Background: Seeding of eukaryotic amyloids is highly specific.
Results: Curli subunits from different bacteria can cross-seed and such interspecies interactions restore surface attachment and biofilm formation.
Conclusion: Curli cross-seeding is relaxed, which promotes interspecies biofilms.
Significance: This is the first study on cross-seeding of bacterial amyloids and will help define the roles of bacterial amyloids in multispecies biofilms.
Keywords: Amyloid, Bacteria, Biofilm, Microbiology, Protein Aggregation, Protein-Protein Interactions, Bacterial Amyloids, Cross-seeding, Interspecies Interactions
Abstract
Amyloids are highly aggregated proteinaceous fibers historically associated with neurodegenerative conditions including Alzheimers, Parkinsons, and prion-based encephalopathies. Polymerization of amyloidogenic proteins into ordered fibers can be accelerated by preformed amyloid aggregates derived from the same protein in a process called seeding. Seeding of disease-associated amyloids and prions is highly specific and cross-seeding is usually limited or prevented. Here we describe the first study on the cross-seeding potential of bacterial functional amyloids. Curli are produced on the surface of many Gram-negative bacteria where they facilitate surface attachment and biofilm development. Curli fibers are composed of the major subunit CsgA and the nucleator CsgB, which templates CsgA into fibers. Our results showed that curli subunit homologs from Escherichia coli, Salmonella typhimurium LT2, and Citrobacter koseri were able to cross-seed in vitro. The polymerization of Escherichia coli CsgA was also accelerated by fibers derived from a distant homolog in Shewanella oneidensis that shares less than 30% identity in primary sequence. Cross-seeding of curli proteins was also observed in mixed colony biofilms with E. coli and S. typhimurium. CsgA was secreted from E. coli csgB− mutants assembled into fibers on adjacent S. typhimurium that presented CsgB on its surfaces. Similarly, CsgA was secreted by S. typhimurium csgB− mutants formed curli on CsgB-presenting E. coli. This interspecies curli assembly enhanced bacterial attachment to agar surfaces and supported pellicle biofilm formation. Collectively, this work suggests that the seeding specificity among curli homologs is relaxed and that heterogeneous curli fibers can facilitate multispecies biofilm development.
Introduction
Amyloids are β-sheet-rich proteinaceous fibrils traditionally associated with protein misfolding and cytotoxicity (1–3). Amyloid formation is the hallmark of many neurodegenerative diseases such as Alzheimer, Parkinson, and prion-based diseases. Recently a rapidly growing class of “functional amyloids” suggests that the amyloid-fold can be utilized to facilitate nondegenerative physiological tasks (4–6). A number of functional amyloids have been described, such as curli produced by Escherichia coli (7), TasA by Bacillus subtilis (8), and Pmel17 by mammalian cells (9). Despite having little similarity in primary structure, amyloids share biochemical and structural propensities. Amyloid fibers are characterized by cross-β-sheet structures, with each β-strand perpendicular to the fiber axis (1, 10–12). These fibers are extraordinarily stable, resistant to most denaturation treatments and protease K digestion (7, 13), and possess the distinct tinctorial ability of binding the dyes Congo red and thioflavin T (ThT)3 (7, 14). Another common feature of amyloids is the nucleation-dependent kinetics of assembly, in which amyloid proteins polymerize into fibers after a lag phase followed by an exponential growth (15–17). Formation of an oligomeric nucleus or seeds is rate-limiting and is associated with amyloid toxicity (18, 19). The self-polymerization of amyloid proteins can be accelerated by the presence of preformed fibers or nucleators in a process called seeding (20, 21).
Most amyloidogenic proteins can be seeded by fibers derived from the same protein. In rare cases, one amyloidogenic protein can be cross-seeded by different amyloid fibers. Cross-seeding is considered a possible mechanism for diverse pathologies of amyloid diseases and prion infections (22–25). Cross-seeding was observed between the Alzheimer-associated peptide Aβ and islet amyloid polypeptide (25), as well as between Aβ and human prion element PrP (24). Additionally, Aβ1–42 fibers have been reported to induce the formation of tau-containing filaments in vivo (26), and in vitro preformed Aβ1–42 oligomers can induce the conversion of tau monomers to β-sheet-rich, toxic oligomers (27). Also, functional amyloids curli and Sup35 can promote amyloid protein A amyloidosis, suggesting that interactions between heterogeneous amyloid proteins may be a risk factor for accelerating the onset of amyloid diseases (23). Cross-seeding is also observed between some of the mammalian and yeast prion species, providing a mechanism for prion transmission and prion-based disease infection (28, 29).
Although limited cross-seeding among diverse amyloids has been reported, these interactions typically occur with reduced efficiency or are often completely prevented by species barriers. Seeding and cross-species transmission of most mammalian prion and yeast prion proteins are highly specific (29–31). Strict species barriers are present among conserved yeast prion domains including closely related Sup35 homologs from the Saccharomyces sensu stricto group (32, 33). A single amino acid mutation can alter the seeding specificity of Sup35 (34). Even the same Sup35 protein polymerizing at different temperatures forms fibers with distinct seeding specificity (35). Cross-seeding is also inefficient among mammalian prions (29), closely related synuclein homologs (36), different immunoglobulin domains (37), and lysozymes from different species (38).
Functional amyloids have been widely described in bacteria including E. coli, Salmonella spp., B. subtilis, Streptomyces coelicolor, and Pseudomonas fluorescens (7, 8, 39–41). Although cross-seeding among amyloid proteins has been extensively studied in disease-associated amyloids and prions, the seeding specificity of bacterial amyloids has not been investigated. To assess cross-seeding among functional bacterial amyloids, as well as the resultant biological consequences, we utilized the well studied bacterial functional amyloid called curli. Curli are amyloid fibers produced on the cell surface of E. coli and other enteric bacteria that facilitate adherence to biotic and abiotic surfaces (42, 43), biofilm development (39, 43–45), and pathogen-host interactions (46–48). Unlike disease-associated amyloids, curli assembly is highly regulated by dedicated pathways (49, 50). At least seven proteins, encoded by the csgBAC and csgDEFG operons (curli specific gene), are involved in curli biogenesis (4, 49). The major subunit of curli is CsgA. In vivo, the polymerization and membrane-localization of CsgA is dependent on the nucleator protein CsgB (51). The secretion of both CsgA and CsgB requires the outer-membrane pore-forming protein CsgG (52) and chaperone proteins CsgE and CsgF (53, 54). In vitro CsgA and CsgB self-assemble into amyloid fibers (51, 55) with β-helix structures (12). The in vitro fibrillization of CsgA can be seeded by its own fibers or by fibers of the nucleator protein CsgB (51, 55).
We report here that CsgA from E. coli, Salmonella enterica serovar Typhimurium LT2, Citrobacter koseri, and even a distant CsgA homolog from Shewanella oneidensis MR-1 are able to cross-seed in vitro. In vivo, both S. typhimurium and E. coli share curli subunits as building blocks to assemble functional fibers in colony biofilms and such interspecies interactions of curli subunits aid in bacterial adherence to abiotic surfaces and restore biofilm formation. Our results suggest that seeding between curli homologs is relaxed and that cross-seeding between different bacteria has an impact on multispecies communities.
EXPERIMENTAL PROCEDURES
Bacterial Growth
Bacteria were grown in LB at 37 °C with overnight shaking. To induce curli expression, bacteria were grown on YESCA agar (1 g/liter of yeast extract, 10 g/liter of casamino acids, and 20 g/liter of agar) or YESCA-CR (50 μg/ml of Congo red and 1 μg/ml of Coomassie Blue) at 26 °C for 48 h. To induce pellicle biofilm formation, bacteria were inoculated in 4 ml of static LB-no salt broth (10 g/liter of tryptone and 5 g/liter of yeast extract) at 26 °C for 3 days. Antibiotics were added at the following concentration: kanamycin, 50 μg/ml, and ampicillin, 100 μg/ml.
Strains and Plasmid
Strains and plasmids used in this study are listed in supplemental Tables S1 and S2. Primer sequences are listed in supplemental Table S3. S. typhimurium curli mutants were constructed according to the methods described by Datsenko and Wanner (56). All the S. typhimurium curli mutants can be complemented by expressing S. typhimurium CsgA or CsgB under the control of S. typhimurium csgBA promoter from the plasmid pACYC177.
Protein Purification
Purification of CsgA/CsgB homologs was adapted from Cegelski et al. (57) and Wang et al. (58). Briefly, expression of C-terminal His6-tagged CsgA or CsgB homologs without the Sec signal sequence in NEB3016 was induced at A600 0.9 by 0.5 mm isopropyl 1-thio-β-d-galactopyranoside at 37 °C for 1 h. Bacteria were lysed in 8 m guanidine hydrochloride in 50 mm potassium phosphate buffer (Pi) overnight. After centrifugation at 10,000 × g for 20 min the supernatant was incubated with nickel-nitrilotriacetic acid resin (Sigma) at room temperature for 1 h and then loaded onto a disposable polypropylene column (Thermo). Proteins were eluted into 50 mm potassium Pi containing 125 mm imidazole. To get monomeric CsgA, fractions with the target protein were combined and loaded onto a 30-kDa centrifugal filter units (Thermo) to remove dimers and other oligomers.
Aβ1–42 Disaggregation
100% Trifluoroacetic acid (TFA) was added to Aβ1–42 peptides to a 1 mg/ml ratio and bath sonicated at room temperature for 10 min. TFA was removed by a SpeedVac at room temperature for 1 h. Residual TFA was removed by dissolving the pellet in 500 μl of 1,1,1,3,3,3-hexafluoro-2-propanol (HFIP) and SpeedVac drying for 1 h; the process was repeated 3 times. The peptides were dissolved in 2 mm cold potassium hydroxide to 62.5 μm and cold 5× potassium Pi buffer was immediately added. Samples were centrifuged at 70,000 × g at 4 °C for 3 h and the top solution was taken carefully for the polymerization assay.
