Background: The periplasmic lipoprotein SpaI protects the subtilin-producing Bacillus subtilis against its own lantibiotic by an unknown mechanism.
Results: The first structure of a lantibiotic immunity protein, SpaI, reveals a novel fold and its membrane-interacting regions.
Conclusion: The membrane interaction is important for SpaI-mediated immunity.
Significance: The SpaI structure will help to understand the immunity of B. subtilis against subtilin on a structural level.
Keywords: Antibiotic Resistance, Bacillus, Lipoprotein, Nuclear Magnetic Resonance, Protein Structure, LanI Protein, Lantibiotic, Lantibiotic Immunity, SpaI, Subtilin
Abstract
Lantibiotics are peptide-derived antibiotics that inhibit the growth of Gram-positive bacteria via interactions with lipid II and lipid II-dependent pore formation in the bacterial membrane. Due to their general mode of action the Gram-positive producer strains need to express immunity proteins (LanI proteins) for protection against their own lantibiotics. Little is known about the immunity mechanism protecting the producer strain against its own lantibiotic on the molecular level. So far, no structures have been reported for any LanI protein. We solved the structure of SpaI, a LanI protein from the subtilin producing strain Bacillus subtilis ATCC 6633. SpaI is a 16.8-kDa lipoprotein that is attached to the outside of the cytoplasmic membrane via a covalent diacylglycerol anchor. SpaI together with the ABC transporter SpaFEG protects the B. subtilis membrane from subtilin insertion. The solution-NMR structure of a 15-kDa biologically active C-terminal fragment reveals a novel fold. We also demonstrate that the first 20 N-terminal amino acids not present in this C-terminal fragment are unstructured in solution and are required for interactions with lipid membranes. Additionally, growth tests reveal that these 20 N-terminal residues are important for the immunity mediated by SpaI but most likely are not part of a possible subtilin binding site. Our findings are the first step on the way of understanding the immunity mechanism of B. subtilis in particular and of other lantibiotic producing strains in general.
Introduction
A number of Gram-positive bacteria including Bacillus subtilis produce a class of bacteriocins called “lantibiotics” (1). They are highly active against Gram-positive bacteria including clinically challenging pathogens such as methicillin-resistant Staphylococcus aureus (2, 3). Lantibiotics are small peptide antibiotics containing characteristic lanthionine and methyllanthionine bridges (Fig. 1A). The biosynthesis of lantibiotics starts with ribosomally synthesized prepeptides. The lanthionine and methyllanthionine bridges are introduced post-translationally by dehydration of serine and threonine residues followed by cyclization reactions with cysteine residues mediated by a membrane-bound multimeric protein complex, which consists of a dehydratase, LanB, and a cyclase, LanC (4). The modified prepeptide is then transported through the membrane whereupon the leader peptide is removed (5–8).
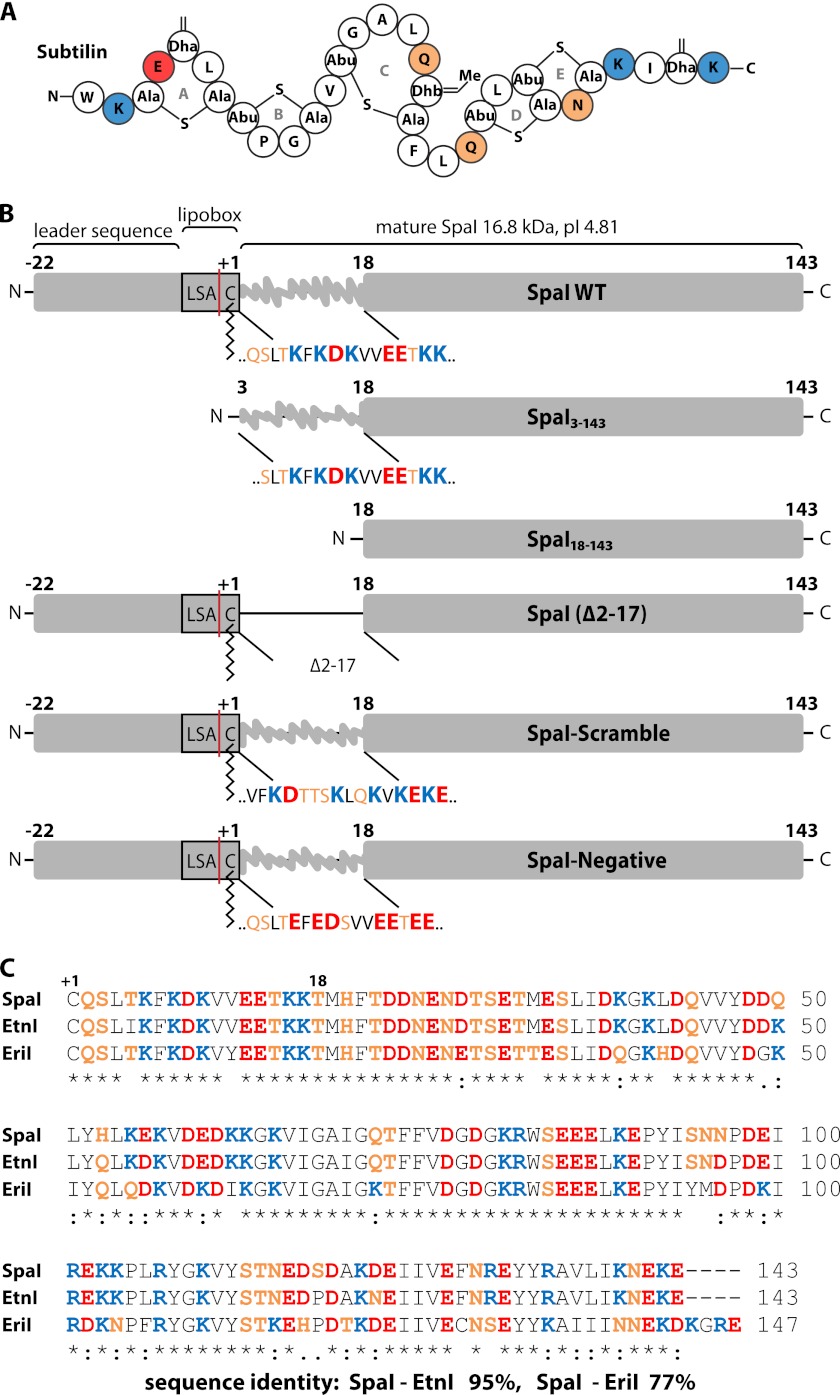
FIGURE 1.
