Background: TIS11 proteins control the degradation of mRNA containing AU-rich elements (ARE).
Results: Drosophila dTIS11 activates the polysomal deadenylation of the ARE mRNA Cecropin A1.
Conclusion: Drosophila dTIS11 controls fewer aspects of ARE mRNA decay as compared with its mammalian homologs.
Significance: TIS11 proteins may have acquired additional functions to control mRNA decay during evolution from invertebrates to mammals.
Keywords: Antimicrobial Peptides, Drosophila, MRNA Decay, Polyadenylation, RNA-binding Protein, AU-rich Element, Deadenylase, TIS11, Tristetraprolin
Abstract
The destabilization of AU-rich element (ARE)-containing mRNAs mediated by proteins of the TIS11 family is conserved among eukaryotes including Drosophila. Previous studies have demonstrated that Tristetraprolin, a human protein of the TIS11 family, induces the degradation of ARE-containing mRNAs through a large variety of mechanisms including deadenylation, decapping, and P-body targeting. We have previously shown that the degradation of the mRNA encoding the antimicrobial peptide Cecropin A1 (CecA1) is controlled by the TIS11 protein (dTIS11) in Drosophila cells. In this study, we used CecA1 mRNA as a model to investigate the molecular mechanism of dTIS11-mediated mRNA decay. We observed that during the biphasic deadenylation and decay process of this mRNA, dTIS11 enhances deadenylation performed by the CCR4-CAF-NOT complex while the mRNA is still associated with ribosomes. Sequencing of mRNA degradation intermediates revealed that the complete deadenylation of the mRNA triggers its decapping and decay in both the 5′-3′ and the 3′-5′ directions. Contrary to the observations made for its mammalian homologs, overexpression of dTIS11 does not promote the localization of ARE-containing mRNAs in P-bodies but rather decreases the accumulation of CecA1 mRNA in these structures by enhancing the degradation process. Therefore, our results suggest that proteins of the TIS11 family may have acquired additional functions in the course of evolution from invertebrates to mammals.
Introduction
The dynamic control of mRNA stability is one of the key strategies used by eukaryotic cells to regulate gene expression. The controlled degradation of mRNAs containing an AU-rich element (ARE)5 in their 3′-untranslated region is a prototypical example of such post-transcriptional control of mRNA degradation. In mammals, ARE-containing mRNAs are targeted by an array of RNA-binding proteins, among which one of the most extensively studied is the Tristetraprolin (TTP) protein (see Ref. 1 for a recent review). TTP belongs to a family of evolutionary conserved zinc finger RNA-binding proteins able to bind AREs, thereby accelerating mRNA degradation. First identified as a regulator of TNF-α production (2), TTP has since been recognized to control the degradation of a large variety of ARE mRNAs including mediators of the inflammatory and the anti-inflammatory responses (3, 4). The TIS11 protein family contains three members in humans and four members in mice. These multifunctional proteins can favor the decay of ARE mRNAs by acting at several levels in the mRNA degradation pathway. Deadenylases are key players in the control of mRNA degradation (5). TTP has been initially shown to stimulate deadenylation of ARE-containing messenger RNAs, suggesting that this protein could favor deadenylase recruitment (6). An in vitro analysis suggested that the poly-A specific ribonuclease (PARN) deadenylase was involved in TTP-mediated deadenylation of mRNAs (7). This hypothesis was sustained by the fact that PARN is required for ARE mediated-decay (AMD) in human HeLa cells (8). More recently, it has been demonstrated that the CCR4-CAF-NOT deadenylation complex participates in the in vivo deadenylation of an ARE-containing reporter mRNA in murine fibroblasts, suggesting that this complex is involved in the ARE-mediated deadenylation of mRNAs (9). Recent in vitro and in vivo experiments confirmed that in mammals, TTP is able to recruit the deadenylase CAF1 through its interaction with the NOT1 adapting factor and targets ARE mRNAs to rapid deadenylation (10–12). The interaction between the deadenylation machinery and TTP is regulated by the MAPK-activated protein (MAPKAP) kinase 2-dependent phosphorylation of TTP (10, 11). Although deadenylation seems to play a major role in TTP-mediated mRNA decay in mammals, the observation that TTP can promote AMD when the poly(A) tail is artificially replaced by a histone 3′ end-processing sequence suggests that TTP can exert its effects through alternative mechanisms (13).
The detection of the ARE-binding proteins TTP and BRF1 in complex with the decapping machinery and the observation that decapping of mammalian ARE mRNAs can be a limiting step in the degradation process suggested a mechanism in which TTP could also induce AMD by promoting mRNA decapping (14). This proposed mechanism is further supported by the fact that TTP activates DCP2 decapping activity in vitro (15).
In eukaryotes, RNA degradation enzymes are found concentrated in P-bodies, which are considered as “factories for mRNA decay” (see Ref. 16) for review). The mammalian members of the TIS11 protein family are all found at least partially localized in P-bodies (17). The observation that ARE mRNAs are specifically targeted to P-bodies (18) and that in some cases inhibition of P-body formation prevents AMD (19) suggested that an additional function of TTP and of related mammalian proteins was to deliver ARE mRNAs to degradation in P-bodies. Alternatively, P-bodies could function as reservoirs to sequester ARE mRNAs from the translational machinery when mRNA decay is delayed (18). ARE mRNA degradation can occur through the 3′-5′ exosome-dependent degradation pathway (20, 21). However, in mammalian cells, the observation that both ARE mRNAs and TTP proteins accumulate in P-bodies when the 5′-3′ degradation pathway is inhibited suggested that this mode of degradation is favored in TTP-mediated ARE mRNA decay (18, 19). Altogether, these observations indicate that in mammals, TTP and the other members of the TIS11 family are multipotent proteins, which favor AMD by enhancing several crucial steps in the mRNA degradation process.
AMD is a conserved mechanism among eukaryotes. In yeast, CTH2p, the homolog of TTP, regulates the stability of mRNAs important for iron metabolism through an ARE-dependent process (22). Recently, we and others have shown that AMD is conserved in Drosophila. In this organism, dTIS11 regulates the stability of ARE-containing mRNAs including messenger RNAs involved in the immune response, such as antimicrobial peptide-coding mRNAs (23–25). We have observed that AMD requires the CAF1 protein in Drosophila. However, the molecular mechanism of mRNA decay mediated by proteins of the TIS11 family remains so far mostly uncharacterized in invertebrates. By using the mRNA encoding the Cecropin A1 (CecA1) antimicrobial peptide as a model substrate of AMD, we explored the involvement of dTIS11 in several steps of mRNA degradation. We observed that the deadenylation and decay of CecA1 mRNA is a biphasic process. The initial deadenylation occurs in the absence of mRNA decay, and the final deadenylation occurs while the messenger is still loaded on polysomes and is the key step controlled by dTIS11 to target the mRNA for degradation. Interestingly, in Drosophila cells, ARE-containing mRNAs are neither specifically addressed to P-bodies nor do they seem to be preferentially targeted for decapping. Our results suggest that the activity of dTIS11 is mainly based on its capacity to enhance deadenylation in Drosophila cells and that proteins of this family may have acquired additional functions later in the course of evolution from invertebrates to mammals.
EXPERIMENTAL PROCEDURES
Reagents
Actinomycin D and DNA oligonucleotides were purchased from Sigma-Aldrich. Hygromycin B and peptidoglycan (Escherichia coli K12) were purchased from InvivoGen. Tobacco acid pyrophosphatase was purchased from Epicenter, and T4 RNA ligase was from New England Biolabs.
Plasmids and dsRNAs
Expression vectors coding for dTIS11 and GFP were prepared by cloning corresponding RT-PCR products in the pMT/V5-His vector (Invitrogen). CAF1 cDNA was amplified with a 3′ primer replacing the natural stop codon by a HA tag and cloned in the pMT vector. The CAF1-D53A/E55A dominant negative form was obtained by site-directed mutagenesis to replace aspartate 53 and glutamate 55 by alanines. dsRNAs were prepared by in vitro transcription (MEGAscript RNAi, Ambion) of either PCR products or cDNAs cloned in the pBluescript vector. Amplified sequences were selected with the E-RNAi software (26).
Cell Culture, Transfection, and RNAi Experiments
S2 cells were maintained at 25 °C in Schneider medium (Invitrogen) supplemented with 10% fetal bovine serum. Highly transfectable S2 subclone (kind gift of N. Silverman, University of Massachusetts, Worcester, MA) were stably transfected with FuGENE® HD (Roche Applied Science) and maintained in Schneider medium supplemented with 300 μg/ml hygromycin B. For RNAi experiments, cells were grown in the presence of dsRNA (5 μg/ml) for 7 days.
