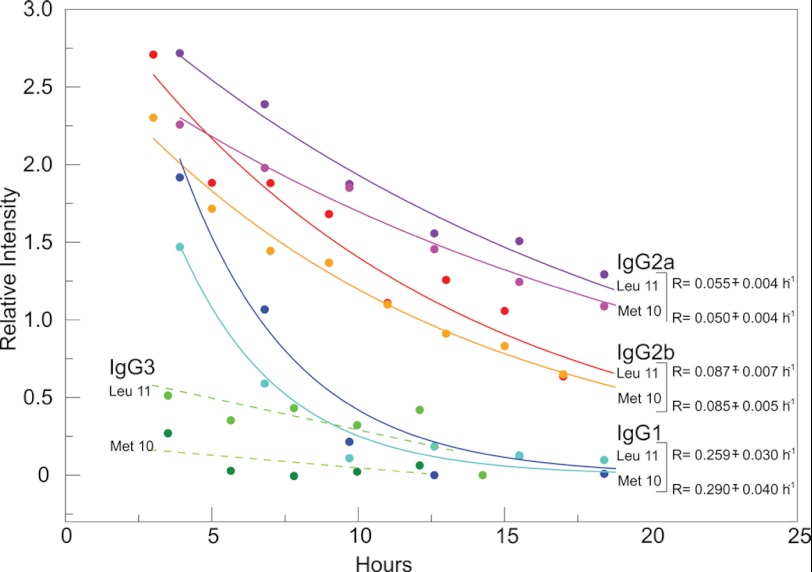
The hydrolysis rates were calculated by fitting the intensities of NMR resonance peaks to an exponential decay function (Fig. 5), and their differences and temperature dependence may be explained by modification of the energy landscape of isotypes by the C regions. Furthermore, the time required for 50% P1 hydrolysis differed for the various isotypes; ∼2.4–2.7 h for IgG1, ∼12.6–13.9 h for IgG2a, and ∼8.0–8.15 h for IgG2b. Hence, IgG2b had proteolytic activity for the peptide at both 25 and 37 °C, IgG1 and IgG2a had observed proteolytic activity at only 37 °C, and IgG3 had no observed proteolytic activity at either temperature despite sharing identical V region sequences. We also measured the binding of all of the 3E5 IgGs to [15N]Met-10–[15N]Leu-11-labeled P1 by ELISA and did not see a significant difference from that of binding to unlabeled P1 (data not shown).
FIGURE 5.
mAb hydrolysis rates derived from NMR. Hydrolysis rates were calculated by fitting the intensities of NMR resonance peaks of [15N]Met-10- and [15N]Leu-11-labeled P1 at 37 °C over a period of 18 h to an exponential decay function (solid lines). The fittings were done for both Met-10 and Leu-11 resonances from Fig. 2. The resonances seen in the IgG3 spectra were significantly broadened because of the tight binding between P1 and IgG3, and they could not be fit accurately (dashed lines) because of the low signal to noise ratio. The errors in rates are S.E.
In addition to the mAb 3E5 set, we studied two other mAbs as positive and negative controls. As a positive control we studied mAb 18B7, another IgG1 mAb against the C. neoformans capsular polysaccharide, which is known to bind peptide P1. However, unlike the mAb 3E5 family, mAb 18B7 has a total of 33 amino acid differences in its V region, of which 12 are in the CDRs (27), and thus, by definition, has a different paratope. mAb 18B7 was tested for binding and hydrolysis of [15N]Met-10–[15N]Leu-11-labeled P1. As expected, 18B7 bound and cleaved P1 but generated a different binding and hydrolysis pattern. Chemical shift perturbations were observed for [15N]-Met-10 and [15N]Leu-11 residues; the resonance positions of the bound form, as well as the resonances of the cleaved products, were different from those of 3E5-IgG1 (see supplemental Fig. S2). As a negative control, we used IgG2b mAb MOPC195. The HSQC spectra of [15N]Met-10–[15N]Leu-11-labeled P1 did not show changes in P1 peak intensities or positions upon MOPC195 mAb addition (data not shown).