Structure of subtilin and overview of SpaI variants used in this study. A, subtilin is a 32-residue type A(I) lantibiotic containing five rings (A–E) closed by lanthionine (Ala-S-Ala) or methyl-lanthionine bridges (Abu-S-Ala). Didehydroalanine (Dha) and didehydrobutyrin (Dhb) are dehydrated serine and threonine residues, respectively. Abu stands for 1-α-aminobutyric acid and the single letter code is used for standard amino acids. Positively charged amino acids are colored in blue, negatively charged amino acids in red, hydrophilic amino acids in orange, and hydrophobic amino acids in white, respectively. B, schematic representation of WT SpaI and SpaI variants used in this study. The proposed cleavage site for the leader sequence following export across the cytoplasmic membrane is indicated by a red line. As a lipoprotein SpaI contains a so called “lipobox” with the sequence LSAC where a diacylglyceryl moiety is attached covalently to the cysteine as indicated. The sequence of the highly basic N-terminal region directly succeeding the lipobox is highlighted with the color coding as in A. This sequence was varied in the different SpaI variants used in this study as shown. C, sequence alignment (ClustalW2 (72)) of the three homologues LanI proteins, SpaI, EtnI, and EriI, starting at the lipid anchored cysteine and color coded as in A.
Lantibiotics of the nisin group (e.g. nisin (9, 10), subtilin (11), ericin S (12), and entianin (13)) share the same elongated structure with five (methyl-) lanthionine bridges and they are characterized by a dual mode of action (14). On one hand they bind to (15) and sequester the essential cell wall precursor lipid II thereby displacing it from its functional locations and inhibiting cell wall synthesis as demonstrated for nisin (16). On the other hand, in complex with lipid II, lantibiotics of the nisin group are able to form pores in the membrane causing a breakdown of the membrane potential (17–19). The solution NMR structure of the nisin-lipid II complex revealed that the two N-terminal lanthionine rings bind to the phosphate moiety of lipid II (20). This so-called phosphate cage binding motif has also been proposed for other nisin group members. In particular, subtilin binding to pyrophosphate moieties was demonstrated by solid-state NMR (19). As shown for nisin, pore formation occurs when the lipid II-bound C terminus of the lantibiotic inserts into the membrane and the lipid II-nisin complex oligomerizes forming a pore consisting of eight lantibiotic and four lipid II molecules (21, 22).
Lantibiotic producing bacteria must protect themselves from the bacteriocidic effects of their own lantibiotics (23, 24). How they achieve this immunity is poorly understood both on a structural and functional level. Although the relevant players have been identified, no structure of any of the proteins involved in lantibiotic immunity has been solved. Subtilin producing B. subtilis and nisin producing Lactococcus lactis strains, for example, possess four immunity genes as an integral part of the subtilin and nisin biosynthetic gene clusters (25–27). In the case of B. subtilis, these four genes encode SpaI as well as the three subunits of the ABC transporter SpaFEG (28). The N-terminal leader sequence of the lipoprotein SpaI marks this protein for extracellular export. After transport across the cytoplasmic membrane SpaI is anchored in the membrane via a diacylglyceryl moiety covalently attached to a cysteine residue of its own lipobox motif (Fig. 1B) (29). Interestingly, the expression of SpaI alone is sufficient to confer subtilin immunity to B. subtilis. The expression of SpaFEG leads to a higher level of immunity than expression of SpaI alone. However, the strongest subtilin immunity is observed upon simultaneous expression of all four genes (28). Another well described example of proteins involved in lantibiotic immunity are the L. lactis proteins NisI, the equivalent to SpaI, and the ABC transporter NisFEG, which confer resistance to the lantibiotic nisin (27, 30–32). Quantitative in vitro peptide release suggested that SpaFEG as well as NisFEG actively expel subtilin and nisin, respectively, into the surrounding medium. Thus, less subtilin or nisin was found in the membrane fraction when the transporter SpaFEG or NisFEG was expressed compared with the expression of the LanI protein alone (28, 32). The exact mechanism of how LanI proteins confer immunity is currently unclear. However, a direct interaction with the lantibiotic appears likely. An interaction between lipid-free NisI and nisin with an affinity in the micromolar range has been demonstrated (33). Furthermore, hexahistidine-mediated cross-linking of His6-tagged SpaI with subtilin indicated a direct interaction between SpaI and subtilin but not between SpaI and nisin in agreement with a high specificity of the involved LanI proteins (28). This is interesting, because subtilin and nisin not only share a very similar overall structure but also a sequence homology of more than 60% (34, 35). However, the observed absence of SpaI and NisI mediated cross-immunity is in line with the very weak sequence homology (23) of these two LanI proteins and their very different molecular sizes.
Currently, four mechanisms for SpaI-mediated immunity can be discussed: (a) binding and sequestering of subtilin and thereby protecting the membrane from subtilin insertion, (b) preventing subtilin oligomerization prior to pore formation, (c) preventing the formation of the subtilin-lipid II complex prior to pore formation, or (d) acting as substrate-binding protein for the ABC transporter SpaFEG. SpaI as well as other LanI proteins do not show any sequence homology to proteins of known function or structure. Thus, an understanding of the immunity mechanism for the nisin group lantibiotics in general and the specificity and mode of action of the immunity protein SpaI in particular will benefit from structural insights at the molecular level.
To this end, we present here the solution NMR structure of the first LanI protein, SpaI, which represents a novel three-dimensional fold. In vivo studies and NMR titration experiments with liposomes reveal determinants for the membrane interaction of SpaI and indicate a functional importance for this interaction. Our results are the first step on the way to understand subtilin immunity of B. subtilis on a structural level at atomic resolution.
EXPERIMENTAL PROCEDURES
Production and Purification of SpaI NMR Constructs
The coding sequences for all SpaI NMR constructs (SpaI3–143, SpaI18–143, SpaI28–143) were inserted into a modified pQE9 vector containing an N-terminal His6 tag followed by a TEV cleavage site. After TEV cleavage the expressed proteins contained two artificial residues at the N terminus, Gly and Ser for SpaI18–143 and SpaI28–143 and Gly and Arg for SpaI3–143 (the numbering scheme is referring to the wild type SpaI sequence, starting at the lipidated cysteine (Fig. 1B and supplemental Table S1).
Expression and purification of all isotopically labeled SpaI constructs for NMR experiments was essentially done as described previously (36). In brief, expression of isotopically labeled SpaI was carried out using standard methods overnight at 37 °C after induction with 1 mm isopropyl 1-thio-β-d-galactopyranoside. The His-tagged SpaI was first purified over a HisTrap HP column (GE Healthcare). After overnight TEV cleavage the His tag and protease were removed via an anion exchange column and SpaI was further purified via gel filtration. Selectively labeled [15N]lysine (250 mg/liter), [13C]threonine (230 mg/liter) SpaI3–143 was produced as described before (36).
Preparation of Liposomes
1,2-Dioleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (DOPG),3 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), and cardiolipin (CL) were purchased from Avanti Polar Lipids and mixed in chloroform in a 3:2:3 molar ratio. The solvent was removed by rotary evaporation overnight and the lipid film was hydrated in NMR buffer. Liposomes were formed by 5 to 6 freeze-thaw cycles.
