Background: The two-dimensional kinetics of receptor-ligand interactions determines cell adherence and release.
Results: Based on kinetic parameters, time- and force-dependent transitions of bimolecular integrin αIIbβ3-fibrinogen complex were revealed.
Conclusion: Coupled force-free binding and forced unbinding kinetics reflects dynamics of surface-attached receptor and ligand molecules.
Significance: A new approach to explore the mechanism and kinetics of bimolecular interactions has been proposed.
Keywords: Cell Adhesion, Fibrinogen, Integrin, Kinetics, Single Molecule Biophysics
Abstract
Using a combined experimental and theoretical approach named binding-unbinding correlation spectroscopy (BUCS), we describe the two-dimensional kinetics of interactions between fibrinogen and the integrin αIIbβ3, the ligand-receptor pair essential for platelet function during hemostasis and thrombosis. The methodology uses the optical trap to probe force-free association of individual surface-attached fibrinogen and αIIbβ3 molecules and forced dissociation of an αIIbβ3-fibrinogen complex. This novel approach combines force clamp measurements of bond lifetimes with the binding mode to quantify the dependence of the binding probability on the interaction time. We found that fibrinogen-reactive αIIbβ3 pre-exists in at least two states that differ in their zero force on-rates (kon1 = 1.4 × 10−4 and kon2 = 2.3 × 10−4 μm2/s), off-rates (koff1 = 2.42 and koff2 = 0.60 s−1), and dissociation constants (Kd1 = 1.7 × 104 and Kd2 = 2.6 × 103 μm−2). The integrin activator Mn2+ changed the on-rates and affinities (Kd1 = 5 × 104 and Kd2 = 0.3 × 103 μm−2) but did not affect the off-rates. The strength of αIIbβ3-fibrinogen interactions was time-dependent due to a progressive increase in the fraction of the high affinity state of the αIIbβ3-fibrinogen complex characterized by a faster on-rate. Upon Mn2+-induced integrin activation, the force-dependent off-rates decrease while the complex undergoes a conformational transition from a lower to higher affinity state. The results obtained provide quantitative estimates of the two-dimensional kinetic rates for the low and high affinity αIIbβ3 and fibrinogen interactions at the single molecule level and offer direct evidence for the time- and force-dependent changes in αIIbβ3 conformation and ligand binding activity, underlying the dynamics of fibrinogen-mediated platelet adhesion and aggregation.
Introduction
Cell-cell and cell-substrate adhesion is mediated by specific interactions between cell surface receptors and ligands (1, 2). In turn, the kinetics of ligand binding has major effects on the rate of cell adherence and release under the influence of external mechanical factors and under physiological and pathophysiological conditions (3–5).
Most often the kinetic parameters of receptor-ligand interactions are determined using methods that measure the binding and unbinding of ensembles of receptors and ligands (6, 7). Thus, measured parameters represent a composite of interactions that could vary substantially because of possible heterogeneity of receptor and ligand conformations. However, state-of-the-art dynamic nanomechanical methods, such as atomic force microscopy (8), biomembrane force probe (9, 10), and magnetic (11) or optical (12, 13) tweezers, enable kinetic measurements at the single molecule level. In these methods, receptor and ligand molecules are firmly attached to apposed surfaces, mimicking cell adhesion where receptors and their ligands interact at an interface. Clamped or ramped piconewton pulling forces are then applied to dissociate bimolecular ligand (L)3-receptor (R) complexes (LR), and bond lifetime and rupture force measurements, respectively, are used to determine the kinetics of forced unbinding (LR → L + R). Information about the equilibrium energy landscape, i.e. the force-free off-rate koff and working distance between the bound and transition states x†, is then obtained indirectly using theoretical models for the force dependence of koff, such as the Bell model (14). Because the statistics of binding events are not analyzed, the reverse association process (L + R → LR), which is governed by the on-rate constant kon and important at small forces, is not characterized.
To account for dynamically coupled competing kinetic pathways, we propose a new method of analysis named binding-unbinding correlation spectroscopy (BUCS) that can be viewed as an analog of two-dimensional spectroscopy in the time domain (15). BUCS is based on a correlation between the kinetics of receptor-ligand complex formation at zero force and the kinetics of forced dissociation of the complex. Similar multidimensional approaches have been devised to explore the influence of relaxation of polypeptide chains on unfolding times for proteins (16) and to probe correlations between protein dynamics and unbinding kinetics for protein-protein complexes (17). The approach proposed here combines the measurements of bond lifetimes (unbinding phase) with measurements of association signals during time-controlled contact between the receptor- and ligand-coated surfaces (binding phase). Subsequent analysis is based on a probabilistic description of reversible association-dissociation transitions (L + R ↔ LR), taking into account the probability of binding and of bond survival per single receptor-ligand contact.
We have used this approach to investigate the interaction of the platelet integrin αIIbβ3 and its major ligand, fibrinogen. αIIbβ3, the major adhesion molecule on platelets, is essential for the hemostatic platelet aggregates that form when fibrinogen bound to αIIbβ3 cross-links adjacent platelets (18, 19). Fibrinogen binding to αIIbβ3 has been quantified at the single molecule level using the optical trap (12, 20, 21) and atomic force microscopy (22) and shown to be a complex multistep process in which the αIIbβ3-fibrinogen complex exists in at least two bound states with different mechanical stabilities. Here, using BUCS, we provide a model-free estimation of the kinetic parameters governing the two-dimensional association and dissociation kinetics of the αIIbβ3-fibrinogen complex. We found that the resistance of the complex to mechanical disruption increases with the duration of contact between the αIIbβ3- and fibrinogen-coated surfaces, a result of a time-dependent force-induced structural rearrangement in the αIIbβ3-fibrinogen complex that favors the formation of a high affinity state.
EXPERIMENTAL PROCEDURES
Optical Trap-based Force Clamp
A custom-built optical trap, described in detail previously (21), was used to measure individual ligand-receptor interactions under constant pulling force. Briefly, the core of the laser tweezer system is a Nikon Diaphot 300 inverted microscope and a 100× 1.3 numerical aperture Fluor lens combined with an FCBar neodymium-doped yttrium aluminum garnet laser (λ = 1,064 nm) with 4 watts of power in continuous TEM-00 mode. A computer-operated two-dimensional acousto-optical deflector is used to control the trap position. The force exerted by the trap on the displaced bead is measured with a quadrant detector. This system enables control of the duration of the compressive contact between interacting surfaces, the magnitude of the compressive force, and the magnitude of the tensile force during bond rupture. All experiments are conducted at an average trap stiffness of 0.10 ± 0.02 pN/nm. Force calibration and trap stiffness are routinely confirmed by the Stokes' force method. LabVIEW® software is used to control and record laser beam deflection, move the piezoelectric stage, and analyze data off-line.
Surfaces and Proteins
Surfaces were coated covalently with the interacting proteins essentially as described previously (20). Briefly, purified human fibrinogen in 20 mm HEPES, 150 mm NaCl, 1 mm CaCl2, pH 7.4 was immobilized to the bottom-anchored 5-μm spherical silica pedestal precoated with glutaraldehyde-activated polyacrylamide. Purified human αIIbβ3 in 20 mm HEPES, 150 mm NaCl, 30 mm n-octyl β-d-glucoside, 1 mm CaCl2, pH 7.4 was covalently coupled to carboxylate-modified 1.75-μm latex beads using water-soluble carbodiimide either in the absence or in the presence of 1 mm MnCl2. Bovine serum albumin (BSA) was used as a blocker for both surfaces. The surface density of the functional αIIbβ3 molecules capable of binding 125I-fibrinogen was determined to be 3,072 ± 412 molecules/μm2 (supplemental Fig. S2). αIIbβ3 was isolated from human platelets (13), and fibrinogen was from HYPHEN BioMed (France). The polypeptide composition, purity, and lack of oligomerization of αIIbβ3 and fibrinogen were assessed by SDS-PAGE and by transmission electron microscopy. The binding probability and rupture force profiles ensure single molecule interactions as inferred from the criteria based on previous experiments performed using the same protein binding protocols (12, 20, 21, 23).
