Background: IPMK is a key enzyme in signaling required for cellular adaptation and organismal development.
Results: We report the structure of IPMK, use structure-based enzyme design and perform complementation analysis in model systems.
Conclusion: We define a basis for substrate selectivity and report that 6-kinase activity is important for signaling.
Significance: Determination of the IPMK structure enabled rewiring of signaling in organisms.
Keywords: Development, Drosophila Metabolism, Inositol Phosphates, Kinetics, Protein Design, Structural Biology, Yeast Metabolism
Abstract
Inositol phosphates (IPs) regulate vital processes in eukaryotes, and their production downstream of phospholipase C activation is controlled through a network of evolutionarily conserved kinases and phosphatases. Inositol phosphate multikinase (IPMK, also called Ipk2 and Arg82) accounts for phosphorylation of IP3 to IP5, as well as production of several other IP molecules. Here, we report the structure of Arabidopsis thaliana IPMKα at 2.9 Å and find it is similar to the yeast homolog Ipk2, despite 17% sequence identity, as well as the active site architecture of human IP3 3-kinase. Structural comparison and substrate modeling were used to identify a putative basis for IPMK selectivity. To test this model, we re-engineered binding site residues predicted to have restricted substrate specificity. Using steady-state kinetics and in vivo metabolic labeling studies in modified yeast strains, we observed that K117W and K117W:K121W mutants exhibited nearly normal 6-kinase function but harbored significantly reduced 3-kinase activity. These mutants complemented conditional nutritional growth defects observed in ipmk null yeast and, remarkably, suppressed lethality observed in ipmk null flies. Our data are consistent with the hypothesis that IPMK 6-kinase activity and production of Ins(1,4,5,6)P4 are critical for cellular signaling. Overall, our studies provide new insights into the structure and function of IPMK and utilize a synthetic biological approach to redesign inositol phosphate signaling pathways.
Introduction
Inositol phosphates (IPs)3 are a large class of distinct chemical codes that regulate various intracellular processes. Classically, IP-mediated signaling begins with the production of IPs in response to cleavage of phosphoinositides by phosphatidylinositol-specific phospholipase C (reviewed in Ref. 1). Additional IP species such as inositol tetrakisphosphate (IP4) and inositol pentakisphosphate (IP5) are generated by subsequent series of phosphorylation and dephosphorylation (2–5). Multiple IP species have been linked to processes, including metabolic regulation and nutrient selection, stress responses, nuclear mRNA export, apoptosis, RNA editing, and organism development (6, 7).
Inositol phosphate multikinase (IPMK) is among the enzymes central to the production of IP species downstream of phospholipase C activation. For clarity, we use IPMK throughout this manuscript, as it most accurately reflects the “promiscuous” 6-/3-/5-kinase functions of the enzyme. As a historical note, IPMK was first designated ArgRIII (later called Arg82) based on a yeast genetic screen designed to identify the components of a nutrient response pathway in which arginine served as the sole nitrogen source (8–11). ArgRIII was then renamed Ipk2 based on the discovery that it functioned as an inositol phosphate kinase (12, 13), and this nomenclature was also used in referring to the plant gene orthologs when they were first identified and characterized as AtIpk2α and AtIpk2β in Arabidopsis (14, 15). IPMK has now been cloned from a variety of species and has been shown to function as a 6/3/5-kinase that catalyzes the phosphorylation of at least seven different IP species. Its loss of function in a variety of organisms is critical for survival (reviewed in Refs. 5 and 6).
To gain insights into the basis of IPMK substrate promiscuity, we initiated a structural approach of the plant enzyme, which possesses three kinase activities capable of phosphorylating the D-6, D-3, and D-5 ring positions of a variety of IP substrates (14). We obtained Arabidopsis thaliana IPMKα crystals, complete data sets of native and seleno-methionine derivatives, and calculated an electron density map from experimental phases. Concomitant to our studies, the yeast IPMK crystal structure was published (16). From these studies and a number of subsequent biological and protein engineering experiments, we now report the crystal structure of A. thaliana IPMKα at 2.9 Å, a basis of IP substrate binding, the kinetic characterization of the wild-type enzyme and selective K117W and K121W mutants, and the yeast and fly IP profiles and phenotypes of the K121W and K117W/K121W mutants. Collectively, our data build upon and extend insights derived from the yeast crystal structure and, for the first time, provide a re-engineering of IPMK to be substrate-selective. We exploit these mutants to address important biological questions related to IPMK function, and our data are consistent with a role for 6-kinase activity in the regulation of cellular and organismal signaling.
EXPERIMENTAL PROCEDURES
Plasmid Constructs
Full-length and N-terminal truncated (residues 16–286; Δ16) A. thaliana IPMK were amplified from the pGEX-KG construct (14) with the restriction sites NdeI or PsiI (N terminus or Δ16 truncation) and BamHI (C terminus) and cloned into the 6xHis-tagged vector pET-15b. The 16–286 construct was designed based on alignment with the Saccharomyces cerevisiae IPMK sequence when crystals of full-length A. thaliana IPMK were found to be twinned. The 5′-sense full-length primer was 5′-CAT AGT GGA CAT ATG CAG CTC AAA GTC CCT GAA CAT CAG-3′ (restriction sites are underlined). The Δ16–286 was 5′-CAG TAT TAT AAA GAC GGG AAG CCT GGT CCT CTC-3′. The 3′-antisense primer was 5′-TAC GGA TCC TCA TCA CTA AGA ATC TGC AGA CTC ATC TGC-3′. The constructs were transformed into XL1-blue chemically competent cells and plasmids extracted from transformants and sequenced at the Duke University DNA sequencing facility.
Generation of Altered Specificity or Kinase-dead Constructs
PCR mutagenesis based on the Stratagene QuikChange kit was used to generate the mutants K100A, K117W, K121W, and K117W:K121W in full-length A. thaliana IPMK in pET-15b. Lys-100 is part of the canonical inositol kinase PxxxDxKxG motif, whereas Lys-117 and Lys-121 were identified from the A. thaliana IPMK structure as potential sites for generating IP substrate-selective mutants.
Expression and Purification of Recombinant A. thaliana IPMK
Constructs were transformed into BL21(DE3) (Invitrogen) cells and protein expression was induced with 1 mm isopropyl-β-d-thiogalactopyranoside at 30 °C for 3 h (seleno-methionine labeled) or overnight (native). Seleno-methionine labeling was performed by the methionine inhibition process (17), and complete incorporation was confirmed by mass spectrometry (supplemental Fig. S1). Cells were suspended in lysis buffer (50 mm Tris-HCl (pH 7.5), 500 mm KCl, 5 mm imidazole, 5 mm β-mercaptoethanol, and 1 tablet “Complete Mini” protease inhibitor (Roche Applied Science) per 10 g of cell pellet) and lysed by passing three times through an M110L microfluidizer (Microfluidics). The soluble fraction was loaded onto a nickel-nitrilotriacetic acid Superflow column (Qiagen), washed, resuspended in one column volume of buffer, and digested with human thrombin (Hematologic Technologies). The resin was repacked and run over a nickel-nitrilotriacetic acid plus benzamidine-Sepharose 4 Fast Flow (GE Healthcare) column to remove any low-affinity proteins, and the thrombin and fractions were collected. Fractions with protein were concentrated and run over a Superdex 200 HiLoad 16/60 gel filtration column (Pharmacia). Final purity was confirmed by SDS-PAGE gels, concentrated to 10 mg/ml, aliquoted, and stored at −80 °C.