In Vitro Polymerization and Seeding Assay
100 μl of freshly purified CsgA homologs were loaded on a 96-well opaque plate and polymerization kinetics were monitored by ThT fluorescence with excitation wavelength at 438 nm and emission at 495 nm (bandwidth 20 nm) on a Tecan plate reader. ThT fluorescence was normalized as described (55). Seeds were prepared by sonication of preformed fibers with three 15-s bursts on ice. ThT was added to a final concentration of 20 μm.
Stability Assay
22 μg Fibers were spun down and resuspended in 50 mm potassium Pi or 50 (v/v), 70, 80, 90, or 100% HFIP for 5 min and immediately dried on a SpeedVac for 3 h. The resulting pellets were boiled in 2× SDS sample buffer for 5 min before being loaded on 15% SDS-PAGE gels.
Biacore Binding Assay
A BIAcore3000 (GE Healthcare) was used to monitor real-time interactions between monomers and fibers. A CM5 sensor chip was activated with 35 μl of 1:1(v/v) mixture of 0.4 m 1-ethyl-3-(3-dimethylpropyl)-carbodiimide and 0.1 m N-hydroxysuccinimide at a flow rate of 5 μl/min. Mature CsgA or CsgB fibers were sonicated (Sonicator XL2020, Misonix) with three 15-s bursts at power 2 and 50-s pauses between. These seeds were then diluted into acetate buffer to final concentrations of 3.5 μm for CsgB fibers and 2.6 μm for CsgA fibers. 55 μl of sonicated fibers were injected at a flow rate of 5 μl/min to allow the immobilization of 2500–3500 resonance units. Blank flow cells on the same chip were used as negative controls. After ligand immobilization, excessive reactive groups were deactivated with 35 μl of 1 m ethanolamine-HCl, pH 8.5, and the sensor chip was primed with 50 μm potassium Pi, pH 7.4. 40 μl of 0.25 μm monomeric E. coli CsgA or S. typhimurium CsgA was injected over the sensor chip surface at a flow rate of 50 μl/min and the response was recorded in resonance units. Elution of a mock purification from NEB3013 harboring an empty vector pET11d and monomeric Aβ1–42 with a concentration higher than 1 μm were used as negative controls.
Western Blot Analysis
Western blot analysis of the whole cell lysates and bacteria with the underlying agar (plug) were performed as described previously (45, 58). For whole cell lysate, bacteria grown on YESCA agar at 26 °C for 48 h were suspended in 50 mm potassium Pi, pH 7.2, normalized by optical density at 600 nm (A600), and pelleted down. The pellets were pre-treated with or without 70 μl of HFIP immediately followed by SpeedVac centrifugation for 30 min at 45 °C. For the plug, overnight cultures of bacteria were normalized by A600 and 4 μl of culture were spotted on thin YESCA agar, incubated at 26 °C for 48 h. 8-Millimeter circular plugs including bacteria colonies and the underlying agar were collected, treated with or without 150 μl of HFIP, and dried immediately by a SpeedVac. Proteins in whole cell lysates or plugs were separated by electrophoresis and transferred onto a polyvinylidene difluoride membrane. CsgA was probed by antiserum raised against purified E. coli curli fibers (Proteintech, Chicago, IL).
Transmission Electron Microscopy
A Philips CM100 transmission electron microscope was used to visualize bacteria samples that were prepared as previously described (58).
Mixed Colony Biofilms
Interspecies curli assembly assay in mixed colony biofilms was adapted from Chapman et al. (7) and White et al. (59). Briefly, overnight cultures of E. coli and S. typhimurium curli mutants were normalized by A600 and mixed at 1:1 (v/v) ratio. 4 μl of each sample were spotted on YESCA agar and incubated at 26 °C for 3 days. Curli formation was analyzed by Western blot and transmission electron microscopy.
Measurement of Bacterial Adhesiveness
Overnight cultures of E. coli and S. typhimurium were normalized by A600 and were mixed at 1:1 (v/v) ratio. Bacterial cultures were spread on 3 ml of YESCA agar on 12-well plates and incubated at 26 °C for 3 days. To determine the bacterial adhesiveness, 1 ml of phosphate-buffered saline (PBS) was added into each well, and plates were rocked vigorously on an orbital titer plate shaker at speed 5 for 30 min at room temperature. Nonadherent bacteria in PBS were removed and the cell densities were determined by A600. Adherent bacteria were suspended in another 1 ml of PBS buffer with an inoculation loop and the A600 was measured. The percentage of adherent bacteria was calculated. To determine the adhesiveness of S. typhimurium, two approaches were used. S. typhimurium WT or mutants were transformed with mCherry-expressing plasmid pAH9 and E. coli cells were transformed with YFP-expressing plasmid pAH16. Both mCherry and YFP were driven by Staphylococcus aureus sarA P1 promoter, which was constitutively on in E. coli (60). Nonadherent S. typhimurium and adherent S. typhimurium were determined by mCherry signal with the excitation wavelength at 600 ± 9 nm and the emission wavelength at 630 ± 20 nm. Alternatively, nonfluorescence labeled E. coli and S. typhimurium were used for adhesiveness as described above. Bacteria collected before and after washing with PBS were diluted and plated out on YESCA-CR plates. E. coli curli mutants formed white colonies, whereas S. typhimurium curli mutants formed pink colonies. Colony forming units of pink colonies before and after washing were counted and the percentage of adhesive S. typhimurium was calculated.
Pellicle Biofilm Assay
Overnight cultures of S. typhimurium WT, csgA− and csgBA− mutants were diluted by 1:1,000 (v/v) into 4 ml of liquid LB-no salt medium. Freshly purified E. coli CsgA, sonicated E. coli CsgA fibers, or mock purification from cells harboring an empty vector was added. Bacteria were incubated at 26 °C without shaking for 3 days. The liquid was removed carefully and pellicles were stained with 0.1% crystal violet for visualization.
RESULTS
Curli Subunits of Different Bacteria Cross-seed in Vitro
To investigate the seeding specificity of curli amyloid, closely related homologs of E. coli CsgA from S. typhimurium and C. koseri (∼70% identity) and a distant homolog from S. oneidensis MR-1 (28% identity) were purified and tested for their ability to cross-seed E. coli CsgA polymerization in vitro. For simplicity, we refer to the CsgA homologs from different organisms as CsgAEC (CsgA from E. coli), CsgAST (S. enterica serovar Typhimiuium), CsgACK (C. koseri), and CsgASO (S. oneidensis).
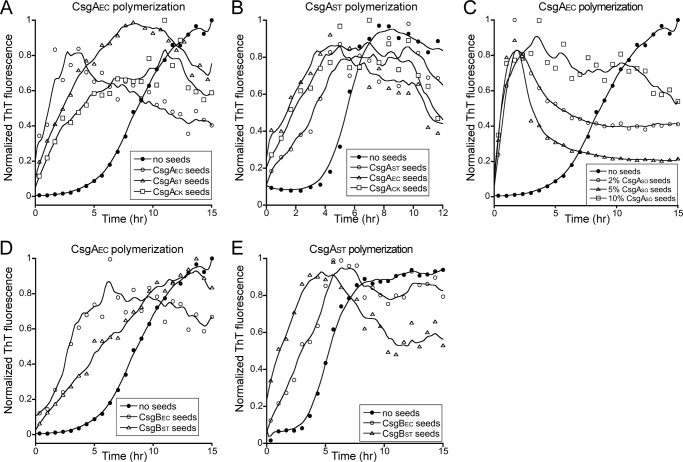
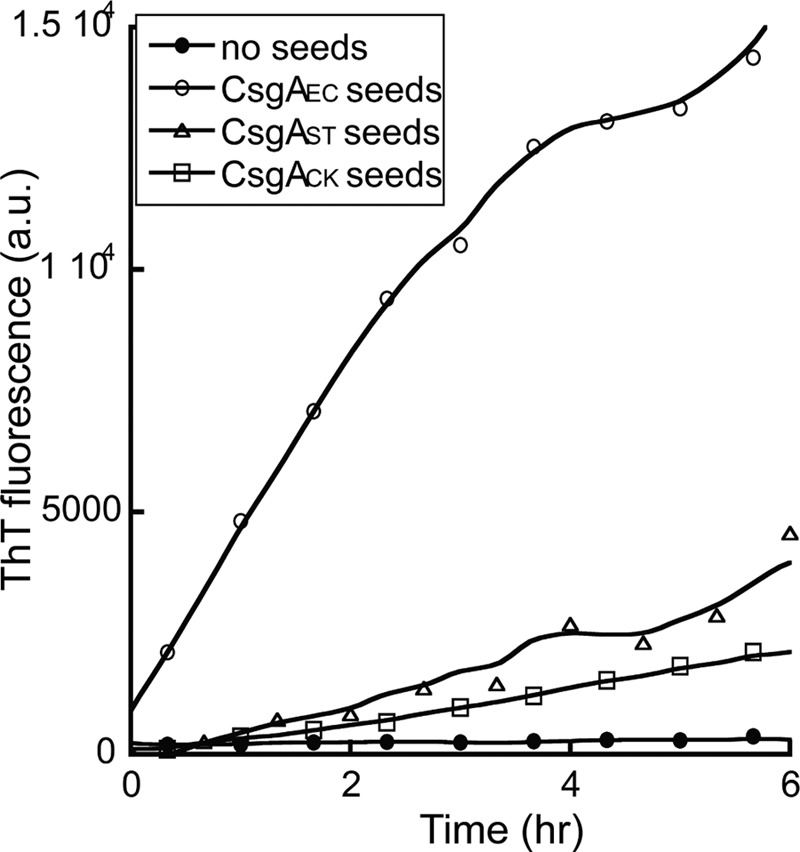
Freshly purified CsgAEC monomers spontaneously assemble into fibers with identifiable lag, exponential and stationary phases that can be followed by thioflavin T fluorescence in real time (55). Addition of preformed CsgAEC fiber seeds completely eliminated the lag phase (Fig. 1A), which was consistent with previous observations (55). In the presence of 5% (w/w) preformed CsgAST or CsgACK seeds, CsgAEC fibrillated with no lag phase, indicating fibers of CsgAST or CsgACK efficiently cross-seeded CsgAEC polymerization (Fig. 1A). Like CsgAEC, monomeric CsgAST assembled into fibers with an approximately 4 h lag phase. The polymerization was accelerated by 5% (w/w) of its own fibers, CsgAEC fibers, or CsgACK fibers (Fig. 1B). The decrease in ThT fluorescence after the exponential phase was possibly due to the adhesion of fiber aggregates to the wall of wells, a phenomenon we often observe. CsgACK also self-polymerized and could be cross-seeded by seeds of CsgAEC or CsgAST (data not shown). Interestingly, the polymerization of CsgAEC was also seeded efficiently by a distant CsgA homolog from S. oneidensis. CsgASO spontaneously assembled into amyloid-like fibers that were morphologically similar to E. coli curli fibers (supplemental Fig. S1A) and were rich in β-sheet secondary structure (supplemental Fig. S1B). The addition of preformed CsgASO fibers effectively eliminated the lag phase of CsgAEC polymerization (Fig. 1C). Taken together, these results suggest that CsgA homologs from different bacteria efficiently cross-seed in vitro.