All Atom Molecular Dynamics (MD) Simulations on 3E5-IgG1 Homolog in Complex with Peptide P1
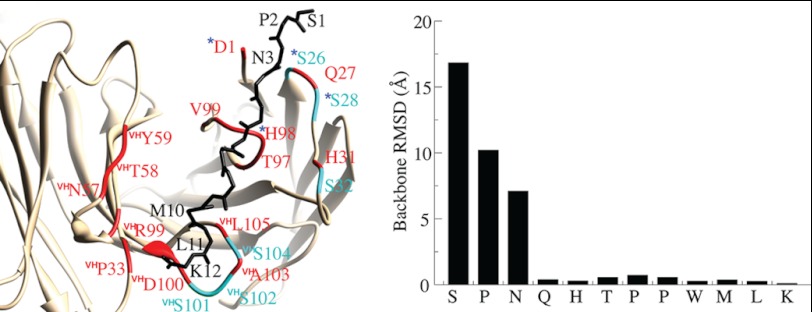
The structure of another IgG1 mAb to GXM (2H1) has been solved bound to a peptide mimetic PA1 that is similar to P1 (34). Because mAb 2H1 uses the same V regions as the 3E5 family and differs by only a few somatic mutations, we were able to use its atomic coordinate data in MD simulations of the 3E5-IgG1 homolog. To explore the binding pocket and protease mechanism, we have modeled 3E5-IgG1+P1 by mutating the 2H1 mAb and peptide PA1 in silico to the corresponding sequences of mAb 3E5-IgG1 and P1, utilizing UCSF-Chimera (39). We then employed AMBER (35) and performed 10 ns of MD simulation on the 3E5-IgG1+P1 model with explicit solvent. The simulation revealed that the interaction is exclusively hydrophobic and that peptide P1 binds to the same surface as peptide PA1 (Fig. 6). The C-terminal peptide residues are highly rigid, whereas the N-terminal residues have a fewer number of contacts, and thus, they are more flexible. The V region residues within contact distance to P1 are colored red. Interestingly, the binding pocket harbors six Ser residues. Of these Ser residues, Ser-26 together with Asp-1 and His-98 form the triad that is canonical in Ser proteases (40). However, these residues are loosely coupled, which may explain the slow protease activity. The C-terminal residues of the peptide is in close proximity of a Ser-rich heavy chain CDR3 loop that harbors an Asp and three Ser residues together with two hydrophobic amino acids that interact with the peptide P1.
FIGURE 6.
MD simulation on the 3E5-IgG1+P1 complex. The P1 binding pocket is shown on the lowest energy structure (left). Peptide P1 is shown as a backbone trace and IgG1 in ribbon representation (tan). The amino acids that belong to the IgG1 V region and that are in contact with peptide P1 are colored red. The serine residues that are in close proximity are highlighted (cyan). The residues that can form an Asp-Ser-His triad are depicted with asterisks. The graph on the right shows the backbone root mean square deviations of P1 residues between the lowest energy structure and the structure with the highest average P1 backbone root mean square deviation (RMSD).
DISCUSSION
The binding response of the four IgG subclasses to all peptides containing alanine substitutions differed significantly for the various isotypes; the binding data consistently indicated that two motifs consisting of residues His-5, Thr-6, and Pro-7 and residues Trp-9, Met-10, and Leu-11 were very important for binding of P1 to all of the isotypes. These six amino acid substitutions decreased binding to the range of 9–41% compared with the original peptide P1 (100%). It is possible that alterations in these amino acids result in steric effects that affect their fit into the Ab paratope. These results are supported by an earlier x-ray crystallographic study using a 3E5 family variable region identical anti-GXM IgG1, 2H1, and a similar peptide, PA1 (34). In the crystal structure of PA1 and mAb 2H1, the parts of the peptide located in the binding pocket of 2H1 correspond to two motifs involving Thr-5 and Pro-6 and Trp-8, Met-9, and Leu-10, which are comparable with two motifs in P1 that show the greatest decreases in binding when mutated to alanine. Because these studies were done with immobilized peptide, some differences in binding observed for IgG1 and IgG3 may reflect a loss of avidity resulting from an inability to form binding complexes given Ab isotype-related differences in hinge angles (41). However, we think this explanation is less likely to apply for all isotypes given the strong reactivity of the IgG2a and IgG2b subclasses, with both GXM and P1 (7).