Measurements of Surface Interactions
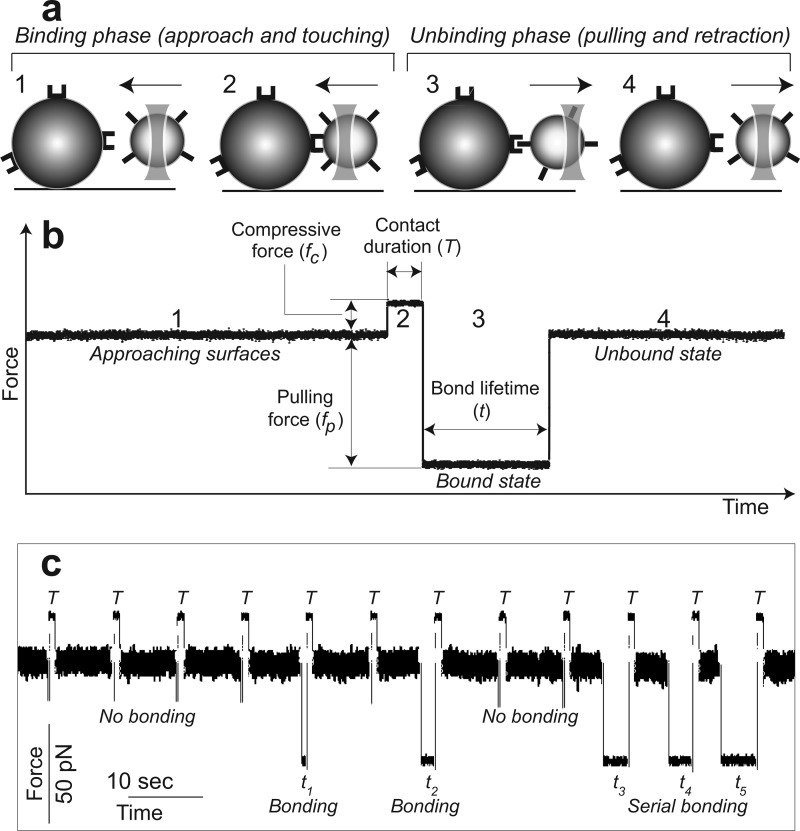
First, a flow chamber was equilibrated with the working buffer (0.1 m HEPES, pH 7.4 buffer with 2 mg/ml BSA, 0.1% Triton X-100, and 1 mm CaCl2 or 1 mm CaCl2, 1 mm MnCl2) at room temperature. 1 μl of the αIIbβ3-coated bead suspension (107 beads/ml) was added to 50 μl of the working buffer and flowed into a chamber containing pedestals with immobilized fibrinogen on their surface. After the chamber was placed on the microscope stage, a single bead was trapped and oscillated, and the stage was moved manually to bring a pedestal within 1–2 μm of the trapped bead. The separation of the pedestal and the bead was then reduced until repeated contacts were observed. The repeated touching occurred with a compressive force of 20 pN and a pulling force of 50 pN. The flexible parameter was the duration of contact, which varied from 0.05 to 2.0 s. The data were recorded at 2,000 scans/s. Several tens of pedestal-bead pairs were analyzed for each experimental condition. The binding-unbinding events from individual files were summarized so that the total number of bond lifetime values observed at each experimental condition varied from ∼400 to ∼4,000. Bond lifetimes <40 ms represented nonspecific interactions and were susceptible neither for inhibition nor activation of the integrin. Accordingly, they were neglected in data presentation and analysis.
Kinetic Model and Probability Measures
Consider L-R interactions where there is only one bound state (LR), i.e. L + R ↔ LR. In the binding phase of the measurements, we probe the reversible association-dissociation kinetics, which is governed by the on-rate kon and off-rate koff. In the unbinding phase, we examine the irreversible dissociation process, LR → L + R, described by the force-dependent dissociation rate k (we assume that the on-rate is negligible). Suppose we now combine the two types of measurements into a single measurement in which we observe the sequential transitions L + R ↔ LR → L + R. The experimental quantity, which describes the likelihood of observing these transitions, is the joint probability (P(T,t)). For the single step kinetics, P(T,t) is given by Equation 1.
 |
In the second line of Equation 1, Pb(T) = kondmax(1 − exp[−(kondmax + koff)T])/(kondmax + koff) and Ps(t) = exp[−kt] are the binding probability Pb(T) and bond survival probability Ps(t), respectively; dmax = max{dL,dR} (22) is the density of the species (L or R) present in excess; kon and koff are the force-free on- and off-rates, respectively; and k is the off-rate under force. In our experiment, integrin receptors are present in excess over fibrinogen ligands, and hence, dmax = dR. The kinetic rates (kon, koff, and k), equilibrium dissociation constant (Kd = koff/kon), and the population of the ligand-bound and unbound form of the receptor (Pb and Pu, respectively) can be obtained by performing a numerical fit of Equation 1 to the experimental data (dR can be obtained by using a radioactively labeled protein).
We analyzed the experimental data for integrin-fibrinogen interactions using the following minimal kinetic model (Scheme 1). The equilibrium kinetic transitions probed in the binding phase of the measurements are shown in Scheme 2. In the unbinding phase of the measurements, we examine the force-dependent transitions shown in Scheme 3. For this model, the binding probability Pb(T) involves contributions from the two pathways, reversible formation of the ligand-bound states LR1 and LR2, as shown in Equation 2.
The joint probability P(T,t), which is the sum of contributions for irreversible dissociation from states LR1 (LR1 → L + R1) and LR2 (LR2 → L + R2), also includes the pathway mixing due to the conformational fluctuations (LR1 ↔ LR2) as shown in Equation 3.
 |
In Equations 2 and 3, the individual binding probabilities Pb1(T) and Pb2(T) and bond survival probabilities Ps1(t) and Ps2(t) are given by Equations 4–7,
 |
 |
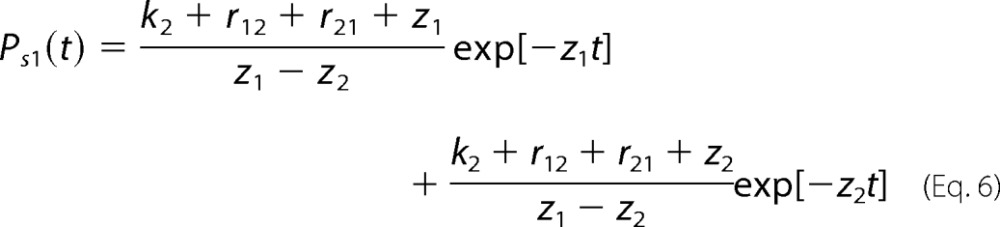 |
 |
where λ1 and λ2 are the initial populations of the receptor states R1 and R2. In Equations 4–7, z1,2 = (k1 + k2 +r12 + r21 ± D)/2 where D = ,b = k1+k2+r12+r21, and a = k1k2 + k1r21 + k2r12.
SCHEME 1.
SCHEME 2.
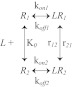
SCHEME 3.