FIGURE 1.
Curli subunits cross-seeded in vitro. A, normalized ThT fluorescence monitoring the polymerization kinetics of 10 μm E. coli CsgA (CsgAEC) alone (●), or in the presence of 5% (w/w) sonicated CsgAEC seeds (○), S. typhimurium CsgA (CsgAST) seeds (Δ), or C. koseri CsgA (CsgACK) seeds (□). B, 10 μm freshly purified CsgAST polymerized with no seeds (●), or in the presence of 5% sonicated CsgAST seeds (○), CsgAEC seeds (Δ), or CsgACK seeds (□). C, 10 μm CsgAEC polymerized alone (●), or in the presence of 2 (○), 5 (Δ), or 10% (□) S. oneidensis CsgA (CsgASO) seeds. D, 10 μm CsgAEC polymerized alone (●), with 5% CsgBEC seeds (○) or with 5% CsgBST seeds (Δ). E, 10 μm CsgAST polymerized alone (●), with 5% CsgBEC seeds (○) or with 5% CsgBST seeds (Δ).
CsgAEC polymerization can also be seeded by E. coli CsgB (CsgBEC) fibers in vitro (Fig. 1D). CsgBEC is proposed to quickly adopt an amyloid-fold and to template CsgAEC fiber formation in vivo (51). The amino acid sequences of CsgAEC and CsgBEC are less than 30% identical. Therefore, the interaction between CsgA and CsgB in E. coli represents a unique example of cross-seeding among amyloids (51, 61). It is unknown if CsgB has species-specific seeding determinants, or if CsgB can seed CsgA homologs from other bacterial species. To determine the seeding specificity of CsgB, CsgBE and CsgB homologs from S. typhimurium (CsgBST) were expressed and purified. 5% (w/w) CsgBST fibers efficiently promoted the polymerization of CsgAEC (Fig. 1D). Similarly, 5% (w/w) CsgBEC seeds cross-seeded the polymerization of CsgAST and CsgACK (Fig. 1E and data not shown), suggesting that CsgB can cross-seed CsgA of a different bacteria.
We also measured the stability of fibers formed by CsgA homologs, CsgB homologs, or fibers formed by CsgAEC in the presence of various seeds in terms of HFIP resistance. HFIP is a strong denaturant that dissociates curli fibers into SDS-soluble monomers that migrate at 17 kDa on a SDS-PAGE gel (57). These fibers showed similar resistance to HFIP treatment: fibers were mostly resistant to 50, 70, or 80% HFIP treatment, and were largely dissociated with 90 or 100% HFIP treatment (supplemental Fig. S2). This result suggests that CsgA/CsgB fibers or those formed in the cross-seeding reaction may adopt similar conformations.
We further tested whether curli subunits can cross-seed with unrelated amyloidogenic peptides and proteins. The peptide amyloid β(1–42) (Aβ1–42) and the prion domain of yeast prion element Sup35 (Sup35 NM, which includes the N-terminal and the middle domain) were analyzed for their ability to cross-seed with E. coli CsgA. Neither Aβ1–42 nor Sup35 NM was able to seed CsgAEC, nor could the fibrillization of Aβ1–42 or Sup35 NM be seeded by CsgBEC fibers (supplemental Fig. S3). The addition of CsgAEC seeds slightly increased the fibrillization of Sup35 NM, although to much lower levels than Sup35 NM seeded by its own fibers (supplemental Fig. S3). Therefore, although cross-seeding occurs between curli homologs, curli cannot cross-seed with unrelated amyloidogenic proteins.
CsgA Monomers Physically Bind Preformed Curli Fibers
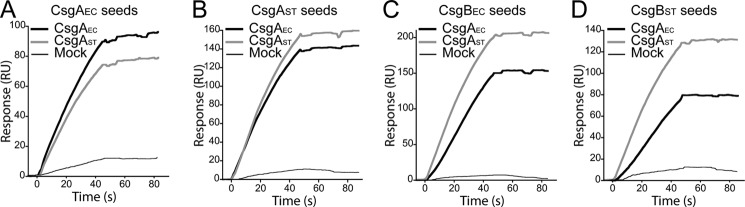
To determine whether monomeric CsgA can physically interact with heterogeneous seeds or whether seed addition allows for adoption of a new fold without direct binding, surface plasmon resonance was performed to monitor interactions between CsgA monomers and preformed fibers in real time. Preformed CsgAEC fibers were immobilized on a sensor chip and 0.25 μm freshly purified CsgAEC or CsgAST was injected over the chip. An increase in resonance units with no obvious signal decay was observed after the injection, suggesting a strong interaction between CsgAEC monomers and CsgAEC seeds, and between CsgAST and CsgAEC seeds (Fig. 2A). The same response was detected when monomeric CsgAEC or CsgAST was flowed over CsgAST seeds, showing a strong association of polymeric CsgAST with CsgAST or CsgAEC monomers (Fig. 2B). Similarly, both freshly purified CsgAEC and CsgAST bound to polymeric CsgBEC or CsgBST (Fig. 2, C and D). As the control, BSA or mock purification from cells harboring an empty vector was flowed over polymeric CsgAEC, CsgAST, CsgBEC, or CsgBST and no significant interaction was observed (Fig. 2 and data not shown). Finally, consistent with results shown in supplemental Fig. S3B, Aβ1–42 only weakly interacted with CsgAEC seeds and quickly disassociated after injection (supplemental Fig. S4A). Aβ1–42 did not interact with CsgBEC seeds at all (supplemental Fig. S4B). Together, these results suggest monomeric CsgA homologs directly bind fibers during cross-seeding reactions.
FIGURE 2.
Freshly purified CsgA bound preformed CsgA or CsgB seeds. Surface plasmon resonance sensorgrams of 0.25 μm freshly purified CsgAEC (black line) and CsgAST (gray line) or products from a mock CsgA purification from strains harboring an empty vector (thin black line) were injected over CsgAEC seeds (A), CsgAST seeds (B), CsgBEC seeds (C), or CsgBST (D) that were immobilized on a CM5 sensor chip.
Mutations of Conserved Gln/Asn Residues Abolish the Cross-seeding of Curli
All the CsgA or CsgB homologs contain a C-terminal domain predicted to have imperfect β-strand-loop-β-strand repeating units and conserved Gln and Asn stacks (62). The first and last repeating units of E. coli CsgA are required for self-seeding and interactions between CsgA and CsgB (58, 63). Mutations of Gln and Asn residues in these repeating units to Ala result in a slow polymerizing variant of CsgA named CsgAslowgo (CsgAQ49A,N54A,Q139A,N144A), which can be seeded by E. coli CsgA fibers, but cannot respond efficiently to CsgB-mediated heteronucleation (64), suggesting that CsgA self-seeding and heteronucleation may be mediated by distinct mechanisms. Interestingly, we found the conserved Gln and Asn residues were also necessary for cross-seeding between E. coli CsgA and other CsgA homologs. The polymerization of freshly purified CsgAslowgo was not efficiently seeded by fibers of CsgAST, CsgACK, or CsgASO (Fig. 3 and data not shown). Consistent with previous results, 5% E. coli CsgA fibers completely eliminated the lag phase and promoted the polymerization of CsgAslowgo (Fig. 3). These results indicated that the conserved Gln and Asn residues in E. coli CsgA help mediate cross-seeding.
FIGURE 3.
Conserved Gln and Asn residues were required for cross-seeding between curli subunits. ThT fluorescence of 30 μm freshly purified E. coli CsgAslowgo alone (●), or in the presence of 5% CsgAEC seeds (○), CsgAST seeds (Δ), or CsgACK seeds (□).
Interspecies Cross-seeding in Vivo
Like E. coli, S. typhimurium, C. koseri, and S. oneidensis all harbor the csgDEFG and csgBA operons required for curli biogenesis, and curli fibers were detected on the surface of S. typhimurium and C. koseri (65) (data not shown). The relaxed seeding of curli subunits in vitro led us to ask if cross-seeding of curli also occurred in vivo between different bacterial species.