NMR Spectroscopy
All NMR samples were prepared in 50 mm sodium phosphate buffer, 100 mm NaCl, pH 6.4, and 10% (v/v) D2O at SpaI concentrations of 200–420 μm for assignment and structure determination experiments and 100 μm for liposome titrations. NMR spectra were recorded at 23 °C on a Bruker DRX 600 MHz spectrometer equipped with a room temperature triple resonance probe or Bruker AVANCE 600, 700, 800, 900, and 950 MHz spectrometers equipped with cryogenic triple resonance probes.
The chemical shift assignment of SpaI18–143 was based on the standard set of triple resonance experiments as described elsewhere (36). Chemical shifts are referenced to 2,2-dimethyl-2-silapentane-5-sulfonic acid (37). To identify the 15 additional backbone amide resonances of SpaI3–143 a three-dimensional HNCA and a three-dimensional HNCACB-experiment were recorded on a uniformly 13C,15N-labeled sample (38). A 15N-HSQC and a one-dimensional HNCO spectrum of a selectively [15N]lysine, [13C]threonine-labeled SpaI3–143 sample were used to verify assignments of the five lysine residues and two threonine-lysine pairs in this N-terminal stretch.
{1H},15N-HetNOE and 15N R1 and R2 relaxation rates were recorded for 200 μm 15N-labeled SpaI18–143 at 23 °C at a 600 MHz spectrometer using standard Bruker pulse sequences (39, 40). Peaks were integrated using Bruker TopSpin 2.1 and peak volumes were plotted against relaxation decay times and fitted to f(t) = ae−bt (b = R1 or R2, t = relaxation decay time) using the program OriginLab 8.1 to obtain R1 and R2 relaxation rates, respectively. Relaxation decay times used for determining the longitudinal and transverse relaxation rates were 0.05, 0.1, 0.2, 0.3, 0.4, 0.6, 0.8, 1, 1.5, 2, 2.5, and 3 s and 15.5, 31, 46.6, 62.1, 77.6, 93.1, 108.6, 124.2, 139.7, and 155.3 ms, respectively.
Residual dipolar couplings (RDC) were measured via recording an IPAP-[1H,15N]-HSQC spectrum (41) with 15N-labeled SpaI18–143 in the absence or presence of pf1 phage (15 mg/ml, Hyglos GmbH) alignment media at 900 MHz (42). 1D(N,H) values were extracted where signals could be tracked reliably and peak maxima could be determined unambiguously by using the peak picking function in the program CcpNmr Analysis (43). RDC of 60 residues with a HetNOE value >0.5 were used for analysis using the programs MODULE (44) and CYANA (45).
For direct identification of hydrogen bonds, a standard TROSY three-dimensional HNcaCO experiment and a long range TROSY three-dimensional HNCO experiment (46) with the NC-transfer delay set to 133 ms (32 scans per increment, 64 and 80 complex points in the indirect 15N and 13C dimension, respectively) were recorded on 2H,13C,15N-labeled SpaI28–143.
For PRE measurements 15N-HSQC spectra of the three SpaI18–143 single point mutants S30C, K62C, and S94C were recorded prior to and after reduction of the spin label by the addition of sodium ascorbate to the final concentration of 500 μm. For tracking of signals 15N-HSQC spectra of SpaI18–143 cysteine mutants not containing any spin label were recorded. All spectra were processed using Bruker TopSpin 2.1 and analyzed using the programs CARA (47) and CcpNmr Analysis (43).
Structural Restraints
Peaks in the NOESY spectra used for structure calculation were picked either manually or automatically with the software ATNOS (48) and inspected manually afterward. NOE distance restraints were obtained using the automated NOE assignment and structure calculation protocol available in CYANA (45, 49). Thus, resonances from 15N-NOESY-HSQC, 15N-NOESY-TROSY, 13C-NOESY-HSQC (in H2O, aliphatic carbons), 13C-HSQC-NOESY (in D2O, aliphatic carbons), and 13C-NOESY-HSQC (in H2O, aromatic carbons) spectra were assigned automatically on the basis of the almost complete manual chemical shift assignments (36) and converted into distance restraints. These automatically obtained NOE assignments were visually inspected and chemical shifts were manually corrected in the case of obvious artifacts or incorrect assignments. For all NOESY spectra an assignment of more than 90% of the NOESY cross-peaks was achieved (Table 1). Dihedral angle restraints were derived from chemical shifts of HN, N, CO, Cα, Cβ, and Hα using the Talos+ webserver (50) and used in the structure calculation only for residues with a HetNOE value >0.5 (Table 1). For all proline residues a trans conformation was detected as described elsewhere (36). 15 hydrogen bond restraints identified by analyzing the long range TROSY three-dimensional HNCO were converted into upper limit distance restraints with a HN-CO distance of 2.2 Å and a N-CO distance of 3.2 Å. In the last round of structure calculations, three additional hydrogen bond restraints were added for amide protons that did not exchange against D2O after 48 h and which formed hydrogen bonds in more than 15 of the 20 lowest energy CYANA structures in preliminary structure calculations. 60 RDC restraints for backbone amide groups were included in the structure calculation for residues with well resolved signals and a HetNOE value >0.5.
TABLE 1.
NOE assignment and structure statistics of the NMR structure of SpaI18–143
| NOE assignmenta | ||
| Total number of peaks | 8228 | |
| Assigned peaks | 7708 | 93.7% |
| 13C-HSQC-NOESY (D2O, aliphatic) | 3312 | 95.7% |
| 13C-NOESY-HSQC (H2O, aliphatic) | 2093 | 93.2% |
| 13C-NOESY-HSQC (H2O, aromatic) | 251 | 90.4% |
| 15N-NOESY-HSQC | 1468 | 90.8% |
| 15N-NOESY-TROSY | 1104 | 93.1% |
| Unassigned | 520 | 6.3% |
| Restraints | ||
| Total NOE distance restraints | 3246 | |
| Intraresidue |i - j| = 0 | 781 | 24.1% |
| Sequential |i - j| = 1 | 741 | 22.8% |
| Short-range |i - j| ≤ 1 | 1522 | 46.9% |
| Medium-range 1 < |i - j|< 5 | 423 | 13.0% |
| Long-range |i - j| ≥ 5 | 1301 | 40.1% |
| Dihedral angle restraints (Talos+) | 144 | |
| Hydrogen bond restraints (long-range HNCO) | 18 | |
| Residual dipolar couplings (DNH) | 60 | |
| Structure statisticsb | ||
| CYANA target function value after energy minimization (Å2) | 3.45 ± 0.38 | |
| Amber energies total (kcal/mol) | −4873.8 ± 151.48 | |
| Restraint violationsc | ||
| Maximal distance restraint violation (Å) | 0.15 ± 0.01 | |
| Number of violated distance restraints > 0.2 Å | 0 | |
| Maximal dihedral angle violation (°) | 9.37 ± 0.56 | |
| Number of violated dihedral angle constrains > 5° | 1 | |
| Maximal RDC restraint violation (Hz) | 2.28 ± 0.24 | |
| Structure validationd | ||
| Non-trivial restraints per residue | 27.6 | |
| Long range restraints per residue | 10.3 | |
| Ramachandran plote | ||
| Residues in most favored regions | 85.1% | |
| Residues in additionally allowed regions | 14.8% | |
| Residues in generously allowed regions | 0.1% | |
| Residues in disallowed regions | 0.0% | |
| Root mean square deviation (residues 32–89, 102–115, 122–139)e | ||
| Average backbone r.m.s. deviation to mean (Å) | 0.30 | |
| Average heavy atom r.m.s. deviation to mean (Å) | 0.60 | |
Structure Calculation
The first structure calculations were done using only NOE restraints. Structures were checked for agreement with experimentally determined hydrogen bond restraints before their explicit incorporation in later rounds of the structure calculation. The correlation between RDC and structures calculated with additional hydrogen bond restraints was determined with CYANA. The resulting alignment tensor magnitude and rhombicity were used in the final structure calculation with an uncertainty of ±3 Hz for RDC values.