mAb binding was then studied with peptides with conserved substitutions, but none of the substitutions restored binding. Each conserved replacement change resulted in a 10-fold decrease in binding, except for the T6S mutation, which resulted in a 3-fold decrease in binding, indicating that the Thr-6 residue may have less interaction with or is not located close enough to side chains in the Ab paratope as the rest of the P1 residues. Given that these mAbs have the same V region sequence but differ in isotype, we interpreted these results as indicating differences in the Ab contact surface or paratope that manifested themselves through binding differences, with the caveat that a contribution from avidity cannot be excluded as noted above.
To investigate the electronic microenvironment of the paratope in the various isotypes, we monitored changes in Trp fluorescence upon GXM binding and found that the wavelength of maximal emission was blue-shifted for all 3E5 isotypes. The fluorescence measurements were done after 1 h of room temperature incubation, and the antigen was GXM; because GXM is a polysaccharide, these results are not expected to be affected by the proteolysis phenomena observed at longer times with the peptide mimetic. The magnitudes of the shifts varied from 1.8 to 4.8 nm, with IgG3 showing the greatest change and IgG1 showing the smallest change. Trp fluorescence is influenced by water molecules and the proximity of charged amino acids to the Trp chromophore. Depending on how the charges near the chromophore shift upon Ag binding, the peak emission wavelength of the Trp shifts. Blue shifting indicates a less polar environment surrounding the Trp molecule (42), arising from conformational changes in the binding pocket residues or from exclusion of water molecules from the binding pocket. The differences observed among the isotypes therefore indicate differences in the movement of charges within their binding pockets. To put the nanometer emission difference values into perspective, we calculated the associated energy changes (1–5 kJ/mol), which were comparable with the free energy required to remove a CH2 group from an aqueous solution (∼3 kJ/mol), an important hydrophobic effect (43). Our recent observation that the V and C domains influence each other allosterically upon GXM binding, (17) strongly suggests that amino acid differences in C regions between the various isotypes create structural constraints that can influence the electronic properties of the mAb-binding pocket.
NMR results show a two-step reaction for IgG1, IgG2a, and IgG2b: first, P1 binding, and second, P1 hydrolysis. As stated previously, when mutated to alanine, both the Met-10 and Leu-11 residues in P1 decreased binding of all 3E5 isotypes by 60–90%; therefore, these residues are important for P1 binding. Proteolytic activity was evident by the appearance of new sharp P1 resonance peaks after IgG binding at 37 °C and has been verified by MS analysis. All of the 3E5 IgGs bound to P1 at both temperatures, although only the IgG2b spectra showed P1 cleavage at 25 °C, suggesting that at 25 °C, when bound to P1, only 3E5-IgG2b adopts a conformation that favors the hydrolysis of P1. At 37 °C, over a period of 17 h, all of the 3E5 mAbs except IgG3 were able to hydrolyze P1 in the same manner as IgG2b at 25 °C. The rates of hydrolysis at 37 °C differed for the mAbs; IgG1 rates were 6-fold larger than those of IgG2b and IgG2a, which were similar, and IgG3 showed no detectable cleavage.
Although the hydrolysis of P1 was quite slow, binding to P1 was immediate on the NMR experimental time scale. The rates and times of 50% P1 proteolysis are distinctly different for the isotypes. Because the new NMR peaks coming from cleaved P1 are at the same place for all of the 3E5 mAbs, they are the same fragments of P1, suggesting an identical cleavage site for all isotypes. Furthermore, when testing a related mAb with a known different V region and paratope, the IgG1 18B7 (27) as a positive control, we found that the bound peak positions of P1 as well as the cleavage peak positions were altered with respect to both free P1 and P1 + 3E5 mAbs. Thus, the 12-amino acid difference between mAb 18B7 heavy chain CDRs and the mAb 3E5 set creates a sufficient difference in the chemical environments of P1 residues that could be detected by NMR. This difference in CDR amino acids was enough to alter the 18B7 paratope from that of the 3E5 mAbs and provides an important control supporting the conclusion that the changes observed among the 3E5 mAb set are consequences of isotype-related paratope differences.