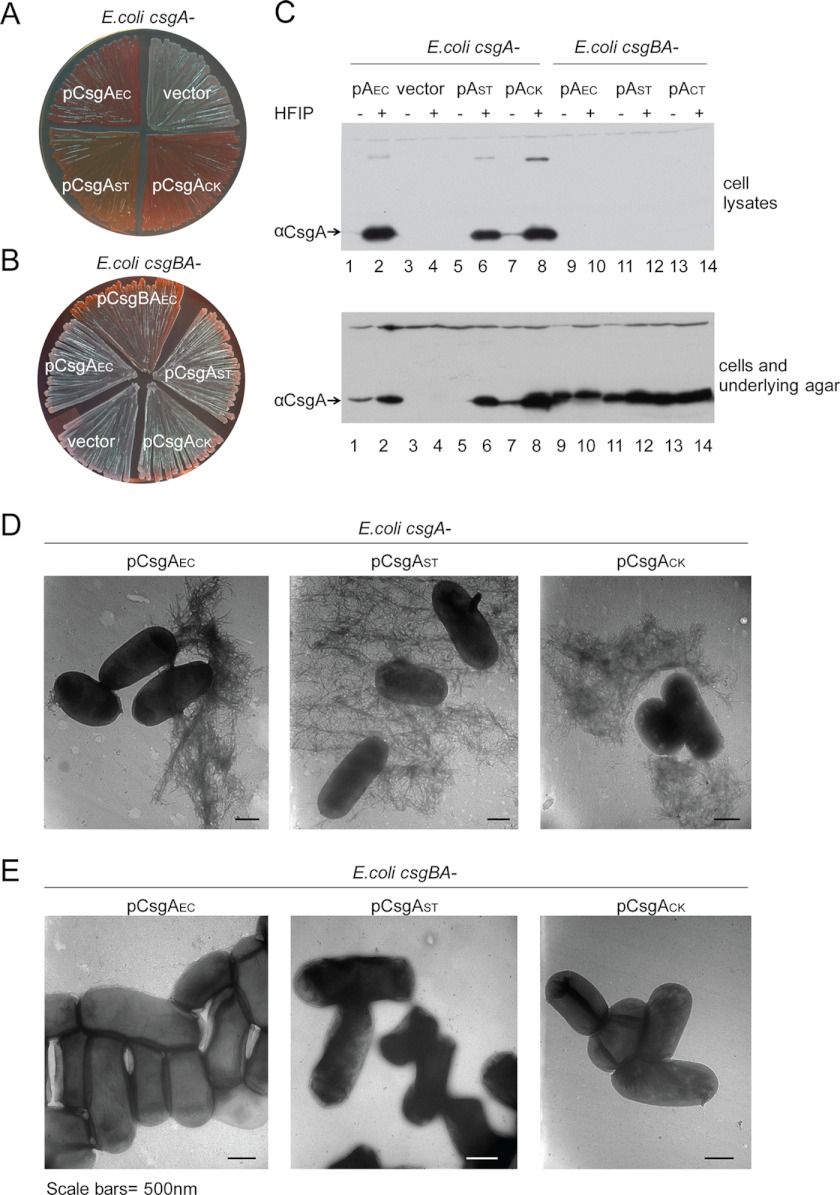
To test if CsgBEC cross-seeds CsgA homologs under physiological conditions, we expressed csgAST, csgACK, and csgASO driven by the E. coli csgBA promoter from a low copy plasmid in an E. coli csgA− mutant. As a control, we cloned the gene encoding CsgAEC into the same plasmid. If expression of CsgA homologs complements curli formation in an E. coli csgA− mutant, it would suggest that these homologs can be cross-seeded by CsgBEC on bacterial surfaces. Colonies formed by an E. coli csgA− mutant expressing CsgAEC, CsgAST, or CsgACK stained red on YESCA-CR plates, whereas the csgA− mutant with the vector control appeared white (Fig. 4A). Bacteria-associated CsgAEC, CsgAST, and CsgACK fibers were detected in whole cell lysates by Western blot after treatment with HFIP. CsgAST and CsgACK in the whole cell lysates were mostly SDS-insoluble, indicating that these CsgA homologs were incorporated into SDS-resistant curli fibers that could not migrate into SDS-PAGE (Fig. 4C). Curli-like fibers were observed on the E. coli csgA− mutant expressing CsgAST and CsgACK by electron microscopy (Fig. 4D). Thus, CsgAST and CsgACK complemented curli assembly in an E. coli csgA− mutant. Moreover, the complementation was CsgB dependent. csgBA−/pCsgAST or csgBA−/pCsgACK did not bind Congo red or produce fibers on the cell surface (Fig. 4, B and E), and no cell-associated CsgAST/CsgACK was detected by Western blot. Instead, SDS-soluble CsgAST/CsgCK was found in agar blocks underneath the colonies (Fig. 4C), indicating that CsgA homologs were secreted without polymerizing into fibers. Collectively, CsgAST and CsgACK can be seeded by CsgBEC on the cell surface. Additionally, an E. coli csgB− mutant could be complemented by expression of CsgBST and CsgBCK (supplemental Fig. S5). Thus, CsgAEC could also be seeded by CsgB homologs from different bacteria. CsgASO and CsgBSO were unable to complement E. coli csgA− or E. coli csgB− mutants (data not shown), possibly indicating that CsgASO and CsgBSO are not properly expressed or localized in E. coli. Consistent with this we were unable to detect expression of His-tagged CsgASO in E. coli (data not shown).
FIGURE 4.
An E. coli csgA− mutant was complemented by CsgA homologs from S. typhimurium or C. koseri in a CsgB-dependent manner. A, the expression of CsgA homologs complemented the Congo red binding of an E. coli csgA− mutant. E. coli csgA− harboring an empty vector control or plasmids encoding CsgAEC (pCsgAEC), CsgAST (pCsgAST), or CsgACK (pCsgACK) were grown on YESCA-CR plates at 26 °C for 48 h. B, CsgB was required for the complementation of Congo red binding. E. coli csgBA− mutant harboring an empty vector or plasmid pCsgBAEC, pCsgAEC, pCsgAST, or pCsgACK were grown on a YESCA-CR agar at 26 °C for 48 h. C, Western blot of whole cell lysates (top panel) and plugs (bottom panel) of an E. coli csgA− mutant transformed with pCsgAEC (lanes 1 and 2), the vector control (lanes 3 and 4), pCsgAST (lanes 5 and 6), or pCsgACK (lanes 7 and 8), and an E. coli csgBA− mutant with pCsgAEC (lanes 9 and 10), pCsgAST (lanes 11 and 12), or pCsgACK (lanes 13 and 14) grown on YESCA agar plates at 26 °C for 48 h. Samples were treated with (+) or without (−) HFIP before electrophoresis and analyzed by aCsgA antibody. D, TEM of E. coli csgA− or csgBA− mutant transformed with pCsgAEC, pCsgAST, or pCsgACK. Scale bars equal 500 nm.
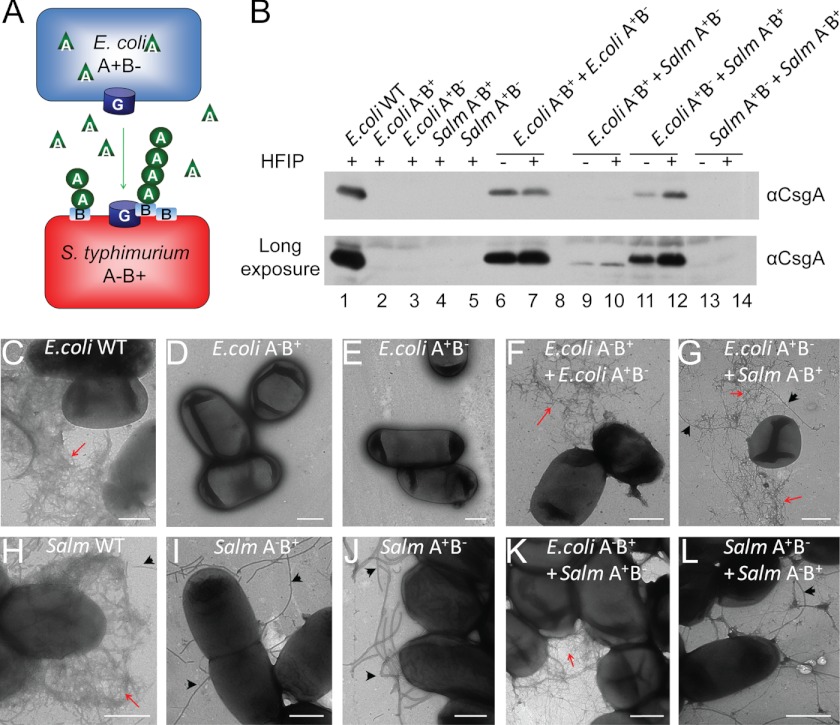
We next asked if different bacteria grown in the same community could exchange curli subunits to assembled interbacterial curli fibers. Here we used E. coli and S. typhimurium because curli biogenesis is well understood in these two organisms and are proposed to frequently share the same ecological niche (65–67). During normal curli assembly in E. coli, CsgA is secreted unpolymerized into the extracellular environment prior to incorporation into fibers (7, 55). Soluble CsgA produced by an E. coli csgB− mutant (A+B−) can assemble into fibers on an adjacent E. coli csgA− mutant (A−B+) (7). In S. enterica serovar Enteritidis, such interbacterial curli assembly was detected only when the lipopolysaccharide O polysaccharide synthesis gene was mutated (59). To determine whether interspecies curli assembly occurred between E. coli and S. typhimurium (schematic in Fig. 5A), E. coli and S. typhimurium csgA−/csgB− mutants were mixed at a 1:1 (A600/A600) ratio and grown into mixed colonies. In a mixed colony with E. coli csgB− (A+B−) and S. typhimurium csgA− (A−B+), bacteria-associated, SDS-insoluble CsgA was readily detected by Western blot (Fig. 5B) and curli fibers were observed by TEM (Fig. 5G). Similar interbacterial curli assembly was detected in a mixed colony with E. coli csgA− (A−B+) and S. typhimurium csgB− (A+B−) by Western analysis (Fig. 5B) and TEM (Fig. 5K). It is interesting to notice that less curli were formed between E. coli csgA− (A−B+) and S. typhimurium csgB− (A+B−) than between E. coli csgB− (A+B−) and S. typhimurium csgA− (A−B+). This was probably because there were less E. coli csgA− (A−B+) cells to template CsgAST into fibers. S. typhimurium csgB− (A+B−) outgrew E. coli csgA− (A−B+) by a ratio of 4:1 in mixed colonies, whereas S. typhimurium csgA− (A−B+) did not outcompete E. coli csgB− (A+B−) (supplemental Fig. S6A). Consistent with this notion, more SDS-soluble CsgA was detected in agar underneath the colony with E. coli csgA− (A−B+) and S. typhimurium csgB− (A+B−) (supplemental Fig. S6B), suggesting that some CsgAST subunits were not incorporated into fibers and were secreted into the agar.
FIGURE 5.