100 conformers were calculated using 20,000 torsion angle dynamics steps and sorted according to their target function values (51). The 20 conformers with the lowest target function, along with the complete restraint dataset, were used as input for the CYANA ”regularized“-macro (52). The single representative structure obtained from this procedure as well as the 19 conformers with the lowest CYANA target function were then subjected to restrained energy refinement with the program OPALp (53), which uses the AMBER94 force field (54). These 20 structures were deposited in the Protein Data Bank under the accession number 2lvl. The structure was validated using Protein Structure Validation Software (PSVS) suite 1.4 (55) by selecting ordered residues with HetNOE values above 0.5 (residues 32–89, 102–115, 122–139). Electrostatic surface potential calculations were prepared with the PDB2PQR webserver (56) using the PARSE force field and visualized with the APBS (57) plug-in for PyMOL (58).
B. subtilis Growth Conditions
B. subtilis cells were grown in Luria-Bertani (LB) medium at 37 °C. For the determination of LanI-mediated immunity, B. subtilis cells were grown in TY medium (0.3 m NaCl, 1% xylose) plus 1% xylose for induction of lanI expression (59). All strains used in this study are listed in supplemental Table S1. Chloramphenicol (5 μg/ml) and erythromycin (1 μg/ml) were used for the selection of B. subtilis transformants.
Determination of LanI-mediated Immunity
B. subtilis BSF 2470 (B2470) strains expressing different lanI variants were used to determine the immunity against the nisin group lantibiotic entianin (13, 59). An overnight culture of the corresponding strain was diluted in TY (0.3 m NaCl, 1% xylose) to a final A600 of 0.1. 200-μl aliquots were transferred into 96-well plates and incubated for 3 h at 37 °C. After addition of different amounts of entianin (in % supernatant of the entianin producing strain B15029) the samples were incubated for 1 h at 37 °C. The A600 was measured with a SpectraMax® M5 MultiMode Microplate Reader (Molecular Devices). The growth after a 1-h incubation with entianin of each culture in every well was normalized as the percent growth to a control without lantibiotic. The strain expressing the wt-spaI gene (B2470.SB3) and the strain expressing no lanI gene (B2470.SD3) were used as positive and negative controls. Protein expression and localization was verified by Western blot analysis.
RESULTS
SpaI Has an Unstructured N Terminus in Aqueous Solution
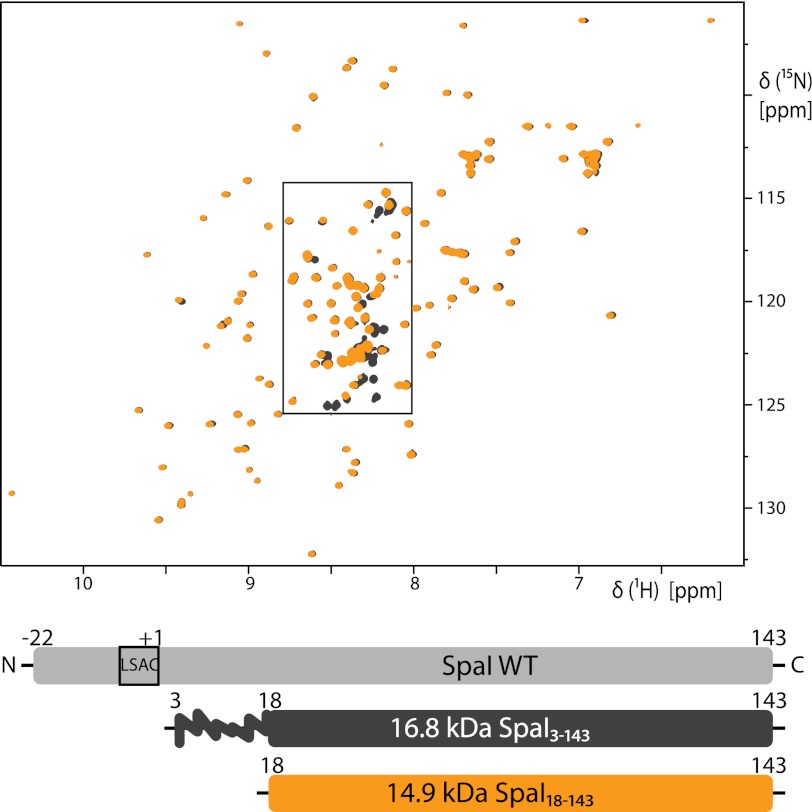
To determine the structure of SpaI with solution NMR spectroscopy we started with a construct lacking the leader sequence and lipobox (SpaI3–143) (Fig. 1B). Unfortunately, under conditions suitable for NMR-based structure determination this SpaI variant was only stable for a limited time and at lower concentrations. A controlled trypsin digest (supplemental Fig. S1) resulted in a stable SpaI fragment with a molecular mass of approximately 15 kDa. Mass spectrometry indicated that this fragment lacked the first 17 N-terminal amino acids following the lipobox (SpaI18–143) (Fig. 1B). The overlay of the 15N-HSQC spectra of these two SpaI variants (Fig. 2) indicates that both have identical structures because the vast majority of NMR signals have the same chemical shifts in both proteins. The NMR signals that correspond to the 15 additional amino acids at the N terminus of SpaI3–143 have chemical shifts and line widths defining them as unstructured in solution (boxed region in Fig. 2). Therefore the shorter variant SpaI18–143 was used for the structure determination in solution due to its superior stability and solubility. For this fragment nearly complete NMR resonance assignments were obtained for the backbone and side chain resonances with standard triple resonance experiments (BMRB accession number 17534 (36)).
FIGURE 2.
SpaI has an unstructured N terminus. An overlay of the [1H,15N]-HSQC spectra of two different soluble SpaI variants (color coded according to the cartoon representation below) is shown. Both lack the leader sequence as well as the amino acids of the lipobox. The two variants adopt identical structures in solution as indicated by their virtually identical spectra. Differences are only found in the center of the spectrum (boxed area, visible black signals). These signals correspond to the first 15 amino acids directly following the lipobox, which are present only in the 16.8-kDa variant. Their chemical shifts indicate that these amino acids are unstructured in aqueous solution. Consequently, for NMR structure determination the 14.9-kDa variant SpaI18–143 (orange) was used.