RESULTS
BUCS
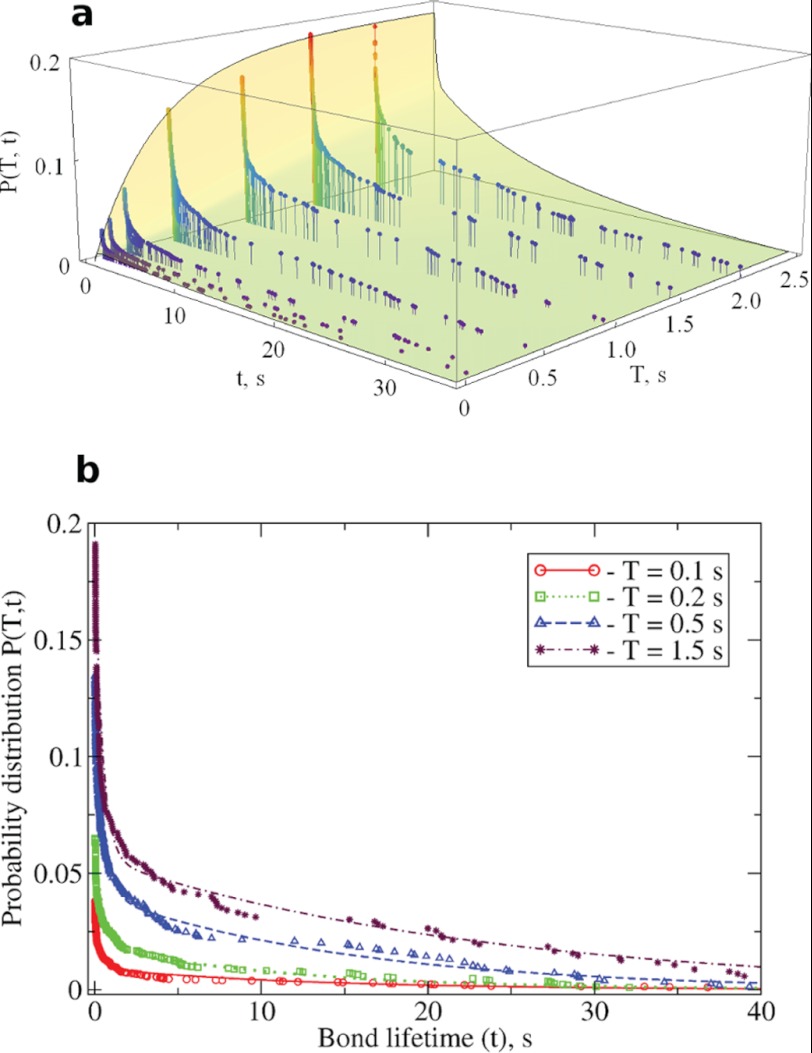
To analyze the kinetics of the reversible association and dissociation of LR using optical trap-based force spectroscopy, we devised a method that combines measurements of binding probability (Pb) as a function of interaction time (T) at low compressive forces (fc) with measurements of the probability of bond survival (Ps) at constant pulling forces (fp). This allows a probabilistic description of association-dissociation transitions. In the binding phase, T is varied to measure the T-dependent ratio of binding events (Nb) to binding attempts (N). When N is large, the ratio Nb(T)/N approaches a stable value equal to the binding probability Pb(T) (supplemental Fig. S1). When T is large, Pb(T) is equal to Pb, the equilibrium population of the bound state LR. Hence, Pb(T) from the binding phase provides the initial conditions for measurements of bond lifetime (t), which can then be used to calculate the probability of bond survival Ps(t). In turn, Pb(T) and Ps(t) can be convoluted to form the joint probability distribution P(T,t), defined as the probability that a bond formed before or at time T will survive until time t. Model calculations of Pb(T), Ps(t), and P(T,t) for the reversible single step kinetics L + R ↔ LR are shown in Fig. 1. We see that when T is large compared with the characteristic time scale for reaching equilibrium ((kondL + koff)−1) the binding probability Pb(T) plateaus, attaining the constant value kondL/(kondL + koff), the equilibrium population of the ligand-receptor bound state Pb.
FIGURE 1.
Results of model calculations of the probability measures Pb(T), Ps(t), and P(T,t) for the single step reversible association-dissociation kinetics, L + R ↔ LR → L + R (see Equation 1 under “Experimental Procedures”). We used the following kinetic parameters: kon = 0.6 × 10−3 μm2/s, koff = 0.5 s−1, k = 0.65 s−1, and ligand surface density dL = 103 μm−2. a, the binding probability Pb(T) as a function of interaction time T. b, the bond survival probability Ps(t) versus the bond lifetime t. Here, Ps(t) was calculated using the initial condition Pb = 1 (only the bonds formed are dissociated over time t). c, the two-dimensional surface of the joint probability distribution P(T,t) as a function of contact duration time T and bond survival time t.
The data needed to calculate the P(T,t) are obtained experimentally using a two-step optical trap protocol. First, by applying a compressive force, fc, a single receptor-ligand pair is approximated and allowed to interact for a fixed time, T1. Then a pulling force, fp, is applied to dissociate the complex (Fig. 2A). Within a single approach-retract cycle, both bond formation and the bond lifetime t are recorded (Fig. 2B). When the approach-retract cycles are repeated N times (Fig. 2C), the number of binding events carries information about the binding probability, and the bond lifetime measurements carry information about probability of bond survival at fixed T = T1. The combined product of binding probability Pb(T1) and bond survival Ps(t) is P(T1,t). Bond lifetime data collected at increasing contact times T = T1 < T2 < … < Tm and the corresponding binding probabilities (Pb(T1), Pb(T2), …, Pb(Tm)) are combined to obtain the joint distribution P(T,t). When there is only one conformational state of R or L and only one bound form of LR, the joint probability P(T,t) can be factorized (P(T,t) = Pb(T)Ps(t)), implying that there are no dynamic correlations between the unbinding data and binding data. Hence, the data from binding and unbinding assays can be analyzed and modeled separately. This approach is used in traditional dynamic force measurements where bond lifetime data are analyzed, but the statistics of binding events are ignored, which is equivalent to setting Pb(T) = 1. For simple kinetics such as L + R ↔ LR, this approach is fully justified. However, when there is dynamic competition among several states of the receptor (R1, R2, …) or ligand (L1, L2, …) or when there are conformational transitions involving several bound forms (LR1, LR2, …), the joint probability distribution P(T,t) cannot be factorized, i.e. P(T,t) ≠ Pb(T)Ps(t). Information about correlations is contained in the difference P(T,t) − Pb(T)Ps(t). Thus, the two data sets must be modeled jointly.
FIGURE 2.
A bead oscillation cycle (a) and a corresponding data trace (b) of αIIbβ3-fibrinogen interactions under a constant compressive force (fc) and pulling force (fp) are shown. Multiple oscillation cycles with alternating bond formation and rupture are shown in c. The bead motion cycle, which has binding and unbinding phases, consists of four parts: 1, a bead approaching a pedestal; 2, bead-pedestal contact; 3, bonding under compressive force fc (if a bond formed); and 4, the bead retraction (application of pulling force fp). When the signal is positive, the ligand-coated bead is stopped for a certain time (T) at a constant compressive force (fc) against a silica pedestal coated with receptor protein. When the signal is negative (no bond was formed during the compression), the trap is stopped at a fixed distance away from the pedestal. However, if a bond was formed, the trap maintains the pulling force (fp) during the entire lifetime (t) of the bond.