Together with the MS data, as well as NMR data using a single [15N]Leu-11-labeled P1 (data not shown), the new NMR resonances seen are all assigned to the [15N]Met-10 within the fragments SPNQHTPPWM, PNQHTPPWM, and, finally, NQHTPPWM. P1 cleavage sites occur between Ser-1 and Pro-2, between Pro-2 and Asn-3, and between Met-10 and Leu-11. Met-10–Leu-11 cleavage releases the Leu-11–Lys-12 fragment, which then gains an NH3+ moiety on the Leu-11 residue that is invisible by HSQC. Most catalytic proteolytic mAbs that have been studied have serine-like protease activity (44), and like them, our mAbs have the same light chain V region catalytic triad of Asp-1, Ser-26, and His-98 (40). MD simulations performed on the 3E5-IgG1+P1 model reveal that in the 3E5 mAbs, this catalytic triad is located proximal to the N terminus of the P1 peptide and could explain the cleavage of the Ser-1 and Pro-2 residues. In addition, the residues forming this triad are loosely coupled, which may explain the low catalysis rates. However, the mechanism for cleavage of the Met-10–Leu-11 amide bond is not as clear. There are Ser residues (Ser-101, Ser-102, and Ser-104), as well as an Asp residue (Asp-100) in close proximity to the Met-10–Leu-11 peptide bond, which may mediate catalysis. Ser can act as a nucleophile, but an efficient catalysis requires the presence of a His residue, as well as water molecules to stabilize the cleaved product. Because the binding pocket at the C terminus of P1 is highly hydrophobic, water accessibility is limited, and peptide hydrolysis would be hindered, which could further explain the slow rates of hydrolysis. The difference in hydrolysis rates observed with different isotypes can be explained by the differences in dynamic restraints imposed by different C regions on their binding pockets. The mAbs investigated here did not evolve to be highly efficient catalysts; rather, they are slow and possibly have broad specificity. Furthermore, incubation of 3E5 mAb preparations with saturating amounts of Ser protease inhibitors blocked the appearance of the new resonances, suggesting that Ser protease activity in the 3E5 IgGs is inhibited, and the hydrolysis of Met-10–Leu-11 peptide bond is impeded. A complete understanding of the proteolysis mechanism requires structural studies on 3E5-IgG+P1 complexes, as well as 3E5 IgGs with P1 fragments.
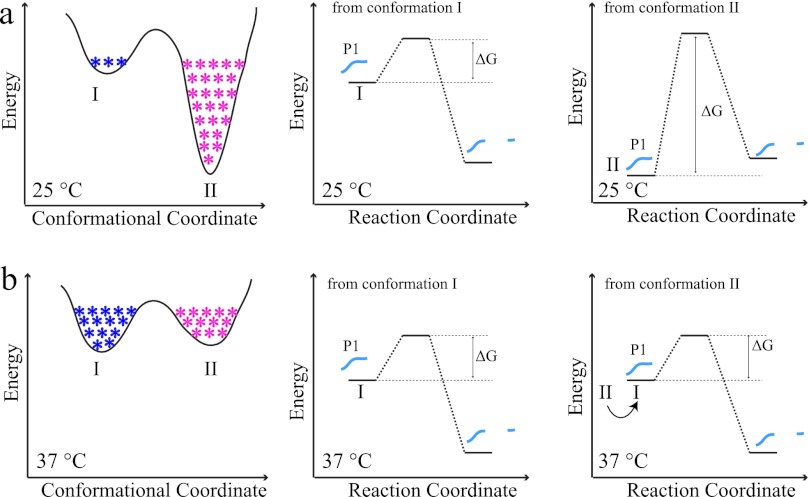
In enzyme kinetics, factor and/or ligand binding and differences in environmental conditions, such as temperature, pH, or ionic strength, affect enzyme function by altering the energy landscape, biasing the conformational coordinate to distinct conformational states and thus affecting the reaction coordinate and causing changes in turnover rates (45). An increasing number of studies have shown the effects of conformational sampling on ligand binding, catalysis, or product formation (46–49). In this view, within the 3E5 isotypes, depending on the temperature, different C regions inhibit or promote Ag hydrolysis by modulating the intrinsic energy landscape to favor active or inactive states. Proteolysis of peptide mimetic P1 is highly dependent on the energy landscape of the Ab isotype. To explain temperature effects, we propose a model using a highly simplified energy landscape (Fig. 7). We note that in reality the landscape is multidimensional, reflecting the effects of other intermolecular and environmental factors, and the energy profile is more rugged because of the existence of conformational substates.