The Structure and Dynamics of SpaI18–143
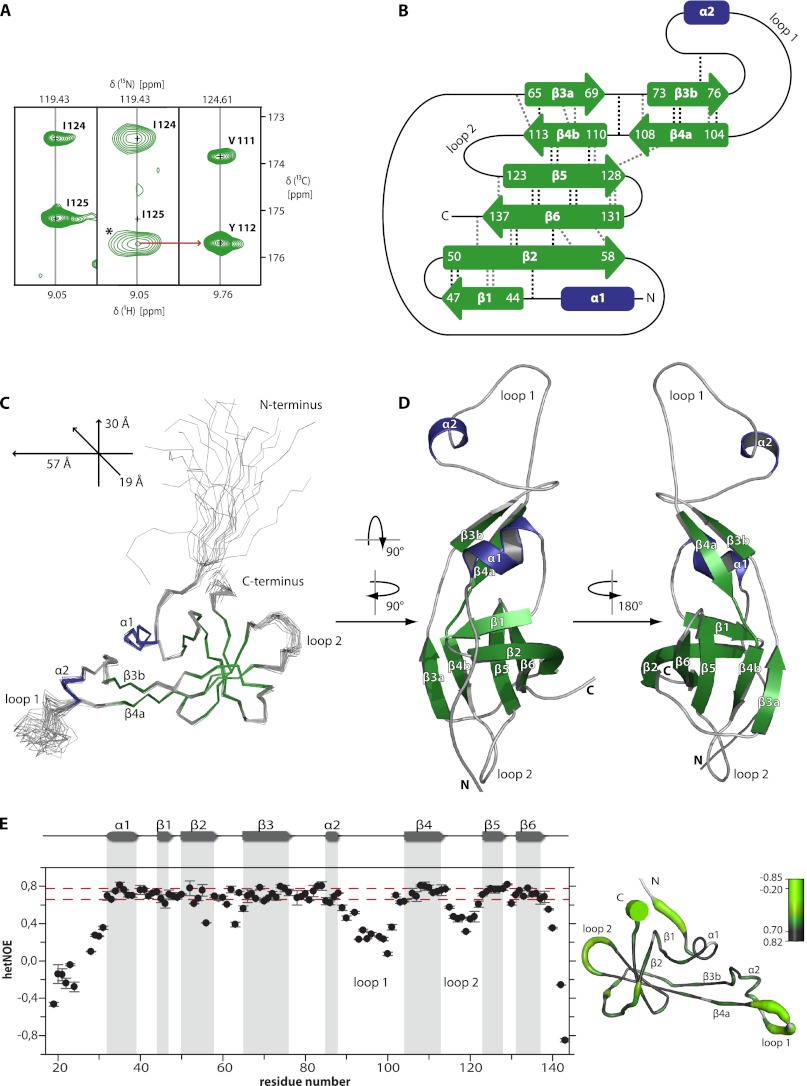
Preliminary structure calculations for SpaI18–143 were based exclusively on NOE and torsion angle restraints. These calculations showed an unusual elongated structure and an unprecedented arrangement of secondary structure elements (see below). Therefore a three-dimensional long range HNCO (46) for the direct detection of hydrogen bonds (Fig. 3A) and experiments for the measurement of residual dipolar couplings for the backbone amides (supplemental Fig. S2) were recorded to complement the conventional NOESY experiments. The analysis of the long range HNCO and a reference HNcaCO experiment (Fig. 3A) allowed the direct identification of 15 hydrogen bonds (Fig. 3B). The presence of three additional hydrogen bonds was deduced because their amide protons did not exchange against D2O after 48 h (supplemental Fig. S3). Accordingly, the final structure calculation included 3246 NOE-derived upper distance restraints, 144 torsion angle restraints from chemical shift information, 18 restraints from the experimentally determined hydrogen bonds, and 60 residual dipolar couplings of backbone amides. The resulting bundle of the 20 lowest energy conformers of SpaI18–143 (Fig. 3C) has a low backbone r.m.s. deviation of 0.3 Å for the rigid parts of the protein and no violated distance restraints larger than 0.2 Å. The statistics for the structure calculation with CYANA (45) are listed in Table 1. The structure has been deposited in the Protein Data Bank under accession number 2lvl wherein the first structure of the bundle represents the energy minimized mean structure.
FIGURE 3.
Solution structure and dynamics of SpaI18–143. A, direct detection of hydrogen bonds by NMR spectroscopy. Shown are the 1H-13C slices of the three-dimensional HNcaCO spectrum (left) and the three-dimensional long-range HNCO spectrum (middle) for residue Ile-125 (hydrogen bond donor) and of the HNcaCO spectrum for Tyr-112 (right, hydrogen bond acceptor). The cross-peak revealing the existence of a hydrogen bond between the amide group of Ile-125 and the backbone carbonyl group of Tyr-112 is marked with an asterisk (*) in the slice from the long-range HNCO experiment (middle). B, NMR-derived secondary structure diagram of SpaI18–143 showing α-helices in blue and β-strands in green. Residue numbers are indicated for β-strands. The black dashed lines show hydrogen bonds in the β-strands detected in the long-range HNCO or in an H/D-exchange experiment. Additional hydrogen bonds found in at least 15 of the 20 final structures are indicated by gray dashed lines. C, backbone trace of the 20 lowest energy conformers after superposition of the Cα atoms of ordered residues representing the solution structure of SpaI, colored as in B. Dimensions of the protein in Å are indicated in the upper left corner. D, ribbon representation of the conformer closest to the mean structure colored as in B. The structure does not show the entire unstructured N terminus. E, {1H},15N-HetNOE values of the backbone amides of SpaI18–143 plotted against the sequence. The secondary structure is shown above and indicated throughout the plots with gray columns. HetNOE values between the red dashed lines indicate the rigid parts of the protein. Flexibility as derived from the HetNOE data is represented by color on the structure of SpaI (residue 25–143) ranging from high HetNOEs and rigid parts of the protein (gray, thin tubes) to low HetNOEs and flexible regions (green, thick tubes). Missing data points in the plot and white thin tubes in the structure indicate prolines or residues for which the HetNOE could not be determined reliably due to resonance overlap.
