Background: The DNA-binding protein CSL regulates transcription from Notch target genes.
Results: We developed assays in cells to characterize CSL mutants with activated forms of Notch and two different transcriptional reporters.
Conclusion: Our analysis of CSL mutants reveals differential responses dependent upon Notch paralog and promoter architecture.
Significance: This study provides important molecular insights into Notch transcription complexes and generates useful reagents for future studies.
Keywords: Isothermal Titration Calorimetry, Notch Pathway, Protein Complexes, Protein-DNA Interaction, Protein-Protein Interactions, Signal Transduction, Transcription Regulation
Abstract
Notch is a conserved signaling pathway that plays essential roles during embryonic development and postnatally in adult tissues; misregulated signaling results in human disease. Notch receptor-ligand interactions trigger cleavage of the Notch receptor and release of its intracellular domain (NICD) from the membrane. NICD localizes to the nucleus where it forms a transcriptionally active complex with the DNA-binding protein CSL and the coactivator Mastermind (MAM) to up-regulate transcription from Notch target genes. Previous studies have determined the structure of the CSL-NICD-MAM ternary complex and characterized mutations that affect complex assembly in functional assays. However, as CSL is expressed in all cell types, these studies have been limited to analyzing mutations in NICD and MAM. Here, we describe a novel set of cellular reagents to characterize how mutations in CSL affect its function as a transcriptional activator. Using retrovirally transduced embryonic fibroblasts from a CSL-null mouse, we generated cell lines that express either wild-type or mutant CSL molecules. We then analyzed these mutants for defects in Notch1- (NICD1) or Notch2 (NICD2)-mediated activation from two different transcriptional reporters (HES-1 or 4×CBS). Our results show that mutations targeted to the different domains of CSL display significant differences in their ability to adversely affect transcription from the two reporters. Additionally, a subset of CSL mutants is sensitive to whether NICD1 or NICD2 was used to activate the reporter. Taken together, these studies provide important molecular insights into how Notch transcription complexes assemble at different target genes and promoter arrangements in vivo.
Introduction
The Notch pathway is a cell-to-cell signaling mechanism that regulates a multitude of developmental and physiological processes, including hematopoiesis (1), organogenesis (2), neurogenesis (3), angiogenesis (4), and the maintenance and self-renewal of stem cells (5). The significance of Notch signaling in human health is underscored by the strong association of misregulated signaling with disease, e.g. in the case of T-cell acute lymphoblastic leukemia (6). Given its clinical importance, there has been much effort in identifying reagents that target Notch signaling for therapeutic purposes (7).
The central components of the Notch pathway are the receptor Notch, the ligand DSL (for Delta, Serrate, Lag-2), and the nuclear effector CSL (for CBF-1, Su(H), Lag-1) (8). Notch and DSL are both transmembrane proteins with a single transmembrane spanning region; CSL is a DNA binding transcription factor that is required for both repression and activation of transcription from genes that are responsive to Notch signals. Mammals have multiple receptors (Notch1–4) and ligands (Delta-like1,3,4 and Jagged1,2) but only one nuclear effector (CSL; also known as RBP-J). Canonical signaling is initiated when Notch receptors and ligands on neighboring cells interact, which triggers proteolytic cleavage of Notch and release of its intracellular domain (NICD)2 from the cell membrane (8). NICD translocates to the nucleus where it forms a transcriptionally active ternary complex with CSL and a member of the Mastermind (MAM) family of coactivators. Assembly of the CSL-NICD-MAM ternary complex at a target gene is the switch for up-regulating transcription from this locus (9).
Structural studies of Notch transcription complexes from our group and others have defined the overall folds, domain organization, and interacting regions for CSL, NICD, and MAM proteins from mammals and nematodes (Fig. 1, A and B) (10). CSL is composed of three domains as follows: N-terminal domain (NTD), β-trefoil domain (BTD), and C-terminal domain (CTD), in which its NTD and BTD interact with DNA (11). NICD binds CSL through its RBP-J-associated molecule (RAM) and ankyrin (ANK) repeats domains, which interact with the BTD and CTD of CSL, respectively (12, 13). MAM forms an elongated helix with a distinctive bend, in which its N-terminal helical region forms a tripartite complex with ANK and CTD, and its C-terminal helical region binds the NTD of CSL (12, 13).
FIGURE 1.
Overview of structure, domain schematics, reporters, and mutations. A, ribbon diagram of transcriptionally active CSL-NICD-MAM ternary complex bound to DNA (Protein Data Bank code 2FO1) (12). The NTD, BTD, and CTD of CSL are colored cyan, green, and orange, respectively. A long β-strand that makes hydrogen-bonding interactions with all three domains is colored magenta. The RAM and ANK domains of NICD are colored blue and yellow, respectively; MAM is colored red; and the DNA is colored gray. B, domain organization of CSL, NICD, and MAM. Coloring matches structure in A. C, Notch-responsive transcriptional reporters used in this study, showing the arrangement of CSL-binding sites. The HES-1 reporter consists of two CSL-binding sites arranged in a conserved head-to-head manner with a 16-bp spacer; the 4×CBS reporter consists of four iterative CSL-binding sites arranged in a head-to-tail manner. D, figure shows the 12 CSL mutants analyzed in this study mapped to the structure of CSL in complex with NICD and MAM. CSL is depicted as a gray surface with mutations in the NTD, BTD, and CTD colored cyan, green, and orange, respectively. NICD (RAM + ANK) and MAM are shown as ribbon diagrams and colored as in A. Two ∼180° views of the CSL-NICD-MAM complex are shown.
CSL binds the consensus DNA sequence -(C/t)GTGGGAA- with moderate affinity (∼200 nm Kd) (14–16); similar sites have been found in vivo at the enhancer and promoter elements of Notch target genes (17). The promoter regions of some, but not all, Notch target genes are composed of two CSL-binding sites arranged in a head-to-head manner with an ∼16-bp spacer sequence separating the two sites (Fig. 1C). This particular binding site arrangement was first identified in flies and termed SPS (Su(H)-paired site) (18), and it is also conserved in mammals, as typified by the HES-1 promoter element (19). More recently, the structural basis for the cooperative assembly of two CSL-NICD-MAM ternary complexes onto the SPS has been elucidated, in which interactions between two ANK molecules mediate the cooperativity (20, 21). Although additional CSL-binding site arrangements will likely be identified in the future, presently the Notch target genes can generally be subdivided into two groups as follows: those targets that contain an SPS element and those target genes that contain CSL-binding sites but do not conform to the strict paired site arrangement.
The x-ray structures of CSL-NICD-MAM complexes have enabled the biochemical, biophysical, and cellular characterization of the domains and residues important for assembly of these nuclear complexes, which has led to considerable molecular insights into these processes (10). However, cellular studies have been limited to only characterizing NICD mutants, because CSL is ubiquitously expressed in all cell types (22). The central role CSL plays in mediating interactions with both transcriptional coactivators and corepressors emphasizes the need for developing new tools to analyze CSL function in cultured cells.