RNA Analysis
In Drosophila cells, negative cross-talks operating in the immunodeficiency (Imd) pathway result in transient activation of the Cecropin A1 promoter in response to peptidoglycan stimulation (Ref. 27 and references therein). This process allowed us to evaluate the stability of CecA1 mRNA for several hours after stimulation of S2 cells with peptidoglycan and without the addition of potentially toxic pharmacological inhibitors of transcription. However, as this kinetic of transcriptional shutdown cannot be precisely controlled, evaluations of CecA1 mRNA stability over short periods of time were performed in the presence of actinomycin D to achieve rapid transcriptional inhibition. Northern blot and RT-PCR were performed as described elsewhere (23). For high resolution Northern blot, RNA samples were separated by electrophoresis on 3% agarose gels. Molecular weight markers were prepared by in vitro transcription of defined size cDNAs using 32P-labeled UTP. In vitro deadenylated CecA1 mRNA was prepared by hybridization of total RNA with oligo(dT) (18–21-mer) at 37 °C for 30 min in the presence of 1 unit of RNase H. Circularized rapid amplification of cDNA ends (cRACE) analysis was performed according to the adaptation of Rissland and Norbury based on previously described methods (Ref. 28 and references therein). Briefly, total RNA from S2 cells was purified with TRI Reagent® (Molecular Research Center). Samples were treated or not with alkaline phosphatase and tobacco acidic phosphatase to remove the cap structure prior to circularization with T4 RNA ligase. cDNA clones obtained by RT-PCR amplification of the CecA1 mRNA using divergent primers (forward, GGCCAGAATGAGAGCGACGAAAAC; reverse, ATCGAACGCGTTGGTCAGCAC) were cloned in the PCRII vector (Invitrogen) and sequenced. The selected primers allowed the detection of mRNA degradation extending 120 nucleotides and 180 nucleotides from the 5′ and 3′ ends, respectively.
RNA Fluorescent in Situ Hybridization
The CecA1 antisense RNA probe was synthesized by in vitro transcription of a 320-bp fragment of the CecA1 cDNA and incorporation of biotinylated UTP. S2 cells were seeded on glass coverslips. Samples were washed twice with phosphate-buffered saline (PBS), fixed with paraformaldehyde 4% for 30 min, rinsed three times in PBS for 10 min, and treated with ethanol at 70, 90, and 100% for 3 min followed by 3 min in ethanol at 90 and 70%, respectively. Coverslips were rinsed in PBS and treated with 0.1% pepsin for 15 min at 37 °C. Final fixation was performed in paraformaldehyde 4% for 10 min before rinsing three times for 10 min in PBS. Samples were hybridized overnight with the CecA1 antisense probe at 42 °C in hybridization buffer (5% formamide, 2.25× saline/sodium phosphate/EDTA, 10% dextran, 2.5× Denhardt's solution, 100 μg/ml tRNA, 5 mm DTT, 100 μg/ml salmon sperm DNA, and 40 units/ml RNase OUT® (Invitrogen)). Samples were rinsed consecutively for 5 min in 3× SSC and NTE buffer (Nad-Tris-EDTA) before treatment with RNase A (in NTE) at 50 μg/ml for 30 min at 37 °C and rinsed again three times in NTE for 5 min and 30 min in 2× SSC. Final wash was performed in 0.1× SSC at 57 °C for 1 h. Signal was amplified by the Tyramide amplification method (TSA) (Molecular Probes) according to the manufacturer's instructions and Oregon green as fluorescent label. Coverslips were rinsed in water and mounted with fluorescent mounting medium (Dako) supplemented with 4′,6-diamidino-2-phenylindole (100 pg/ml) and sealed with nail polish before imaging on a Zeiss observer inverted microscope with a 100× objective. In combined FISH/immunofluorescent experiments, samples were not treated with pepsin, and GFP was detected with an anti-GFP antibody and Alexa Fluor 594-labeled secondary antibody due to loss of intrinsic fluorescent properties of the GFP during the FISH procedure.
Polysome Fractionation
S2 cells were treated by cycloheximide (100 μg/ml) for 5 min and harvested in buffer containing 25 mm Hepes, 100 mm KCl, 5 mm MgCl2, 0.5% Nonidet P-40, 2 μg/ml heparin, and 10 μg/ml cycloheximide. Cell extracts were cleared by centrifugation at 12,000 × g for 10 min and loaded onto 15–50% linear sucrose gradients. Samples were ultracentrifuged at 200,000 × g for 3 h in a SW40.1 rotor and subsequently fractionated with a gradient fractionator (Brandel).
RESULTS
Overexpression of dTIS11 Accelerates the Degradation of CecA1 mRNA but Does Not Alter the Biphasic Profile of mRNA Deadenylation and Decay
General mRNA deadenylation and decay is a biphasic process in eukaryotes. The first slow and synchronous deadenylation shortens the poly(A) tail without detectable decay of the mRNA. The second phase of deadenylation is less synchronous and is accompanied by mRNA decay (recently reviewed in Ref. 29). In Drosophila S2 cells, the mRNA encoding the antimicrobial peptide Cecropin A1 is rapidly accumulated in response to the activation of the Imd pathway by Gram-negative peptidoglycan. Due to the ARE present in its 3′-UTR, this messenger RNA is also rapidly degraded. The analysis of the deadenylation process of the CecA1 mRNA is facilitated by its small RNA body size (353 nucleotides) and can be directly evaluated by Northern blot. Therefore, the degradation of CecA1 mRNA is a useful model to study the mechanism of AMD in Drosophila cells. We have previously shown that this mRNA is deadenylated by a two-step mechanism (23). Our RNAi experiments indicated that both dTIS11 and CAF1 were necessary for the second phase of deadenylation, whereas depletion of these proteins alone had no detectable influence on the first phase of deadenylation (23).
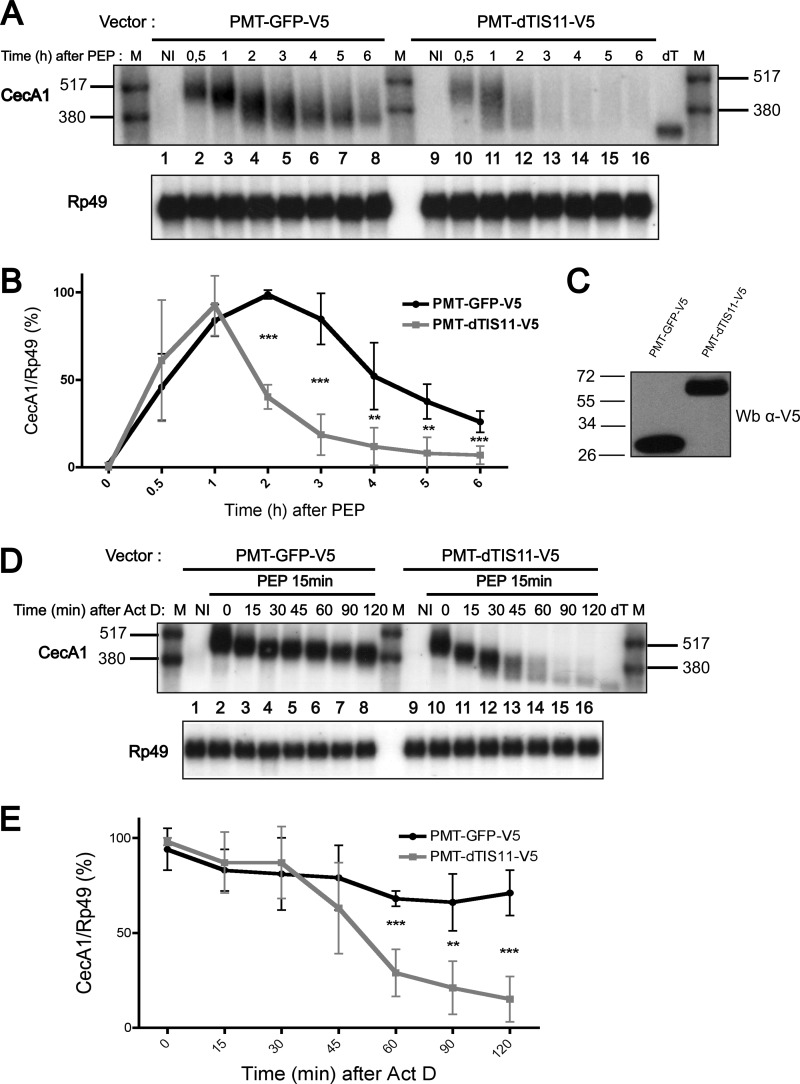
To better delineate the influence of dTIS11 on the deadenylation and decay mechanism of Drosophila ARE-containing mRNAs, we established S2 cell lines stably transfected with an expression vector where dTIS11 or GFP coding sequences were placed under the control of the Metallothionein gene promoter. S2 cells were treated with copper sulfate to induce the accumulation of GFP or dTIS11 (Fig. 1C) and stimulated with peptidoglycan to transiently activate CecA1 gene expression (27). We observed that CecA1 mRNA accumulation over time is significantly reduced in cells overexpressing dTIS11 as compared with cells expressing GFP (Fig. 1A, compare left and right panels and Fig. 1B), demonstrating that the accumulation of dTIS11 is sufficient to enhance CecA1 mRNA degradation. In GFP control cells, the average length of the CecA1 mRNA is markedly decreased during the initial 2 h of stimulation (Fig. 1A, lanes 2–4), suggesting a rapid deadenylation of the mRNA after synthesis, whereas a decrease in the total amount of mRNA is initiated only after 2 h of stimulation (Fig. 1B). As described previously (23), this degradation phase of the mRNA is concomitant with a further decrease in length of the remaining pool of messenger RNA (Fig. 1A, lanes 4–8). In dTIS11-overexpressing cells, CecA1 mRNA degradation is detectable as soon as 1 h after stimulation (Fig. 1B), whereas this mRNA is already partially deadenylated as indicated by the smear detectable in Fig. 1A (lane 11). Transcriptional blockade by actinomycin D during peptidoglycan stimulation further confirmed the absence of mRNA degradation during the initial phase of CecA1 mRNA deadenylation in control cells (Fig. 1D, lanes 2–8, and Fig. 1E). Interestingly, in dTIS11-overexpressing cells, this initial deadenylation of ∼70–80 nucleotides occurs more rapidly as it is completed after 15–30 min of actinomycin D treatment (Fig. 1D, compare lane 8 with lanes 11–12). However, the total amount of CecA1 mRNA remains constant and not significantly different from the level observed in control cells during this period of time (0–30 min after actinomycin D addition, Fig. 1E). After 30 min of actinomycin D treatment, we observed a processive deadenylation and decay of the CecA1 mRNA pool (Fig. 1D, lanes 12–16). Therefore, although dTIS11 is able to accelerate the deadenylation and the decay process of CecA1 mRNA, our observations suggest that this factor is only able to trigger mRNA decay when the poly(A) tail has been markedly reduced from its initial size.