The structured core of SpaI18–143, which includes amino acids 32–139 (see below), exhibits an overall elongated three-dimensional shape with one long axis of about 57 Å and two short axes of ∼30 and ∼19 Å (axes are indicated in Fig. 3C). SpaI is a mainly β-sheet protein with a core of 6 antiparallel twisted β-strands with the strand order: β1 β2 β6 β5 β4b β3a (Fig. 3B) and an unusual long β-hairpin comprised of strands β3b and β4a flanked by two short α-helices (Fig. 3D). The extended β-hairpin is stabilized by hydrophobic packing interactions with helix α1 as indicated by NOEs between side chain hydrogens of residue Ile-37 of helix α1 to side chain hydrogens of residue Phe-74 and Val-76 of β3b and Leu-106 of β4a. The β-strands β3 and β4 are disrupted by glycine residues that in general are thought to destabilize β-strands. However, a tight hydrogen bonding network that deviates from the canonical β-sheet hydrogen bonding pattern (Fig. 3B) stabilizes these strands. Thus, Gly-68 of β-strand β3a is in a hydrogen bond with the amide hydrogen of Val-111 of β4b. β3a and β3b are interrupted by Gly-71 and β4 is disrupted by Gly-109 dividing this strand in β4a and β4b. Gly-109 is in a hydrogen bond with the amide hydrogen of residue Ile-70 as evidenced by the long range HNCO experiment.
The N terminus of SpaI18–143 (residues 18–32) is unstructured and flexible in solution as indicated by the lack of medium and long-range NOEs for these residues and low heteronuclear NOE values (Fig. 3E). In general, the heteronuclear NOE (HetNOE) values are in good agreement with the secondary structure of SpaI18–143 given that essentially all amides in secondary structure elements have HetNOE values > ∼0.6 (red dashed lines in Fig. 3E). Additionally, the measured R1 and R2 relaxation rates show the same pattern as the heteronuclear NOE values (supplemental Fig. S4). Thus residues 32–139 represent the stable structured core of SpaI in solution. Furthermore, the complete deletion of the flexible N terminus in SpaI28–143 (a construct starting at amino acid 28) does not alter the structure of the folded core region (supplemental Fig. S5). There is not even evidence for transient interactions between N-terminal amino acids 3–27 and the structured core of SpaI because virtually identical chemical shifts and signal line widths are observed for residues 30–143 in all tested SpaI variants (supplemental Fig. S5).
In addition to the flexible N-terminal region, all three relaxation parameters reveal that SpaI18–143 has two flexible internal loops, a large loop 1 between α2 and β4a and a shorter loop 2 between β4b and β5 (Fig. 3, C and E, and supplemental Fig. S4). The other loops between α1/β1, β2/β3a, and β3b/α2 as well as the β-hairpins between β1/β2 and β5/β6 show HetNOE values of ∼0.6 or higher indicating their structural rigidity. The measured rotational correlation time of SpaI18–143 of τC = 11.6 ± 0.9 ns (for backbone amides with HetNOE larger than 0.6) is in good agreement with the τC calculated by the program HYDRONMR (60) from the structure yielding τC = 11.3 ns. In contrast, according to the rule of thumb for globular proteins (τC (ns) = 0.6 × kDa) SpaI18–143 should exhibit a τC of ∼8.9 ns. Thus, the higher τC of SpaI18–143 is in good agreement with its elongated tertiary structure.
Structure Validation Using Paramagnetic Relaxation Enhancement (PREs)
Based on our NMR structure (Fig. 3, C and D), three individual sites for spin label attachment were chosen for the validation of the SpaI18–143 structure (single point mutants S30C, K62C, and S94C, for positions see supplemental Fig. S6B). K62C was selected to probe the relative orientation between loop β2/β3a and β-strands β2, β3a, and β6, whereas S30C is close to the unstructured N terminus and S94C is placed in the large flexible loop 1. 15N-HSQC spectra were recorded prior and after reduction of the spin label to identify signals of amides affected by the presence of the spin label (supplemental Fig. S6A). After visual inspection of the spectra the peaks affected by the presence of the spin label were grouped into four intensity groups and visualized on the calculated structure of SpaI18–143. With the spin label located in position K62C (supplemental Fig. S6) the peaks that broadened out completely are assigned not only to amides close in sequence to the spin-labeled site but also in β-strands β2 and β6 and loop 2. These structure elements are close in space to the spin label as predicted from our structure (distance <16 Å). The results of the other two spin label mutants (S30C and S94C) fit equally well to the structure thereby validating the NMR solution structure of SpaI18–143. As expected for these two spin labels only amino acids close in sequence are affected by the presence of the spin label (data not shown).
SpaI18–143 Defines a Novel Fold Class
To identify possible structural homologs of SpaI a search using the DALI server (61) for identification of structurally similar proteins was performed. However, even the best match has a Z-score of only 2.5, which is only slightly above the threshold of being significant (Z > 2). The two best matches are the structures of a pyruvate carboxylase and a 165-amino acid long hypothetical protein with Z-scores of 2.5 and 2.4, respectively (see supplemental Table S2). However, the pyruvate carboxylase structures 3hbl-D and 3bg5-D fit only with an r.m.s. deviation of 11.7 and 11.2 Å to SpaI18–143, respectively. In addition, only the three β-strands β4b, β6, and β5 of SpaI including only 64 of its 107 Cα-atoms in the structured core region align weakly with three of the β-strands of the pyruvate carboxylase. The hypothetical protein (2dc4-A) has a Z-score of 2.4 but fits with a r.m.s. deviation of 4.5 Å to SpaI18–143 and shows the highest number of aligned residues (78 residues to 107 residues of the structured core of SpaI). This hypothetical protein has a β-barrel structure and aligns with β-strands β3–6 of SpaI18–143 because these twisted antiparallel strands also form a β-barrel-like structure. Even so, the hypothetical protein does not show similarities to the unusual long β-hairpin and the two α-helices. Hence we conclude that SpaI18–143 defines a novel three-dimensional fold.