Here, we develop a set of cellular reagents to characterize the transcriptional outputs from CSL mutants. Transcription from these CSL mutants was assayed using the activated forms of Notch1 (NICD1) or Notch2 (NICD2) with two different luciferase reporters (Fig. 1C) as follows: 1) the HES-1 reporter, which corresponds to the SPS found at the HES-1 gene; and 2) the 4×CBS reporter, which consists of four tandem CSL-binding sites. Our findings suggest that mutations in the BTD of CSL significantly affect transcription from the 4×CBS reporter with NICD1 but have little to no effect on transcription from the HES-1 reporter; however, this effect was concentration-ependent and diminished when NICD2 was used to activate the reporter. The effect of mutations in the NTD was dependent on both the reporter and Notch paralog used, with NICD1–4×CBS and NICD2-HES-1 most adversely affected. Charge-reversal mutations in the CTD severely blunted transcription from both reporters and with both NICD1 and NICD2. These mutants can be rescued by making the corresponding charge-reversal mutation in NICD1. Taken together, these studies provide molecular insights into the role certain domains of CSL play in the assembly of Notch pathway transcription complexes at different promoter arrangements with different Notch paralogs.
EXPERIMENTAL PROCEDURES
Cell Lines
OT11 and OT13 cell lines (23), which correspond to rbp-j null and wild-type mouse embryonic fibroblasts (MEFs), respectively, were a generous gift from Drs. Tasuku Honjo, Jae Jung, and Heesoon Chang. MEFs were maintained at 37 °C in 5% CO2 in Dulbecco's modified essential medium (DMEM) supplemented with 10% fetal bovine serum and penicillin/streptomycin. All subsequent MEF cell lines expressing either wild-type or mutant CSL molecules were maintained in a similar manner.
Retroviral Transduction of OT11 Cells
The coding region for murine rbp-j, which corresponds to amino acids 1–526, and a C-terminal FLAG tag was PCR-cloned into the MigR1 retroviral vector (24), using the BglII and EcoRI restriction sites. This wild-type CSL construct with a C-terminal FLAG tag was used as a template to make the site-directed CSL mutants, using QuikChange mutagenesis protocol (Stratagene). As shown in Table 1 and Fig. 1D, 12 single-site mutations were generated that targeted all three domains of CSL. Wild-type and mutant CSL MigRI constructs were cotransfected with pVSV-G into the retrovirus packaging cell line HEK GP2-293. Supernatants containing the retroviruses were collected 96 h post-transfection. Retroviral transduction of OT11 MEFs was performed by plating 1.1 × 105 cells in a 12-well plate with 0.5 ml of the appropriate retrovirus-containing supernatant. Successful transduction was monitored by flow cytometry, which examined GFP expression from an internal ribosome entry site contained within MigR1, and immunoblots for the FLAG epitope. GFP-negative cells were removed from the population by fluorescence-activated cell sorting, which resulted in homogeneous cell lines used for subsequent experiments.
TABLE 1.
CSL interfacial mutants
Table shows amino acids, numbering, and interfaces targeted for murine CSL residues mutated in this study. In addition, residues and numbering are shown for human, worm, and fly CSL orthologs. 3BRG, 2F8X, and 2FO1 represent Protein Data Bank (PDB) codes for mouse, human, and worm CSL x-ray structures.
| Mutant | Mouse | Human | Worm | Fly | Interface |
|---|---|---|---|---|---|
| PDB 3BRG | PDB 2F8X | PDB 2FO1 | |||
| 1 | F128R | Phe-88 | Tyr-289 | Phe-176 | NTD-MAM |
| 2 | F172R | Phe-132 | Phe-348 | Phe-220 | NTD-MAM |
| 3 | R186E | Arg-146 | Arg-362 | Arg-234 | NTD-ANK |
| 4 | F261R | Phe-221 | Phe-442 | Phe-309 | BTD-RAM |
| 5 | V263R | Val-223 | Val-444 | Val-311 | BTD-RAM |
| 6 | A284R | Ala-244 | Ala-465 | Ala-332 | BTD-RAM |
| 7 | Q333R | Gln-293 | Gln-517 | Gln-381 | BTD-RAM |
| 8 | E398R | Glu-358 | Glu-580 | Glu-446 | CTD-ANK |
| 9 | N389R | Asn-349 | Asp-571 | Asn-437 | CTD-ANK |
| 10 | E425R | Glu-385 | Glu-607 | Glu-473 | CTD-ANK |
| 11 | N407R | Asn-367 | Asn-589 | His-455 | CTD-MAM |
| 12 | R422E | Arg-382 | Arg-604 | Arg-470 | CTD-ANK-MAM |
Luciferase Reporter Assays
Wild-type or mutant CSL cell lines were grown to ∼80% confluence in 6-well plates and transiently transfected with a construct that expresses either NICD1 or NICD2 to activate Notch signaling. As described by Ong et al. (25), NICD1 corresponds to mouse Notch1 residues 1744–2531 and contains an N-terminal 3×FLAG tag; NICD2 corresponds to murine Notch2 residues 1669–2470 and also contains an N-terminal 3×FLAG tag. NICD1 or NICD2 was cotransfected with either the HES-1 or 4×CBS luciferase reporter constructs and phRL, which expresses Renilla luciferase to normalize for transfection efficiency. Residues 8–67 of murine Mastermind1 (Maml1) were cloned into pcDNA3.1C to create a dominant-negative Mastermind construct. The SatisFection (Stratagene) reagent was used for the transfections, following the manufacturer's protocol, and the amount of transfected DNA was normalized using pBluescript. 48 h post-transfection, the cells were harvested and assayed for luciferase activity using the Dual-Luciferase kit (Promega). Firefly luciferase expression from either the HES-1 or 4×CBS reporter was first normalized to Renilla luciferase expression and reported as fold-activation by comparing cells transfected with and without NICD1/2. Percent activity was determined by comparing fold-activation for wild-type and mutant CSL cell lines. Average values, errors, and standard deviations were determined from three individual experiments performed in duplicate. GraphPad Prism was used to perform paired Student's t tests.
Immunoblots
MEFs were grown in 100-mm dishes to ∼80% confluency. Cells were then isolated, and extracts were prepared using the NE-PER nuclear and cytoplasmic extraction kit (Thermo Scientific, Pierce). Extracts were separated by SDS-PAGE and subjected to Western blotting with antibodies directed against RBP-J (2ARBP2-T6709, Institute of Immunology, kindly provided by Nadean Brown), FLAG M2 (Sigma), or β-actin (kindly provided by Bill Miller and James Lessard). Reactive proteins were detected by using the appropriate secondary antibodies and enhanced chemiluminescence (Thermo Scientific, Pierce). Using the Bradford reagent, cellular extracts were normalized to total protein concentration levels.
ITC
As described previously (26), recombinant mouse CSL (RBP-J, residues 53–474) and RAM (Notch1, residues 1744–1771) proteins were overexpressed and purified from bacteria using a combination of affinity, ion exchange, and size exclusion column chromatography. CSL mutants were purified in a similar manner. A 21-residue peptide that corresponds to the RAM domain of Notch2 (residues 1702–1722) was chemically synthesized and purified to homogeneity using reverse phase HPLC. A MicroCal VP-ITC calorimeter was used for CSL-RAM binding studies. Experiments were carried out at 25 °C in a buffer consisting of 50 mm sodium phosphate, pH 6.5, and 150 mm NaCl. Proteins were degassed and buffer-matched using size exclusion chromatography and/or dialysis. A typical experiment consisted of 10 μm CSL in the cell and 100 μm RAM in the syringe. Protein concentrations were determined by both UV absorbance at 280 nm and BCA assay (Pierce). The binding data reported are the average of at least three individual experiments (n = 3); the c value (c = Ka[M]N) for all experiments was between 1 and 500; and the N value (stoichiometry ligand/macromolecule (M)) for all experiments was between 0.8 and 1.4. The data were analyzed using the ORIGIN software and fit to a one-site binding model.