FIGURE 1.
dTIS11 accelerates the degradation of CecA1 mRNA but does not alter the biphasic profile of mRNA deadenylation and decay. A, S2 cells stably transfected with GFP or dTIS11 expression vectors were incubated in the presence of peptidoglycan for the indicated periods of time. Total RNA was extracted, and 8 μg were analyzed by Northern blot using the indicated riboprobes for hybridization. In vitro deadenylated mRNA sample (dT) and radiolabeled RNAs (M) were added for size reference. NI, non induced. A high exposure time autoradiography is reproduced to reveal the differences between control and transfected cells upon long stimulation with peptidoglycan. B, four independent Northern blots performed as in A were quantified by PhosphorImager, and the ratio of CecA1/Rp49 signal was plotted as a function of time. PEP, peptidoglycan. C, analysis of GFP and dTIS11 protein levels by Western blot (Wb). Protein extracts from cells transfected with GFP or dTIS11 expression vectors were analyzed by Western blot using a primary antibody directed against the V5 tag fused to GFP and dTIS11. D, S2 cells were stimulated with peptidoglycan for 15 min before the addition of actinomycin D (Act D) for the indicated periods of time before analysis as in A. E, three independent experiments performed as in D were quantified by PhosphorImager, and the ratio of CecA1/Rp49 signal was plotted as a function of time. In B and E, error bars represent S.D. for each time point. Mean values were tested for statistical significance by a one-tailed Student's t test. **, p < 0.01, ***, p < 0.001.
The Initial Deadenylation of CecA1 mRNA Is Indifferently Performed by the PAN2-PAN3 and CCR4-CAF1-NOT Complexes, whereas the Late Deadenylation Phase Relies Solely on the CCR4-CAF1-NOT Complex and on CAF1 Catalytic Activity
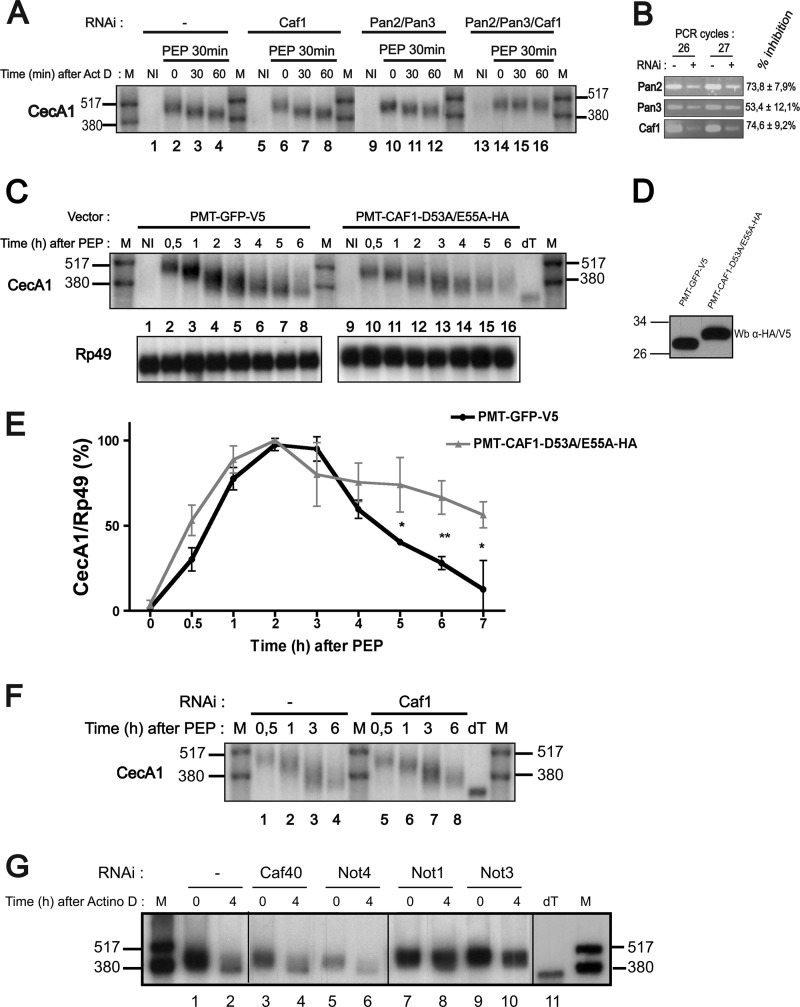
To identify the enzymatic activities involved in the different deadenylation steps of CecA1 mRNA, S2 Drosophila cells were treated with dsRNA targeting messenger RNAs encoding all the Drosophila deadenylation complexes described so far (5), including the PAN2-PAN3 complex, which was reported in mammals to be responsible for the initiation of general mRNA deadenylation (30). Following incubation with dsRNA, cells were stimulated with peptidoglycan. After 30 min, actinomycin D was added for short periods of time to observe the initial phase of CecA1 mRNA deadenylation. As shown in Fig. 2A (left panel), the size of CecA1 mRNA is rapidly reduced after actinomycin D treatment (compare lanes 2 and 4). Pretreatment of cells with dsRNAs targeting mRNAs encoding CAF1 or PAN2 and PAN3 proteins efficiently reduces the accumulation of these mRNAs (Fig. 2B) but has no effect on the shortening of CecA1 mRNA (Fig. 2A, second and third panels). Similarly, knockdown of mRNAs encoding CCR4, Nocturnin, and Angel deadenylases does not affect the initial deadenylation of the mRNA (not shown). However, the combined knockdown of PAN2, PAN3, and CAF1 leads to the accumulation of the full-size CecA1 mRNA (Fig. 2A, right panel), thereby indicating that the PAN2-PAN3 complex and CAF1 protein play redundant functions in controlling the initial phase of CecA1 mRNA deadenylation.
FIGURE 2.