To clarify the latter point, consider the case where a receptor exists in both low affinity (R1) and high affinity (R2) states, forming two bound states, LR1 and LR2 (see Scheme 1). Using the bond survival probability alone and ignoring the binding probabilities for these states would result in an erroneous estimation of the forced dissociation rate and an incorrect interpretation of the interaction mechanism. Indeed, when conformational transitions (LR1 ↔ LR2) are slow compared with the rates of unbinding (r12, r21 ≪ k1, k2), forced dissociation from the states LR1 and LR2 is decoupled, and the bond survival probability is bimodal (21). In this regime, forced dissociation is dominated by the fastest pathway at shorter times and by the slowest pathway at longer times. At the opposite extreme (r12, r21 ≫ k1, k2), the dissociation pathways are strongly coupled, and single exponential unbinding kinetics, Ps(t) ≈ exp[−(k1 + k2)t], is determined by the sum of the rate constants k1 and k2 (24, 25). When conformational relaxation and unbinding transitions occur on the same time scale (r12, r21 ≈ k1, k2), the bond survival probability Ps(t) interpolates between these extremes. Hence, the lack of precise knowledge about the initial populations of receptor states R1 and R2 precludes meaningful interpretation of bond lifetime data.
The Likelihood of Forming Stable αIIbβ3-Fibrinogen Bonds Depends on Contact Duration
We used BUCS to analyze optical trap-based measurements of the association and dissociation of the complex formed by the platelet integrin αIIbβ3 and its principal ligand, fibrinogen. We found that at fixed values of compressive force and pulling force the probability of αIIbβ3-fibrinogen interactions was time-dependent (Fig. 3A). Thus, when the contact duration was varied between 0.05 and 2.0 s, the cumulative probability of forming an αIIbβ3-fibrinogen bond increased up to ∼20%, reaching a plateau at T ≈ 1.5 s. To exclude the contribution of nonspecific αIIbβ3-fibrinogen interactions, we counted only signals that lasted for >0.04 s in the unbinding phase (21). Because only a small number of interface contacts resulted in the formation of αIIbβ3-fibrinogen bonds within this range of T, the majority of bond rupture events were due to formation and subsequent dissociation of single bonds (26). To ensure the specificity of the interactions between fibrinogen- and αIIbβ3-coated surfaces, measurements were performed in the presence of the αIIbβ3 antagonist abciximab (27). Abciximab reduced the binding probability by ∼70–80%, indicating that the vast majority of the measured binding signals resulted from the specific interaction of αIIbβ3 with fibrinogen (Fig. 3A).
FIGURE 3.
Binding probability Pb(T) as a function of contact duration T for formation of the integrin-fibrinogen complex in the absence (a) and presence (b) of Mn2+. Experimental data points are shown as diamonds, and error bars represent S.D. a also shows the results of control experiments (squares) performed in the presence of a specific inhibitor mAb for which the binding probability is substantially reduced. The numerical fits of the theoretical binding probability curve Pb(T) = Pb1(T) + Pb2(T) (Equations 2 and 4–5) to the experimental data points are shown as the solid curves. Insets, the contributions to binding probability from the low affinity state LR1, Pb1(T) (dashed curves), and the high affinity state LR2, Pb2(T) (dash-dotted curves). Numerical values of the kinetic parameters (see Scheme 1 and Equation 2) obtained from the fits of the theoretical curve of Pb(T) to the experimental data points are summarized in Table 1.
When αIIbβ3 was pretreated with 1 mm Mn2+, a potent allosteric integrin activator, the slope of Pb(T) became steeper, implying faster binding (Fig. 3B). On the other hand, the plateau level did not change. These results imply that the surface density of fibrinogen-reactive αIIbβ3 molecules and the population of the αIIbβ3-fibrinogen complex remained the same before and after treatment with Mn2+. Because Mn2+ is expected to recruit active αIIbβ3 molecules from the available pool (23, 28), αIIbβ3 should have been present in excess initially compared with fibrinogen.
Average Bond Lifetimes Are Dependent on the Duration of αIIbβ3-Fibrinogen Contact (T)
Next, we tested whether precisely controlling the duration of the contact between the interacting surfaces coated with αIIbβ3 and fibrinogen would affect the mechanical stability of the αIIbβ3-fibrinogen complex. Varying the time of contact (T) between αIIbβ3 and fibrinogen from 0.05 to 2 s at a fixed compressive force of 20 pN and a pulling force of 50 pN revealed a positive correlation between the average αIIbβ3-fibrinogen bond lifetimes and the duration of αIIbβ3-fibrinogen contact (Fig. 4). Shorter lifetimes (2–4 s) collected at T = 0.05–0.2 s corresponded mainly to force-driven dissociation from a lower affinity state (LR1), whereas longer lifetimes (≈5–7 s) collected at T = 1.0–2.0 s reflected the growing contribution from a higher affinity state (LR2; Fig. 3, insets). A plateau was reached at T ≈ 1 s in Ca2+-containing medium, but in the presence of Mn2+, average bond lifetime increased at a faster rate and reached a plateau at T = 0.5 s. These results suggest that the strength of αIIbβ3-fibrinogen interactions, reflected by the average bond lifetime, is time-dependent. The gradual strengthening of the αIIbβ3-fibrinogen complex is due to the increasing contribution of a higher affinity population of αIIbβ3-fibrinogen complexes, a phenomenon that is more pronounced in the presence of Mn2+.
FIGURE 4.
The ensemble average bond lifetimes of the αIIbβ3-fibrinogen complex collected at constant pulling force fp = 50 pN as a function of the contact duration T in the absence of Mn2+ (black squares) and presence of Mn2+ (black circles). The experimental data points were fitted with the exponential function tmax(1 − exp[−t/τ]) where tmax is the largest bond lifetime and τ is the characteristic time.
Bond Survival Probability Shows a Bimodal Dependence on Bond Lifetimes
Under force clamp conditions, we observed a broad range of bond lifetimes varying from milliseconds to tens of seconds. Based on their sensitivity to αIIbβ3 antagonists, the bond lifetimes >0.04 s were considered to result from specific αIIbβ3-fibrinogen interactions (21). Unbinding data were then gathered into bond survival curves in which the probability of bond survival Ps(t) was plotted against actual bond lifetimes (t). As we showed above, the slope of the bond survival curves as displayed on a semilogarithmic scale was proportional to the off-rate (k) for a particular bound state. Moreover, when measurements were performed at different contact duration times, the probability of bond survival Ps(t) showed a bimodal exponential decay, indicating the presence of two unbinding pathways. Thus, as shown in Fig. 5, at T = 0.5 s and fp = 50 pN and in the absence or presence of Mn2+, plots of Ps(t) versus bond lifetime (t) have two distinctly different slopes, corresponding to rapid and slow modes of forced unbinding. This agrees with the results of our previously studies, namely bimodal forms of histograms of rupture forces (20) and bond lifetimes (21), and confirms the existence of at least two bound states, the lower and higher affinity forms of the αIIbβ3-fibrinogen complexes, LR1 and LR2.
FIGURE 5.
Bond survival curves for the integrin-fibrinogen complex subject to pulling force fp = 50 pN in the absence (a) and presence (b) of 1 mm Mn2+. The bond lifetimes are collected for single integrin-fibrinogen bonds formed over time T = 0.5 s. The curves display two distinct probability slopes due to the bimodal nature of the bond lifetimes. Slopes 1 and 2 correspond to the two bound states of the αIIbβ3-fibrinogen complex, LR1 and LR2, characterized by markedly different mechanical stability. Note that Slope 2 in a is larger than Slope 2 in b, indicating that the forced dissociation rate for the bound state LR2, k2, is smaller in the presence of Mn2+ (see Tables 2 and 3).