Circular Dichroism
CD measurements were taken in triplicate using an Aviv circular dichroism spectrometer model 215. Measurements were collected in a 0.02-cm cuvette at 25 °C using 1.0 nm wavelength steps between 190 and 290 nm. Native and mutant mouse CSL proteins (RBP-J, residues 53–474) were characterized in a buffer containing 50 mm sodium phosphate, pH 6.5, and 75 mm NaCl with protein concentrations ranging from 20 to 50 μm. CD data were analyzed on Dichroweb using CDSSTR with reference set four (27, 28).
RESULTS
Generation of CSL Mutant Cell Lines
To characterize the effects CSL mutants have on cellular transcription, we cultured mouse embryonic fibroblasts (MEFs) derived from a rbp-j (−/−) mouse embryo (23, 29). These MEFs were transduced with a retrovirus that encodes either a wild-type or mutant version of murine CSL with a C-terminal FLAG tag, as well as an internal ribosome entry site that allows for green fluorescent protein (GFP) expression. Fluorescence-activated cell sorting was used to separate GFP-expressing cells from nonexpressing cells, giving a homogeneous population of cells that express CSL. As shown in Fig. 1D and Table 1, we designed 12 single-site CSL mutants, which targeted disruptive mutations to the different protein-protein interfaces that compose the CSL-NICD-MAM ternary complex (12, 13). Similar protein expression levels and nuclear localization for native and mutant CSL proteins were confirmed by immunoblots (Fig. 2). In addition, recombinant CSL proteins were purified from bacteria and analyzed by circular dichroism, demonstrating that the mutations had no effect on the secondary-structure content of CSL (supplemental Fig. S1).
FIGURE 2.
Cellular characterization of CSL-null MEFs. A, figure shows immunoblots for the cytoplasmic and nuclear fractions derived from CSL-null (OT11), wild-type (OT13), and retrovirally transduced (OT11-MIG or OT11-MIG-CSLwt) MEFs. Blots were probed with either anti-RBP-J (CSL) or anti-actin (control). Note that CSL is not detected in the null MEFs (lanes 1 and 5); the CSL expression level of the retrovirally transduced MEFs is similar to that of the wild-type MEFs (compare lanes 6 and 8), and CSL is primarily localized to the nucleus in both the wild-type and transduced MEFs (compare lanes 2, 3, 6, and 8). B and C, immunoblots show expression levels and cellular localization for null MEFs transduced with retroviruses that express either wild-type or mutant forms of CSL. Blots were probed with either anti-RBP-J or anti-actin (control). Note that the mutations neither affect the expression levels nor the nuclear localization of CSL. D, immunoblot shows relative expression levels of NICD1 and NICD2 in retrovirally transduced MEFs following transient transfection. Immunoblots were probed with either anti-FLAG M2 (NICD) or anti-actin (loading control). Expected molecular weights for NICD1 and NICD2 are 87,470 and 85,334 Da, respectively. Asterisk denotes a nonspecific band that was observed in untransfected cells (data not shown).
ITC Binding Assays of CSL Mutants with NICD RAM Domain
To confirm that our targeted mutations in CSL disrupt the protein-protein interfaces of the CSL-NICD-MAM transcriptional activation complex, we performed ITC binding assays with purified recombinant preparations of mutant CSL molecules and the RAM domains of NICD1 and NICD2. Previously, we and others have shown by ITC that the RAM domain forms a high affinity (∼20 nm Kd) interaction with the BTD of CSL (26, 30–32). Thus, we performed ITC binding assays with the BTD mutants (F261R, V263R, A284R, and Q333R) and compared these findings with our previous characterization of complexes formed between wild-type CSL and RAM. As shown in Table 2 and Fig. 3, all four BTD mutants displayed significantly reduced affinity for the RAM domains of NICD1 and NICD2; mutant F261R was the most affected with an approximate 700- and 300-fold reduction in binding for the RAM domains of NICD1 and NICD2, respectively. The other three BTD mutants (V263R, A284R, and Q333R) were also significantly affected, ranging from ∼5- to 100-fold reductions in binding; however, different binding trends were observed for NICD1 and NICD2 with the mutants V263R and A284R, in which the binding of NICD2 RAM was affected to a much greater extent than NICD1 (Table 2).
TABLE 2.
Calorimetric data for the RAM domains of NICD1 and NICD2 binding to wild-type and BTD mutant CSL proteins
All experiments were performed at 25 °C. Values are the mean of at least three independent experiments, and errors represent the standard deviations of multiple experiments.
| CSL | RAM | K | Kd | ΔG0 | ΔH0 | −TΔS0 |
|---|---|---|---|---|---|---|
| m−1 | μm | kcal/mol | kcal/mol | kcal/mol | ||
| Wild type | NICD1 | 5.2 ± 2.1 × 107 | 0.022 | −10.4 ± 0.3 | −17.1 ± 0.3 | 6.6 ± 0.1 |
| Wild type | NICD2 | 3.8 ± 0.2 × 107 | 0.032 | −10.2 ± 0.03 | −12.4 ± 0.9 | 2.2 ± 1.0 |
| F261R | NICD1 | 6.9 ± 1.6 × 104 | 15 | −6.6 ± 0.1 | −5.5 ± 0.1 | −1.1 ± 1.2 |
| F261R | NICD2 | 1.1 ± 0.1 × 105 | 9.54 | −6.9 ± 0.1 | −5.2 ± 0.3 | −1.7 ± 0.2 |
| V263R | NICD1 | 2.4 ± 0.5 × 106 | 0.43 | −8.7 ± 0.1 | −17.4 ± 0.7 | 8.7 ± 0.7 |
| V263R | NICD2 | 2.9 ± 0.7 × 105 | 3.59 | −7.5 ± 0.1 | −11.5 ± 0.9 | 4.0 ± 0.9 |
| A284R | NICD1 | 1.4 ± 0.3 × 106 | 0.74 | −8.5 ± 0.1 | −16.7 ± 0.4 | 8.2 ± 4.5 |
| A284R | NICD2 | 4.6 ± 2.3 × 105 | 2.51 | −7.7 ± 0.3 | −13.5 ± 1.3 | 5.8 ± 1.0 |
| Q333R | NICD1 | 2.7 ± 0.6 × 106 | 0.38 | −8.8 ± 0.6 | −16.7 ± 0.6 | 7.9 ± 0.6 |
| Q333R | NICD2 | 5.4 ± 1.3 × 106 | 0.2 | −9.1 ± 0.2 | −10.5 ± 0.5 | 1.4 ± 0.4 |
FIGURE 3.