FIGURE 7.
Simplified mAb energy landscape model. Different C regions result in differences in conformational coordinates, which alters the reaction coordinate of proteolysis. a, conformational coordinate of an arbitrary mAb. State I is the active state, which is poorly populated (blue asterisks), whereas state II is the inactive state (highly populated; magenta asterisks). b, there is a large energy penalty for this mAb to digest its substrate (right panel) at 25 °C. At high temperatures (e.g., 37 °C), the mAb avoids the local minima by virtue of cooperative transitions; the low energy gap is eliminated, causing an increase in active state population; thus the rate of catalysis increases. The model explains 3E5 mAbs behavior seen in this study. IgG2a, IgG1, and IgG3 isotypes are catalytically inactive at 25 °C because of their unique energy landscapes with highly populated, deep inactive states, which do not favor catalysis. At 37 °C, the energy landscape is remodeled, and all mAbs adopt their unique active or inactive states with different activity profiles.
Together, we interpret these findings as implying that the attachment of the same V region to different C regions results in subtle structural changes in the V domain that alter the immunoglobulin paratope to cause profound effects on GXM and peptide binding and peptide hydrolysis. In the complexes with different mAbs, the shifts seen in the amide resonances in NMR experiments prior to proteolysis were negligible, indicating that the amide bonds of the two labeled residues were in a similar chemical environment as they were in their Ab-free form. Future uniform 13C,15N labeling of P1 may give more insight into the chemical environments and subtle differences of the mAb-binding pockets imposed by different C regions.
These studies provide a mechanism for prior observations that V region identical mAbs differing in isotype manifest Ag specificity differences. We show that the presence of different C regions attached to the same V region confers new properties to the IgG in electronic emission spectra and proteolytic capacity of its Ag. Furthermore, these studies provide a new example of Ab catalysis as an intrinsic characteristic of Ab function and suggest a possible explanation why peptide mimetics have not been very effective as vaccines (50). These results are interpreted as indicating that C region-mediated changes in the V region structure result in isotype-related differences in Ab paratope. Subtle changes in the energy landscape of an Ab molecule translate into changes in the stabilities of different Ab conformations that may have significant consequences at the systems level. Fine-tuning the immunoglobulin energy landscape by switching the C regions in accordance with a specific immune response might be crucial in facilitating quick and specific responses to changes in external stimuli.
Supplementary Material
Acknowledgments
We thank Dr. Matthew D. Scharff for critical reading of this manuscript. We thank Mary Ann Gawinowicz at the Columbia University Protein Core Facility for the Mass Spectrometry analysis. Molecular graphics and analyses were performed with the UCSF Chimera package, which was developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco.
This work was supported, in whole or in part, by National Institutes of Health Grant 2P41RR001081. This work was also supported by National Institute of General Medical Sciences Grant 9P41GM103311.

This article contains supplemental Figs. S1 and S2.
- Ab
- antibody
- C region
- constant region
- V region
- variable region
- Ag
- antigen
- GXM
- glucuronoxylomannan
- HSQC
- heteronuclear single quantum coherence
- MD
- molecular dynamics.