Extrapolating from the Joint Probability Distribution P(T,t) to Kinetic Parameters
To derive kinetic parameters for the interaction of αIIbβ3 with fibrinogen using the results presented above, we used a minimal kinetic model based on the following principles. First, in its free form, αIIbβ3 exists in at least two conformations, R1 and R2, that are in dynamic equilibrium with an equilibrium constant, K0. Accordingly, there are at least two pools of αIIbβ3 that are defined by their equilibrium populations, λ1 = 1/(K0 + 1) and λ2 = K0/(K0 + 1), and that associate with fibrinogen with the on-rates kon1 and kon2. Second, fibrinogen binding to αIIbβ3 is reversible. Thus, dissociation of fibrinogen from αIIbβ3 is governed by the off-rates koff1 and koff2 in the absence of force fp. Third, the lifetime of αIIbβ3-fibrinogen bonds depends strongly on the duration of contact between αIIbβ3 and fibrinogen. Thus, the probability of fibrinogen binding to αIIbβ3 is affected progressively by the presence of more mechanically stable high affinity complexes (LR2). Fourth, there is a force-induced redistribution of αIIbβ3-fibrinogen complexes that is described by the equilibrium constant K = r12/r21 where r12 and r21 are the kinetic rates for the forward transition LR1 → LR2 and reverse transition LR2 → LR1, respectively. Lastly, the external pulling force fp can modulate off-rates so that k1 ≠ koff1 and k2 ≠ koff2. This minimal kinetic model is summarized by Scheme 1, and the kinetic transitions of the binding and unbinding phases are shown by Schemes 2 and 3, respectively. Corresponding to the kinetic model, theoretical treatments for Pb(T) and P(T,t) are given by Equations 2–7.
We then fit the experimental data for αIIbβ3-fibrinogen complex formation in the absence and presence of Mn2+ as a function of contact duration time T using Equations 2 and 4–5. This enabled us to extract the zero force two-dimensional kinetic parameters kon and koff in the absence of external mechanical factors. The apparent force-free on-rates were converted into true kinetic on-rates by factoring them for the surface density of reactive αIIbβ3 molecules, ≈3,000 molecules/μm2 as measured by their ability to specifically bind 125I-fibrinogen with high affinity (Kd ≈ 18 nm) (supplemental Fig. S2). As summarized in Table 1, we found that active αIIbβ3 exists in at least two states (R1 and R2) that differ in their zero force on-rates, off-rates, and corresponding equilibrium dissociation constants (Kd = koff/kon). In the absence of αIIbβ3 activators and inhibitors, the ratio of low affinity state LR1 (population λ1) to high affinity state LR2 (population λ2) is 10.1 with a zero force equilibrium constant K0 = λ2/λ1 ≈ 0.098. Thus, the low affinity αIIbβ3 conformation (R1) makes the major contribution to the probability of αIIbβ3-fibrinogen bond formation (Fig. 3A, inset). For the low affinity state, the on-rate was ∼2-fold slower, and the off-rate was ∼4-fold faster than for the high affinity state, accounting for the ∼7-fold difference in the equilibrium dissociation constant (Kd1 = 1.7 × 104 μm−2 for LR1 versus Kd2 = 2.6 × 103 μm−2 for LR2). In the presence of Mn2+, the ratio of the low affinity to the high affinity populations decreased to 5.25, and K0 increased to ∼0.19. Mn2+ had a dual effect on the equilibrium on-rates: it decreased the association rate for the low affinity state by ∼2.5-fold but increased the on-rate for the high affinity state ∼9-fold, whereas the off-rates for both the low and high affinity states were unchanged. Hence, the net effect of Mn2+ on αIIbβ3-fibrinogen interactions is a change in the distributions of lower affinity and higher affinity populations and in the on-rate of fibrinogen binding.
TABLE 1.
Force-free kinetics of the αIIbβ3-fibrinogen interactions
Initial populations of the integrin-fibrinogen complexes in the low affinity state LR1 (λ1) and high affinity state LR2 (λ2), the force-free apparent and true kinetic on-rates (kon′ and kon), and off-rates (koff) are shown. These parameters were obtained from the fit of the theoretical binding probability curve Pb(T) (Equations 2 and 4–5; also see Fig. 3) to the experimental data from binding measurements collected at compressive force fc = 20 pN in the absence and presence of the integrin activator Mn2+.
| Parameter |
|||||
|---|---|---|---|---|---|
| kon1′, kon1 × 10−3 | kon2′, kon2 × 10−3 | koff1 | koff2 | λ1, λ2 | |
| s−1 | s−1 | ||||
| Ca2+ | 0.42 ± 0.05 s−1, 0.14 ± 0.01 μm2/s | 0.70 ± 0.08 s−1, 0.23 ± 0.02 μm2/s | 2.42 ± 0.29 | 0.60 ± 0.29 | 0.91 ± 0.11, 0.09 ± 0.01 |
| Ca2+/Mn2+ | 0.16 ± 0.02 s−1, 0.05 ± 0.006 μm2/s | 6.10 ± 0.79 s−1, 1.98 ± 0.26 μm2/s | 2.50 ± 0.32 | 0.65 ± 0.08 | 0.84 ± 0.11, 0.16 ± 0.03 |
Next, we calculated the joint probability distribution P(T,t) of αIIbβ3-fibrinogen complexes as a function of contact duration time T and bond lifetime t. In theoretical modeling, we used the numerical values of the force-free two-dimensional kinetic rates shown in Table 1 to extract additional kinetic parameters, the force-dependent dissociation rates (k1 and k2) and the transition rates (r12 and r21), for the conformational interconversion LR1 ↔ LR2. The results are displayed in Fig. 6 for measurements made in the absence of Mn2+ and in supplemental Fig. S3 for measurements made in the presence of Mn2+ and are summarized in Tables 2 and 3. We found that in the absence and presence of Mn2+ the force-dependent dissociation rate for the high affinity state LR2 (k2 = 0.106 s−1) is ∼44–48-times slower than the force-dependent dissociation rate for the low affinity state LR1 (k1 = 5.1 s−1). This is an indication of the markedly different mechanical stabilities of the high and low affinity forms of the αIIbβ3-fibrinogen complex. In the presence of Mn2+, the average k1 and k2 are only slightly lower than their Mn2+-free counterparts. This indicates that Mn2+ has a limited effect on the stability of the αIIbβ3-fibrinogen complex and correlates with the equilibrium off-rates, which also revealed a marginal dependence on the presence of Mn2+ (Table 1). There is a relatively slow force-induced conformational transition from the low affinity to the high affinity states in the absence of Mn2+ (r12 = 0.012 s−1) that increases slightly following the addition of Mn2+ (r12 = 0.026 s−1). The reverse process (LR2 → LR1) is very slow both in the absence and presence of Mn2+ (r21 = 0.001 s−1), implying that the backward transition did not occur in the experimental time scale (∼40 s). Nonetheless, the forced equilibrium constant increases from K1 = 11.6 (without Mn2+) to K1 = 25.0 (with Mn2+), evidence that fibrinogen-bound αIIbβ3 can be activated mechanically. Moreover, comparison of the force-free off-rates (koff1 and koff2) with the force-dependent dissociation rates (k1 and k2) indicates that mechanical force has a dual effect on the strength of αIIbβ3-fibrinogen bonds, increasing the dissociation rate for the low affinity state LR1 (k1 = 4.1–5.1 s−1 versus koff1 = 2.4–2.5 s−1) while at the same time decreasing the dissociation rate for the high affinity state LR2 (k1 = 0.092–0.106 s−1 versus koff1 = 0.60–0.65 s−1).
FIGURE 6.
Dynamics of association-dissociation interactions between single integrin and fibrinogen molecules in the absence of Mn2+. a, the joint probability distribution P(T,t) as a function of the contact duration T and bond lifetime t. The theoretical probability surface is compared with the experimental data points obtained for different contact duration times T = 0.05, 0.1, 0.2, 0.5, 1.0, 1.5, and 2.0 s. b, the theoretical profiles of P(T,t) versus bond lifetime t for fixed T = 0.1, 0.2, 0.5, and 1.5 s (curves) are compared against the experimental data points (symbols). The theoretical probability measures were obtained using the kinetic parameters summarized in Tables 1 and 2.
TABLE 2.