CSL-RAM ITC binding assays. Figure shows representative thermograms (raw heat signal and nonlinear least squares fit to the integrated data) for wild-type and mutant CSL molecules binding the RAM domains of NICD1 (A) and NICD2 (B). Forty titrations were performed per experiment, consisting of 7-μl injections that were spaced 120 s apart.
Transcriptional Analysis of Retrovirally Transduced MEFS
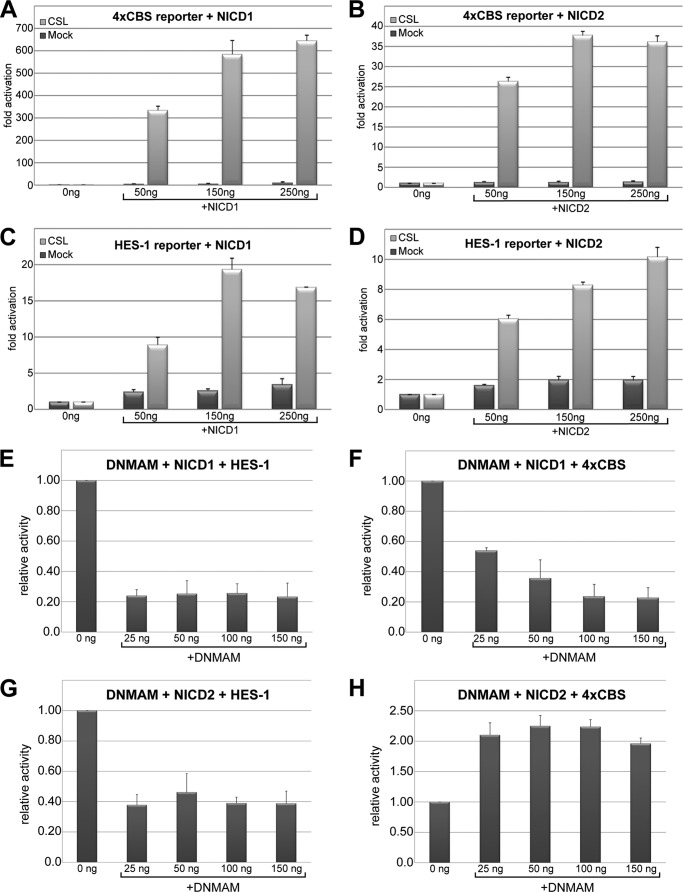
To validate that our transduced MEFs were responsive to Notch signaling, due to the retroviral expression of CSL, we transiently transfected the MEFs with either the luciferase reporter HES-1 or 4×CBS. Notch signaling was activated in these cells by cotransfection with truncated forms of either the Notch1 (NICD1) or Notch2 (NICD2) receptor, which has been shown previously to be constitutively active in cellular assays and in vivo (19, 25, 33). As shown in Fig. 4, MEFs transduced with wild-type CSL, but not an empty retrovirus (mock), activated transcription from both reporters in a dose-dependent manner. NICD1 provided much stronger activation from the reporters than NICD2, which was not due to different levels of expression (Fig. 2D). Similar differences in the transcriptional potency of NICD1 and NICD2 have been reported elsewhere (25). Unless otherwise noted, in all subsequent experiments 150 ng of NICD1 or NICD2 was used to activate transcription from the luciferase reporters. We also attempted transcriptional reporter assays with the other Notch paralogs NICD3 and NICD4; however, NICD3 and NICD4 produced little activity from these reporters in our MEFs (data not shown), making analysis of our CSL mutants intractable, and therefore were not pursued further.
FIGURE 4.
Activation of HES-1 and 4×CBS reporters by NICD1 and NICD2 in transduced MEFs. A–D show reporter activity from CSL-null (rbpj−/−) MEFs that have been transduced with either an empty retrovirus (mock) or a retrovirus that encodes wild-type CSL. Notch signaling was activated in the MEFs by transiently expressing increasing amounts (50, 150, and 250 ng) of the intracellular domains of Notch1 (NICD1) or Notch2 (NICD2). Notch activity was monitored by observing the luciferase activity from the co-transfection of the HES-1 or 4×CBS transcriptional reporters. Luciferase activity was normalized to MEFs that were not transfected with NICD1 or NICD2 and reported as fold-activation. A and B show the luciferase activity from 4×CBS with NICD1 and NICD2, respectively; C and D show the activity from HES-1 with NICD1 and NICD2, respectively. E–H show the effect increasing concentrations of DNMAM have on activity from the HES-1 and 4×CBS reporters with NICD1 (E and F) and NICD2 (G and H). Relative activity for cells transfected with 150 ng of NICD1 or NICD2 is plotted on the y axis, and amounts of transfected DNMAM DNA (nanograms) is plotted on the x axis. Plotted data represent average values and means ± S.E. from three independent experiments performed in duplicate.
To provide a base line for analyzing the negative effects our CSL mutants have on transcription, we used the well established Notch signaling inhibitor dominant-negative Mastermind (DNMAM). DNMAM is a truncated form of the transcriptional coactivator MAM that can form ternary complexes with CSL and NICD but cannot activate transcription (34). We characterized the effect DNMAM had on reporter activity (HES-1 or 4×CBS) with either NICD1 or NICD2. As shown in Fig. 4, E and F, DNMAM reduced activity from the HES-1 and 4×CBS reporters with NICD1 by ∼80%. Unexpectedly for NICD2, DNMAM only reduced transcription from the HES-1 reporter by ∼60% and was completely ineffective at inhibiting activity from the 4×CBS reporter for all concentrations of DNMAM tested (Fig. 4, G and H). Although DNMAM has been shown to be an effective inhibitor of signaling mediated by Notch2 in previous studies (35), these data may suggest that NICD2 is functioning independently of MAM in this specific cellular context. Certainly, additional studies are needed to validate this hypothesis; however, the molecular basis for the lack of NICD2 inhibition by DNMAM was not pursued further in this study.
Characterization of NTD Mutants
Next, we analyzed how our 12 mutant CSL cell lines in conjunction with NICD1 or NICD2 activated transcription from the HES-1 and 4×CBS reporters (Figs. 5 and 6). Mutations in the NTD (F128R and F172R), which target one of the MAM-binding sites on CSL (Fig. 1D), reduce transcription from the HES-1 and 4×CBS reporters ∼40 and 70%, respectively, when compared with wild-type CSL with NICD1 (Figs. 5A and 6A); the third mutation in the NTD (R186E), which targets a contact between NTD and ANK, similarly decreases transcription by ∼40% for the HES-1 reporter and ∼70% for the 4×CBS reporter (Figs. 5A and 6A). Interestingly, this interaction was more prominent in the worm CSL-NICD-MAM ternary complex structure than the human complex (12, 13); nonetheless, this mutant significantly affects transcription from the reporter in mammalian cells. When NICD2 was used to activate the reporters, a stronger reduction in transcription was observed from the NTD mutants with the HES-1 reporter (60–70%) (Fig. 5A). This was particularly evident for mutant F128R, in which the difference between NICD1 and NICD2 was found to be statistically significant (p < 0.05). (Table 3). However, in contrast to NICD1, activity from the 4×CBS reporter with NICD2 was less affected by the NTD mutants (F128R and F172R), which resulted in approximately twice as much activity as observed for NICD1 (Fig. 6A).