Crystallization and Data Collection
Native Δ16 AtIPK2α crystallized in 2.0–2.15 m ammonium sulfate and 0.1 m Tris-HCl, pH 8.5, at 16–20 mg/ml. Full-length SeMet AtIPK2α crystallized in 2.10 m ammonium sulfate and 0.1 m Tris-HCl, pH 7.0, at 18 mg/ml. All crystals were grown by hanging-drop vapor diffusion using Hampton trays. Crystals were cryo-protected by successive transfer into 30% glycerol and saturated ammonium sulfate at the crystallization pH in 5% glycerol increments. They were equilibrated overnight in the final protectant and then flash-frozen with liquid nitrogen. Data were collected at the Advanced Photon Source, Argonne National Laboratory (Argonne, IL), beamlines 22ID (SeMet) and 22BM (native) on MAR300 or MAR225 CCD detectors, respectively. The data were processed with XDS (18).
Structure Determination and Model Refinement
The SHELXC/D/E suite (19) was used to solve the structure by single-wavelength anomalous dispersion. Experimental synthesis maps were calculated to 3.10 Å for both enantiomers in space groups P3221 and P3121. Comparison clearly revealed the correct hand and space group (P3221). The initial phases were improved by solvent flattening and density modification with CNS (version 1.1) (20) using the 2.9 Å Δ16 AtIPK2α native amplitudes. A polyalanine search model was generated in PyMOL (21) using the structures of S. cerevisiae IPMK and Homo sapiens IP3K (Protein Data Bank codes 2IEW, 1TZD, and 2AQX) (16, 22–24). Molrep (25) was used to fit the search model into the experimental density and provide a framework to build the remaining model. Model building was performed in iterative rounds using O (26), Coot (27), and KiNG (28). The initial poly-A model was refined by simulated annealing in CNS (20), and subsequent refinement rounds were carried out with Refmac5 (25). Translation-libration-screw refinement (29) was applied after completion of the backbone. Experimental density was observed equivalent to the β1 strand of S. cerevisiae IPK2 and IP3K but could not be satisfactorily modeled. Structure validation was performed using MolProbity (30). Data and refinement statistics are reported in Table 1.
TABLE 1.
Crystallographic data, phasing, and refinement statistics
| Data collection and phasing | 16-286 | SeMet |
|---|---|---|
| Space group | P3221 | P3221 |
| Unit cell | a = b = 130.8 Å, c = 129.9 Å; α = β = 90°, γ = 120° | a = b = 130.2 Å, c = 129.4 Å; α = β = 90°, γ = 120° |
| Wavelength (Å) | 1.000 | 0.97166 |
| Resolution (Å) | 46-2.9 | 50-3.1 |
| Unique reflections | 28,738 | 44,459 |
| Completeness (%) | 100 (100)a | 100 (100) |
| Rmergeb | 8.5 (52.4) | 9.2 (52.6) |
| FOM (SAD) | 1.31 | |
| Refinement | ||
| Resolution range (Å) | 46–2.9 (2.975–2.90) | |
| No. of reflections | 314,752 (43,621) | |
| R (%) | 23.64 (31.5) | |
| Rfree (%) | 24.61 (37.0) | |
| r.m.s.d.c | ||
| Bond length (Å) | 0.015 | |
| Bond angle | 1.496° | |
| B factor (Å2) main chain bonds | 0.917 | |
| B factor (Å2) side chain bonds | 2.725 | |
| Protein atoms, number | 3444 | |
| Ligand atoms, number | 30 | |
| Residues in allowed ϕ-ψ region | 99.5% | |
a Data in parentheses are for highest resolution shells.
b Rsym = Σhkl√(N/(N − 1))Σi|Ii(hkl) − 〈I(hkl)〉|/ΣiIi(hkl).
c r.m.s.d., root mean square deviation.
Design of Substrate-selective A. thaliana IPMK Mutants
To test the substrate selectivity motif and design substrate-selective mutants for in vivo signal probing, constructs were designed to block specific phosphate-binding pockets. The structures of A. thaliana IPMK, ADP-bound S. cerevisiae IPMK (Protein Data Bank code 2IF8), and substrate-bound H. sapiens IP3K (Protein Data Bank code 1W2C) were overlaid with LSQMAN (31), and the coordinates of Ins(1,4,5)P3 from H. sapiens IP3K were transferred into the structure of A. thaliana IPMK. Using PyMOL (21), a model IP binding library was generated for IP6 (from the structure of ADAR2, Protein Data Bank code 1ZY7) in the 6-/3-/5-kinase reactions while conserving the original ring orientation. Phosphate groups were deleted as necessary to model the appropriate IP3 or IP4 species. Phosphate pockets were defined as the volume of space occupied by an equatorial phosphate group. Residues within 5 Å of the putative γ- or δ-phosphate pockets were identified as potential candidates for mutation. Lys-117 and Lys-121 were mutated in silico using KiNG (28, 30) to residues able to occupy the corresponding phosphate pockets. Candidates were identified by selecting rotamer conformations without protein main-chain steric conflicts.
Substrate-selective Mutant Product Identification
Radiolabeled assays were performed to test the selectivity and identify the product of the mutant IPMKs. Ins(1,4,5)P3 was phosphorylated by the K121W or K117W/K121W mutants in the presence of [γ-32P]ATP, and the products were separated via HPLC. Products were identified by treatment of [32P]IP4 with 5-ptase (selective for Ins(1,3,4,5)P4 but not Ins(1,4,5,6)P4) in the presence of [3H]Ins(1,3,4,5)P4 and separated with a 4.6 × 125 mm Partisphere SAX anion exchange column over a 10 mm to 1.7 m NH4H2PO4 gradient, whereas 3H and 32P activity was observed.
Generation and Purification of [32P]IP4 Products
[32P]Ins(1,4,5,6)P4 and [32P]Ins(1,3,4,5)P4 were generated as follows. 10 μm Ins(1,4,5)P3 and ATP with trace [γ-32P]ATP were treated with either K121W A. thaliana IPMK to generate Ins(1,4,5,6)P4 or D. melanogaster IP3K to generate Ins(1,3,4,5)P4. The reactions were performed in 50 mm Hepes, pH 6.6, 50 mm KCl, 10 μm ATP, 1 mm MgCl2, at 30 °C using 1 μg of IPMK or IP3K and terminated by heat inactivation. [32P]-IP4 products were purified over a 0.2 mm HCl to 0.5 m HCl, 2 mm EDTA, 30-min gradient on a Partisphere SAX column, and 1-ml fractions were collected. Fractions containing [32P]IP4 were identified, and activity was quantitated by scintillation counting with ScintiSafe Econo2 (Fisher Chemicals) scintillation fluid in a Beckman LS6500 multipurpose scintillation counter. Samples were neutralized by the addition of 5 m KOH, until pH 7.0 was reached and brought to 50 mm Hepes, pH 7.0, aliquoted, and stored at −20 °C.
Inositol Phosphate Kinase Assays
All unlabeled IPs were purchased from Cell Signals, Inc. Tritiated IPs were purchased from American Radiolabeled Chemicals. Inositol phosphate steady-state kinetic reactions were performed in 50 mm Hepes, pH 6.6, 50 mm KCl, 1.5 mm ATP, 5 mm MgCl2, and 0.1 mg/ml bovine serum albumin at 30 °C and 9.8–1500 μm Ins(1,4,5)P3, Ins(1,4,5,6)P4, or Ins(1,3,4,5)P4, with trace amounts of the matching species of [3H]IP3 or [32P]IP4. Reactions were halted with 2 μl of 2.5 m HCl and separated by HPLC (3H) or TLC (32P). 3H traces were quantitated with the Karat32TM program, and 32P TLC plates were quantitated with the ImageQuantTM program. All reactions were performed in at least triplicate, and data were analyzed with the Graphpad PrismTM program.