Interbacterial curli assembly between E. coli and S. typhimurium curli mutants. A, a schematic describing the interaction between an E. coli csgB− mutant (A+B−) and S. typhimurium csgA− mutant (A−B+). Unpolymerized E. coli CsgA secreted by E. coli csgB− (A+B−) is templated by CsgB on the surface of S. typhimurium csgA− (A−B+). B, Western blots of HFIP-treated (+) or nontreated (−) whole cell lysates of the indicated strains or strain mixtures. The blots were probed with aCsgA antibody. The bottom panel is a longer exposure of the same blot to visualize the faint bands. C–L, TEM of E. coli WT (C), E. coli csgA− (A−B+) (D), E. coli csgB− (A+B−) (E), a mixed colony with E. coli csgA− (A−B+) and E. coli csgB− (A+B−) (F), a mixture of S. typhimurium csgA− (A−B+) and E. coli csgB− (A+B−) (G), S. typhimurium WT (H), S. typhimurium csgA− (A−B+) (I), S. typhimurium csgB− (A+B−) (J), a mixture of E. coli csgA− (A−B+) and S. typhimurium csgB− (A+B−) (K), and a mixture of S. typhimurium csgA− (A−B+) and S. typhimurium csgB− (A+B−) (L). Curli fibers are indicated by red arrows and flagella are indicated by black arrows. Scale bars equal to 500 nm.
As controls, none of the single mutants produced curli on their own (Fig. 4B), and TEM (Fig. 5, D, E, I, and J) and curli were not detected in a mixture of E. coli csgB− (A+B−) and S. typhimurium csgBA− (A−B−) or S. typhimurium csgB− (A+B−) and E. coli csgBA− (A−B−), demonstrating that the nucleator CsgB was required to mediate interspecies curli assembly (date not shown). Consistent with previous reports (7, 59), interbacterial curli formed between E. coli csgA− and E. coli csgB− mutants (Fig. 5, B and F) but not between S. typhimurium csgA− and csgB− mutants (Fig. 5, B and L). However, curli formed by the two E. coli mutants was less resistant to SDS, suggesting either that those fibers may adopt a distinct conformation or that polymerization was less efficient. Together, both E. coli and S. typhimurium are capable of sharing curli subunits to build interspecies curli.
Interbacterial Complementation between E. coli Curli Mutants and S. typhimurium Curli Mutants Restore Bacterial Surface Attachment
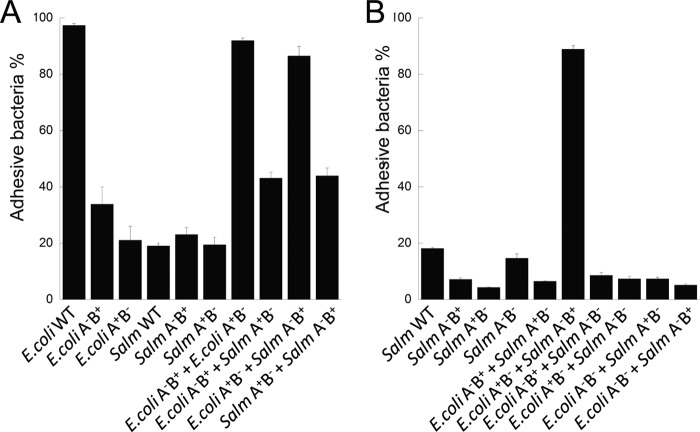
The ability of E. coli and S. typhimurium to share curli subunits (Fig. 5) led us to further investigate the impact of such interspecies interactions on multispecies communities. Curli mediate surface attachment (42, 43) and bacteria-bacteria interactions (44). Wild-type E. coli formed a colony biofilm that tightly adhered to the agar surface even after vigorous shaking in PBS (Fig. 6A). The adhesiveness was dependent on curli production. Colonies of an E. coli csgA− mutant (A−B+) or an E. coli csgB− mutant (A+B−) did not strongly adhere to the agar surface; less than 40% of the curli-defective bacteria remained on agar after washing compared with 95% of WT (Fig. 6A). A csgA− mutant (A−B+) or a csgB− mutant (A+B−) of S. typhimurium also showed low adherence to the agar surface (Fig. 6A). Wild-type S. typhimurium has low surface adherence as well (Fig. 6, A and B), possibly due to the production of cellulose and other extracellular polysaccharides that counteract curli-mediated adherence (59, 68). However, over 80% of a mixed colony with E. coli csgB− (A+B−) and S. typhimurium csgA− (A−B+) stayed attached to the agar surface after vigorous washing with PBS (Fig. 6A). The increase in surface attachment was dependent on interspecies curli assembly, as mixed colonies with E. coli csgB− (A+B−) and S. typhimurium csgBA− (A−B−), or S. typhimurium csgA− (A−B+) and E. coli csgBA− (A−B−) mutants did not attach to the surface well (supplemental Fig. S7A). Consistent with the low level of interspecies curli production, we did not observe a significant increase in adherence when E. coli csgA− (A−B+) and S. typhimurium csgB− (A+B−) was mixed (Fig. 6A). Collectively, these results demonstrate that curli assembled between E. coli and S. typhimurium are effective in restoring bacterial surface attachment.
FIGURE 6.
Interbacterial curli formation between E. coli and S. typhimurium curli mutants restored bacteria adherence to agar surface. A, overnight cultures of E. coli, S. typhimurium, or a 1:1 (A600/A600) mixture of E. coli and S. typhimurium curli mutants were spread on YESCA agar in 12-well tissue culture plates, then incubated at 26 °C for 3 days. Bacterial adhesiveness was determined by the percentage of adhered bacteria after vigorous washing as described under ”Experimental Procedures.“ B, overnight cultures of E. coli constitutively expressing YFP from plasmid pAH16, S. typhimurium expressing constitutively expressing mCherry from pAH9 (60), or the mixed culture of E. coli/pAH16 and S. typhimurium/pAH9 were spread on YESCA agar on 12-well tissue culture plates, incubated at 26 °C for 3 days, and washed in PBS with vigorous shaking. Percentage of adhesive S. typhimurium was determined by mCherry signal with the excitation wavelength at 600 ± 9 nm and the emission wavelength at 630 ± 20 nm.
Because S. typhimurium did not adhere to the agar surface as well as E. coli, we modified the assay to determine whether interspecies curli assembly between E. coli and S. typhimurium also facilitated surface attachment of S. typhimurium. To differentiate between the two species, a S. typhimurium strain carrying a plasmid that constitutively expresses mCherry was used. The adhesiveness of S. typhimurium was determined by mCherry fluorescence before and after the PBS wash. When mixed with E. coli csgB− (A+B−) mutant, the percentage of S. typhimurium csgA− (A−B+) mutant adhering to the agar surface increased by more than 60% (Fig. 6B). The increase in attachment was dependent on the nucleator CsgB; an E. coli csgBA− (A−B−) did not promote S. typhimurium adherence (Fig. 6B). These results were confirmed by the colony forming units of S. typhimurium in the mixed colony (supplemental Fig. S7B). Together, these results demonstrate that interspecies curli assembly between E. coli and S. typhimurium restores bacterial adhesiveness of the whole population and also promotes the attachment of S. typhimurium to agar.
Cross-seeding of Curli Restores Pellicle Biofilm Formation of S. typhimurium
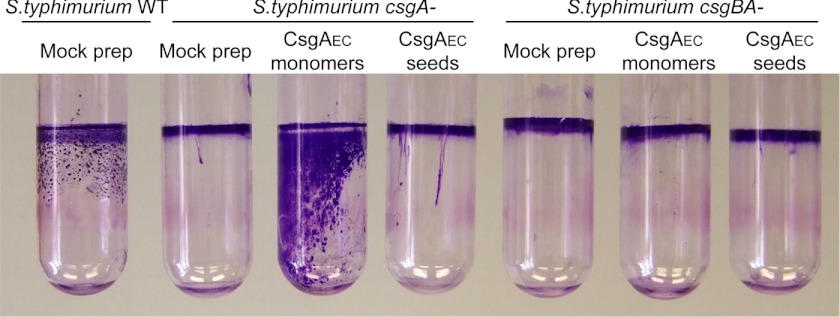
Curli are important for the development of a pellicle biofilm, a type of biofilm that grows at the air-liquid interface (8, 45, 57). In static LB-no salt medium, S. typhimurium formed a pellicle biofilm (Fig. 7). The development of pellicle biofilms requires curli production (45), and indeed S. typhimurium csgA− or csgBA− mutants were unable to make pellicles (Fig. 7). We asked if S. typhimurium csgA− could utilize and incorporate CsgAEC into fibers to support a pellicle biofilm. The addition of freshly purified CsgAEC monomers restored the pellicle development of the S. typhimurium csgA− mutant (Fig. 7). However, a S. typhimurium csgBA− mutant mixed with E. coli CsgA monomers did not form a pellicle (Fig. 7). Furthermore, sonicated, preformed E. coli CsgA seeds did not restore pellicle formation of the S. typhimurium csgA− mutant. This is probably because exogenously added fibers were not associated with cells and thus could not function as a scaffold to support pellicle biofilm formation. Thus, the cross-seeding of CsgAEC by CsgBST on S. typhimurium cell surfaces facilitates pellicle biofilm development.
FIGURE 7.
Pellicle formation of S. typhimurium csgA− was complemented by exogenous E. coli CsgA subunits. S. typhimurium WT, csgA−, or csgBA− mutants were incubated in static LB-no salt for 3 days at 26 °C in glass tubes. 10 μg/ml of freshly purified CsgAEC or CsgAEC seeds, or products from a mock CsgA purification from strains harboring an empty vector were added to each tube. The liquid culture underneath pellicles was removed and tubes were stained with crystal violet. When the liquid media was removed, pellicles fell against the wall of the glass tubes.
DISCUSSION
Amyloid proteins polymerize into fibers with nucleation-dependent kinetics. The conversion of monomers to an oligomeric nucleus contributes to the lag phase, but subsequently preformed fibers or nucleus can act as templates that accelerate amyloid formation. This process is known as seeding. Seeding is a critical step of amyloid propagation and disease development (1, 69). The work presented here demonstrates that bacterial functional amyloids have relatively relaxed seeding specificities. Amyloid cross-seeding of curli subunits was observed both in vitro between E. coli, S. typhimurium, C. koseri, and S. oneidensis as well as in vivo between E. coli and S. typhimurium. Remarkably this interspecies interaction facilitated bacterial surface attachment and biofilm development.