FIGURE 5.
Analysis of CSL mutants with the HES-1 reporter. Figure shows the effects mutations targeted to the individual domains of CSL have on the activity from the HES-1 reporter with either NICD1 or NICD2. The data are normalized to cells expressing wild-type CSL and reported as relative activity. Results for NTD, BTD, and CTD mutants are shown in A–C, respectively. Activation of the HES-1 reporter with NICD1 or NICD2 is depicted as dark gray or light gray bars, respectively. The data shown are derived from three independent experiments performed in duplicate and represent the means ± S.E. Statistical significance was assessed using paired Student's t tests with *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; and ns, not significant.
FIGURE 6.

Analysis of CSL mutants with 4×CBS reporter. Figure shows the effects mutations targeted to the individual domains of CSL have on the activity from the 4×CBS reporter with either NICD1 or NICD2. The data are normalized to cells expressing wild-type CSL and reported as relative activity. Results for NTD, BTD, and CTD mutants are shown in A–C, respectively. Activation of the 4×CBS reporter with NICD1 or NICD2 is depicted as dark gray or light gray bars, respectively. The data shown are derived from three independent experiments performed in duplicate and represent the means ± S.E. Statistical significance was assessed using paired Student's t tests with *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; and ns, not significant.
TABLE 3.
Statistical analysis of the differences observed for NICD1 and NICD2 with the CSL mutants and the HES-1 and 4×CBS reporters
Statistical significance for the pairwise comparison of activity from NICD1 and NICD2 with the HES-1 and 4×CBS reporters and the various CSL mutants was assessed using paired Student's t tests. *, p ≤0.05; **, p ≤0.01; ***, p ≤0.001; and NS, not significant.
| Reporter | NTD mutants |
BTD mutants |
CTD mutants |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F128R | F172R | R186E | F261R | V263R | A284R | Q333R | E398R | N389R | E425R | N407R | R422E | |
| HES-1 | * | NS | NS | * | NS | NS | NS | NS | NS | * | NS | NS |
| 4×CBS | * | NS | NS | *** | NS | * | NS | NS | NS | * | ** | * |
Characterization of BTD Mutants
Mutations within the BTD of CSL target the interaction with the RAM domain of NICD (Fig. 1D). Strikingly, all four BTD mutants have little to no effect on transcriptional activity from the HES-1 reporter with NICD1 (Fig. 5B). The slight increases in transcription observed for F261R and Q333R were not found to be statistically significant. Moreover, three double mutants and one triple mutant within the BTD also do not significantly affect transcription from the HES-1 reporter with NICD1 (supplemental Fig. S2). However, when less NICD1 was transfected into the cells (50 versus 150 ng), the BTD mutants displayed a stronger reduction in HES-1 reporter activity, in particular F261R (supplemental Fig. S2).
Interestingly, a different trend is observed when NICD2 is used with the HES-1 reporter and the BTD mutants (Fig. 5B). For two of the BTD mutants (F261R and Q333R), only a 20% reduction in activity was observed, which is modest but significantly different from the response observed with NICD1. Mutant V263R, on the one hand, has no appreciable effect on reporter activity, but on the other hand, the BTD mutant A284R displays an ∼60% reduction in activity from the HES-1 reporter that was specific for NICD2 but not NICD1 (Fig. 5B). When less NICD2 was transfected into the cells (50 versus 150 ng), this had the strongest effect on mutant V263R, which displayed ∼40% reduction from the HES-1 reporter comparable with the other BTD mutants with NICD2.
In contrast to the HES-1 reporter, the BTD mutants had a much stronger effect on transcription from the 4×CBS reporter with NICD1 but, curiously, to a much lesser extent with NICD2 (Fig. 6B). The BTD mutant F261R with NICD1 decreased transcription from the reporter by at least 75% compared with wild-type. The other BTD mutations (V263R, A284R, and Q333R) with NICD1 decreased transcription by ∼60%. Moreover, the effects of double and triple mutants within the BTD were generally additive in their reduction of transcription from the 4×CBS reporter with NICD1 (supplemental Fig. S2); the double mutants V263R/Q333R and A284R/Q333R had only 30% activity when compared with wild-type CSL; and the triple mutant V263R/A284R/Q333R had only 10% of the transcriptional output as compared with wild-type CSL. Again, NICD2 displayed differential effects from NICD1 with regard to how the BTD mutants affected transcription from the 4×CBS reporter (Fig. 6B). BTD mutants with NICD2 had either minimal (F261R; ∼20%), modest (V263R and A284R; ∼40%), or considerable (Q333R; ∼60%) reductions in activity from the 4×CBS reporter. The differences between NICD1 and NICD2 activating the 4×CBS reporter were the most striking for the CSL mutant F261R (p < 0.001, Table 3).
Characterization of CTD Mutants
To characterize how mutations within the CTD of CSL, which target interactions with MAM and the ANK domain of NICD (Fig. 1D), affect transcription, we performed reporter assays with the CSL mutant cell lines E398R, N389R, E425R, N407R, and R422E. The E398R, N389R, and E425R mutations lie at the CTD-ANK interface; N407R exclusively lies at the CTD-MAM interface; and the mutation R422E lies at the interface of CTD with both ANK and MAM. Two mutations within the CTD of CSL (E398R and R422E) strikingly reduced transcription (∼80%) from both reporters and with both NICD1 and NICD2 (Figs. 5C and 6C). For NICD1, other mutations within the CTD had only a moderate effect on transcription from both the HES-1 and 4×CBS reporters (∼20–60% reduction). For NICD2, the effects were more varied, but similar for both reporters (Figs. 5C and 6C); the CTD mutant E425R was the most affected with ∼80% reduction in activity from both reporters, which interestingly is at least 2-fold stronger than the effect observed for NICD1 with this CSL mutant. The mutant N389R was modestly affected, and N407R was only marginally affected with both reporters. In general, CTD double and triple mutations were additive with their effects in reducing activity from the reporters with NICD1 (supplemental Fig. S2).
CTD-ANK Charge Reversal Mutations
Given that the CTD mutant E398R had the most dramatic reduction in transcription from both reporters with NICD1 and NICD2, we next wanted to test whether a charge reversal mutation in NICD1 could rescue the transcriptional output from mutant E398R. Interestingly, the corresponding glutamate residues in the worm and human CSL-NICD-MAM ternary complex structures form ion pairs with different arginine residues in NICD (12, 13, 36). Based on the interactions from the worm complex, Glu-398 would form a salt bridge with Arg-1952 in the ANK domain of NICD1; however, based on the human complex structure, Glu-398 would form a salt bridge with Arg-1994. Therefore, we made two charge reversal mutations in NICD1-R1952E and -R1994E and performed luciferase reporter assays with the CSL mutant E398R to determine which mutation would rescue activity from the reporter. As shown in Fig. 7, the R1994E mutation in NICD1, but not R1952E, rescued transcription from the HES-1 reporter.
FIGURE 7.