Identification of factors involved in the two phases of CecA1 mRNA deadenylation. A, S2 cells were pretreated or not with double-stranded RNAs directed against the indicated mRNAs for 4 days (Caf1) or 7 days (Pan2/Pan3). Stimulation with peptidoglycan (PEP) was performed before the addition of actinomycin D (Act D) for the indicated periods of time. Total RNA was extracted, and 8 μg were analyzed by Northern blot using the indicated riboprobes for hybridization. The results are representative of three independent experiments. B, knockdown of Caf1, Pan2, and Pan3 genes by RNAi was confirmed by RT-PCR stopped after 26 and 27 cycles. The inhibition efficiencies were determined by quantification of three independent experiments, and S.D. were calculated. C, S2 cells transfected with plasmids encoding GFP or a dominant negative form of CAF1 were incubated in the presence of peptidoglycan for the indicated periods of time. Total RNA was extracted, and 8 μg were analyzed by Northern blot using the indicated riboprobes for hybridization. dT designates in vitro deadenylated mRNA generated by incubation of RNA with oligo(dT) and RNase H, and M designates radiolabeled RNAs. D, protein levels of GFP and the dominant negative form of CAF1 were evaluated by Western blot using a primary antibody directed against the V5 or HA tags fused to the expressed proteins. E, three independent Northern blots performed as in C were quantified by PhosphorImager, and the ratio of CecA1/Rp49 signal was plotted as a function of time. Error bars represent S.D. for each time point. Mean values were tested for statistical significance by a one-tailed Student's t test. *, p < 0.05; **, p < 0.01. F, cells were pretreated or not with double-stranded RNA directed against the Caf1 mRNA and stimulated with peptidoglycan as indicated. CecA1 mRNA was analyzed as in C. G. S2 cells were pretreated with double-stranded RNAs directed against the indicated mRNAs for 7 days. Cells were stimulated with peptidoglycan for 3 h before actinomycin D (Actino D) treatment for 4 h to evaluate the effect of dsRNA treatments on the late phase of deadenylation of the CecA1 mRNA as visualized by high resolution Northern blot.
The CAF1 protein belongs to the CCR4-CAF1-NOT complex involved in general mRNA deadenylation in eukaryotes. Both the CCR4 and the CAF1 components of this complex contain deadenylase domains. Although only CCR4 seems to be catalytically involved in deadenylation in yeast, both proteins are active in mammals (9). In Drosophila, CAF1 has been identified as the main enzyme involved in the general deadenylation of mRNAs (Ref. 31 and references therein). Because we previously described that CAF1 was necessary for the second deadenylation phase and for the decay of ARE mRNAs in Drosophila (23), we determined whether the intrinsic catalytic activity of CAF1 was required for the deadenylation of CecA1 mRNA. S2 cell lines were transfected with vectors encoding the GFP or a dominant negative form of CAF1 (Fig. 2D). We compared the accumulation of CecA1 mRNA in response to prolonged peptidoglycan stimulation of these cell lines. The expression of the catalytic mutant form of CAF1 has little effect on the accumulation of CecA1 during the initial 3 h of peptidoglycan stimulation corresponding to the initial phase of mRNA deadenylation but significantly increases the amount of CecA1 mRNA detected during the later phase of deadenylation and decay occurring after 3 h of stimulation (Fig. 2, C and E). The ability of the dominant negative form of CAF1 to inhibit the secondary deadenylation of the mRNA was further confirmed by the broader band detected on Northern blot after 4 h of stimulation with peptidoglycan (Fig. 2C).
We tested whether subunits of the CCR4-CAF1-NOT complex were required for the second phase of CecA1 mRNA deadenylation. We confirmed that the depletion of CAF1 inhibits the last deadenylation step of CecA1 mRNA (Fig. 2F). Inhibition of Not1 and Not3 expression also induces the accumulation of a partially deadenylated form of CecA1 mRNA. In contrast, knockdown of Not4 and Caf40 had no effect on CecA1 mRNA deadenylation (Fig. 2G). Altogether, our results show that the catalytic activities of CAF1 as well as the NOT1 and NOT3 subunits are required to induce the second phase of CecA1 deadenylation and to target this mRNA to degradation, suggesting that CAF1 deadenylase is active on CecA1 mRNA in the context of the CCR4-CAF1-NOT complex. Because total depletion of NOT subunits cannot be achieved by RNAi, we cannot determine at this point whether all subunits of this complex are required for the deadenylation of this ARE-containing mRNA, but our results suggest that a decrease in the abundance of NOT4 and CAF40 does not affect the deadenylating activity of the complex toward ARE mRNAs.
Decapping Conditions CecA1 mRNA Degradation and Occurs Only on Fully Deadenylated Messenger RNA Molecules
In mammals, proteins of the TIS11 family are able to enhance the decay of ARE mRNAs by stimulating mRNA decapping. TTP and BRF-1 can interact with the Dcp1-Dcp2 decapping complex. TTP has also been shown to activate decapping in vitro (15), and the decapping activator Lsm1 is essential for AMD in mammalian cells (19). Alternatively, decapping of mammalian ARE mRNAs can also be achieved by the Nudt16 decapping factor (32). Analysis of the general mRNA degradation pathway in mammals indicates that decapping can take place after the initial phase of deadenylation or later when the mRNA has reached a fully deadenylated state (30).
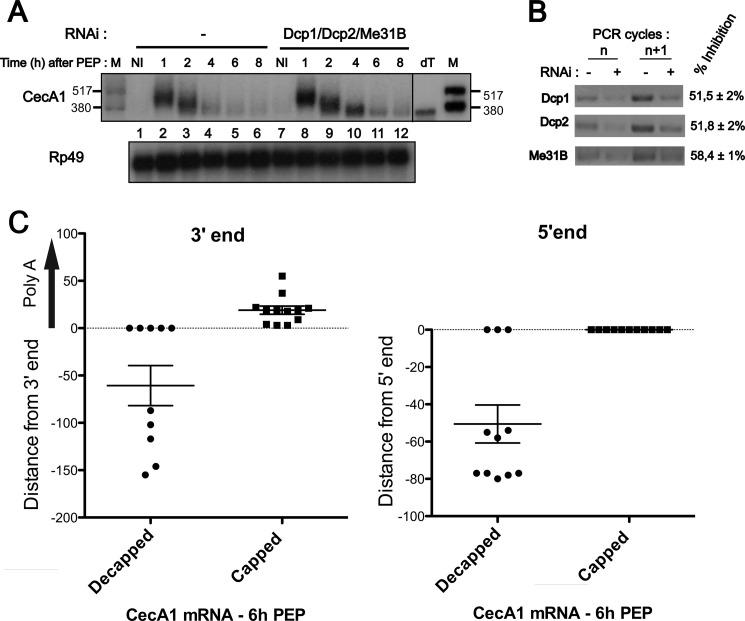
To analyze the influence of decapping on AMD in Drosophila cells, we assessed the degradation of CecA1 mRNA in S2 cells where decapping has been inhibited by depletion of the DCP1/2 components of the decapping complex and of the Me31b decapping co-activator (Fig. 3 B). Cells were stimulated with peptidoglycan for increasing periods of time, and CecA1 mRNA decay was evaluated by Northern blot (Fig. 3A). As described previously for Figs. 1A and 2C, signal quantification shows that the peak concentration of CecA1 mRNA is reached after 2 h and reduced by 48% after 4 h of stimulation with peptidoglycan. However, the inhibition of the decapping activity by depleting Me31B and Dcp1/2 favors the accumulation of a short form of CecA1 messenger RNA co-migrating with in vitro deadenylated molecules (Fig. 3A, right panel). Therefore, our data suggest that decapping favors the subsequent degradation of this messenger.
FIGURE 3.
Cap structure controls CecA1 mRNA degradation but not deadenylation. A, S2 cells were pretreated or not with double-stranded RNAs directed against the indicated mRNAs for 7 days before induction with peptidoglycan (PEP), and in the absence of actinomycin D, for the indicated times. Total RNA was extracted, and 8 μg were analyzed by Northern blot using the indicated riboprobes for hybridization. dT designates in vitro deadenylated mRNA generated by incubation with oligo(dT) and RNase H, and M designates radiolabeled RNAs. Results are representative of three independent experiments. B, knockdown of indicated genes by RNAi was confirmed by RT-PCR stopped after 26 and 27 cycles. The inhibition efficiencies were determined by quantification of three independent experiments, and S.D. were calculated. C, individual clones generated by cRACE were sequenced and compared with a reference full-length deadenylated CecA1 mRNA sequence. Results were plotted on a vertical scattered plot, and means ± S.E. are indicated for each experimental condition.
In mammals, decapping can occur after the first or second phase of mRNA deadenylation (Ref. 29 and references therein). We evaluated the polyadenylation state of capped and decapped CecA1 mRNA molecules by cRACE methods (Ref. 28 and references therein). We isolated total mRNA from S2 cells induced by peptidoglycan for 6 h. As described previously (23), under these conditions, the initial deadenylation phase of CecA1 mRNA has been completed, and the remaining mRNA is engaged in the second phase of deadenylation and decay. To distinguish capped and decapped mRNAs, total RNA was either directly self-ligated with T4 RNA ligase or ligated after treatment with shrimp alkaline phosphatase followed by treatment with tobacco acid pyrophosphatase. Under direct ligation conditions, only mRNA devoid of cap structure and presenting a 5′-monophosphate end can be self-ligated. Removal of this 5′-terminal phosphate with alkaline phosphatase impairs self-ligation of decapped mRNA, whereas subsequent treatment with pyrophosphatase generates 5′-phosphate from initially capped mRNA, allowing the self-ligation of these mRNA molecules. The region containing the junction of the 5′ and 3′ ends of CecA1 mRNA was specifically amplified by RT-PCR on both populations of self-ligated RNA. PCR products were ligated into the pCRII plasmid, and individual clones were sequenced for each experimental condition. Analysis of cDNA sequences obtained from a pool of decapped mRNA shows that under this condition, the 3′ end of CecA1 mRNA is shortened on average from 61 nucleotides downstream from the cleavage and polyadenylation site of the full-length CecA1 mRNA (Fig. 3C, left panel, left column). The same analysis performed on cDNA sequences generated from capped mRNA indicates that the 3′end is intact and carries a short poly(A) tail of 20 nucleotides on average (Fig. 3C, left panel, right column).