REFERENCES
- 1. Gilliland G. L., Luo J., Vafa O., Almagro J. C. (2012) Leveraging SBDD in protein therapeutic development. Antibody engineering. Methods Mol. Biol. 841, 321–349 [DOI] [PubMed] [Google Scholar]
- 2. Cooper L. J., Robertson D., Granzow R., Greenspan N. S. (1994) Variable domain-identical antibodies exhibit IgG subclass-related differences in affinity and kinetic constants as determined by surface plasmon resonance. Mol. Immunol. 31, 577–584 [DOI] [PubMed] [Google Scholar]
- 3. Dam T. K., Torres M., Brewer C. F., Casadevall A. (2008) Isothermal titration calorimetry reveals differential binding thermodynamics of variable region-identical antibodies differing in constant region for a univalent ligand. J. Biol. Chem. 283, 31366–31370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Torres M., Casadevall A. (2008) The immunoglobulin constant region contributes to affinity and specificity. Trends Immunol. 29, 91–97 [DOI] [PubMed] [Google Scholar]
- 5. Torres M., Fernández-Fuentes N., Fiser A., Casadevall A. (2007) The immunoglobulin heavy chain constant region affects kinetic and thermodynamic parameters of antibody variable region interactions with antigen. J. Biol. Chem. 282, 13917–13927 [DOI] [PubMed] [Google Scholar]
- 6. Torres M., Fernandez-Fuentes N., Fiser A., Casadevall A. (2007) Exchanging murine and human immunoglobulin constant chains affects the kinetics and thermodynamics of antigen binding and chimeric antibody autoreactivity. PLoS One 2, e1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Torres M., May R., Scharff M. D., Casadevall A. (2005) Variable-region-identical antibodies differing in isotype demonstrate differences in fine specificity and idiotype. J. Immunol. 174, 2132–2142 [DOI] [PubMed] [Google Scholar]
- 8. Cooper L. J., Shikhman A. R., Glass D. D., Kangisser D., Cunningham M. W., Greenspan N. S. (1993) Role of heavy chain constant domains in antibody-antigen interaction. Apparent specificity differences among streptococcal IgG antibodies expressing identical variable domains. J. Immunol. 150, 2231–2242 [PubMed] [Google Scholar]
- 9. McLean G. R., Torres M., Elguezabal N., Nakouzi A., Casadevall A. (2002) Isotype can affect the fine specificity of an antibody for a polysaccharide antigen. J. Immunol. 169, 1379–1386 [DOI] [PubMed] [Google Scholar]
- 10. Pritsch O., Magnac C., Dumas G., Bouvet J. P., Alzari P., Dighiero G. (2000) Can isotype switch modulate antigen-binding affinity and influence clonal selection? Eur. J. Immunol. 30, 3387–3395 [DOI] [PubMed] [Google Scholar]
- 11. Kato K., Matsunaga C., Odaka A., Yamato S., Takaha W., Shimada I., Arata Y. (1991) Carbon-13 NMR study of switch variant anti-dansyl antibodies. Antigen binding and domain-domain interactions. Biochemistry 30, 6604–6610 [DOI] [PubMed] [Google Scholar]
- 12. Tudor D., Yu H., Maupetit J., Drillet A. S., Bouceba T., Schwartz-Cornil I., Lopalco L., Tuffery P., Bomsel M. (2012) Isotype modulates epitope specificity, affinity, and antiviral activities of anti-HIV-1 human broadly neutralizing 2F5 antibody. Proc. Natl. Acad. Sci. U.S.A. 109, 12680–12685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Casadevall A., Janda A. (2012) Immunoglobulin isotype influences affinity and specificity. Proc. Natl. Acad. Sci. U.S.A. 109, 12272–12273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Elkon K., Casali P. (2008) Nature and functions of autoantibodies. Nat. Clin. Pract. Rheumatol. 4, 491–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Casadevall A., Mukherjee J., Devi S. J., Schneerson R., Robbins J. B., Scharff M. D. (1992) Antibodies elicited by a Cryptococcus neoformans-tetanus toxoid conjugate vaccine have the same specificity as those elicited in infection. J. Infect. Dis. 165, 1086–1093 [DOI] [PubMed] [Google Scholar]
- 16. Spira G., Scharff M. D. (1992) Identification of rare immunoglobulin switch variants using the ELISA spot assay. J. Immunol. Methods 148, 121–129 [DOI] [PubMed] [Google Scholar]