Charge reversal mutation rescues CSL mutant E398R. Mutation of glutamate 398 to arginine (E398R) in the CTD of CSL results in a dramatic reduction in transcription from the HES-1 reporter. In the structures of both human and worm, CSL-NICD-MAM Glu-398 forms a buried salt bridge with a conserved arginine residue from NICD; however, based on the human complex, it would form an ion pair with Arg-1994 of murine NICD1, and based on the worm complex it would form an ion pair with Arg-1952. Thus, two charge reversal mutations were made in murine NICD1 (R1994E and R1952E) to test which mutation would rescue the CSL mutant E398R. The figure shows that NICD1 mutant R1994E, but not R1952E, rescues the CSL mutant E398R. Reporter activity for wild-type and E398R CSL expressing cells are shown in dark and light gray bars, respectively. The data shown are derived from three independent experiments performed in duplicate and represent the means ± S.E.
DISCUSSION
Canonical Notch signaling results in the up-regulation of transcription from Notch target genes (8). The focal point of this regulation is the DNA-binding protein CSL, which binds both corepressors and coactivators to regulate transcriptional repression and activation, respectively (10). In mammals, CSL activates transcription by forming a ternary complex with one of four Notch receptor paralogs (Notch1–4) and a member of the Mastermind family of coactivators (Maml1–3) (10). In turn, these transcriptionally active ternary complexes bind at the promoter and enhancer elements of Notch-responsive genes, which often have conserved binding site arrangements, e.g. the paired CSL-binding sites that compose the hes-1 promoter (19, 21).
The previous structures of CSL and CSL-NICD-MAM activator complexes have afforded detailed biochemical and cellular studies, in conjunction with site-directed mutagenesis, to characterize ternary complex assembly and to define the roles that certain domains play in this process (21, 25, 26, 37). The centrality of CSL in mediating protein-protein interactions with NICD and MAM, as well as with transcriptional corepressors, underscores the usefulness of characterizing mutations in CSL and how these mutants affect function. However, due to the ubiquitous expression of CSL in all cell types, previous cellular studies have been unable to analyze mutations in CSL but rather have focused on mutations in NICD and MAM. To circumvent this problem, we have developed cellular assays in MEFs that were derived from a CSL null mouse (23), in which either wild-type or mutant CSL was stably expressed by retroviral transduction.
Our initial study focused on designing single site mutations in CSL that targeted the protein-protein interfaces of the CSL-NICD-MAM ternary complex. These mutations were predicted to disrupt the assembly of the ternary complex, which would be monitored as a loss of activity from the transcriptional reporter. These studies would provide molecular insights into the importance of particular interactions that compose the CSL-NICD-MAM activator complex. Additionally, our studies had two other aims as follows: 1) to determine whether the different CSL-binding site arrangements in 4×CBS and HES-1 responded similarly or differently to each respective CSL mutant; 2) to determine whether different Notch receptor paralogs (NICD1 and NICD2) activated the reporters similarly or differently with respect to each CSL mutant.
Several trends emerge from our studies that suggest both the receptor paralog and reporter used to activate transcription can affect the responses observed with the different CSL mutants. Overall, mutations in CSL have a greater effect on transcription from the 4×CBS reporter than the HES-1 reporter (Figs. 5 and 6). This is particularly evident for mutations within the BTD, which are much more deleterious to activity from the 4×CBS reporter than HES-1 (Figs. 5B and 6B). However, this effect for the BTD mutants with the HES-1 reporter displayed a significant concentration dependence (supplemental Fig. S2). Taken together, these data suggest that dimeric CSL-NICD-MAM transcription complexes at SPS elements are relatively refractory to the destabilizing effects of mutations in CSL, compared with CSL-NICD-MAM transcription complexes bound at individual sites. We interpret these data as dimers being more resistant to disassembly due to cooperative interactions between the complexes (20, 21), which would have the effect of increasing the occupancy of these complexes at the HES-1 promoter, resulting in higher transcriptional activity from the HES-1 reporter. In contrast, the four ternary complexes that bind the 4×CBS promoter do not form cooperative dimers, or any other higher order complex, and therefore do not experience this increased stability and residency time. This would likely result in faster disassembly kinetics for each of these individual complexes, which would be accelerated by mutations in CSL, ultimately resulting in less relative activity from the 4×CBS reporter.
A closer examination of our binding data for CSL-RAM complexes and how these data correlate with our transcriptional reporter assays reveals interesting differences in Notch paralog function. For the RAM domain of NICD1, our ITC binding studies of CSL-RAM complexes mirror the relative reductions in reporter activity observed with 4×CBS (Fig. 6B and Table 2), e.g. the CSL mutant F261R has the strongest reduction in NICD1 RAM binding in vitro (∼700-fold) and the strongest reduction of 4×CBS reporter activity in cells (∼75%); similar trends are observed for the other BTD mutants with NICD1 RAM and the 4×CBS reporter. Curiously, this trend does not hold true for the RAM domain of NICD2 with the BTD mutants and the 4×CBS reporter (Fig. 6B and Table 2). In fact, a completely opposite trend is observed, e.g. the CSL mutant F261R has the strongest reduction in NICD2 RAM binding in vitro (∼300-fold) but only suffers an ∼25% reduction in 4×CBS reporter activity in cells, whereas the BTD mutant Q333R has a 6-fold reduction in binding NICD2 RAM in vitro but has a 60% reduction in activity from the 4×CBS reporter. A detailed explanation for this observation is not immediately evident; however, a closer inspection of our CSL-RAM binding data may provide some molecular insights into these differences.
Our binding data show that although the RAM domains of NICD1 and NICD2 bind CSL with similar overall affinities, the enthalpic and entropic contributions to binding are substantially different; the enthalpic contribution of NICD1 RAM binding to CSL is ∼4.4 kcal/mol greater than for NICD2 RAM, and correspondingly the entropic penalty for NICD2 RAM binding to CSL is ∼4.4 kcal/mol less than for NICD1 RAM. Lubman et al. (32) observed similar enthalpic/entropic contributions to binding for complexes formed between NICD1 and NICD2 RAM with the isolated BTD of CSL. Because of this differential enthalpic/entropic partitioning to the overall free energy of binding, this suggests that at the thermodynamic level, the balance of forces that facilitate complex assembly and hinder complex disassembly are different for NICD1 and NICD2. This in part may help explain why NICD1 and NICD2 are more or less sensitive to activating the reporter when paired with particular mutations in the BTD (Figs. 5B and 6B) or CTD (Figs. 5C and 6C) of CSL.
One last notable differential response of NICD1 and NICD2 with the CSL mutants is observed with the NTD mutant F128R. On the one hand, activation of the HES-1 reporter by NICD2 is affected significantly more by this mutant than NICD1, but on the other hand, NICD1 is more severely affected than NICD2 in the context of the 4×CBS reporter. This is a curious result given that the NTD mutant F128R lies at the CSL-MAM interface and therefore neither directly contacts NICD1 nor NICD2. One possibility is that NICD1 and NICD2 have different preferences for which MAM paralog they pair with to form CSL-NICD-MAM activator complexes. Consistent with this notion is our reporter data with DNMAM, which showed that DNMAM was much more effective at inhibiting transcription mediated by NICD1 than NICD2 (Fig. 4, E–H). Nevertheless, to our knowledge there is no other experimental evidence to either support or refute this possibility. However, it should also be mentioned that there is modest sequence divergence between the MAM paralogs in this contact region with the NTD of CSL (12, 13), which may help explain the divergent effects the mutant F128R has on NICD1 and NICD2 activation of the reporters.