The presence of a short poly(A) tail on all cDNA clones generated from capped mRNA and the absence of poly(A) tail on all clones generated from decapped mRNA suggest that decapping occurs after the last phase of deadenylation when the mRNA is fully deadenylated. Moreover, these data show that the degradation of decapped CecA1 mRNA molecules can occur in the 3′ to 5′ direction as demonstrated by the 3′ truncation of 50% of cDNA clones derived from decapped mRNA. A parallel analysis of the 5′ sequence shows that decapped CecA1 mRNA can also be degraded in the 5′ to 3′ direction, whereas as expected, capped mRNA are protected from 5′ exonucleolytic degradation (Fig. 3C, right panel). An analysis of the 5′ region by the RNAfold algorithm (33) predicted two hairpin structures centered at position 48 and 83 (not shown). These structures could contribute to the enrichment of intermediate degradation products observed around these positions in the mRNA (Fig. 3C, right panel).
The ability of dTIS11 to stimulate the decapping mechanism of ARE mRNAs could not be evaluated by this approach because the presence of long poly(A) tails on CecA1 mRNA molecules from dTIS11-depleted cells disrupts the cRACE procedure.6 However, because we previously observed that depletion of dTIS11 blocks the secondary deadenylation and decay phase of ARE-containing mRNAs (23), we can conclude that dTIS11 is active on ARE-containing mRNA before the decapping step.
Accumulation of CecA1 mRNA in P-bodies Is Not Required for Its Degradation
In mammals, TTP and BRF1, two proteins of the TIS11 family, specifically deliver ARE mRNAs to P-bodies (18). Indeed, in human cells, the accumulation of ARE-mRNAs in P-bodies correlates with cellular levels of TIS11 proteins. Depleting TTP and BRF1 by siRNA reduces the accumulation of ARE-mRNAs in P-bodies, whereas overexpression of TTP enhances the accumulation of an ARE mRNA to P-bodies (18). These observations suggest that one function of TIS11 proteins in mammals is to stimulate mRNA decay by enhancing the contact between ARE mRNAs and degradation factors concentrated in P-bodies. Alternatively, these factors could silence gene expression by localizing ARE mRNAs away from the translation machinery (18).
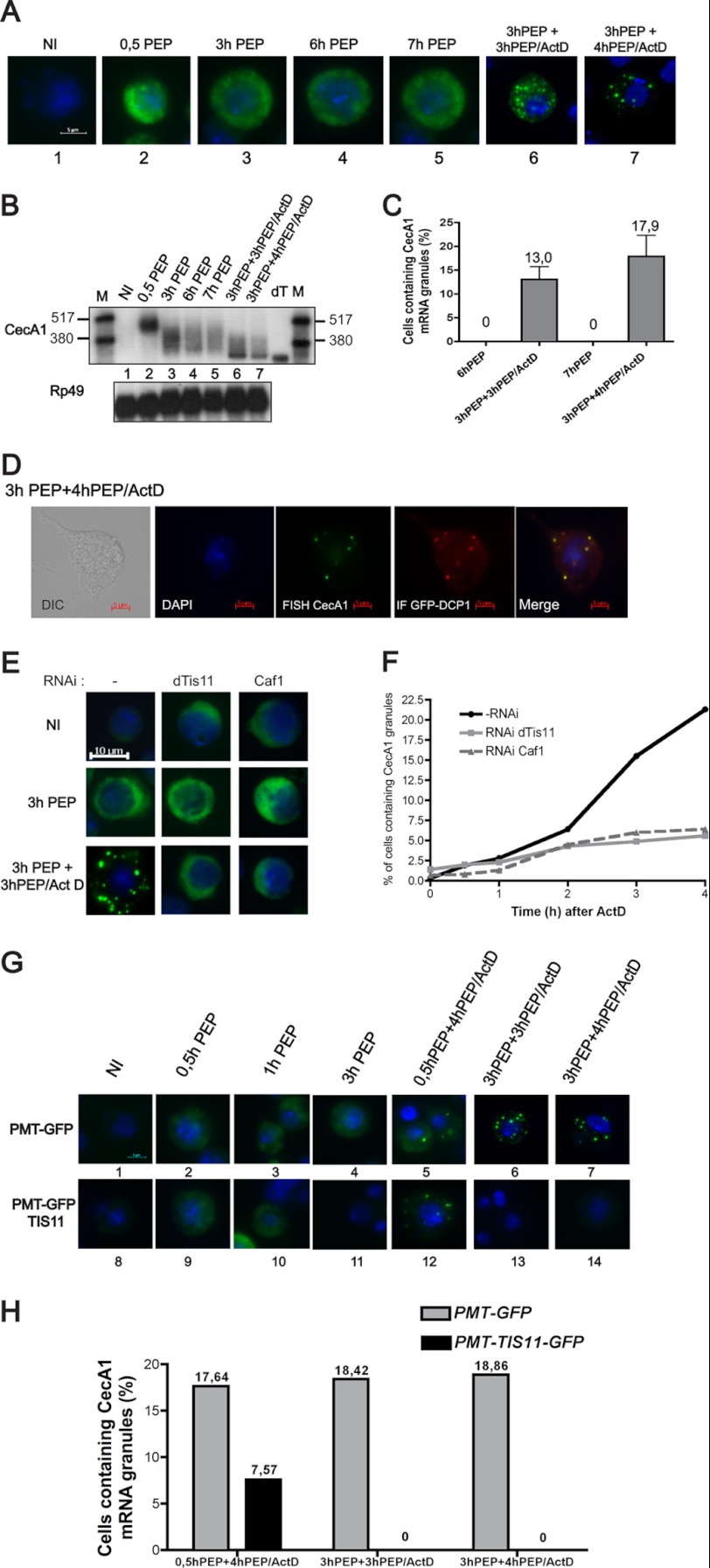
To determine whether this function of TIS11 proteins is conserved in invertebrates, we evaluated the localization of CecA1 mRNA in the course of degradation in Drosophila cells by RNA FISH experiments. As shown in Fig. 4A, CecA1 mRNA is barely detectable in the cytoplasm of unstimulated S2 cells. After 30 min of stimulation by peptidoglycan, CecA1 mRNA accumulates in the cytoplasm. The localization of the messenger remains unchanged upon prolonged exposure of the cells to peptidoglycan. CecA1 mRNA is homogeneously distributed in the cytoplasm during the deadenylation and decay process and does not concentrate in cytoplasmic foci (Fig. 4A, pictures 2–5, and Fig. 4B, lanes 2–5). Interestingly, in cells treated with actinomycin D, CecA1 mRNA is detectable in cytoplasmic granules in ∼15% of the cells (Fig. 4A, pictures 6–7, and Fig. 4C) concomitantly with the accumulation of a fully deadenylated form of this molecule (Fig. 4B, lanes 6–7). Co-localization of these granules with overexpressed DCP1 proteins identifies them as P-bodies (Fig. 4D). In mammals, the formation of P-bodies is stimulated by depletion of the mRNA degradation machinery (18). Therefore, accumulation of CecA1 mRNA in P-bodies could result from a depletion of the RNA degradation machinery induced by actinomycin D in S2 cells.
FIGURE 4.
Differential localization of CecA1 mRNA in S2 cells treated or not with actinomycin D. A, S2 cells were stimulated with peptidoglycan (PEP) alone or peptidoglycan with the addition of actinomycin D (ActD) for the indicated periods of time. Cells were fixed and hybridized with an in vitro transcribed antisense biotinylated CecA1 riboprobe. Fluorescent labeling was performed with the TSA-Oregon green labeling kit (Molecular Probes). Samples were mounted in Dako fluorescent mounting medium in the presence of DAPI and observed at 1000× magnification. B, total RNA extracted from cells treated as in A were analyzed by Northern blot as described in the legend for Fig. 1. dT designates in vitro deadenylated mRNA generated by incubation with oligo(dT) and RNase H, and M designates radiolabeled RNAs. C, three independent experiments were performed as in A. For each experimental condition in every experiment, 300 cells were observed individually at 630× magnification to count CecA1 mRNA-positive granules. D, cells were stimulated with peptidoglycan and actinomycin D as indicated, and combined FISH/immunofluorescence experiments were performed as described under “Experimental Procedures.” E, S2 cells were treated with dsRNA directed against dTis11 or Caf1 mRNA for 7 days before a FISH experiment performed as in A. F, cells treated as in E and incubated in the presence of actinomycin D for increasing periods of time. For each experimental condition, the presence of CecA1 mRNA-containing granules was evaluated on a total of 300 independent cells. G, S2 cells transfected with a GFP or dTIS11-GFP expression vector were stimulated with peptidoglycan alone or peptidoglycan and actinomycin D for the indicated periods of time. FISH experiments were performed as described under “Experimental Procedures.” H, the presence of CecA1 mRNA-containing granules was evaluated on 300 transfected cells for each experimental condition.