Yeast Strains and Media
Yeast strains (12, 14, 32) were grown in complete minimal media with the appropriate amino acids and 2% sugar. Yeast lines were transformed using the lithium acetate method (33).
Yeast Construct Generation
Full-length A. thaliana IPMK was PCR-amplified from the pET-15 construct with primers for SalI and EcoRI N- and C-terminal sites. It was then ligated into pRS426 Cup1 and transformed into XL1-blue cells. An HA tag was included at the C terminus. Plasmids were sequenced and point mutants were generated, as with the pET-15b constructs.
Metabolic Labeling
Labeling and analysis were performed as described previously (34). Strains were inoculated from single colonies into 3 ml of CSM-selective media, containing 20 μCi/ml [3H]d-myo-inositol and 50–150 μm CuSO4 and grown to saturation at 30 °C and 250 rpm. The cultures were harvested by centrifugation, and soluble inositol phosphates were extracted. Organic and aqueous phases were separated by centrifugation, and soluble IPs in the aqueous phase were resolved by HPLC on a 4.6 × 125 mm Partisphere SAX anion exchange column. Individual IP isomers were assigned on the basis of co-elution with IP standards.
Western Blots
Yeast extract blots were performed using mouse anti β-actin (mAbcam 8224) or α-HA (primary) and goat anti-mouse (Odyssey no. 926-32210) and quantitated on an Odyssey Li-Cor thermal imager running software (version 2.1).
Plate Growth Assays
Single colonies were picked from streaks on selective media, suspended in sterile water, and water bath sonicated for 3 × 5 s. 5 μl of 10-fold serial dilutions were spotted on CSM-Ura+Dex, Glu-Ura+Dex, or Orn-Ura+Dex agar media and incubated at 30 °C.
Generation of A. thaliana IPMK Transgenic D. melanogaster Lines
Wild-type and K121W A. thaliana IPMK in pET-15b were used as the templates to generate BamHI-XbaI fragments ligated into enhanced GFP and pUAST. The resultant p-element plasmids were injected into W1118 embryos, and the resultant transgenic lines were crossed with the ipk220B-3 allele on chromosome II.
Measurement of D. melanogaster Wing Morphology
Wings from 6-day-old adult males were dissected and mounted in glycerol. Pictures of the mounted wings were taken using a Nikon dissecting microscope and camera at 10× magnification. The relative sizes of the wings were measured by the Metamorph program.
RESULTS
Crystallization and Structure Determination
The structures of full-length and 16–286 (Δ16) A. thaliana IPMK were solved in space group P3221 by single wavelength anomalous dispersion at 3.1 Å, and the structure of Δ16 A. thaliana IPMK refined to 2.9 Å with an R/Rfree of 23.64/24.61. Each asymmetric unit contains two copies of A. thaliana IPMK, mirroring its elution as a dimer in gel chromatography (data not shown). 215 of 286 residues are modeled in each monomer, consisting of residues 41–70, 81–175, 183–230, and 238–279. Anomalous diffraction density was observed for five of six seleno-methionine residues, the remaining residue in the disordered N terminus. 95.7/99.0% of the residues were in the favored/allowed regions of the Ramachandran plot. The solvent content was abnormally high at 76.5% with Vm = 5.22 (35), but experimental maps reveal no density for additional molecules, and none can be modeled within the constraints of the asymmetric unit. Additional statistics are presented in Table 1.
Structural Description
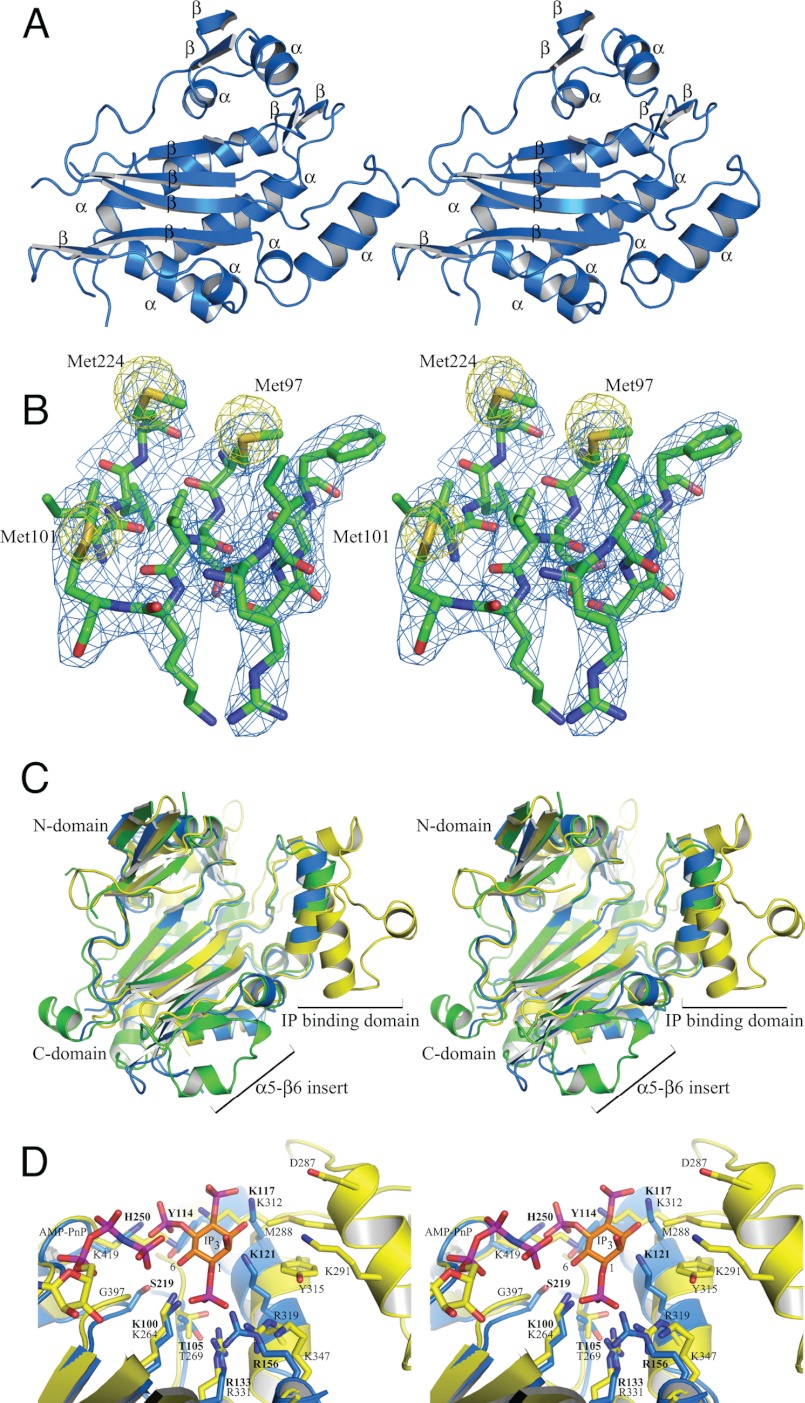
A. thaliana IPMK has a similar secondary and tertiary structure to other members of the ATP grasp fold family (36, 37). It consists of eight α-helices and 10 β-strands organized around a central β-sheet, backed by slightly off-parallel α-helices organized into three domains (Fig. 1A). The N- and C-terminal subdomains are α+β groupings, whereas the inositol binding domain consists of two α-helices and connecting loop regions. To facilitate comparisons, we have followed the secondary structure assignment as used by Holms and Jogl (16) for the yeast structure (supplemental Fig. S2). The larger C-terminal subdomain makes up the majority of the structure of A. thaliana IPMK. Residues 94–102 and 132–277 are organized into five anti-parallel β-strands in a 3-2-1-4-5 order, flanked by helix α6 and backed by helices α7, α8, α10, and two smaller anti-parallel β-strands. Yeast helices α5 and α9 are not observed in the plant structure. α5 is part of an insert in the yeast sequence, whereas α9 is in the residues 231–237 disordered region. Similarly, the β6 strand in the same insert as helix α5 is not present. This may be an insert unique to S. cerevisiae IPMK as it is not present in H. sapiens IP3K, which has the same structure in this region as A. thaliana IPMK (Figs. 2 and 3A). The inositol binding domain is inserted between strands β4 and β5. Helices α6, α7, and α8 connect β6 and β7, whereas the small sheet strands β8 and β11 are between α8 and β9, and β10 and α10, respectively. Packing between non-crystallographic copies is minimal, limited to parts of the N-terminal domain (residues 54–58 and 68), with a few residues in the C-terminal domain (195–196, 203, 274, 277). Crystallographic packing is much more extensive, with contacts throughout the α-helix-packed side of the central β-sheet in a back-to-back arrangement, in which half of the helices interact with one monomer and half with the other monomer. A second set of contacts exist at the edge of the central β-sheet, where the β6 strand of chain A packs anti-parallel to the β6 strand of the chain B symmetry mate. The two sets of symmetry contacts are on nearly orthogonal axes to each other; this may explain the ability of the A. thaliana IPMK crystals to grow to nearly 1-mm dimensions despite their high (75%) solvent content. Large solvent channels are evident when symmetry mates are generated but are not large enough to fit a third monomer.