- 17. Janda A., Casadevall A. (2010) Circular Dichroism reveals evidence of coupling between immunoglobulin constant and variable region secondary structure. Mol. Immunol. 47, 1421–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dadachova E., Bryan R. A., Apostolidis C., Morgenstern A., Zhang T., Moadel T., Torres M., Huang X., Revskaya E., Casadevall A. (2006) Interaction of radiolabeled antibodies with fungal cells and components of the immune system in vitro and during radioimmunotherapy for experimental fungal infection. J. Infect. Dis. 193, 1427–1436 [DOI] [PubMed] [Google Scholar]
- 19. Morahan G., Berek C., Miller J. F. (1983) An idiotypic determinant formed by both immunoglobulin constant and variable regions. Nature 301, 720–722 [DOI] [PubMed] [Google Scholar]
- 20. Lange H., Solterbeck M., Berek C., Lemke H. (1996) Correlation between immune maturation and idiotypic network recognition. Eur. J. Immunol. 26, 2234–2242 [DOI] [PubMed] [Google Scholar]
- 21. Reitan S. K., Hannestad K. (1995) A syngeneic idiotype is immunogenic when borne by IgM but tolerogenic when joined to IgG. Eur. J. Immunol. 25, 1601–1608 [DOI] [PubMed] [Google Scholar]
- 22. Huang X., Yang X., Luft B. J., Koide S. (1998) NMR identification of epitopes of Lyme disease antigen OspA to monoclonal antibodies. J. Mol. Biol. 281, 61–67 [DOI] [PubMed] [Google Scholar]
- 23. Tsang P., Rance M., Fieser T. M., Ostresh J. M., Houghten R. A., Lerner R. A., Wright P. E. (1992) Conformation and dynamics of an Fab'-bound peptide by isotope-edited NMR spectroscopy. Biochemistry 31, 3862–3871 [DOI] [PubMed] [Google Scholar]
- 24. Zilber B., Scherf T., Levitt M., Anglister J. (1990) NMR-derived model for a peptide-antibody complex. Biochemistry 29, 10032–10041 [DOI] [PubMed] [Google Scholar]
- 25. Eryilmaz E., Benach J., Su M., Seetharaman J., Dutta K., Wei H., Gottlieb P., Hunt J. F., Ghose R. (2008) Structure and dynamics of the P7 protein from the bacteriophage phi 12. J. Mol. Biol. 382, 402–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nicholas M. P., Eryilmaz E., Ferrage F., Cowburn D., Ghose R. (2010) Nuclear spin relaxation in isotropic and anisotropic media. Prog. Nucl. Magn Reson. Spectrosc. 57, 111–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Casadevall A., Cleare W., Feldmesser M., Glatman-Freedman A., Goldman D. L., Kozel T. R., Lendvai N., Mukherjee J., Pirofski L. A., Rivera J., Rosas A. L., Scharff M. D., Valadon P., Westin K., Zhong Z. (1998) Characterization of a murine monoclonal antibody to Cryptococcus neoformans polysaccharide that is a candidate for human therapeutic studies. Antimicrob. Agents Chemother. 42, 1437–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nimrichter L., Frases S., Cinelli L. P., Viana N. B., Nakouzi A., Travassos L. R., Casadevall A., Rodrigues M. L. (2007) Self-aggregation of Cryptococcus neoformans capsular glucuronoxylomannan is dependent on divalent cations. Eukaryot. Cell 6, 1400–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McFadden D. C., De Jesus M., Casadevall A. (2006) The physical properties of the capsular polysaccharides from Cryptococcus neoformans suggest features for capsule construction. J. Biol. Chem. 281, 1868–1875 [DOI] [PubMed] [Google Scholar]
- 30. Bodenhausen G., Ruben D. J. (1980) Natural abundance nitrogen-15 NMR by enhanced heteronuclear spectroscopy. Chem. Phys. Lett. 69, 185–189 [Google Scholar]
- 31. Palmer A. G., III, Cavanagh J., Wright P. E., Rance M. (1991) Sensitivity improvement in proton-detected two-dimensional heteronuclear correlation NMR spectroscopy. J. Magn. Reson. 93, 151–170 [Google Scholar]
- 32. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) NMRPipe. A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 33. Johnson B. A. (2004) Using NMRView to Visualize and Analyze the NMR Spectra of Macromolecules Protein NMR Techniques (Downing A. K., ed) pp. 313–352, Humana Press, Totowa, NJ: [DOI] [PubMed] [Google Scholar]