We have previously observed that dTIS11 does not localize in P-bodies in Drosophila S2 cells (23). However, to determine whether dTIS11 could favor the accumulation of CecA1 mRNA in P-bodies as described in mammals, we analyzed the localization of CecA1 mRNA in cells treated with peptidoglycan and actinomycin D after inhibition of dTIS11 expression by RNAi. This experiment revealed that depletion of dTIS11 inhibited the accumulation of CecA1 mRNA in P-bodies (Fig. 4E). The number of cells containing detectable CecA1 mRNA in P-bodies increased with the duration of actinomycin D treatment, and after 4 h, this accumulation was approximately four times lower in cells treated with a dsRNA directed against dTIS11 as compared with untreated cells (Fig. 4F). Similarly, depletion of CAF1 decreased CecA1 mRNA accumulation in P-bodies. In mammals, it has been reported that the formation of P-bodies requires active deadenylation (9). Because we have shown that dTIS11 induces CecA1 mRNA deadenylation, our observations that dTIS11 and CAF1 depletion inhibits the accumulation of CecA1 mRNA in P-bodies could be the consequence of the inhibition of the deadenylation process rather than the ability of dTIS11 to actively target ARE mRNAs to P-bodies.
We further tested the ability of dTIS11 to directly address CecA1 mRNA to P-bodies by comparing the localization of CecA1 mRNA in cells overexpressing a GFP-dTIS11 fusion protein or GFP alone. In both conditions, no CecA1 mRNA granule is observed in the absence of actinomycin D (Fig. 4G, pictures 1–4 and 8–11). Upon treatment with actinomycin D after peptidoglycan stimulation, the number of cells containing detectable CecA1 mRNA in cytoplasmic foci is markedly reduced in cells transfected with GFP-dTIS11 as compared with cells transfected with GFP alone (Fig. 4, G and H), correlating with the highest degradation rate of the CecA1 mRNA in the presence of high amount of dTIS11 (Fig. 1A). Therefore, contrary to the observations made for TTP in mammalian cells (18), overexpression of dTIS11 in Drosophila cells is not able to induce the accumulation of an ARE mRNA in P-bodies. The ability of ARE mRNAs to localize in P-bodies in the presence of actinomycin D in S2 cells could be a consequence of the deadenylation induced by dTIS11 protein and the inhibition of degradation potentially induced by actinomycin D.
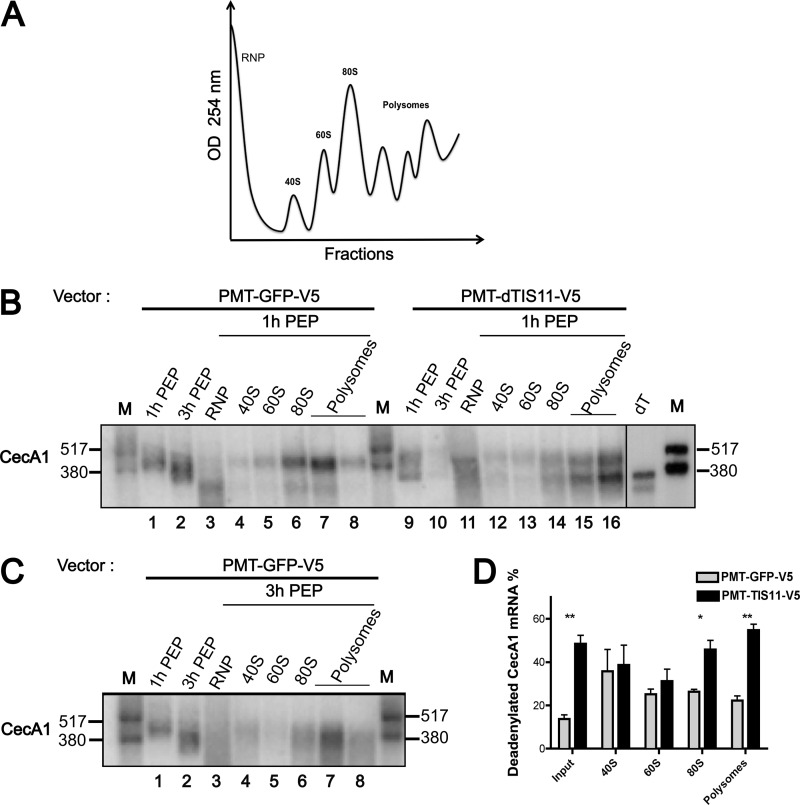
dTIS11 Stimulates the Polysomal Deadenylation of ARE-containing mRNA
Because dTIS11 does not induce CecA1 mRNA accumulation in specialized degradation structures such as P-bodies, we determined whether it promotes mRNA deadenylation once these are released in the cytoplasm or whether this protein targets polysome-associated mRNA molecules. Therefore, we analyzed the polysome distribution of CecA1 mRNA in S2 cells transfected with GFP or dTIS11 expression constructs. Cells were stimulated with peptidoglycan for 1 or 3 h, and cytoplasmic extracts were separated by ultracentrifugation on sucrose gradients. In cells overexpressing a GFP control construct and stimulated with peptidoglycan for 1 h, CecA1 mRNA is mainly distributed in 80 S and polysomal fractions, whereas partially degraded mRNAs are found in the ribosome-free fraction (Fig. 5B, left panel). Interestingly, a minor shortened form of CecA1 mRNA is also present in the ribosomal fractions. This minor form co-migrates with an in vitro deadenylated CecA1 mRNA, suggesting that the deadenylated form of CecA1 mRNA can be found in association with the translational machinery. The ectopic expression of dTIS11 strongly enhances the proportion of this fully deadenylated form of CecA1 mRNA in the 80 S and polysomal fractions (Fig. 5B, right panel, and Fig. 5D). Upon dissociation of polysomes by puromycin, all CecA1 mRNA molecules were shifted toward the top of the gradient (data not shown), further confirming that both adenylated and deadenylated CecA1 mRNA molecules can be found in association with polysomes. Stimulation of the cells with peptidoglycan for 3 h further enhances the deadenylated form of CecA1 mRNA associated with polysomes (Fig. 5, B and C, compare left panels). As described in Fig. 1, dTIS11 overexpression greatly increases the degradation of CecA1 mRNA after 3 h of stimulation with peptidoglycan, and this mRNA is no more detectable in polysomes in such experimental conditions (not shown). Altogether, our data indicate that dTIS11 promotes the deadenylation of CecA1 mRNA while still associated into polysomes and accelerates its degradation.
FIGURE 5.
dTIS11 induces the deadenylation of polysome-associated CecA1 mRNA. A, representative graph of continuous A254 nm recording of sucrose gradient fractionation. OD, optical density. B, cell extracts from S2 cells stably transfected with a GFP or dTIS11 expression vector and stimulated with peptidoglycan (PEP) for 1 h were separated on 15–30% sucrose gradients and fractionated as described under “Experimental Procedures.” Total RNA was extracted from all fractions. Three fractions representative of the 40 S, 60 S, and 80 S peak regions of the gradient were pooled together. The first three and last three fractions of the polysome region were also pooled together. Equal volumes of pooled fractions were analyzed by Northern blot using a CecA1 antisense riboprobe. dT designates an in vitro deadenylated mRNA generated by incubation with oligo(dT) and RNase H, and M designates radiolabeled RNAs. C, cell extracts from S2 cells stably transfected with GFP expression vector and stimulated with peptidoglycan for 3 h were analyzed as in B. D, three independent Northern blots performed as in B were quantified with a PhosphorImager. The ratio of deadenylated/total CecA1 signal was calculated for each experimental point. The deadenylated fractions were defined as the bands co-migrating with the in vitro deadenylated form of CecA1 mRNA. Error bars represent S.D. for each experimental condition. Mean values were tested for statistical significance by a one-tailed Student's t test. *, p < 0.05; **, p < 0.01.