The SpaI N Terminus Interacts with the Membrane Even in the Absence of a Lipid Anchor
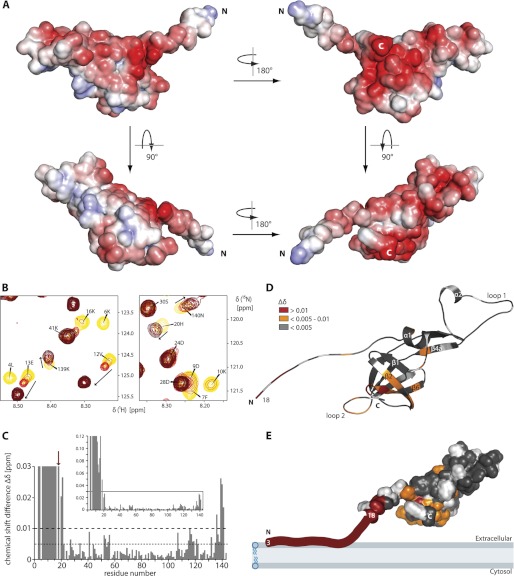
SpaI18–143 is a highly negatively charged protein with 32 negatively and only 20 positively charged amino acids. The electrostatic surface potential map (Fig. 4A) reveals that the negative charges are clustered at the two long sides of SpaI (upper panels in Fig. 4A), whereas the bottom (lower left in Fig. 4A) and top (lower right in Fig. 4A) show positive or hydrophobic amino acids on the surface. In contrast, the unstructured N terminus comprised of amino acids 2–17 is much more basic and contains 5 positively charged lysines and only 3 negatively charged amino acids (Fig. 1B). Furthermore, the N terminus has a sequence with the propensity to form an almost ideal amphipathic α-helix (supplemental Fig. S7) and is predicted to have an α-helical structure by a number of secondary structure prediction programs in contrast to what is observed in aqueous solution. This suggests that the N terminus might be able to interact with negatively charged phospholipid membranes as an amphipathic α-helix. To elucidate whether this highly positively charged N terminus is indeed mediating a membrane interaction of SpaI, the two variants, SpaI3–143 and SpaI18–143, were tested for their interaction with multilamellar liposomes prepared from DOPG, DOPE, and CL (molar ratio 3:2:3) using NMR spectroscopy. The composition and the molar ratios of the three lipids were chosen to represent the major lipids and their ratios found in B. subtilis membranes (62, 63). The shorter variant SpaI18–143, which lacks the basic N terminus, showed no changes in chemical shifts upon titration with these liposomes (supplemental Fig. S8) indicating that this fragment is not interacting with the liposomes. However, for the longer construct, SpaI3–143, significant changes in the appearance of the 15N-HSQC spectrum upon addition of lipids and therefore interaction with the liposomes are observed (Fig. 4B and supplemental Fig. S8). The peaks for residues 3–10 are disappearing completely (Fig. 4, B and C). This is most likely due to a direct and strong interaction with the liposomes, which increases the rotational correlation time of the large complex. In addition, peaks of residues 11–21, 136, and 139–141 show significant chemical shift changes (larger than 0.1 ppm) upon titration with lipids indicating a transient interaction with the large liposomes (Fig. 4, B and C). Plotting chemical shift changes for all residues whose peaks show chemical shift changes larger than 0.05 ppm onto the structure of SpaI18–143 reveals a defined surface (Fig. 4D) affected by the presence of liposomes, which indicates a preferred orientation of SpaI3–143 toward the membrane leading to the model shown in Fig. 4E. In this model the N terminus of SpaI3–143 is directly interacting with the plasma membrane even in the absence of a lipid anchor. This is followed by a flexible linker of ∼13 amino acids. The structured core of SpaI (amino acids 32–139) is oriented with a defined strongly negatively charged surface toward the membrane.
FIGURE 4.
Electrostatic surface potential and membrane interaction mapping of SpaI. A, electrostatic surface potential mapped on the solvent accessible surface of SpaI18–143 in different orientations showing from red (−5 kT/e) to blue (+5 kT/e) negatively to positively charged surface areas, respectively (k = Boltzmann's constant, T = absolute temperature, and e = electron charge). White areas correspond to hydrophobic surfaces. The lower right panel shows the same orientation of the protein as in panels D and E. B, two detailed views of overlays of the [1H,15N]-HSQCs of 0.1 mm SpaI3–143 (yellow) after addition of 0.5 mm (orange), 1.5 mm (light red), 3 mm (red), and 5 mm (dark red) lipids as multilamellar liposomes prepared from DOPG, DOPE, and CL in the molar ratio of 3:2:3. Signals for which chemical shifts could be tracked unambiguously are indicated by arrows. C, histogram showing the chemical shift differences (73) induced in SpaI3–143 (0.1 mm) after the addition of multilamellar liposomes (5 mm). Signals that disappeared completely were set to an arbitrary value of 0.12 (inset). Missing data points indicate prolines or residues whose Δδ could not be traced reliably (colored white in D and E). The inset represents the full plot, whereas the larger plot corresponds to the boxed area of the inset. Δδ values above the dotted or dashed line are colored orange or red in D and E, respectively. The red arrow indicates the position of residue 18, the N terminus of the structures shown in A and D. D, chemical shift differences plotted onto the structure of SpaI18–143 in a ribbon presentation. The orientation of the protein is the same as in the lower right panel of A. E, model of the membrane interaction of SpaI3–143, showing the solvent accessible surface of SpaI18–143 in the same orientation as in the lower right panel of A and the color coding as in D.
Homology Models of the LanI Proteins from Entianin and Ericin S Producing Strains
The subtilin class lantibiotics entianin and ericin produced by B. subtilis DSM 15029T (13) and B. subtilis A1/3 (12), respectively, show high similarity in sequence and structure to subtilin (supplemental Fig. S9). In the case of entianin only three hydrophobic amino acids are exchanged for other hydrophobic amino acids. In analogy, EtnI (the LanI protein of B. subtilis DSM 15029T) and SpaI have a sequence similarity of 95% (Fig. 1C) and SpaI is able to mediate immunity against entianin (Fig. 5). A comparison of the electrostatic surface potential maps of SpaI18–143 and a homology model of EtnI18–143 indicates that both proteins are similarly negatively charged, which is consistent with the fact that subtilin and entianin have the same charge distributions (supplemental Fig. S9). Ericin S differs from subtilin in the same amino acid positions as entianin except for one additional lysine, which is replaced by a histidine making this lantibiotic less positively charged. In addition, the homology model of the corresponding LanI protein EriI18–147 also shows fewer negative charges on one side of the surface compared with SpaI18–143 (supplemental Fig. S9). EriI and SpaI have a sequence homology of 77% (Fig. 1C). However, EriI is still able to mediate immunity against entianin to some extent (data not shown). Interestingly, when comparing the sequences of all three LanI proteins the first 27 N-terminal amino acids of EtnI and EriI differ only in one amino acid from SpaI, whereas all charges are preserved (Fig. 1C). This suggests a conservation of the membrane interaction observed for SpaI in EriI and EtnI.
FIGURE 5.
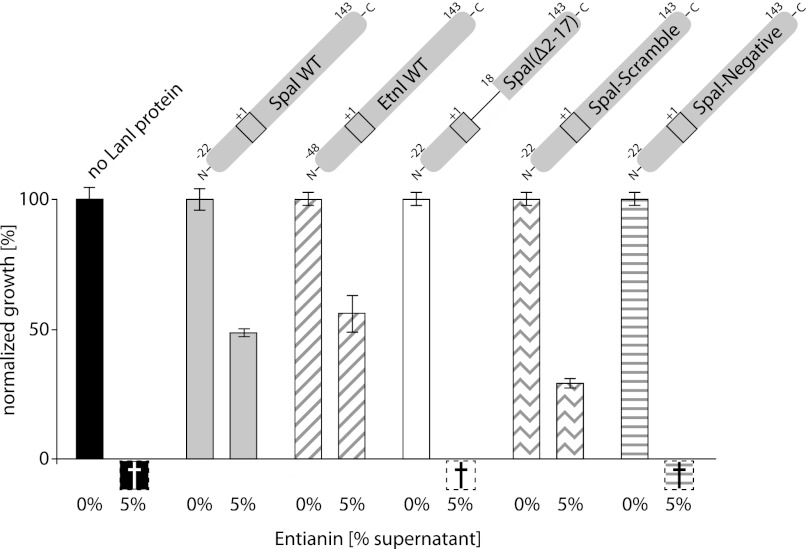
Growth tests of different B. subtilis strains expressing SpaI variants. Percental growth increase after a 1-h incubation without and with entianin (5% supernatant) normalized to the control without lantibiotic. Black bars, strain B2470.SD3 expressing no LanI protein; gray bars, strain B2470.SB3 expressing SpaIWT; fasciated bars, strain B2470.SB8 expressing EtnI WT; white bars, strain B2470.SB2 expressing SpaI(Δ2–17); wavey bars, strain B2470.SB7 expressing SpaI-Scramble; striped bar, strain B2470.SB17 expressing SpaI-Negative. Negative bars indicate loss of optical density presumably due to cell lysis and their height is set to an arbitrary value. The schematics above the bars represent the respective LanI variant.