FIGURE 1.
Structure and comparison of A. thaliana IPMK. A, stereo schematic of secondary structure. B, representative stereo density of core of A. thaliana IPMK and anomalous peaks. Single coefficient-weighted density (calculated by FFT in CCP4 4. 6.1) is shown at 1.5σ (blue), and anomalous density is shown at 12σ around three of the five seleno-methionine residues used to solve the structure. C, stereo overlay of A. thaliana IPMK (blue), S. cerevisiae IPMK (green), and H. sapiens IP3K (yellow) structures. D, active site comparison of A. thaliana IPMK and H. sapiens IP3K with Ins(1,4,5)P3. Stereo view of the active sites of A. thaliana IPMK and H. sapiens IP3K with AMP-PnP from H. sapiens IP3K and Ins(1,4,5)P3 modeled in the proposed 6-kinase orientation. Note the poor structural conservation on the opposite side of the inositol ring and much more extensive structure and contacts of H. sapiens IP3K.
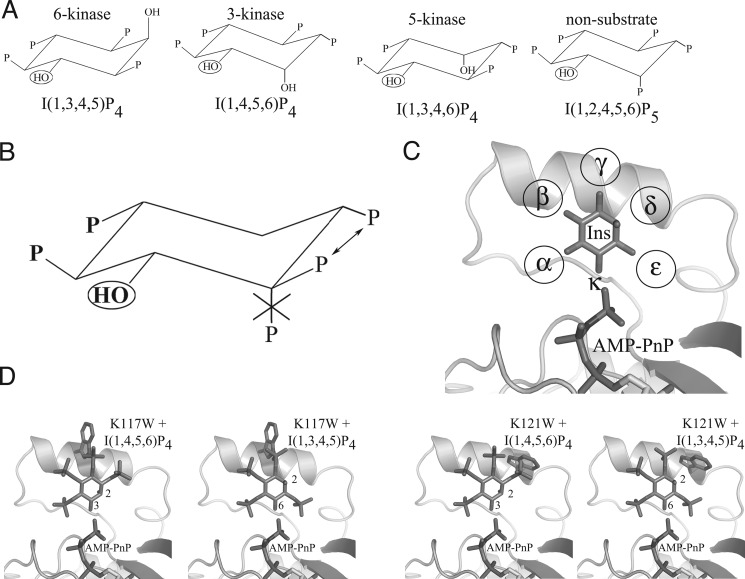
FIGURE 2.
Inositol phosphate species, binding motif, and A. thaliana IPMK selective designs. A, substrate and non-substrate IP species. Three A. thaliana IPMK IP substrate species, one for each kinase activity, and one non-substrate species are shown. All are orientated to preserve the position of the target hydroxyl (circled) and ring orientation. B, proposed IP substrate motif. Also shown are proposed rules for IP substrate selectivity by A. thaliana IPMK. Two phosphate groups are absolutely required (boldface type), the ring orientation must be maintained, one of two phosphate groups is required (arrows), and an axial phosphate cannot be tolerated if it occupies the first position clockwise of the target hydroxyl. C, phosphate binding pockets. Inositol in the active site of A. thaliana IPMK is shown conserving ring-carbon orientation. The phosphate pockets are designated α-ϵ clockwise from the kinase target hydroxyl κ. Surrounding residues are omitted for clarity. D, proposed binding of IP4 species with K117W and K121W mutants. Ins(1,3,4,5)P4 and Ins(1,4,5,6)P4 are modeled with either K117W or K121W mutant models. K117W is predicted to have steric clashes with both IP4 species, whereas K121W is predicted to clash with only Ins(1,4,5,6)P4.
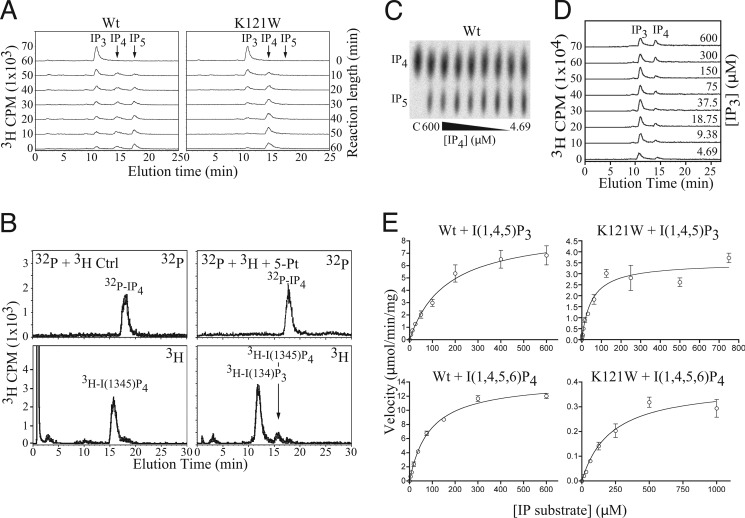
FIGURE 3.
IP production, identity, and kinetics. A, production of IP4 and IP5 over time. Time courses of generation of IP4 and IP5 by wild-type and K121W A. thaliana IPMK from [3H]Ins(1,4,5)P3. Note the simultaneous appearance of IP4 and IP5 in the wild-type reactions, but no IP5 signal for the K121W reactions, even after 60 min. B, identification of the K121W IP4 product as Ins(1,4,5,6)P4. Samples containing [32P]IP4 produced by K121W and [3H]Ins(1,3,4,5)P4 as an internal standard treated or not treated with 5-ptas and separated by HPLC, and radioactivity was counted on 3H and 32P channels simultaneously. 5-Ptase product is observed in only for [3H]IP4. C, representative TLC plate of [32P]IP4 assay. D, representative HPLC trace of [3H]Ins(1,4,5)P3 assay. E, Michaelis-Menten steady-state kinetic curves for Ins(1,4,5)P3 and Ins(1,4,5,6)P4 with wild-type and K121W A. thaliana IPMK.