Amyloid seeding is typically highly specific. For instance, lack of cross-seeding is reported between the Parkinson disease-associated amyloid, α-synuclein, and closely related amyloidogenic homologs with more than 78% similarity (36). Species barriers are also commonly observed between different mammalian prion species, and between Sup35 homologs from different yeast species with up to 95% identity (29, 30, 32, 33). Even a single amino acid mutation could alter the seeding specificity of Aβ1–42 and prions (25, 29). However, our results suggest that the cross-seeding specificity between curli subunits is relaxed. Efficient seeding was observed in vitro between curli subunits from a range of bacteria including E. coli, S. typhimurium, C. koseri, and S. oneidensi. Strikingly, although the CsgASO is only 28% identical to E. coli CsgA in primary structure, it efficiently seeded CsgAEC amyloid formation with only 2% (w/w) seed concentration (Fig. 1C). We have also shown that during seeding and cross-seeding reactions, monomers physically interact with and bind fibers, as indicated by surface plasmon resonance (Fig. 2).
Curli assembly on bacterial surfaces is mediated by the nucleator protein CsgB. A longstanding model of curli biogenesis suggests that, once secreted, CsgB quickly adopts an amyloid fold that serves as a template to direct the assembly of CsgA into amyloid polymers (61, 70). The interaction between CsgA and CsgB in E. coli represents an example of heterogeneous seeding of amyloid proteins because E. coli CsgA and CsgB are less than 30% identical in amino acid sequence (51, 71). In vitro, both CsgAEC and CsgBEC spontaneously polymerize into amyloid fibers with a similar structure measured by CD and EM (51, 55). Shewmaker et al. (12) have also shown that both CsgA and CsgB adopt similar β-helix structure. Thus we hypothesize that CsgB-mediated templating is analogous to CsgA-mediated templating. CsgB-dependent heteronucleation could be mediated either by species-specific recognition sequences or by a promiscuous seeding mechanism. We have shown both in vitro and in vivo that CsgBEC can cross-seed CsgAST and CsgACK, and in turn CsgBST can cross-seed CsgAEC (Figs. 1, 2, and 4, and supplemental Fig. S5). Although cross-seeding between CsgAEC and other CsgB species was not tested due to the technical challenges in protein purification, curli formation by an E. coli csgB− mutant can be complemented by expressing CsgBCK in trans (supplemental Fig. S5), suggesting CsgBCK can also cross-seed the fibrillization of CsgAEC. Therefore, CsgB-mediated seeding is likely to be a result of relaxed seeding of curli subunits.
Despite low sequence identity, both E. coli CsgAEC and CsgBEC are composed of an N-terminal Sec signaling sequence and a C-terminal domain with five 19–24 amino acid imperfect repeating units. Each repeating unit is predicted to adopt a β-strand-loop-β-strand motif and contains conserved Gln and Asn, which are critical for amyloid fiber formation (64). The same primary structure arrangement with regularly spaced Gln and Asn residues was also found in CsgA and CsgB homologs from other Gram-negative bacteria (62). Our previous results have shown that Gln and Asn residues in the N- and C-terminal repeating units (Gln-49, Asn-54, Gln-139, Asn-144) of E. coli CsgA are essential for efficient amyloid formation and interaction with E. coli CsgB, as a CsgAslowgo mutant with those four Gln/Asn residues mutated cannot be cross-seeded by CsgB (64). In this study we further demonstrated that this mutant was also defective in cross-seeding with other CsgA homologs. Therefore these conserved Gln and Asn residues are important sequence determinants that mediate cross-seeding. An emerging consensus from studies of prion seeding specificity suggests that compatibility in protein or fiber conformations governs seeding specificity (35, 72–75). Not surprisingly, small changes in amino acid sequences can dictate conformational variability; therefore seeding specificity is tightly correlated with primary sequence (29, 64, 76). Specific side chain interactions also play an important role in amyloid formation and seeding (77). Gln and Asn residues in amyloid proteins form intra-molecular hydrogen bonds and help to stabilize the cross-β structure (77, 78). It is possible that the hydrogen bonds formed between Gln and Asn mediate interaction and seeding between different curli proteins. CsgAslowgo seeding with E. coli CsgA may be facilitated by other strain-specific side chain interactions.
The spatial arrangement of Gln and Asn within an amyloidogenic protein is proposed to play an important role in amyloid assembly (64). This could explain why the yeast prion Sup35, another Gln/Asn-rich amyloid protein, did not cross-seed with E. coli CsgA (supplemental Fig. S3), as the spacing between Gln and Asp in CsgA and in Sup35 are different. Moreover, circular dichroism spectra revealed that the secondary structures of CsgA fibers and Sup35-NM fibers are different, NM fibers have more random coil structure (7, 79) (data not shown), indicating that CsgA and Sup35 fibers may adopt incompatible conformations that limit the cross-seeding efficiency.
Although no cross-seeding was observed between curli and Sup35 or curli and Aβ1–42 (supplemental Fig. S3), other eukaryotic amyloidogenic peptides including the human antimicrobial peptide LL-37 and amyloid protein A have been suggested to interact with curli directly or indirectly (23, 68). LL-37 is produced by epithelial cells of the urinary track to protect against infections with uropathogenic E. coli (80, 81). It is recently shown that LL-37 strongly bound to polymeric CsgA fibers (68), suggesting that curli may have the potential to promote the aggregation of LL-37 and bacteria might use cross-seeding as a strategy to counteract the antimicrobial effects. Amyloid protein A deposition is a result of chronic inflammations (82). Lundmark et al. (23) has shown that amyloid protein A deposition can be accelerated by amyloids including curli. Thus, cross-seeding may provide a mechanism for amyloidogenesis.
The highly specific seeding between disease-associated amyloids and prions is proposed to be a mechanism to prevent amyloid disease propagation and transmission. As a class of functional amyloids, the relaxed seeding propensity of curli may have an impact on biological events. Curli are an important component of enteric bacterial biofilm communities; mediating surface attachment and cell-cell interactions (43, 44, 57). Cross-seeding of curli subunits was observed in vivo in a mixed-species community, as interspecies curli assembly was found between E. coli and S. typhimurium curli mutants (Fig. 5). Moreover, interspecies assembled curli fibers were biologically functional. Intercellular curli assembly between E. coli and S. typhimurium curli mutants restored bacterial adherence to agar surfaces (Fig. 6).
Curli are also required for pellicle biofilm formation (57). A S. typhimurium csgA− mutant that cannot make a pellicle biofilm was able to utilize E. coli CsgA monomers to form a robust pellicle (Fig. 7). Romero et al. (8) recently showed that addition of B. subtilis amyloids TasA fibers restored the pellicle development of a B. subtilis tasA mutant. The addition of polymerized E. coli CsgA fibers did not restore pellicle formation by S. typhimurium csgA−, indicating that S. typhimurium cannot simply utilize curli fibers as a scaffold to support pellicle formation (Fig. 7). E. coli CsgA monomers or fibers were also unable to restore pellicle formation of a S. typhimurium csgBA− mutant that lacks the nucleator protein on its surface. Thus, the complementation of S. typhimurium pellicle formation by E. coli CsgA is dependent on the nucleation process.
In nature, bacterial communities are composed of multiple species (83, 84), and bacterial amyloids are abundant in natural biofilms (85). Because E. coli and S. typhimurium are proposed to share ecological niches, it is likely that these two species interact in biofilm environments (65, 67). Because curli subunits are secreted into the extracellular environment prior to assembly into polymers, and because, as an amyloid, curli can be cross-seeded, it is plausible that E. coli and S. typhimurium coexisting in natural communities can share curli subunits to build a heterogeneous matrix. Curli-like structures are also found in Enterobacter spp. and Citrobacter spp. (65, 79), and curli homologs are prevalent among Pseudomonas spp. and Shewanella spp. Thus the promiscuous seeding of curli may have a broad impact on the multispecies communities.
Supplementary Material
Acknowledgments
We thank members of the Chapman laboratory for helpful discussions and review of this manuscript. We thank Dr. Blaise Boles and Dr. John Roth for providing the S. typhimurium LT2 strains and technical support for Salmonella genetics. We thank Dr. Susan Lindquist and Dr. Kendra Frederick for providing Sup35.
This work was supported, in whole or in part, by National Institutes of Health Grant AI073847 and the Umeå University Linnaeus Foundation.

This article contains supplemental Figs. S1–S7 and Tables S1–S3.
- ThT
- thioflavin T
- Aβ
- amyloid β
- TEM
- transmission electron microscopy
- HFIP
- 1,1,1,3,3,3-hexafluoro-2-propanol.