Force-dependent kinetics of the αIIbβ3-fibrinogen interactions
Force-dependent dissociation rates k1 and k2 for the low affinity state LR1 and high affinity state LR2, respectively; conformational transition rates r12 (transition LR1 → LR2) and r21 (transition LR2 → LR1); and equilibrium constant K1 = r12/r21 (see kinetic model in Schemes 1–3) are shown. These parameters were obtained from the fit of the theoretical curves for the joint probability distribution P(T,t) at fixed interaction times T = 0.1, 0.2, 0.5, 1.0, and 1.5 s to the experimental data points (see Fig. 6).
| T | k1 | k2 | r12 | r21 | K1 |
|---|---|---|---|---|---|
| s | s−1 | s−1 | s−1 | s−1 | |
| 0.1 | 6.00 | 0.10 | 0.011 | 0.0011 | 10.0 |
| 0.2 | 5.50 | 0.10 | 0.011 | 0.0010 | 11.0 |
| 0.5 | 4.00 | 0.15 | 0.010 | 0.0010 | 10.0 |
| 1.0 | 5.50 | 0.09 | 0.015 | 0.0010 | 15.0 |
| 1.5 | 4.50 | 0.09 | 0.012 | 0.0010 | 12.0 |
| Average | 5.10 ± 0.82 | 0.106 ± 0.025 | 0.012 ± 0.002 | 0.0010 ± 0.0001 | 11.6 ± 2.1 |
TABLE 3.
Force-dependent kinetics of the αIIbβ3-fibrinogen interactions in the presence of Mn2+
The parameters are the same as those shown in Table 2 but for the experimental data obtained in the presence of Mn2+ (see supplemental Fig. S3).
| T | k1 | k2 | r12 | r21 | K1 |
|---|---|---|---|---|---|
| s | s−1 | s−1 | s−1 | s−1 | |
| 0.1 | 3.0 | 0.09 | 0.020 | 0.0010 | 20.0 |
| 0.2 | 4.5 | 0.09 | 0.020 | 0.0010 | 20.0 |
| 0.5 | 5.5 | 0.08 | 0.030 | 0.0011 | 27.3 |
| 1.0 | 4.0 | 0.08 | 0.035 | 0.0010 | 35.0 |
| 1.5 | 3.5 | 0.12 | 0.025 | 0.0011 | 22.7 |
| Average | 4.10 ± 0.96 | 0.092 ± 0.016 | 0.026 ± 0.006 | 0.0010 ± 0.0001 | 25.0 ± 6.3 |
DISCUSSION
Advantages of Binding-Unbinding Correlation Spectroscopy
We developed a new approach named BUCS that combines novel experimental methodology and theoretical modeling to describe a joint probability distribution P(T,t) for the two-dimensional reversible association-dissociation kinetics of receptor-ligand interactions. This higher order statistical measure, which can be obtained by convoluting the binding probability as a function of interaction time T with the probability of bond survival as a function of bond lifetime t, contains complete information about the kinetics of biomolecular interactions. We tested the validity of this approach using an optical trap-based model system capable of making single molecule force clamp or force ramp measurements. However, the approach can be applied to other nanomechanical single molecule techniques such as atomic force microscopy and the biomembrane force probe.
Because many receptor-ligand pairs exhibit structural heterogeneity and flexibility, there can be uncertainty about possible forms of these complexes. Furthermore, different receptor or ligand conformations may interconvert in the experimental time scale. To obtain meaningful models of these biomolecular transitions in forced unbinding studies, it is necessary to gather accurate information about the initial (bound) states of the system, but unbinding data alone do not provide this information. Our approach, based on binding-unbinding correlations, overcomes these limitations. Force-free formation of the receptor-ligand bond precedes force-driven bond dissociation. Hence, equilibrium information can be used to describe the results of unbinding measurements. This can be accomplished by examining the entire joint probability surface P(T,t) and its projections constructed using the bond lifetimes collected for different fixed values of contact duration. Taken together, this approach enables modeling of receptor-ligand interactions at equilibrium as well as under the influence of mechanical factors. Importantly, force-free on-rates and off-rates are obtained directly from experimental measurements without using simplifying assumptions or empirical models. A small compressive force helps to hold the reactive surfaces together, providing the conditions necessary for biomolecules to establish intermolecular contacts at the interface.
Measurements of binding probability as a function of contact duration have previously been used to derive two-dimensional kinetics for complexes composed of the Fcγ receptor IIIA and IgG (9), adhesins of Staphylococcus epidermidis and fibronectin (29), the integrin αLβ2 and intercellular adhesion molecule-1 (30), and the T-cell receptor and peptide-major histocompatibility complex (31, 32). In addition to force-free interactions, the dynamic behavior of αLβ2-intercellular adhesion molecule-1 interactions has been studied by measuring bond lifetimes under ramped tension (33). However, these measurements by themselves could not determine joint probabilities for consecutive association and dissociation transitions and could not be used to analyze the association-dissociation kinetics coupled to internal protein motion. Although the existence of several states of the receptors or ligands can be suggested by the multimodal shape of the histogram of bond lifetimes, different binding pathways may compete, and the shape of the distribution could depend on the duration of receptor-ligand contact. Moreover, binding measurements alone do not capture possible force-induced conformational transitions for large protein biomolecules that can be captured using BUCS.
Two-dimensional Kinetics of Fibrinogen Binding to αIIbβ3 in the Absence and Presence of Mn2+
In the absence of Mn2+, we obtained force-free on-rates (kon1 and kon2), off-rates (koff1 and koff2), force-dependent dissociation rates (k1 and k2), and equilibrium dissociation constants (Kd1 and Kd2) for the non-covalent complex between the platelet integrin αIIbβ3 and it principal ligand, fibrinogen. Previously, we studied αIIbβ3-fibrinogen interactions in a ramp force mode and analyzed binding probability as a function of contact duration and found that that they were heterogeneous (20). Furthermore, using a force clamp mode, we found that the αIIbβ3-fibrinogen complex exists at least in two states that differ in mechanical stability (21). The studies reported here confirm this conclusion and more precisely quantify these states and their dynamics (see supplemental Table S1). These findings are consistent with the “conformational selection” model of protein-ligand interactions in which the unbound protein exists as an ensemble of conformations in dynamic equilibrium (34, 35). Importantly, we found that αIIbβ3 undergoes a slow force-induced reversible transition from a lower to a higher affinity state, and we were able to calculate kinetic rates (r12 and r21) and equilibrium constant (K1) for this process.
Previously, we found that the presence of Mn2+ favored the formation of higher affinity αIIbβ3-fibrinogen complexes characterized by a higher binding energy and reduced off-rate (20, 21, 23). Here, we found that Mn2+ shifts αIIbβ3 to a higher activation state by modulating both the on-rate and the rate at which αIIbβ3 transitions from a lower to a higher affinity state. Thus, the equilibrium constant for αIIbβ3 activation, K0 = λ2/λ1, increased ∼2-fold in the presence of Mn2+; however, force-free off-rates were unchanged, indicating that Mn2+ does not alter the kinetics of the dissociation of the αIIbβ3-fibrinogen complex. By contrast, the on-rate for the formation of the lower affinity state (LR1) decreased, and the on-rate for the higher affinity state (LR2) increased, decreasing the probability of forming the lower affinity state and increasing the probability of forming the high affinity state. Furthermore, the results of forced unbinding experiments indicated that in the presence of Mn2+ the higher affinity state was more stable mechanically with a forced off-rate ∼50-fold less than that of the lower affinity state. With reasonable precautions, the effects of Mn2+ could potentially be extended to αIIbβ3 activation induced by other natural and artificial stimuli.