The Structural Core of A. thaliana IPMK Is Homologous to Those of S. cerevisiae IPMK and H. sapiens IP3K
To examine the structural basis of substrate selectivity, the models of S. cerevisiae IPMK and H. sapiens IP3K were overlaid onto A. thaliana IPMK. As expected, the cores of each protein were nearly identical to one another (root mean square deviation = 1.067 and 1.118 Å, Fig. 1C). Similarly, the active sites of all three proteins were well conserved, with the majority of conserved residues in either the central β-sheet or secondary structure elements that support it. The structure of the IP-binding domain was highly conserved between both IPMK species and diverges from H. sapiens IP3K, with the additional IP-binding domain inserts (Fig. 1C), which provide IP3K with its greater substrate selectivity (23, 24). Outside of the conserved core, there were several differences, concentrated in the external loops and species-specific inserts, and in the expanded inositol binding domain of IP3K.
Comparison of All Known Substrate and Non-substrate IP Species Suggests a Steric-based Binding Motif
To identify an IP substrate binding motif, we compared all known substrate and non-substrate IP species based on a common ring orientation and target hydroxyl position. Two phosphate groups were conserved in all substrate species and were identified as those with the most contacts with the IP3K substrate-bound structure (4′- and 5′-phosphates). Orientated with the target hydroxyl pointed down and away from the plane of view, these phosphates are the first two located clock-wise from the target hydroxyl. No IP species with a 2′-phosphate group that was adjacent to the target hydroxyl and pointed in the same direction relative to the plane of the inositol ring was a substrate. A phosphate was also present for all substrates except Ins(4,5)P2 at one of the first two positions immediately counter-clockwise from the target hydroxyl. Three IP4 substrates of A. thaliana IPMK and a non-substrate species are shown in Fig. 2A, the common motif in Fig. 2B. The presence of a phosphate opposite the target hydroxyl does not appear to play any role and was predicted to be solvent-exposed or interact with residues able to adopt accommodating conformations such as lysine. During the course of this work, Chang and Majerus (38) independently proposed a similar theory for H. sapiens IPMK. Their model addresses the 5-/3-kinase activities of human IPMK and does not include a role for the 2′-hydroxyl and does not account for the 6-kinase activity of the yeast or plant homologues, which is severely reduced in the human form.
Putative Binding Modes of Ins(1,4,5)P3, Ins(1,4,5,6)P4, Ins(1,3,4,5)P4, and Ins(1,3,4,6)P4
All known substrate and non-substrate IP species were modeled into the active site of A. thaliana IPMK, following the proposed motif. As expected, the A. thaliana IPMK structure was able to accommodate all substrate species, and no non-substrate species except Ins(1,4)P2 and Ins(1,3,4)P3. These species lacked one of the two phosphate groups conserved in all substrate species. As expected, the ring positions that did not appear to require or exclude phosphate groups proved to be either solvent-exposed or had potential favorable interactions with residues Lys-117, Lys-121, Lys-153, or Arg-156. All possessed highly flexible side chains with minimal adjacent packing, presumably allowing the conformational flexibility required to tolerate the presence or absence of an IP phosphate group. IP species with a 2′-phosphate group were modeled into this presumed active site and were found to have extensive steric clashes. Fig. 2C shows inositol modeled in the proposed 6-kinase orientation in the active site of A. thaliana IPMK, with each phosphate pocket designated α-ϵ located clockwise from the kinase-target hydroxyl κ.
Substrate-selective Design and Product Identification
To test the substrate-selective motif, mutants were designed that were predicted to exclude specific classes of IP species from binding. Mutant A. thaliana IPMK constructs were treated with [γ-32P]ATP and Ins(1,4,5)P3. The K117W mutant was predicted to partially inhibit both the 6′-phosphorylation of Ins(1,3,4,5)P4 and the 3′-phosphorylation of Ins(1,4,5,6)P4 by blocking the γ-pocket, presumably occupied by a (different) phosphate for both IP4 species. Full inhibition was not expected due to the existence of a clash-relieving near-rotamer. K121W was predicted to severely inhibit the 3′-phosphorylation of Ins(1,4,5,6)P4 but have little or lesser effect on the 6′-phosphorylation of Ins(1,3,4,5)P4, as the blocked δ-pocket would not be occupied by a phosphate for the latter IP4 species (Fig. 2D). Treatment of Ins(1,4,5)P3 with [γ-32P]ATP and K117W resulted in IP4 and IP5 formation, with reduced IP5 levels compared with wild-type, whereas K121W generated IP4 and no or trace IP5 (data not shown). Time courses of phosphorylation of [3H]IP3 by wild-type and K121W confirmed the 32P-labeled observations (Fig. 3A). Treatment of the purified [32P]IP4 product from K121W with 5-ptase identified it as Ins(1,4,5,6)P4 (Fig. 3B), as the internal control [3H]Ins(1,3,4,5)P4 was dephosphorylated to Ins(1,3,4)P3; the [32P]IP4 product was not and eluted at a different time point, matching the control [3H]Ins(1,4,5,6)P4 (data not shown).
K117W and K121W Mutants Possess Different Substrate Profiles
Michaelis-Menten steady-state kinetics experiments were performed to determine the changes in mutant specificity. The kinetic parameters Km and Vmax were measured for the following reactions for wild-type IPMK and both selective mutants: Ins(1,4,5)P3 → Ins(1,4,5,6)P4 (6-kinase), Ins(1,4,5,6)P4 → Ins(1,3,4,5,6)P5 (3-kinase), and Ins(1,3,4,5)P4 → Ins(1,3,4,5,6)P5 (6-kinase) (Fig. 3E and Table 2). The following pattern in kcat/Km was predicted based on the number of blocked sites for each combination of mutant and IP4 species, the presence of adjacent phosphate groups that would restrict the ability of the blocked phosphate to adopt a relieving conformation, and the possibility that alternate residue conformations existed to reduce clash: WT ≥ K121W + IP3 > K117W + Ins(1,3,4,5)P4 > K121W + Ins(1,3,4,5)P4 > K117W + Ins(1,4,5,6)P4 > K121W + Ins(1,4,5,6)P4. This pattern was observed, supporting the inositol selectivity hypothesis. If loss of activity were due to a general inhibition of IP binding such as steric exclusion of the inositol ring, similar losses of activity would have been expected for all IP species for a given mutant. The conservation of the 1′- and 4′-phosphates in two conserved pockets proposed by Ongusaha (39) and reviewed by Irvine (3) is not consistent with the single active site nor the pattern of activity loss.
TABLE 2.