DISCUSSION
The destabilization of ARE-containing mRNAs mediated by proteins of the TIS11 family is conserved among eukaryotes. Previous studies have demonstrated that TTP, a human protein of the TIS11 family, induces the degradation of ARE-containing mRNAs through a large variety of mechanisms including deadenylation, decapping, and P-body targeting. Sequence alignment of TIS11 family members reveals a high degree of conservation in the CCCH zing finger RNA-binding region (34), whereas other regions of the protein are poorly conserved between invertebrates and mammals, suggesting that the molecular mechanism controlling the activity of TIS11 proteins may have evolved in the course of eukaryotic evolution.
We have investigated the molecular mechanism mediating the action of the Drosophila dTIS11 protein on the ARE-containing mRNA coding for the antimicrobial peptide CecA1. We observed that the degradation of CecA1 mRNA is initiated by a two-step deadenylation mechanism. We have shown that CAF1 is required for the late deadenylation of the CecA1 mRNA. Moreover, CAF1 requires the presence of at least the NOT1 and NOT3 proteins to deadenylate this mRNA. Our data also suggest that the PAN2-PAN3 and CCR4-CAF1-NOT deadenylation complexes play a redundant role at the beginning of the deadenylation process because the initial deadenylation phase is inhibited solely when proteins from both complexes are depleted. Our results corroborate previous data indicating that both the PAN2-PAN3 and the CCR4-CAF-NOT complexes were involved in mRNA deadenylation in Drosophila (35).
The ability of TIS11 family members to enhance the deadenylation of ARE-containing mRNAs has been clearly established in mammals (6, 9). TTP is able to recruit CAF1 deadenylase, and this interaction relies on the recruitment of NOT1 by the C-terminal region of TTP (10–12). We have also observed a co-immunoprecipitation of dTIS11 with the CCR4-CAF1-NOT complex (data not shown), suggesting that the ability of TIS11 proteins to recruit the CCR4-CAF1-NOT complex on ARE-containing mRNAs has been conserved from invertebrates to mammals. By depletion of CCR4-CAF1-NOT complex subunits, we observed that NOT1 and NOT3 are required for the second deadenylation phase of CecA1 mRNA, whereas depletion of NOT4 and CAF40 does not alter this process. Although we cannot exclude the requirement of NOT4 and CAF40 on the basis of RNAi depletion experiments, our results confirm previous observations that CAF1, NOT1, NOT2, and NOT3 are required for the general deadenylation mechanism of mRNA in Drosophila and that NOT4 and CAF40 do not seem necessary for this process. Moreover, NOT4 was not detected as a stable component of the CCR4-CAF1-NOT complex (31).
We observed that overexpression of TIS11 accelerates the decay of CecA1 mRNA (Fig. 1), whereas we have previously shown that dTIS11 is essential for the late, CAF1-dependent deadenylation but is not required for the early deadenylation of ARE mRNAs (23). Therefore, the ability of TIS11 to stimulate the CAF1-dependent deadenylation of ARE mRNAs might be timely controlled. Phosphorylation events could transiently inactivate the ability of TIS11 to stimulate CAF1-induced deadenylation as observed with the TTP protein in mammals (10, 11). Alternatively, when both deadenylation complexes are present, early deadenylation of ARE mRNAs could be preferentially carried out by PAN2-PAN3 and therefore would prevent TIS11 from modulating the early steps of deadenylation in a CAF1-dependent manner. Our observations that a dominant negative form of CAF1 preferentially inhibits the late deadenylation of the CecA1 mRNA but does not alter the early deadenylation step (Fig. 2C) are in accordance with this hypothesis. However, these models are not mutually exclusive, and both mechanisms could coexist in vivo. Overall, our results suggest that dTIS11 enhances a general mechanism of mRNA deadenylation.
In mammals, evidence suggests that other deadenylases could mediate TTP function. Poly-A specific ribonuclease (PARN) deadenylase has been involved in AMD and was shown to interact with TTP in vitro (7, 8), although this interaction is not detected in cells overexpressing both factors (11). TTP is also able to recruit PAN2 deadenylase in a phosphorylation-dependent manner, suggesting that this enzyme could also participate in TTP-induced mRNA deadenylation (11). In Drosophila, the absence of PARN ortholog and the implication of the PAN2-PAN3 complex in the initial dTIS11-independent deadenylation phase of CecA1 mRNA suggest that the activity of dTIS11 protein is concentrated on the second deadenylation phase of ARE-containing mRNAs, where it enhances the CAF1-induced deadenylation of ARE-containing mRNA.
In eukaryotes, the removal of the cap structure is believed to occur on transcripts with short poly(A) tails but not necessarily on fully deadenylated mRNAs (30). In our cRACE analysis, the absence of residual poly(A) tail on decapped CecA1 mRNA and the systematic presence of a poly(A) tail on capped mRNA suggest that decapping occurs only on fully deadenylated CecA1 mRNA molecules. These observations could reflect the fact that the accelerated deadenylation induced by dTIS11 reduces the probability for the decapping machinery to trap an mRNA molecule before completion of the deadenylation process. Alternatively, partially deadenylated CecA1 mRNA could remain inaccessible to the decapping machinery. Because depletion of dTIS11 leads to the accumulation of partially deadenylated CecA1 mRNA (23), whereas decapping occurs on fully deadenylated messenger RNAs, it is unlikely that dTIS11 influences the decapping of ARE-containing mRNA in Drosophila, contrary to what has been observed with TTP (15).
By depleting components of the decapping machinery, we have observed the accumulation of a fully deadenylated form of CecA1 mRNA. Moreover, by the cRACE approach, we have observed the degradation of CecA1 mRNA core only of decapped mRNA, suggesting that decapping is required for mRNA degradation. Sequencing of cRACE cDNA clones also revealed that the core of CecA1 mRNA can be degraded both from the 5′ and from the 3′ ends. In mammals, ARE mRNA silencing involves the delivery of transcripts to P-bodies by TTP or by the related protein BRF-1 (18). In S2 cells, we detected CecA1 mRNA in P-bodies at late stages of mRNA degradation only when cells were treated with the transcription inhibitor actinomycin D. Moreover, this actinomycin D-dependent localization of CecA1 mRNA in P bodies is markedly reduced by dTIS11 overexpression (Fig. 4, G and H), suggesting that in contrast to its mammalian counterparts, dTIS11 is devoid of the ability to address ARE-containing mRNA to P-bodies. We also observed that preserving CecA1 mRNA from deadenylation by depletion of dTIS11 or CAF1 also abolished its migration into P-bodies in response to actinomycin D (Fig. 4E). Therefore, the migration of CecA1 mRNA into P-bodies in these conditions might be caused by the accumulation of deadenylated CecA1 mRNA in waiting for further degradation due to the saturation of the degradation machinery. In mammals, ARE mRNAs migrate into P-bodies in normal degradation conditions, but the amount of ARE mRNA localized in these structures is enhanced by depleting components of the RNA degradation machinery. Altogether, these observations could reflect the fact that the degradation machinery is more rapidly saturated in mammalian cells than in Drosophila S2 cells.
The general model of mRNA degradation suggests that ribosome dissociation is necessary for mRNA decapping and degradation (36). However, recent studies suggest that in yeast, mRNA degradation can occur in polysomes (37). In the major eukaryotic mRNA degradation pathway, mRNA decay is concomitant with the second phase of mRNA deadenylation (reviewed in Ref. 29). Our observation that dTIS11 enhances the second phase of CecA1 mRNA deadenylation before decapping without further addressing to P-bodies suggests that dTIS11 could target CecA1 mRNA in polysomes. This hypothesis is confirmed by our observation that the second phase of CecA1 mRNA deadenylation occurs in polysomes and is accelerated by overexpressing dTIS11. In agreement with this observation, both the CCR4-CAF1-NOT complex and TTP have been detected in polysomes (38, 39). Therefore, the polysomal deadenylation of ARE-containing mRNAs might represent the ancestral function of TIS11 family members, whereas new functions could have emerged in the course of evolution leading to the optimization of AMD.
Acknowledgments
We thank Neil Silverman and Nicholas Paquette for providing the highly transfectable S2 cell line and useful technical advices. We also thank Suzana Valane and Elisa Izaurralde for providing plasmid constructs.
This work was supported by the Fund for Medical Scientific Research (Belgium, Grant 2.4.511.00.F), the Fonds Brachet, the Fonds Van Buuren, and the “Actions de Recherches Concertées” (Grant AV.06/11-345).
C. Vindry, unpublished observations.
- ARE
- AU-rich element
- TTP
- Tristetraprolin
- AMD
- ARE-mediated decay
- cRACE
- circularized rapid amplification of cDNA ends
- CCR4
- carbon catabolite repression 4
- CAF1
- CCR4 associated factor 1.