For all of the CSL mutants analyzed in this study, two mutations within the CTD (E398R and R422E) had the most striking reductions in activity (∼80%, Figs. 5C and 6C), which were as effective as the well established Notch signaling inhibitor DNMAM. Moreover, these effects were consistent for both reporters and with both Notch paralogs, emphasizing the importance of these interactions at the CTD-ANK interface. However, the structural basis for the detrimental effects of these two mutants stems from different reasons. Based on the human and worm CSL-NICD-MAM structures (12, 13), Glu-398 of CSL forms a salt bridge with an arginine residue in the ANK domain of NICD that is buried within the CTD-ANK interface. Glu-398 likely forms a similar interaction with the ANK domain of NICD2, based on homology modeling (data not shown). In fact, of all the CSL residues mutated in this study, Glu-398 has one of the smallest solvent-accessible surface areas when in complex with the ANK domain of NICD. Moreover, Glu-398 is stereochemically restrained by surrounding residues in CSL and NICD, such that when it is mutated to arginine (E398R), there are no other conformations that it can access to avoid the charge repulsion from the incoming arginine interaction with NICD. Other mutations within the CTD (e.g. Asn-389, Glu-425, and Asn-407) are more solvent-exposed and therefore can more easily access side chain conformations that would avoid the steric and repulsive effects of the mutation, thereby limiting their detrimental effects on activity from the reporters.
In contrast to Glu-398, Arg-422 is one of the most solvent-exposed residues at the CTD-ANK interface, having approximately three times more solvent-accessible surface area than Glu-398, which raises the question as to why mutation of Arg-422 to glutamate has such a significant effect on activity from the transcriptional reporters. Arg-422 lies at the interface with ANK and MAM, and interestingly, based on the CSL-NICD-MAM structures (12, 13), Arg-422 does not appear to form ionic interactions with ANK or MAM but rather hydrogen bonding and cation-π interactions. These interactions are incompatible with a glutamate side chain at this position. Moreover, despite the relatively high solvent accessibility of this residue, there appear to be few side chain conformations of residue 422 that would accommodate a negatively charged glutamate residue at this position. Thus, steric clashes or charge repulsion with either ANK or MAM are unavoidable when Arg-422 is mutated to glutamate. It should also be mentioned that Arg-422 likely forms a similar interface with the ANK domain of NICD2 and MAM based on homology modeling, which provides a similar explanation for why the R422E mutant dramatically reduces the activities from both reporters with NICD2.
Previously, we noted that although the overall architecture of the worm and human CSL-NICD-MAM activator complexes are remarkably similar, upon closer examination of the side chain interactions that compose the two ternary complexes, striking differences are observed between the worm and human orthologous structures (36). However, to date, it has been unclear whether these differences are species-specific or represent two different functional conformations of the activator complex. Because our cell culture system can analyze mutations in both CSL and NICD, this afforded us the opportunity to directly analyze whether the molecular interactions that were solely observed in the worm ternary complex affect transcriptional activation by the mammalian CSL-NICD-MAM complex. One of the most striking differences in side chain interactions between the worm and human complexes occurs at residue Glu-398, which, as mentioned previously, resides at the CTD-ANK interface and forms an ion pair with an arginine residue from NICD. However, based on the two orthologous activator complex structures (12, 13), the arginine residue with which Glu-398 interacts is different, and based on the worm and human complexes, it would pair with either Arg-1952 or Arg-1994, respectively. Thus, we made two charge reversal mutations in NICD1 (R1952E and R1994E) to test which one would rescue the severe reduction in transcription associated with the CSL mutant E398R. As conclusively shown in Fig. 7, only the NICD1 mutation R1994E, which is consistent with the human interactions, rescued the CSL mutant E398R in transcriptional reporter assays. Taken together, this strongly suggests that the structural differences observed in the worm and human CSL-NICD-MAM activator complexes are not conserved, but rather indicate that species-specific differences have arisen through evolution that are likely tailored for the overall benefit of the organism. This raises a cautionary note when comparing the phenotype of a particular mutant in one model organism and predicting what its phenotype would be in a divergent organism.
In summary, we have generated a set of cellular reagents that were specifically designed to characterize mutations in CSL in order to determine how mutations in CSL would affect transcriptional regulation in the Notch pathway. Although our present study analyzed the effects these mutations had on activation of cellular reporters with NICD1 and NICD2, certainly these reagents will be beneficial for future studies to analyze how these CSL mutants interact with corepressors and affect the function of CSL as a transcriptional repressor. In particular, these reagents will be invaluable for determining whether corepressors utilize similar binding surfaces to contact CSL as coactivators and whether these CSL mutants have defects in transcriptional repression in cells.
Supplementary Material
Acknowledgments
We thank Bill Miller and Rod Dekoter for technical advice and reagents regarding culturing and transduction of MEFs. We thank Raphi Kopan and Yoshio Hamada for plasmids and Tasuku Honjo for OT11/13 MEFs. We also thank Nadean Brown for the RBP-J antibody.
This work was supported, in whole or in part, by National Institutes of Health Grant CA120199. This work was also supported by Muscular Dystrophy Association and Leukemia and Lymphoma Society (to R. A. K.) and predoctoral fellowships from the American Heart Association (to K. J. R. and D. R. F.).

This article contains supplemental Figs. 1 and 2.
- NICD
- Notch intracellular domain
- NTD
- N-terminal domain
- BTD
- β-trefoil domain
- CTD
- C-terminal domain
- RAM
- RBP-J-associated molecule
- ANK
- ankyrin repeats
- ITC
- isothermal titration calorimetry
- MAM
- Mastermind
- CSL
- CBF1, Su(H), Lag-1
- DSL
- Delta, Serrate, Lag-2
- SPS
- Su(H)-paired site
- MEF
- mouse embryonic fibroblast
- DNMAM
- dominant-negative Mastermind.