Kinetics
| IP Species | Wild-type | K117W | K121W |
|---|---|---|---|
| I(1,4,5)P3 | |||
| Vmax (μmol/min/mg) | 8.96 | NPa | 3.52 |
| Km (μm) | 162.2 | 50.05 | |
| kcat/Km (s−1 m−1) | 2.97 × 104 | 3.78 × 104 | |
| I(1,3,4,5)P4 | |||
| Vmax (μmol/min/mg) | 6.06 | 6.11 | 6.61 |
| Km (μm) | 30.82 | 568 | 490 |
| kcat/Km (s−1 m−1) | 1.06 × 105 | 5.79 × 103 | 7.27 × 103 |
| I(1,4,5,6)P4 | |||
| Vmax (μmol/min/mg) | 11.04 | 0.96 | 0.39 |
| Km (μm) | 74.97 | 254 | 216 |
| kcat/Km (s−1 m−1) | 7.91 × 104 | 2.04 × 103 | 9.73 × 102 |
a NP, not performed.
In Vivo Different Inositol Phosphate Populations Are Altered by K121W and K117W/K121W
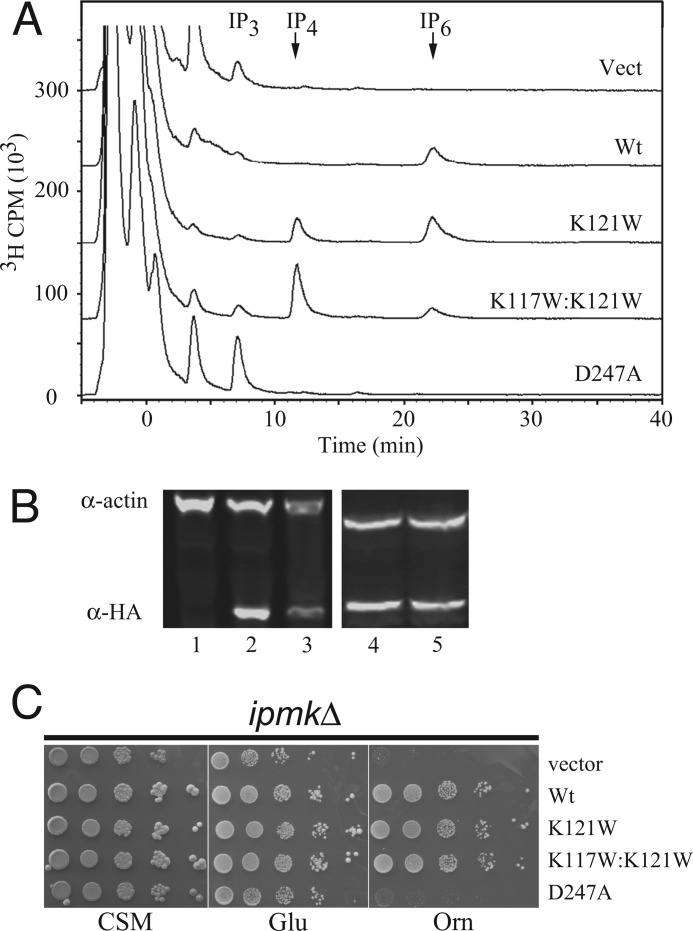
To determine whether changes in the in vitro selectivity of the mutants affected in vivo IP production, the inositol profiles of ipk2Δ S. cerevisiae with various A. thaliana IPMK constructs were measured by HPLC (Fig. 4A). In addition to the K121W and K117W/K121W double-mutants, empty vector, wild-type, and D247A kinase-dead constructs were also used. Similar levels of Ins, IP1, and IP2 were measured for all constructs. IP3 levels were highest in the control and kinase-dead constructs, and progressively less in double mutant, K121W, and wild-type (negligible). Slightly elevated levels of IP3 observed in D247A mutant as compared with vector were not found to be statistically significant. IP4 levels were negligible in all constructs except the K121W and K117W/K121W mutants, approaching the control and kinase-dead IP3 levels. IP6 (produced from IP5 by IPK1(40)) levels were equivalent in K121W and wild-type but greatly reduced in the double mutant. The IP4 to IP6 ratios changed dramatically with the different selective mutants; although essentially, no IP4 is present with wild-type IPMK, levels of IP4 and IP6 are equivalent in K121W, and IP4 is 6-fold higher or greater than IP6 in K117W/K121W.
FIGURE 4.
In vivo inositol phosphate production and rescue of ipmkΔ. A, HPLC traces of soluble inositol phosphates. HPLC traces of soluble [3H]IP species from ipmkΔ transformed with indicated A. thaliana IPMK constructs. B, Western blot of IPMK construct expression. Soluble A. thaliana IPMK expression detected with α-HA and normalized to α-actin. K117W/K121W and D247A blots are from a second series. C, growth on ornithine. 10-Fold serial dilution spot assays on CSM-Ura, Glu-Ura, or Orn-Ura media. Vect, vector.
Mutants Have Different Rescue of Nitrogen Growth Defects Than Wild-type
Rescue of the ipmkΔ ornithine growth defect was compared between wild-type A. thaliana IPMK and the selective constructs (Fig. 4C). Yeast, plant, and fly IPMK have previously been shown to fully rescue growth with ornithine as the sole source of nitrogen (12, 14). Growth on CSM and glutamate media was unaffected by the deletion of IPMK but was lost when plated onto media containing ornithine as the sole nitrogen source. Both IPMK mutants were able to fully rescue growth on ornithine, with similar colony number and size as wild-type A. thaliana IPMK, whereas the catalytic knock-out D247A mutant failed to rescue.
Nitrogen Metabolism Control Does Not Appear to Require a Polyaspartate Region
It has been proposed that the polyaspartate region of S. cerevisiae IPMK, rather than the kinase functionality, is responsible for rescue of ornithine growth (41). Although kinase-dead mutants of yeast IPMK were reported to partially rescue growth on ornithine media, we observed that growth could also be rescued by plant and fly IPMK. Not only does A. thaliana IPMK clearly lack the polyaspartate insert or any sequence similarity to it (supplemental Fig. S3A), but the structure reveals that short of a complete unfolding of strands β9 and β10, there is no way a similar conformation could be adopted. Although residues 231–236 are disordered, any possible loop would be far too small to imitate the size of the 28-residue disordered region in yeast that includes the polyaspartate insert. Furthermore, the polyaspartate region in yeast begins pointed in the opposite direction of the beginning of the disordered region in plant (supplemental Fig. S3B). With only six residues, the plant disordered region would be unable to imitate even the residues of the yeast insert that are observable, much less the disordered region.
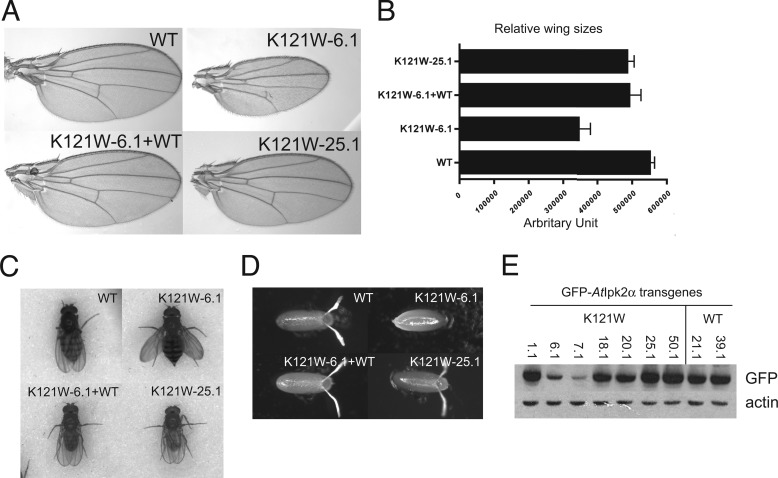
K121W Flies Have Rescued Growth but Exhibit Expression-dependent Defects in Wing Morphology
ipmkΔ flies with transgenes for the K121W A. thaliana IPMK construct are able to mature, unlike ipmkΔ flies, which fail to progress beyond the larval stage. However, at lower expression levels, defects are observed in wing morphology, size, and posture (Fig. 5, A–C). Wing veins appear to be malformed, and the average wing size is reduced, whereas mature flies are unable to fully fold their wings along their back. At the L3 stage of development, wing discs are not observed for the K121W-6.1 transgene construct but are observed for the wild-type, wild-type plus K121W-6.1, and K121W-25.1 construct, which is a stronger promoter. Western blots confirm that expression of K121W is significantly higher in the K121W-25.1 construct (Fig. 5E).