Implications of the Application of BUCS to αIIbβ3-Fibrinogen Interactions
Because applied tensile force facilitates dissociation of receptor-ligand complexes, it was expected that forced off-rates for the lower affinity form of αIIbβ3 in the absence or presence of Mn2+ would be larger than their force-free counterparts. Paradoxically, however, forced off-rates for the higher affinity form were significantly smaller that those of their force-free counterpart. One possible explanation for this finding might be that the extended conformation of higher affinity αIIbβ3 provides more contacts at the binding interface. Thus, whereas the application of a pulling force on fibrinogen weakens the lower affinity complex, mechanical deformation of the higher affinity complex leads to the formation of a tighter binding pocket, more side chain contacts, or both. This type of behavior resembles the catch bonds where bond stability increases with increasing mechanical tension (36, 37). Nonetheless, the existence of multiple bound states is essential but not sufficient for the emergence of the “catch regime” (24, 25), and we showed previously that in the force range of up to 50 pN the αIIbβ3-fibrinogen interactions display classical slip bonds (21).
We found that the application of pulling force induces a slow reversible conformational transition from lower to higher affinity αIIbβ3-fibrinogen complexes. The similarity of the rate constants for the forward transition r12 (LR1 → LR2) and reverse reaction r21 (LR2 → LR1) in the absence and presence of Mn2+ indicates that this transition is a purely mechanical effect. Indeed, we found that the transition rate constants r12 and r21 and corresponding equilibrium constants for the conformational transitions K1 (LR1 ↔ LR2) with and without Mn2+ are very similar. Moreover, comparison of the equilibrium constants for αIIbβ3 activation in its ligand-free form both in the absence and presence of Mn2+ (K0 ≈ 0.1 and ≈0.2, respectively) with the equilibrium constants for the ligand-bound form under the same conditions (K1 ≈ 11 without Mn2+ and K0 ≈ 25 with Mn2+) indicates that mechanical activation is more profound than biochemical activation by Mn2+.
Kinetic parameters derived from two-dimensional single molecule assays and from three-dimensional bulk experiments are substantially different and cannot be compared directly (38). Indeed, kinetic parameters derived from the forced dissociation of surface-bound molecules depend on surface density, steric limitations due to surface confinement, and the relative orientation of the molecules. Nonetheless, two-dimensional kinetic parameters may be more relevant to cell-cell and cell-substrate adhesion because in vivo both the receptor and the ligand are usually attached to surfaces (39). In the case of platelet adhesion and aggregation mediated by fibrinogen binding to αIIbβ3, processes that occur in vivo under dynamic conditions of continuous hydrodynamic shear, our results demonstrate a time-dependent increase in the stability of the αIIbβ3-fibrinogen complex, implying ligand-induced binding site remodeling in αIIbβ3 (40). Forced extension of two other integrins, αvβ3 and αLβ2, has been recently demonstrated in silico (41–43), and this is a possible mechanism for the observed force-induced activation of αIIbβ3. If this is the case, then the enhanced activation of αIIbβ3 under the extremely high shear stress in atherosclerotically narrowed arteries may be an important component of the arterial thrombosis.
In conclusion, we have devised an experimental protocol and theoretical framework entitled BUCS that enables the characterization of two-dimensional receptor-ligand interactions. In a broader perspective, this approach, which can be used with existing experimental instrumentation, provides a convenient method to explore the kinetics and mechanism of bimolecular coupling, including protein-protein and protein-DNA interactions, when they are affected by the internal dynamics of one or both of the biomolecules involved. We then applied this approach to the interaction between the platelet integrin αIIbβ3 and its principal ligand, fibrinogen. Using an optical trap-based force clamp system in conjunction with the probabilistic description of the kinetics of receptor-ligand interactions, we found that fibrinogen-reactive αIIbβ3 exists in at least two conformational forms. These states differ in their zero force on-rates, off-rates, and corresponding equilibrium dissociation constants by 1 order of magnitude. The applied pulling force favors the higher affinity form of αIIbβ3 by speeding up the dissociation from the lower affinity state while increasing the population of and slowing down the dissociation from the higher affinity state. Hence, the application of mechanical force is accompanied by a chemical change, i.e. the increased stability and prevalence of a particular form of αIIbβ3. This indicates that there is interrelation or linkage between the mechanics (conformational motion) and kinetics (association and dissociation) of the αIIbβ3-fibrinogen pair that can be further explored using linked functions and reciprocal effects for ligand binding and linkage in polyfunctional biological macromolecules (44–46). The strength of αIIbβ3-fibrinogen interactions gradually increases with duration of contact between αIIbβ3 and fibrinogen due to progressive growth in the fraction of the faster forming higher affinity complexes, a phenomenon that is driven mechanically and is accelerated in the presence of Mn2+, a potent integrin activator. Taken together, our results provide kinetic parameters for the interaction of αIIbβ3 under both force-free and forced unbinding conditions and offer new mechanistic insights into the αIIbβ3-mediated interactions that underlie platelet function.
Supplementary Material
Acknowledgments
We thank Drs. Douglas Cines and Serge Yarovoi (University of Pennsylvania) for help in performing the experiments with radioactivity and Olga Kononova (University of Massachusetts, Lowell) for help in the manuscript preparation.
This work was supported, in whole or in part, by National Institutes of Health Grants HL030954/HL090774 (to J. W. W.) and HL40387/HL81012 (to J. S. B.). This work was also supported by American Heart Association Grant 09SDG2460023 (to V. B.).

This article contains supplemental Figs. S1–S3 and Table S1.
- L
- ligand
- R
- receptor
- RL
- receptor-ligand complex(es)
- BUCS
- binding-unbinding correlation spectroscopy
- Pb(T)
- binding probability
- Ps(t)
- bond survival probability
- P(T,t)
- joint probability
- pN
- piconewtons
- T
- interaction time
- t
- bond lifetime
- fc
- compressive force
- fp
- pulling force.