REFERENCES
- 1. Chiti F., Dobson C. M. (2006) Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 75, 333–366 [DOI] [PubMed] [Google Scholar]
- 2. Glenner G. G., Wong C. W. (1984) Alzheimer disease. Initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem. Biophys. Res. Commun. 120, 885–890 [DOI] [PubMed] [Google Scholar]
- 3. Prusiner S. B. (1996) Molecular biology and pathogenesis of prion diseases. Trends Biochem. Sci. 21, 482–487 [DOI] [PubMed] [Google Scholar]
- 4. Blanco L. P., Evans M. L., Smith D. R., Badtke M. P., Chapman M. R. (2012) Diversity, biogenesis and function of microbial amyloids. Trends Microbiol. 20, 66–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Badtke M. P., Hammer N. D., Chapman M. R. (2009) Functional amyloids signal their arrival. Sci. Signal. 2, pe43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fowler D. M., Koulov A. V., Balch W. E., Kelly J. W. (2007) Functional amyloid. From bacteria to humans. Trends Biochem. Sci. 32, 217–224 [DOI] [PubMed] [Google Scholar]
- 7. Chapman M. R., Robinson L. S., Pinkner J. S., Roth R., Heuser J., Hammar M., Normark S., Hultgren S. J. (2002) Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 295, 851–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Romero D., Aguilar C., Losick R., Kolter R. (2010) Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc. Natl. Acad. Sci. U.S.A. 107, 2230–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fowler DM, K. A., Alory-Jost C., Marks M. S., Balch W. E., Kelly J. W. (2006) Functional amyloid formation within mammalian tissue. PLoS Biol. 4, 100–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sunde M., Blake C. (1997) The structure of amyloid fibrils by electron microscopy and x-ray diffraction. Adv. Protein Chem. 50, 123–159 [DOI] [PubMed] [Google Scholar]
- 11. Sunde M., Serpell L. C., Bartlam M., Fraser P. E., Pepys M. B., Blake C. C. (1997) Common core structure of amyloid fibrils by synchrotron x-ray diffraction. J. Mol. Biol. 273, 729–739 [DOI] [PubMed] [Google Scholar]
- 12. Shewmaker F., McGlinchey R. P., Thurber K. R., McPhie P., Dyda F., Tycko R., Wickner R. B. (2009) The functional curli amyloid is not based on in-register parallel β-sheet structure. J. Biol. Chem. 284, 25065–25076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nordstedt C., Näslund J., Tjernberg L. O., Karlström A. R., Thyberg J., Terenius L. (1994) The Alzheimer A-β-peptide develops protease resistance in association with its polymerization into fibrils. J. Biol. Chem. 269, 30773–30776 [PubMed] [Google Scholar]
- 14. Ban T., Hamada D., Hasegawa K., Naiki H., Goto Y. (2003) Direct observation of amyloid fibril growth monitored by thioflavin T fluorescence. J. Biol. Chem. 278, 16462–16465 [DOI] [PubMed] [Google Scholar]
- 15. Serio T. R., Cashikar A. G., Kowal A. S., Sawicki G. J., Moslehi J. J., Serpell L., Arnsdorf M. F., Lindquist S. L. (2000) Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science 289, 1317–1321 [DOI] [PubMed] [Google Scholar]
- 16. Harper J. D., Lieber C. M., Lansbury P. T., Jr. (1997) Atomic force microscopic imaging of seeded fibril formation and fibril branching by the Alzheimer disease amyloid-β protein. Chem. Biol. 4, 951–959 [DOI] [PubMed] [Google Scholar]
- 17. Walsh D. M., Hartley D. M., Kusumoto Y., Fezoui Y., Condron M. M., Lomakin A., Benedek G. B., Selkoe D. J., Teplow D. B. (1999) Amyloid β-protein fibrillogenesis. Structure and biological activity of protofibrillar intermediates. J. Biol. Chem. 274, 25945–25952 [DOI] [PubMed] [Google Scholar]
- 18. Demuro A., Mina E., Kayed R., Milton S. C., Parker I., Glabe C. G. (2005) Calcium dysregulation and membrane disruption as a ubiquitous neurotoxic mechanism of soluble amyloid oligomers. J. Biol. Chem. 280, 17294–17300 [DOI] [PubMed] [Google Scholar]
- 19. Kayed R., Sokolov Y., Edmonds B., McIntire T. M., Milton S. C., Hall J. E., Glabe C. G. (2004) Permeabilization of lipid bilayers is a common conformation-dependent activity of soluble amyloid oligomers in protein misfolding diseases. J. Biol. Chem. 279, 46363–46366 [DOI] [PubMed] [Google Scholar]
- 20. Glover J. R., Kowal A. S., Schirmer E. C., Patino M. M., Liu J. J., Lindquist S. (1997) Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell 89, 811–819 [DOI] [PubMed] [Google Scholar]
- 21. Jarrett J. T., Berger E. P., Lansbury P. T., Jr. (1993) The carboxyl terminus of the β-amyloid protein is critical for the seeding of amyloid formation. Implications for the pathogenesis of Alzheimers disease. Biochemistry 32, 4693–4697 [DOI] [PubMed] [Google Scholar]
- 22. Furukawa Y., Kaneko K., Matsumoto G., Kurosawa M., Nukina N. (2009) Cross-seeding fibrillation of Q/N-rich proteins offers new pathomechanism of polyglutamine diseases. J. Neurosci. 29, 5153–5162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lundmark K., Westermark G. T., Olsén A., Westermark P. (2005) Protein fibrils in nature can enhance amyloid protein A amyloidosis in mice. Cross-seeding as a disease mechanism. Proc. Natl. Acad. Sci. U.S.A. 102, 6098–6102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Morales R., Estrada L. D., Diaz-Espinoza R., Morales-Scheihing D., Jara M. C., Castilla J., Soto C. (2010) Molecular cross-talk between misfolded proteins in animal models of Alzheimers and prion diseases. J. Neurosci. 30, 4528–4535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. O'Nuallain B., Williams A. D., Westermark P., Wetzel R. (2004) Seeding specificity in amyloid growth induced by heterologous fibrils. J. Biol. Chem. 279, 17490–17499 [DOI] [PubMed] [Google Scholar]
- 26. Götz J., Chen F., van Dorpe J., Nitsch R. M. (2001) Formation of neurofibrillary tangles in P301l tau transgenic mice induced by Aβ42 fibrils. Science 293, 1491–1495 [DOI] [PubMed] [Google Scholar]
- 27. Lasagna-Reeves C. A., Castillo-Carranza D. L., Guerrero-Muoz M. J., Jackson G. R., Kayed R. (2010) Preparation and characterization of neurotoxic tau oligomers. Biochemistry 49, 10039–10041 [DOI] [PubMed] [Google Scholar]
- 28. Derkatch I. L., Uptain S. M., Outeiro T. F., Krishnan R., Lindquist S. L., Liebman S. W. (2004) Effects of Q/N-rich, poly(Q), and nonpoly(Q) amyloids on the de novo formation of the [PSI+] prion in yeast and aggregation of Sup35 in vitro. Proc. Natl. Acad. Sci. U.S.A. 101, 12934–12939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vanik D. L., Surewicz K. A., Surewicz W. K. (2004) Molecular basis of barriers for interspecies transmissibility of mammalian prions. Mol. Cell 14, 139–145 [DOI] [PubMed] [Google Scholar]
- 30. Kocisko D. A., Priola S. A., Raymond G. J., Chesebro B., Lansbury P. T., Jr., Caughey B. (1995) Species specificity in the cell-free conversion of prion protein to protease-resistant forms. A model for the scrapie species barrier. Proc. Natl. Acad. Sci. U.S.A. 92, 3923–3927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Prusiner S. B., Scott M., Foster D., Pan K. M., Groth D., Mirenda C., Torchia M., Yang S. L., Serban D., Carlson G. A., Hoppe P. C., Westaway D., Dearmond S. J. (1990) Transgenetic studies implicate interactions between homologous Prp isoforms in scrapie prion replication. Cell 63, 673–686 [DOI] [PubMed] [Google Scholar]
- 32. Chen B., Newnam G. P., Chernoff Y. O. (2007) Prion species barrier between the closely related yeast proteins is detected despite coaggregation. Proc. Natl. Acad. Sci. U.S.A. 104, 2791–2796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Santoso A., Chien P., Osherovich L. Z., Weissman J. S. (2000) Molecular basis of a yeast prion species barrier. Cell 100, 277–288 [DOI] [PubMed] [Google Scholar]
- 34. DePace A. H., Santoso A., Hillner P., Weissman J. S. (1998) A critical role for amino-terminal glutamine/asparagine repeats in the formation and propagation of a yeast prion. Cell 93, 1241–1252 [DOI] [PubMed] [Google Scholar]
- 35. Tanaka M., Chien P., Yonekura K., Weissman J. S. (2005) Mechanism of cross-species prion transmission. An infectious conformation compatible with two highly divergent yeast prion proteins. Cell 121, 49–62 [DOI] [PubMed] [Google Scholar]
- 36. Biere A. L., Wood S. J., Wypych J., Steavenson S., Jiang Y., Anafi D., Jacobsen F. W., Jarosinski M. A., Wu G. M., Louis J. C., Martin F., Narhi L. O., Citron M. (2000) Parkinson disease-associated α-synuclein is more fibrillogenic than β- and γ-synuclein and cannot cross-seed its homologs. J. Biol. Chem. 275, 34574–34579 [DOI] [PubMed] [Google Scholar]
- 37. Wright C. F., Teichmann S. A., Clarke J., Dobson C. M. (2005) The importance of sequence diversity in the aggregation and evolution of proteins. Nature 438, 878–881 [DOI] [PubMed] [Google Scholar]
- 38. Krebs M. R., Morozova-Roche L. A., Daniel K., Robinson C. V., Dobson C. M. (2004) Observation of sequence specificity in the seeding of protein amyloid fibrils. Protein Sci. 13, 1933–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Austin J. W., Sanders G., Kay W. W., Collinson S. K. (1998) Thin aggregative fimbriae enhance Salmonella enteritidis biofilm formation. FEMS Microbiol. Lett. 162, 295–301 [DOI] [PubMed] [Google Scholar]
- 40. Dueholm M. S., Petersen S. V., Sonderkaer M., Larsen P., Christiansen G., Hein K. L., Enghild J. J., Nielsen J. L., Nielsen K. L., Nielsen P. H., Otzen D. E. (2010) Functional amyloid in Pseudomonas. Mol. Microbiol. 77, 1009–1020 [DOI] [PubMed] [Google Scholar]
- 41. Elliot M. A., Karoonuthaisiri N., Huang J., Bibb M. J., Cohen S. N., Kao C. M., Buttner M. J. (2003) The chaplins. A family of hydrophobic cell-surface proteins involved in aerial mycelium formation in Streptomyces coelicolor. Genes Dev. 17, 1727–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Olsén A., Jonsson A., Normark S. (1989) Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature 338, 652–655 [DOI] [PubMed] [Google Scholar]
- 43. Kikuchi T., Mizunoe Y., Takade A., Naito S., Yoshida S. (2005) Curli fibers are required for development of biofilm architecture in Escherichia coli K-12 and enhance bacterial adherence to human uroepithelial cells. Microbiol. Immunol. 49, 875–884 [DOI] [PubMed] [Google Scholar]