FIGURE 5.
Fly complementation analysis and wing morphology. A and B, representative wings and size distributions. Mean size ± S.D., n ≥ 19. C and D, wing postures and eggs laid by indicated genotypes. WT, WT + K121W, ipmkΔ + K121W-6.1, and ipmkΔ + K121W-25.1. E, Western blot of GFP-A. thaliana IPMK expression. Expression of various GFP-A. thaliana IPMK transgenes in D. melanogaster L3 larvae using α-GFP and α-actin as loading controls.
DISCUSSION
Our work reveals the structure of plant IPMK, a basis of its substrate selectivity, and, through selective mutants, a linkage between specific IP species and phenotypes in yeast and flies. IPMK discriminates between the myriad different IP species through a combination of steric exclusion and requiring the occupation of specific phosphate binding pockets. It remains unclear whether or not there are accessory factors or post-translational modifications that regulate IPMK 6-, 3-, or 5-kinase activities. However, recombinant protein produced in bacteria appears to perform phosphorylation of a multitude of substrates depending on their availability in kinetically similar fashion. With respect to proposed lipid kinase activity of mammalian IPMK, the structure of the plant enzyme and modeling of the active site suggests that 3-kinase activity toward phosphatidylinositol 4,5-bisphosphate may be prevented or greatly reduced by depth of the active site and steric incompatibilities. Indeed, we have been unsuccessful in measuring lipid kinase activity for recombinant A. thaliana IPMK in vitro or in vivo (data not shown).
The ability to alter the kinetic profiles of IPMK through single- and double-residue substitution confirms insights gleaned from substrate modeling. The restriction of the specificity of A. thaliana IPMK through mutations to both Lys-117 and Lys-121 also allowed us to rewire inositol signaling pathways in two model systems. The combination mutant K117W/K121W achieved the best outcome and harbored normal Ins(1,4,5)P3 6-kinase, but greatly reduced Ins(1,4,5,6)P4 3-kinase activity. Our phenotypic studies in yeast indicate that the utilization of ornithine as a sole nitrogen source requires 6-kinase activity and that greatly reduced 3-kinase activity does not impair cellular responses.
Remarkably, we were also able to use a synthetic biology approach to probe inositol signaling specificity in the fruit fly D. melanogaster. Our recent work has defined an essential role for IPMK in fly development, proliferation, and cell death pathways.4 Interestingly, the low level expression of K121W A. thaliana IPMK mutant (promoter 6.1) is able to rescue the viability and imaginal disc development of ipmk null mutants; however, we noticed that proper wing patterning and position in adult flies remained perturbed, whereas high copy expression of the mutant (promoter 25.1) fully rescued viability and wing development. Our in vitro kinetics and yeast in vivo metabolic labeling of the K121W mutant indicates IP3 → IP4 production is normal, but conversion to IP5 is reduced but not abolished. We favor a model in which 6-kinase activity is critical; however, we cannot rule out that the residual 3-kinase, and possible untested 5-kinase, activities also contribute to the IMPK function in fly development. Further optimization of selective IPMK designs and testing of the K117W/K121W mutant in flies may help resolve these hypotheses in the fly system.
Our structure also provided suggestions that non-catalytic regions of IPMK may participate in cellular signaling and regulation. Of interest, the polyaspartate loop of S. cerevisiae IPMK, which was disordered in the crystal structure, was not present in A. thaliana IPMK. We also found that the loop of six residues between the β9 and β10 strands was disordered. Given that the plant protein does not harbor a polyaspartate region, the fact that the protein is fully capable of restoring growth with ornithine as the sole nitrogen source contradicts previous reports that the polyaspartate region is required for transcriptional responses mediated by IMPK (41). The fact that the rescue of growth by the plant enzyme depends on catalysis further emphasizes a role for IPMK and production of Ins(1,4,5,6)P4 in regulating transcriptional responses through the ArgR-Mcm1 complex. We also note that recent reports of the structural basis for Ins(1,4,5,6)P4 in binding to and regulating the histone deacetylase/co-repressor complex (HDAC3-NCOR2) provides an exciting new target for inositol signaling through IPMK (42).
Overall, our study provides insights into both the structure and biology of IPMK through a synthetic biology approach. Insights into substrate binding, kinase specificity and non-catalytic regions of IMPK have been used to answer unique biological questions in two model organisms, budding yeast and fruit flies. These have helped resolve relevance of signaling molecules and provide a foundation for future studies aimed at understanding signaling specificity and biology.
Supplementary Material
Acknowledgments
We thank the members of the York laboratory for helpful discussions, in particular Dr. D. Eric Dollins for discussions during phase determination, model building, and kinetic assays; Dr. Jim Otto and Peter Fridy for HPLC analysis, and Ace Hatch and Trang Pham for protocols and advice. We thank Drs. Jane and David Richardson and Dr. Bruce R. Donald (R01-GM-078031) for helpful support. We finally thank the x-ray crystallographic structure facility at Duke University and the Advanced Photon Source at Argonne National Laboratories and Advanced Light Source (Berkeley) where diffraction data were collected.
This work was supported by the Howard Hughes Medical Institute and National Institutes of Health Grant RO1 HL-55672 (both to J. D. Y.).

This article contains supplemental Figs. S1–S3.
A. M. Seeds, M. M. Tsui, C. Sunu, E. P. Spana, and J. D. York, submitted for publication.
- IP
- inositol phosphate
- IP4
- tetrakisphosphate
- IP5
- inositol pentakisphosphate
- IPMK
- inositol phosphate multikinase
- FOM
- figure of merit
- SAD
- single anomalous dispersion
- CSM
- complete synthetic medium.