REFERENCES
- 1. Hynes R. O. (2009) The extracellular matrix: not just pretty fibrils. Science 326, 1216–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Weber G. F., Bjerke M. A., DeSimone D. W. (2011) Integrins and cadherins join forces to form adhesive networks. J. Cell Sci. 124, 1183–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hynes R. O. (2002) Integrins: bidirectional, allosteric signaling machines. Cell 110, 673–687 [DOI] [PubMed] [Google Scholar]
- 4. Hynes R. O. (2007) Cell-matrix adhesion in vascular development. J. Thromb. Haemost. 5, Suppl. 1, 32–40 [DOI] [PubMed] [Google Scholar]
- 5. Bergmeier W., Hynes R. O. (2012) Extracellular matrix proteins in hemostasis and thrombosis. Cold Spring Harb. Perspect. Biol. 4, a005132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Frojmovic M., Wong T., van de Ven T. (1991) Dynamic measurements of the platelet membrane glycoprotein IIb-IIIa receptor for fibrinogen by flow cytometry. I. Methodology, theory and results for two distinct activators. Biophys. J. 59, 815–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kuroki K., Maenaka K. (2011) Analysis of receptor-ligand interactions by surface plasmon resonance. Methods Mol. Biol. 748, 83–106 [DOI] [PubMed] [Google Scholar]
- 8. Rico F., Chu C., Moy V. T. (2011) Force-clamp measurements of receptor-ligand interactions. Methods Mol. Biol. 736, 331–353 [DOI] [PubMed] [Google Scholar]
- 9. Chesla S. E., Selvaraj P., Zhu C. (1998) Measuring two-dimensional receptor-ligand binding kinetics by micropipette. Biophys. J. 75, 1553–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kinoshita K., Leung A., Simon S., Evans E. (2010) Long-lived, high-strength states of ICAM-1 bonds to β2 integrin, II: lifetimes of LFA-1 bonds under force in leukocyte signaling. Biophys. J. 98, 1467–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Danilowicz C., Greenfield D., Prentiss M. (2005) Dissociation of ligand-receptor complexes using magnetic tweezers. Anal. Chem. 77, 3023–3028 [DOI] [PubMed] [Google Scholar]
- 12. Litvinov R. I., Shuman H., Bennett J. S., Weisel J. W. (2002) Binding strength and activation state of single fibrinogen-integrin pairs on living cells. Proc. Natl. Acad. Sci. U.S.A. 99, 7426–7431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sun J. E., Vranic J., Composto R. J., Streu C., Billings P. C., Bennett J. S., Weisel J. W., Litvinov R. I. (2012) Bimolecular integrin-ligand interactions quantified using peptide-functionalized dextran-coated microparticles. Integr. Biol. 4, 84–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bell G. I. (1978) Models for the specific adhesion of cells to cells. Science 200, 618–627 [DOI] [PubMed] [Google Scholar]
- 15. Mukamel S. (1995) Principles of Nonlinear Optical Spectroscopy, Oxford University Press, New York [Google Scholar]
- 16. Barsegov V., Thirumalai D. (2005) Probing protein-protein interactions by dynamic force correlation spectroscopy. Phys. Rev. Lett. 95, 168302. [DOI] [PubMed] [Google Scholar]
- 17. Barsegov V., Klimov D. K., Thirumalai D. (2006) Mapping the energy landscape of biomolecules using single molecule force correlation spectroscopy: theory and applications. Biophys. J. 90, 3827–3841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bennett J. S. (2005) Structure and function of the platelet integrin αIIbβ3. J. Clin. Investig. 115, 3363–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bennett J. S., Berger B. W., Billings P. C. (2009) The structure and function of platelet integrins. J. Thromb. Haemost. 7, Suppl. 1, 200–205 [DOI] [PubMed] [Google Scholar]
- 20. Litvinov R. I., Bennett J. S., Weisel J. W., Shuman H. (2005) Multi-step fibrinogen binding to the integrin αIIbβ3 detected using force spectroscopy. Biophys. J. 89, 2824–2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Litvinov R. I., Barsegov V., Schissler A. J., Fisher A. R., Bennett J. S., Weisel J. W., Shuman H. (2011) Dissociation of bimolecular αIIbβ3-fibrinogen complex under a constant tensile force. Biophys. J. 100, 165–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Agnihotri A., Soman P., Siedlecki C. A. (2009) AFM measurements of interactions between the platelet integrin receptor GPIIbIIIa and fibrinogen. Colloids Surf. B Biointerfaces 71, 138–147 [DOI] [PubMed] [Google Scholar]
- 23. Litvinov R. I., Nagaswami C., Vilaire G., Shuman H., Bennett J. S., Weisel J. W. (2004) Functional and structural correlations of individual αIIbβ3 molecules. Blood 104, 3979–3985 [DOI] [PubMed] [Google Scholar]
- 24. Barsegov V., Thirumalai D. (2005) Dynamics of unbinding of cell adhesion molecules: transition from catch to slip bonds. Proc. Natl. Acad. Sci. U.S.A. 102, 1835–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Barsegov V., Thirumalai D. (2006) Dynamic competition between catch and slip bonds in selectins bound to ligands. J. Phys. Chem. B 110, 26403–26412 [DOI] [PubMed] [Google Scholar]
- 26. Zhu C., Long M., Chesla S. E., Bongrand P. (2002) Measuring receptor/ligand interaction at the single-bond level: experimental and interpretative issues. Ann. Biomed. Eng. 30, 305–314 [DOI] [PubMed] [Google Scholar]
- 27. Coller B. S. (1985) A new murine monoclonal antibody reports an activation-dependent change in the conformation and/or microenvironment of the platelet glycoprotein IIb/IIIa complex. J. Clin. Investig. 76, 101–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Eng E. T., Smagghe B. J., Walz T., Springer T. A. (2011) Intact αIIbβ3 integrin is extended after activation as measured by solution x-ray scattering and electron microscopy. J. Biol. Chem. 286, 35218–35226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bustanji Y., Arciola C. R., Conti M., Mandello E., Montanaro L., Samorí B. (2003) Dynamics of the interaction between a fibronectin molecule and a living bacterium under mechanical force. Proc. Natl. Acad. Sci. U.S.A. 100, 13292–13297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang F., Marcus W. D., Goyal N. H., Selvaraj P., Springer T. A., Zhu C. (2005) Two-dimensional kinetics regulation of αLβ2-ICAM-1 interaction by conformational changes of the αL-inserted domain. J. Biol. Chem. 280, 42207–42218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huang J., Zarnitsyna V. I., Liu B., Edwards L. J., Jiang N., Evavold B. D., Zhu C. (2010) The kinetics of two-dimensional TCR and pMHC interactions determine T-cell responsiveness. Nature 464, 932–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Edwards L. J., Zarnitsyna V. I., Hood J. D., Evavold B. D., Zhu C. (2012) Insights into T cell recognition of antigen: significance of two-dimensional kinetic parameters. Front. Immunol. 3, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Evans E., Kinoshita K., Simon S., Leung A. (2010) Long-lived, high-strength states of ICAM-1 bonds to β2 integrin, I: lifetimes of bonds to recombinant αLβ2 under force. Biophys. J. 98, 1458–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Boehr D. D., Nussinov R., Wright P. E. (2009) The role of dynamic conformational ensembles in biomolecular recognition. Nat. Chem. Biol. 5, 789–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Csermely P., Palotai R., Nussinov R. (2010) Induced fit, conformational selection and independent dynamic segments: an extended view of binding events. Trends Biochem. Sci. 35, 539–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Marshall B. T., Long M., Piper J. W., Yago T., McEver R. P., Zhu C. (2003) Direct observation of catch bonds involving cell-adhesion molecules. Nature 423, 190–193 [DOI] [PubMed] [Google Scholar]
- 37. Zhu C., Lou J., McEver R. P. (2005) Catch bonds: physical models, structural bases, biological function and rheological relevance. Biorheology 42, 443–462 [PubMed] [Google Scholar]
- 38. Huang J., Meyer C., Zhu C. (2012) T cell antigen recognition at the cell membrane. Mol. Immunol. 52, 155–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Long M., Lü S., Sun G. (2006) Kinetics of receptor-ligand interactions in immune responses. Cell. Mol. Immunol. 3, 79–86 [PubMed] [Google Scholar]
- 40. Hantgan R. R., Stahle M. C., Connor J. H., Horita D. A., Rocco M., McLane M. A., Yakovlev S., Medved L. (2006) Integrin αIIbβ3:ligand interactions are linked to binding-site remodeling. Protein Sci. 15, 1893–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen W., Lou J., Zhu C. (2010) Forcing switch from short- to intermediate- and long-lived states of the αA domain generates LFA-1/ICAM-1 catch bonds. J. Biol. Chem. 285, 35967–35978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen W., Lou J., Hsin J., Schulten K., Harvey S. C., Zhu C. (2011) Molecular dynamics simulations of forced unbending of integrin αvβ3. PLoS Comput. Biol. 7, e1001086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xiang X., Lee C. Y., Li T., Chen W., Lou J., Zhu C. (2011) Structural basis and kinetics of force-induced conformational changes of an αA domain-containing integrin. PLoS One 6, e27946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wyman J., Jr. (1964) Linked functions and reciprocal effects in hemoglobin: a second look. Adv. Protein Chem. 19, 223–286 [DOI] [PubMed] [Google Scholar]
- 45. Di Cera E., Gill S. J., Wyman J. (1988) Canonical formulation of linkage thermodynamics. Proc. Natl. Acad. Sci. U.S.A. 85, 5077–5081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Di Cera E. (1990) Thermodynamics of local linkage effects. Contracted partition functions and the analysis of site-specific energetics. Biophys. Chem. 37, 147–164 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.