- 34. Young A. C., Valadon P., Casadevall A., Scharff M. D., Sacchettini J. C. (1997) The three-dimensional structures of a polysaccharide binding antibody to Cryptococcus neoformans and its complex with a peptide from a phage display library. Implications for the identification of peptide mimotopes. J. Mol. Biol. 274, 622–634 [DOI] [PubMed] [Google Scholar]
- 35. Case D. A., Cheatham T. E., III, Simmerling C. L., Wang J., Duke R. E., Luo R., Zhang W., Merz K. M., Roberts B., Wang B., Hayik S., Roitberg A., Seabra G., Wong K. F., Paesani F., Vanicek J., Liu J., Wu X., Brozell S. R., Steinbrecher T., Cai Q., Ye X., Wang J., Hsieh M. J., Cui G., Roe D. R., Mathews D. H., Sagui C., Babin V., Luchko T., Gusarov S., Kovalenko A., Kollman P. A. (2010) AMBER11, University of California, San Francisco [Google Scholar]
- 36. Hornak V., Abel R., Okur A., Strockbine B., Roitberg A., Simmerling C. (2006) Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 65, 712–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L. (1983) Comparison of simple potential functions for simulating liquid water. J. Chem. Physics 79, 926 [Google Scholar]
- 38. Valadon P., Nussbaum G., Boyd L. F., Margulies D. H., Scharff M. D. (1996) Peptide libraries define the fine specificity of anti-polysaccharide antibodies to Cryptococcus neoformans. J. Mol. Biol. 261, 11–22 [DOI] [PubMed] [Google Scholar]
- 39. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera. A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 40. Okochi N., Kato-Murai M., Kadonosono T., Ueda M. (2007) Design of a serine protease-like catalytic triad on an antibody light chain displayed on the yeast cell surface. Appl. Microbiol. Biotechnol. 77, 597–603 [DOI] [PubMed] [Google Scholar]
- 41. Saphire E. O., Stanfield R. L., Crispin M. D., Parren P. W., Rudd P. M., Dwek R. A., Burton D. R., Wilson I. A. (2002) Contrasting IgG structures reveal extreme asymmetry and flexibility. J. Mol. Biol. 319, 9–18 [DOI] [PubMed] [Google Scholar]
- 42. Vivian J. T., Callis P. R. (2001) Mechanisms of tryptophan fluorescence shifts in proteins. Biophys. J. 80, 2093–2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tanford C. (1997) How protein chemists learned about the hydrophobic factor. Protein Sci. 6, 1358–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Erhan S., Greller L. D. (1974) Do immunoglobulins have proteolytic activity? Nature 251, 353–355 [DOI] [PubMed] [Google Scholar]
- 45. Messina T. C., Talaga D. S. (2007) Protein free energy landscapes remodeled by ligand binding. Biophys. J. 93, 579–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Baldwin A. J., Kay L. E. (2009) NMR spectroscopy brings invisible protein states into focus. Nat. Chem. Biol. 5, 808–814 [DOI] [PubMed] [Google Scholar]
- 47. Benkovic S. J., Hammes G. G., Hammes-Schiffer S. (2008) Free-energy landscape of enzyme catalysis. Biochemistry 47, 3317–3321 [DOI] [PubMed] [Google Scholar]
- 48. Fraser J. S., Clarkson M. W., Degnan S. C., Erion R., Kern D., Alber T. (2009) Hidden alternative structures of proline isomerase essential for catalysis. Nature 462, 669–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Henzler-Wildman K. A., Thai V., Lei M., Ott M., Wolf-Watz M., Fenn T., Pozharski E., Wilson M. A., Petsko G. A., Karplus M., Hübner C. G., Kern D. (2007) Intrinsic motions along an enzymatic reaction trajectory. Nature 450, 838–844 [DOI] [PubMed] [Google Scholar]
- 50. Bianchi E., Ingallinella P., Finotto M., Joyce J., Liang X., Miller M. D., Kinney G. G., Ciliberto G., Shiver J. W., Pessi A. (2009) Synthetic peptide vaccines. The quest to develop peptide vaccines for influenza, HIV and Alzheimer's disease. Adv. Exp. Med. Biol. 611, 121–123 [DOI] [PubMed] [Google Scholar]
- 51. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.