REFERENCES
- 1. Drin G., Scarlata S. (2007) Stimulation of phospholipase Cβ by membrane interactions, interdomain movement, and G protein binding–how many ways can you activate an enzyme? Cell Signal. 19, 1383–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gonzales M. L., Anderson R. A. (2006) Nuclear phosphoinositide kinases and inositol phospholipids. J. Cell Biochem. 97, 252–260 [DOI] [PubMed] [Google Scholar]
- 3. Irvine R. F. (2005) Inositide evolution–towards turtle domination? J. Physiol. 566, 295–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Blazer-Yost B. L., Nofziger C. (2005) Phosphoinositide lipid second messengers: New paradigms for transepithelial signal transduction. Pflugers Arch. 450, 75–82 [DOI] [PubMed] [Google Scholar]
- 5. Hatch A. J., York J. D. (2010) SnapShot: Inositol phosphates. Cell 143, 1030–1030.e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. York J. D. (2006) Regulation of nuclear processes by inositol polyphosphates. Biochim. Biophys. Acta 1761, 552–559 [DOI] [PubMed] [Google Scholar]
- 7. Seeds A. M., Bastidas R. J., York J. D. (2005) Molecular definition of a novel inositol polyphosphate metabolic pathway initiated by inositol 1,4,5-trisphosphate 3-kinase activity in Saccharomyces cerevisiae. J. Biol. Chem. 280, 27654–27661 [DOI] [PubMed] [Google Scholar]
- 8. Bechet J., Greenson M., Wiame J. M. (1970) Mutations affecting the repressibility of arginine biosynthetic enzymes in Saccharomyces cerevisiae. Eur. J. Biochem. 12, 31–39 [DOI] [PubMed] [Google Scholar]
- 9. Dubois E., Messenguy F. (1994) Pleiotropic function of ArgRIIIp (Arg82p), one of the regulators of arginine metabolism in Saccharomyces cerevisiae. Role in expression of cell-type-specific genes. Mol. Gen. Genet. 243, 315–324 [DOI] [PubMed] [Google Scholar]
- 10. Messenguy F., Dubois E., Boonchird C. (1991) Determination of the DNA-binding sequences of ARGR proteins to arginine anabolic and catabolic promoters. Mol. Cell Biol. 11, 2852–2863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dubois E., Messenguy F. (1985) Isolation and characterization of the yeast ARGRII gene involved in regulating both anabolism and catabolism of arginine. Mol. Gen. Genet. 198, 283–289 [DOI] [PubMed] [Google Scholar]
- 12. Odom A. R., Stahlberg A., Wente S. R., York J. D. (2000) A role for nuclear inositol 1,4,5-trisphosphate kinase in transcriptional control. Science 287, 2026–2029 [DOI] [PubMed] [Google Scholar]
- 13. York J. D., Odom A. R., Murphy R., Ives E. B., Wente S. R. (1999) A phospholipase C-dependent inositol polyphosphate kinase pathway required for efficient messenger RNA export. Science 285, 96–100 [DOI] [PubMed] [Google Scholar]
- 14. Stevenson-Paulik J., Odom A. R., York J. D. (2002) Molecular and biochemical characterization of two plant inositol polyphosphate 6-/3-/5-kinases. J. Biol. Chem. 277, 42711–42718 [DOI] [PubMed] [Google Scholar]
- 15. Stevenson-Paulik J., Bastidas R. J., Chiou S. T., Frye R. A., York J. D. (2005) Generation of phytate-free seeds in Arabidopsis through disruption of inositol polyphosphate kinases. Proc. Natl. Acad. Sci. U.S.A. 102, 12612–12617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Holmes W., Jogl G. (2006) Crystal structure of inositol phosphate multikinase 2 and implications for substrate specificity. J. Biol. Chem. 281, 38109–38116 [DOI] [PubMed] [Google Scholar]
- 17. Van Duyne G. D., Standaert R. F., Karplus P. A., Schreiber S. L., Clardy J. (1993) XDS atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol. 229, 105–124 [DOI] [PubMed] [Google Scholar]
- 18. Kabsch W. (1993) XDS automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 26, 795–800 [Google Scholar]
- 19. Sheldrick G. M. (1997) SHELX97 Manual, University of Gottingen, Germany [Google Scholar]
- 20. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 21. DeLano W. L. (2010) The PyMOL Molecular Graphics System, version 1.3r1, Schrödinger, LLC, New York [Google Scholar]
- 22. Chamberlain P. P., Sandberg M. L., Sauer K., Cooke M. P., Lesley S. A., Spraggon G. (2005) Structural insights into enzyme regulation for inositol 1,4,5-trisphosphate 3-kinase B. Biochemistry 44, 14486–14493 [DOI] [PubMed] [Google Scholar]
- 23. Miller G. J., Hurley J. H. (2004) Crystal structure of the catalytic core of inositol 1,4,5-trisphosphate 3-kinase. Mol. Cell 15, 703–711 [DOI] [PubMed] [Google Scholar]
- 24. González B., Schell M. J., Letcher A. J., Veprintsev D. B., Irvine R. F., Williams R. L. (2004) Structure of a human inositol 1,4,5-trisphosphate 3-kinase: Substrate binding reveals why it is not a phosphoinositide 3-kinase. Mol. Cell 15, 689–701 [DOI] [PubMed] [Google Scholar]
- 25. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 26. Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 27. Emsley P., Cowtan K. (2004) Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 28. Lovell S. C., Davis I. W., Arendall W. B., 3rd, de Bakker P. I., Word J. M., Prisant M. G., Richardson J. S., Richardson D. C. (2003) Structure validation by Cα geometry: φ, ψ, and Cβ deviation. Proteins 50, 437–450 [DOI] [PubMed] [Google Scholar]
- 29. Winn M. D., Isupov M. N., Murshudov G. N. (2001) Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D Biol. Crystallogr. 57, 122–133 [DOI] [PubMed] [Google Scholar]
- 30. Davis I. W., Murray L. W., Richardson J. S., Richardson D. C. (2004) MOLPROBITY: Structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 32, W615–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kleywegt G. J. (1996) Use of non-crystallographic symmetry in protein structure refinement. Acta Crystallogr. D Biol. Crystallogr. 52, 842–857 [DOI] [PubMed] [Google Scholar]
- 32. Seeds A. M., Sandquist J. C., Spana E. P., York J. D. (2004) A molecular basis for inositol polyphosphate synthesis in Drosophila melanogaster. J. Biol. Chem. 279, 47222–47232 [DOI] [PubMed] [Google Scholar]
- 33. Ito H., Fukuda Y., Murata K., Kimura A. (1983) Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153, 163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stolz L. E., Huynh C. V., Thorner J., York J. D. (1998) Identification and characterization of an essential family of inositol polyphosphate 5-phosphatases (INP51, INP52, and INP53 gene products) in the yeast Saccharomyces cerevisiae. Genetics 148, 1715–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Matthews B. W. (1968) Solvent content of protein crystals. J. Mol. Biol. 33, 491–497 [DOI] [PubMed] [Google Scholar]
- 36. Murzin A. G. (1996) Structural classification of proteins: New superfamilies. Curr. Opin. Struct. Biol. 6, 386–394 [DOI] [PubMed] [Google Scholar]
- 37. Fan C., Moews P. C., Shi Y., Walsh C. T., Knox J. R. (1995) A common fold for peptide synthetases cleaving ATP to ADP: Glutathione synthetase and d-alanine:d-alanine ligase of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 92, 1172–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chang S. C., Majerus P. W. (2006) Inositol polyphosphate multikinase regulates inositol 1,4,5,6-tetrakisphosphate. Biochem. Biophys. Res. Commun. 339, 209–216 [DOI] [PubMed] [Google Scholar]
- 39. Ongusaha P. P., Hughes P. J., Davey J., Michell R. H. (1998) Inositol hexakisphosphate in Schizosaccharomyces pombe: Synthesis from Ins(1,4,5)P3 and osmotic regulation. Biochem. J. 335, 671–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ives E. B., Nichols J., Wente S. R., York J. D. (2000) Biochemical and functional characterization of inositol 1,3,4,5,6-pentakisphosphate 2-kinases. J. Biol. Chem. 275, 36575–36583 [DOI] [PubMed] [Google Scholar]
- 41. Dubois E., Dewaste V., Erneux C., Messenguy F. (2000) Inositol polyphosphate kinase activity of Arg82/ArgRIII is not required for the regulation of the arginine metabolism in yeast. FEBS Lett. 486, 300–304 [DOI] [PubMed] [Google Scholar]
- 42. Watson P. J., Fairall L., Santos G. M., Schwabe J. W. (2012) Structure of HDAC3 bound to co-repressor and inositol tetraphosphate. Nature 481, 335–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.