- 44. Prigent-Combaret C., Prensier G., Le Thi T. T., Vidal O., Lejeune P., Dorel C. (2000) Developmental pathway for biofilm formation in curli-producing Escherichia coli strains. Role of flagella, curli, and colanic acid. Environ. Microbiol. 2, 450–464 [DOI] [PubMed] [Google Scholar]
- 45. Weiss-Muszkat M., Shakh D., Zhou Y., Pinto R., Belausov E., Chapman M. R., Sela S. (2010) Biofilm formation by and multicellular behavior of Escherichia coli O55:H7, an atypical enteropathogenic strain. Appl. Environ. Microbiol. 76, 1545–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tukel C., Nishimori J. H., Wilson R. P., Winter M. G., Keestra A. M., van Putten J. P., Baumler A. J. (2010) Toll-like receptors 1 and 2 cooperatively mediate immune responses to curli, a common amyloid from enterobacterial biofilms. Cell Microbiol. 12, 1495–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tükel C., Wilson R. P., Nishimori J. H., Pezeshki M., Chromy B. A., Bäumler A. J. (2009) Responses to amyloids of microbial and host origin are mediated through Toll-like receptor 2. Cell Host Microbe 6, 45–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tükel C., Raffatellu M., Humphries A. D., Wilson R. P., Andrews-Polymenis H. L., Gull T., Figueiredo J. F., Wong M. H., Michelsen K. S., Akçelik M., Adams L. G., Bäumler A. J. (2005) CsgA is a pathogen-associated molecular pattern of Salmonella enterica serotype Typhimurium that is recognized by Toll-like receptor 2. Mol. Microbiol. 58, 289–304 [DOI] [PubMed] [Google Scholar]
- 49. Barnhart M. M., Chapman M. R. (2006) Curli biogenesis and function. Annu. Rev. Microbiol. 60, 131–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gerstel U., Römling U. (2003) The csgD promoter, a control unit for biofilm formation in Salmonella typhimurium. Res. Microbiol. 154, 659–667 [DOI] [PubMed] [Google Scholar]
- 51. Hammer N. D., Schmidt J. C., Chapman M. R. (2007) The curli nucleator protein, CsgB, contains an amyloidogenic domain that directs CsgA polymerization. Proc. Natl. Acad. Sci. U.S.A. 104, 12494–12499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Robinson L. S., Ashman E. M., Hultgren S. J., Chapman M. R. (2006) Secretion of curli fiber subunits is mediated by the outer membrane-localized CsgG protein. Mol. Microbiol. 59, 870–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nenninger A. A., Robinson L. S., Hammer N. D., Epstein E. A., Badtke M. P., Hultgren S. J., Chapman M. R. (2011) CsgE is a curli secretion specificity factor that prevents amyloid fiber aggregation. Mol. Microbiol. 81, 486–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nenninger A. A., Robinson L. S., Hultgren S. J. (2009) Localized and efficient curli nucleation requires the chaperone-like amyloid assembly protein CsgF. Proc. Natl. Acad. Sci. U.S.A. 106, 900–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang X., Smith D. R., Jones J. W., Chapman M. R. (2007) In vitro polymerization of a functional Escherichia coli amyloid protein. J. Biol. Chem. 282, 3713–3719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Datsenko K. A., Wanner B. L. (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cegelski L., Pinkner J. S., Hammer N. D., Cusumano C. K., Hung C. S., Chorell E., Aberg V., Walker J. N., Seed P. C., Almqvist F., Chapman M. R., Hultgren S. J. (2009) Small-molecule inhibitors target Escherichia coli amyloid biogenesis and biofilm formation. Nat. Chem. Biol. 5, 913–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang X., Zhou Y., Ren J. J., Hammer N. D., Chapman M. R. (2010) Gatekeeper residues in the major curlin subunit modulate bacterial amyloid fiber biogenesis. Proc. Natl. Acad. Sci. U.S.A. 107, 163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. White A. P., Gibson D. L., Collinson S. K., Banser P. A., Kay W. W. (2003) Extracellular polysaccharides associated with thin aggregative fimbriae of Salmonella enterica serovar enteritidis. J. Bacteriol. 185, 5398–5407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Malone C. L., Boles B. R., Lauderdale K. J., Thoendel M., Kavanaugh J. S., Horswill A. R. (2009) Fluorescent reporters for Staphylococcus aureus. J. Microbiol. Methods 77, 251–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hammer N. D., McGuffie B. A., Zhou Y., Badtke M. P., Reinke A. A., Brännström K., Gestwicki J. E., Olofsson A., Almqvist F., Chapman M. R. (2012) The C-terminal repeating units of CsgB direct bacterial functional amyloid nucleation. J. Mol. Biol. 422, 376–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Morten S., Dueholm P. H. N., Chapman M., Otzen D. (2012) Functional Amyloid in Bacteria, Wiley, New York, in press [Google Scholar]
- 63. Wang X., Hammer N. D., Chapman M. R. (2008) The molecular basis of functional bacterial amyloid polymerization and nucleation. J. Biol. Chem. 283, 21530–21539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wang X., Chapman M. R. (2008) Sequence determinants of bacterial amyloid formation. J. Mol. Biol. 380, 570–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zogaj X., Bokranz W., Nimtz M., Römling U. (2003) Production of cellulose and curli fimbriae by members of the family Enterobacteriaceae isolated from the human gastrointestinal tract. Infect. Immun. 71, 4151–4158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Römling U. (2005) Characterization of the rdar morphotype, a multicellular behavior in Enterobacteriaceae. Cell Mol. Life Sci. 62, 1234–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Römling U., Bian Z., Hammar M., Sierralta W. D., Normark S. (1998) Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J. Bacteriol. 180, 722–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kai-Larsen Y., Lüthje P., Chromek M., Peters V., Wang X., Holm A., Kádas L., Hedlund K. O., Johansson J., Chapman M. R., Jacobson S. H., Römling U., Agerberth B., Brauner A. (2010) Uropathogenic Escherichia coli modulates immune responses and its curli fimbriae interact with the antimicrobial peptide LL-37. PLoS Pathog. 6, e1001010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Jarrett J. T., L. P. J. (1993) Seeding “one-dimensional crystallization” of amyloid. A pathogenic mechanism in Alzheimer disease and scrapie? Cell 18, 1055–1058 [DOI] [PubMed] [Google Scholar]
- 70. Shu Q., Crick S. L., Pinkner J. S., Ford B., Hultgren S. J., Frieden C. (2012) The E. coli CsgB nucleator of curli assembles to β-sheet oligomers that alter the CsgA fibrillization mechanism. Proc. Natl. Acad. Sci. U.S.A. 109, 6502–6507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. White A. P., Collinson S. K., Banser P. A., Gibson D. L., Paetzel M., Strynadka N. C., Kay W. W. (2001) Structure and characterization of AgfB from Salmonella enteritidis thin aggregative fimbriae. J. Mol. Biol. 311, 735–749 [DOI] [PubMed] [Google Scholar]
- 72. Chien P., Weissman J. S. (2001) Conformational diversity in a yeast prion dictates its seeding specificity. Nature 410, 223–227 [DOI] [PubMed] [Google Scholar]
- 73. Jones E. M., Surewicz W. K. (2005) Fibril conformation as the basis of species- and strain-dependent seeding specificity of mammalian prion amyloids. Cell 121, 63–72 [DOI] [PubMed] [Google Scholar]
- 74. Ma B., Nussinov R. (2012) Selective molecular recognition in amyloid growth and transmission and cross-species barriers. J. Mol. Biol. 421, 172–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Tanaka M., Chien P., Naber N., Cooke R., Weissman J. S. (2004) Conformational variations in an infectious protein determine prion strain differences. Nature 428, 323–328 [DOI] [PubMed] [Google Scholar]
- 76. Tessier P. M., Lindquist S. (2007) Prion recognition elements govern nucleation, strain specificity, and species barriers. Nature 447, 556–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Nelson R., Sawaya M. R., Balbirnie M., Madsen A. Ø., Riekel C., Grothe R., Eisenberg D. (2005) Structure of the cross-β spine of amyloid-like fibrils. Nature 435, 773–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Lipfert J., Franklin J., Wu F., Doniach S. (2005) Protein misfolding and amyloid formation for the peptide GNNQQNY from yeast prion protein Sup35. Simulation by reaction path annealing. J. Mol. Biol. 349, 648–658 [DOI] [PubMed] [Google Scholar]
- 79. Kim S. M., Lee H. W., Choi Y. W., Kim S. H., Lee J. C., Lee Y. C., Seol S. Y., Cho D. T., Kim J. (2012) Involvement of curli fimbriae in the biofilm formation of Enterobacter cloacae. J. Microbiol. 50, 175–178 [DOI] [PubMed] [Google Scholar]
- 80. Gudmundsson G. H., Agerberth B., Odeberg J., Bergman T., Olsson B., Salcedo R. (1996) The human gene FALL39 and processing of the cathelin precursor to the antibacterial peptide LL-37 in granulocytes. Eur. J. Biochem. 238, 325–332 [DOI] [PubMed] [Google Scholar]
- 81. Frohm Nilsson M., Sandstedt B., Sørensen O., Weber G., Borregaard N., Ståhle-Bäckdahl M. (1999) The human cationic antimicrobial protein (hCAP18), a peptide antibiotic, is widely expressed in human squamous epithelia and colocalizes with interleukin-6. Infect. Immun. 67, 2561–2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Kobayashi H., Tada S., Fuchigami T., Okuda Y., Takasugi K., Matsumoto T., Iida M., Aoyagi K., Iwashita A., Daimaru Y., Fujishima M. (1996) Secondary amyloidosis in patients with rheumatoid arthritis. Diagnostic and prognostic value of gastroduodenal biopsy. Br J. Rheumatol. 35, 44–49 [DOI] [PubMed] [Google Scholar]
- 83. Rickard A. H., Gilbert P., High N. J., Kolenbrander P. E., Handley P. S. (2003) Bacterial coaggregation. An integral process in the development of multispecies biofilms. Trends Microbiol. 11, 94–100 [DOI] [PubMed] [Google Scholar]
- 84. Peters B. M., Jabra-Rizk M. A., O'May G. A., Costerton J. W., Shirtliff M. E. (2012) Polymicrobial interactions. Impact on pathogenesis and human disease. Clin. Microbiol. Rev. 25, 193–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Jordal P. B., Dueholm M. S., Larsen P., Petersen S. V., Enghild J. J., Christiansen G., Hojrup P., Nielsen P. H., Otzen D. E. (2009) Widespread abundance of functional bacterial amyloid in mycolata and other Gram-positive bacteria. Appl. Environ. Microb. 75, 4101–4110 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.