REFERENCES
- 1. Baou M., Jewell A., Murphy J. J. (2009) TIS11 family proteins and their roles in post-transcriptional gene regulation. J. Biomed. Biotechnol. 2009, 634520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Carballo E., Lai W. S., Blackshear P. J. (1998) Feedback inhibition of macrophage tumor necrosis factor-α production by tristetraprolin. Science 281, 1001–1005 [DOI] [PubMed] [Google Scholar]
- 3. Lai W. S., Parker J. S., Grissom S. F., Stumpo D. J., Blackshear P. J. (2006) Novel mRNA targets for tristetraprolin (TTP) identified by global analysis of stabilized transcripts in TTP-deficient fibroblasts. Mol. Cell. Biol. 26, 9196–9208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Stoecklin G., Tenenbaum S. A., Mayo T., Chittur S. V., George A. D., Baroni T. E., Blackshear P. J., Anderson P. (2008) Genome-wide analysis identifies interleukin-10 mRNA as target of tristetraprolin. J. Biol. Chem. 283, 11689–11699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Goldstrohm A. C., Wickens M. (2008) Multifunctional deadenylase complexes diversify mRNA control. Nat. Rev. Mol. Cell Biol. 9, 337–344 [DOI] [PubMed] [Google Scholar]
- 6. Lai W. S., Carballo E., Strum J. R., Kennington E. A., Phillips R. S., Blackshear P. J. (1999) Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor-α mRNA. Mol. Cell. Biol. 19, 4311–4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lai W. S., Kennington E. A., Blackshear P. J. (2003) Tristetraprolin and its family members can promote the cell-free deadenylation of AU-rich element-containing mRNAs by poly(A) ribonuclease. Mol. Cell. Biol. 23, 3798–3812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lin W. J., Duffy A., Chen C. Y. (2007) Localization of AU-rich element-containing mRNA in cytoplasmic granules containing exosome subunits. J. Biol. Chem. 282, 19958–19968 [DOI] [PubMed] [Google Scholar]
- 9. Zheng D., Ezzeddine N., Chen C. Y., Zhu W., He X., Shyu A. B. (2008) Deadenylation is prerequisite for P-body formation and mRNA decay in mammalian cells. J. Cell Biol. 182, 89–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Marchese F. P., Aubareda A., Tudor C., Saklatvala J., Clark A. R., Dean J. L. (2010) MAPKAP kinase 2 blocks tristetraprolin-directed mRNA decay by inhibiting CAF1 deadenylase recruitment. J. Biol. Chem. 285, 27590–27600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clement S. L., Scheckel C., Stoecklin G., Lykke-Andersen J. (2011) Phosphorylation of tristetraprolin by MK2 impairs AU-rich element mRNA decay by preventing deadenylase recruitment. Mol. Cell. Biol. 31, 256–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sandler H., Kreth J., Timmers H. T., Stoecklin G. (2011) Not1 mediates recruitment of the deadenylase Caf1 to mRNAs targeted for degradation by tristetraprolin. Nucleic Acids Res. 39, 4373–4386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lai W. S., Blackshear P. J. (2001) Interactions of CCCH zinc finger proteins with mRNA: tristetraprolin-mediated AU-rich element-dependent mRNA degradation can occur in the absence of a poly(A) tail. J. Biol. Chem. 276, 23144–23154 [DOI] [PubMed] [Google Scholar]
- 14. Lykke-Andersen J., Wagner E. (2005) Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev. 19, 351–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fenger-Grøn M., Fillman C., Norrild B., Lykke-Andersen J. (2005) Multiple processing body factors and the ARE-binding protein TTP activate mRNA decapping. Mol. Cell. 20, 905–915 [DOI] [PubMed] [Google Scholar]
- 16. Kulkarni M., Ozgur S., Stoecklin G. (2010) On track with P-bodies. Biochem. Soc. Trans. 38, 242–251 [DOI] [PubMed] [Google Scholar]
- 17. Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke-Andersen J., Fritzler M. J., Scheuner D., Kaufman R. J., Golan D. E., Anderson P. (2005) Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169, 871–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Franks T. M., Lykke-Andersen J. (2007) TTP and BRF proteins nucleate processing body formation to silence mRNAs with AU-rich elements. Genes Dev. 21, 719–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stoecklin G., Mayo T., Anderson P. (2006) ARE-mRNA degradation requires the 5′-3′ decay pathway. EMBO Rep. 7, 72–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen C. Y., Gherzi R., Ong S. E., Chan E. L., Raijmakers R., Pruijn G. J., Stoecklin G., Moroni C., Mann M., Karin M. (2001) AU-binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107, 451–464 [DOI] [PubMed] [Google Scholar]
- 21. Mukherjee D., Gao M., O'Connor J. P., Raijmakers R., Pruijn G., Lutz C. S., Wilusz J. (2002) The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J. 21, 165–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vergara S. V., Thiele D. J. (2008) Post-transcriptional regulation of gene expression in response to iron deficiency: coordinated metabolic reprogramming by yeast mRNA-binding proteins. Biochem. Soc. Trans. 36, 1088–1090 [DOI] [PubMed] [Google Scholar]
- 23. Lauwers A., Twyffels L., Soin R., Wauquier C., Kruys V., Gueydan C. (2009) Post-transcriptional regulation of genes encoding anti-microbial peptides in Drosophila. J. Biol. Chem. 284, 8973–8983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cairrao F., Halees A. S., Khabar K. S., Morello D., Vanzo N. (2009) AU-rich elements regulate Drosophila gene expression. Mol. Cell. Biol. 29, 2636–2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Spasic M., Friedel C. C., Schott J., Kreth J., Leppek K., Hofmann S., Ozgur S., Stoecklin G. (2012) Genome-wide assessment of AU-rich elements by the AREScore algorithm. PLoS Genet. 8, e1002433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Horn T., Boutros M. (2010) E-RNAi: a web application for the multispecies design of RNAi reagents: 2010 update. Nucleic Acids Res. 38, W332–W339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim T., Yoon J., Cho H., Lee W. B., Kim J., Song Y. H., Kim S. N., Yoon J. H., Kim-Ha J., Kim Y. J. (2005) Down-regulation of lipopolysaccharide response in Drosophila by negative cross-talk between the AP1 and NF-κB signaling modules. Nat. Immunol. 6, 211–218 [DOI] [PubMed] [Google Scholar]
- 28. Rissland O. S., Norbury C. J. (2009) Decapping is preceded by 3′ uridylation in a novel pathway of bulk mRNA turnover. Nat. Struct. Mol. Biol. 16, 616–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen C. Y., Shyu A. B. (2011) Mechanisms of deadenylation-dependent decay. Wiley Interdiscip. Rev. RNA 2, 167–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yamashita A., Chang T. C., Yamashita Y., Zhu W., Zhong Z., Chen C. Y., Shyu A. B. (2005) Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat. Struct. Mol. Biol. 12, 1054–1063 [DOI] [PubMed] [Google Scholar]
- 31. Temme C., Zhang L., Kremmer E., Ihling C., Chartier A., Sinz A., Simonelig M., Wahle E. (2010) Subunits of the Drosophila CCR4-NOT complex and their roles in mRNA deadenylation. RNA 16, 1356–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li Y., Song M., Kiledjian M. (2011) Differential utilization of decapping enzymes in mammalian mRNA decay pathways. RNA 17, 419–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lorenz R., Bernhart S. H., Höner Zu Siederdissen C., Tafer H., Flamm C., Stadler P. F., Hofacker I. L. (2011) ViennaRNA Package 2.0. Algorithms Mol. Biol. 6, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cao H., Deterding L. J., Blackshear P. J. (2007) Phosphorylation site analysis of the anti-inflammatory and mRNA-destabilizing protein tristetraprolin. Expert Rev. Proteomics 4, 711–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bönisch C., Temme C., Moritz B., Wahle E. (2007) Degradation of hsp70 and other mRNAs in Drosophila via the 5′-3′ pathway and its regulation by heat shock. J. Biol. Chem. 282, 21818–21828 [DOI] [PubMed] [Google Scholar]
- 36. Coller J., Parker R. (2004) Eukaryotic mRNA decapping. Annu. Rev. Biochem. 73, 861–890 [DOI] [PubMed] [Google Scholar]
- 37. Hu W., Sweet T. J., Chamnongpol S., Baker K. E., Coller J. (2009) Co-translational mRNA decay in Saccharomyces cerevisiae. Nature 461, 225–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Collart M. A., Panasenko O. O. (2012) The Ccr4: not complex. Gene 492, 42–53 [DOI] [PubMed] [Google Scholar]
- 39. Pfeiffer J. R., McAvoy B. L., Fecteau R. E., Deleault K. M., Brooks S. A. (2011) CARHSP1 is required for effective tumor necrosis factor-α mRNA stabilization and localizes to processing bodies and exosomes. Mol. Cell. Biol. 31, 277–286 [DOI] [PMC free article] [PubMed] [Google Scholar]