REFERENCES
- 1. Radtke F., Fasnacht N., Macdonald H. R. (2010) Notch signaling in the immune system. Immunity 32, 14–27 [DOI] [PubMed] [Google Scholar]
- 2. Chiba S. (2006) Notch signaling in stem cell systems. Stem Cells 24, 2437–2447 [DOI] [PubMed] [Google Scholar]
- 3. Pierfelice T., Alberi L., Gaiano N. (2011) Notch in the vertebrate nervous system. An old dog with new tricks. Neuron 69, 840–855 [DOI] [PubMed] [Google Scholar]
- 4. Gridley T. (2010) Notch signaling in the vasculature. Curr. Top. Dev. Biol. 92, 277–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liu J., Sato C., Cerletti M., Wagers A. (2010) Notch signaling in the regulation of stem cell self-renewal and differentiation. Curr. Top. Dev. Biol. 92, 367–409 [DOI] [PubMed] [Google Scholar]
- 6. Aster J. C., Blacklow S. C., Pear W. S. (2011) Notch signaling in T-cell lymphoblastic leukemia/lymphoma and other hematological malignancies. J. Pathol. 223, 262–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Koch U., Radtke F. (2010) Notch signaling in solid tumors. Curr. Top. Dev. Biol. 92, 411–455 [DOI] [PubMed] [Google Scholar]
- 8. Kopan R., Ilagan M. X. (2009) The canonical Notch signaling pathway. Unfolding the activation mechanism. Cell 137, 216–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Borggrefe T., Oswald F. (2009) The Notch signaling pathway: transcriptional regulation at Notch target genes. Cell. Mol. Life Sci. 66, 1631–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kovall R. A., Blacklow S. C. (2010) Mechanistic insights into Notch receptor signaling from structural and biochemical studies. Curr. Top. Dev. Biol. 92, 31–71 [DOI] [PubMed] [Google Scholar]
- 11. Kovall R. A., Hendrickson W. A. (2004) Crystal structure of the nuclear effector of Notch signaling, CSL, bound to DNA. EMBO J. 23, 3441–3451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wilson J. J., Kovall R. A. (2006) Crystal structure of the CSL-Notch-Mastermind ternary complex bound to DNA. Cell 124, 985–996 [DOI] [PubMed] [Google Scholar]
- 13. Nam Y., Sliz P., Song L., Aster J. C., Blacklow S. C. (2006) Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell 124, 973–983 [DOI] [PubMed] [Google Scholar]
- 14. Friedmann D. R., Kovall R. A. (2010) Thermodynamic and structural insights into CSL-DNA complexes. Protein Sci. 19, 34–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Meng X., Brodsky M. H., Wolfe S. A. (2005) A bacterial one-hybrid system for determining the DNA-binding specificity of transcription factors. Nat. Biotechnol. 23, 988–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tun T., Hamaguchi Y., Matsunami N., Furukawa T., Honjo T., Kawaichi M. (1994) Recognition sequence of a highly conserved DNA-binding protein RBP-Jκ. Nucleic Acids Res. 22, 965–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang H., Zou J., Zhao B., Johannsen E., Ashworth T., Wong H., Pear W. S., Schug J., Blacklow S. C., Arnett K. L., Bernstein B. E., Kieff E., Aster J. C. (2011) Genome-wide analysis reveals conserved and divergent features of Notch1/RBPJ binding in human and murine T-lymphoblastic leukemia cells. Proc. Natl. Acad. Sci. U.S.A. 108, 14908–14913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bailey A. M., Posakony J. W. (1995) Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes Dev. 9, 2609–2622 [DOI] [PubMed] [Google Scholar]
- 19. Jarriault S., Brou C., Logeat F., Schroeter E. H., Kopan R., Israel A. (1995) Signaling downstream of activated mammalian Notch. Nature 377, 355–358 [DOI] [PubMed] [Google Scholar]
- 20. Arnett K. L., Hass M., McArthur D. G., Ilagan M. X., Aster J. C., Kopan R., Blacklow S. C. (2010) Structural and mechanistic insights into cooperative assembly of dimeric Notch transcription complexes. Nat. Struct. Mol. Biol. 17, 1312–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nam Y., Sliz P., Pear W. S., Aster J. C., Blacklow S. C. (2007) Cooperative assembly of higher order Notch complexes functions as a switch to induce transcription. Proc. Natl. Acad. Sci. U.S.A. 104, 2103–2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hamaguchi Y., Yamamoto Y., Iwanari H., Maruyama S., Furukawa T., Matsunami N., Honjo T. (1992) Biochemical and immunological characterization of the DNA-binding protein (RBP-Jκ) to mouse Jκ recombination signal sequence. J. Biochem. 112, 314–320 [DOI] [PubMed] [Google Scholar]
- 23. Kato H., Taniguchi Y., Kurooka H., Minoguchi S., Sakai T., Nomura-Okazaki S., Tamura K., Honjo T. (1997) Involvement of RBP-J in biological functions of mouse Notch1 and its derivatives. Development 124, 4133–4141 [DOI] [PubMed] [Google Scholar]
- 24. Pear W. S., Nolan G. P., Scott M. L., Baltimore D. (1993) Production of high titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. U.S.A. 90, 8392–8396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ong C. T., Cheng H. T., Chang L. W., Ohtsuka T., Kageyama R., Stormo G. D., Kopan R. (2006) Target selectivity of vertebrate notch proteins. Collaboration between discrete domains and CSL-binding site architecture determines activation probability. J. Biol. Chem. 281, 5106–5119 [DOI] [PubMed] [Google Scholar]
- 26. Friedmann D. R., Wilson J. J., Kovall R. A. (2008) RAM-induced allostery facilitates assembly of a notch pathway active transcription complex. J. Biol. Chem. 283, 14781–14791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Whitmore L., Wallace B. A. (2004) DICHROWEB, an on-line server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 32, W668–W673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sreerama N., Woody R. W. (2000) Estimation of protein secondary structure from circular dichroism spectra. Comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 287, 252–260 [DOI] [PubMed] [Google Scholar]
- 29. Oka C., Nakano T., Wakeham A., de la Pompa J. L., Mori C., Sakai T., Okazaki S., Kawaichi M., Shiota K., Mak T. W., Honjo T. (1995) Disruption of the mouse RBP-Jκ gene results in early embryonic death. Development 121, 3291–3301 [DOI] [PubMed] [Google Scholar]
- 30. VanderWielen B. D., Yuan Z., Friedmann D. R., Kovall R. A. (2011) Transcriptional repression in the Notch pathway. Thermodynamic characterization of CSL-MINT (Msx2-interacting nuclear target protein) complexes. J. Biol. Chem. 286, 14892–14902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Johnson S. E., Ilagan M. X., Kopan R., Barrick D. (2010) Thermodynamic analysis of the CSL × Notch interaction. Distribution of binding energy of the Notch RAM region to the CSL β-trefoil domain and the mode of competition with the viral transactivator EBNA2. J. Biol. Chem. 285, 6681–6692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lubman O. Y., Ilagan M. X., Kopan R., Barrick D. (2007) Quantitative dissection of the Notch/CSL interaction. Insights into the Notch-mediated transcriptional switch. J. Mol. Biol. 365, 577–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Murtaugh L. C., Stanger B. Z., Kwan K. M., Melton D. A. (2003) Notch signaling controls multiple steps of pancreatic differentiation. Proc. Natl. Acad. Sci. U.S.A. 100, 14920–14925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wu L., Aster J. C., Blacklow S. C., Lake R., Artavanis-Tsakonas S., Griffin J. D. (2000) MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nat. Genet. 26, 484–489 [DOI] [PubMed] [Google Scholar]
- 35. Maillard I., Weng A. P., Carpenter A. C., Rodriguez C. G., Sai H., Xu L., Allman D., Aster J. C., Pear W. S. (2004) Mastermind critically regulates Notch-mediated lymphoid cell fate decisions. Blood 104, 1696–1702 [DOI] [PubMed] [Google Scholar]
- 36. Kovall R. A. (2007) Structures of CSL, Notch, and Mastermind proteins. Piecing together an active transcription complex. Curr. Opin. Struct. Biol. 17, 117–127 [DOI] [PubMed] [Google Scholar]
- 37. Del Bianco C., Aster J. C., Blacklow S. C. (2008) Mutational and energetic studies of Notch 1 transcription complexes. J. Mol. Biol. 376, 131–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.