The Positive Charge and the Length of the N Terminus Is Required for Mediating Immunity but Not Its Sequence
To test the role of the N terminus, which is absent in the construct used for NMR structure determination in mediating immunity in vivo we performed growth tests with B. subtilis strains expressing different variants of SpaI in the presence of increasing concentrations of entianin. Supernatant from B. subtilis DSM 15029T expressing entianin was used for growth tests because of its significantly higher lantibiotic concentration in the supernatant compared with the subtilin expressing strain B. subtilis ATCC 6633. As shown in Fig. 5 (gray and fasciated bar), SpaI and EtnI likewise mediate immunity against entianin, which therefore can be used for the immunity tests instead of the experimentally less accessible subtilin. We have previously shown that there is no cross-immunity between SpaI and nisin or NisI and subtilin, respectively (28, 32). Hence we tested the specificity of our growth tests with B. subtilis strains expressing SpaI and NisI using different entianin concentrations (supplemental Fig. S10). Cell lysis, resulting in a complete loss of growth, is already seen for the NisI expressing strain B2470.SD4 at 5% of the supernatant entianin, whereas for the SpaI expressing strain B2470.SB3 an approximately five times higher entianin concentration is needed to induce cell lysis. For the growth tests with different SpaI variants the effects at 5% entianin containing supernatant are plotted because definite growth differences can be seen best at this concentration (Fig. 5).
The growth test with B. subtilis strain B2470.SB2 expressing the gene coding for SpaI(Δ2–17) (Figs. 1B and 5, white bar) indicates that the N-terminally truncated SpaI18–143 used for NMR analysis is not sufficient to mediate immunity against entianin. Compared with the wild type SpaI (Figs. 1B and 5, grey bar) expressing strain B2470.SB3 complete cell lysis occurs with 5% of entianin containing supernatant. The strain B2470.SB7 expresses SpaI-Scramble, a variant including amino acids 2–17 arranged in a scrambled sequential order (Fig. 1B) but conserving the potential amphipathic α-helix character. This strain is able to grow in the presence of entianin at a level comparable with the wild type SpaI expressing strain (Fig. 5, wavey bar). This indicates that the length and the charge distribution of the unstructured N-terminal region are important to mediate immunity but not the specific amino acid sequence. This is in agreement with a functional role for the positively charged N terminus in membrane binding but argues against a direct role of the unstructured 2–17 amino acid stretch in the formation of a possible entianin or subtilin binding site. To more directly test the importance of the positive charge of the 5 lysines in the N-terminal stretch of SpaI for mediating immunity the positive amino acids were replaced against negatively charged or hydrophilic residues (Figs. 1B and 5, striped bar). The strain B2470.SB17 expressing the gene encoding for SpaI-Negative where the 5 lysines were mutated to glutamate or serine is not able to mediate immunity against entianin as expected when these positive charges are important for mediating a functional membrane interaction in SpaI.
DISCUSSION
Previous studies of the immunity proteins SpaI and NisI indicated that full immunity is conferred to the producer strain only if the LanI protein is present in addition to its corresponding ABC transporter (28, 32). For SpaI and NisI, binding to their corresponding lantibiotic has been suggested by surface plasmon resonance (33) or cross-linking (28, 32) experiments. However, the molecular mechanism for mediation of immunity by LanI proteins is still unknown. We approached this question from a structural point of view and solved the first structure of a LanI protein, SpaI, with solution NMR spectroscopy.
SpaI is a mainly β-sheet protein with an unstructured and flexible N terminus in solution followed by a 13.6-kDa structured core consisting of six β-strands and two α-helices (residues 32–139). The six β-strands form a twisted antiparallel β-sheet flanked by an unusual long β-hairpin and the two helices. The twisted antiparallel β-sheet shows similarities to β-barrel structures but together with the unusual long β-hairpin SpaI defines a novel three-dimensional fold. Accordingly, no definite homologs were found by the DALI server. The previously published secondary structure prediction of SpaI suggested a mainly α-helical fold for SpaI (64), which is not confirmed by our structure.
An unstructured region prior to or after the residue that is prenylated or acetylated for membrane anchoring is a common feature of lipoproteins, e.g. as seen for the Ras protein family where ∼10 amino acids prior to the carboxyl-terminal CAAX motif and the palmitoylation sites form a flexible linker between the structured protein core and the membrane anchoring site (65). It is assumed that this linker enables Ras to experience a large conformational freedom (66). Other bacterial lipoproteins like BamE, which is part of the β-barrel assembly machine complex and RcsF, a part of the Rcs phosphorelay, a stress-induced defense mechanism, also have an unstructured and flexible N terminus of about 20 and 30 amino acids following the lipidated cysteine, respectively (67, 68). Both lipoproteins are assembled in the outer membrane of Gram-negative bacteria facing the periplasm.
Likewise, the 30 N-terminal amino acids of SpaI following the lipidated cysteine are unstructured and flexible in the absence of a membrane. Interestingly, the stretch of the first 17 amino acids contains 5 positively charged lysines and only 3 negatively charged amino acids. This resembles, for instance, the membrane interactions of the K-ras protein where a polybasic domain containing 6 lysine residues complements the prenyl-membrane anchor (65). When we mapped the membrane interaction site of SpaI with NMR titration experiments we found that the ∼15 N-terminal amino acids interact directly with the membrane even in the absence of a lipobox and a lipid anchor (Fig. 4E). Because the NMR signals of the corresponding residues disappear we cannot exclude that this unstructured stretch adopts a structure such as an α-helix in the presence of a membrane. This is predicted for amino acids 4–17 by several secondary structure prediction programs and interestingly such a putative helix would be amphipathic as typical for a membrane-associated helix. Additionally, the length and charge distribution of this unstructured N-terminal region seems to be important for mediating immunity as shown by the B. subtilis growth test with strains B2470.SB7 and B2470.SB17 expressing SpaI-Scramble and SpaI-Negative, respectively (Figs. 1B and 5). We propose that the 5 lysines in the N-terminal stretch play a major role in the functional membrane interaction, because mutating them to negatively charged or hydrophilic residues abolishes the immunity mediated by SpaI. Interestingly, these 5 lysines are conserved between SpaI, EtnI, and EriI (Fig. 1C). Furthermore, NisI also shows 4 positive charged residues within the first 30 N-terminal amino acids.
In our model the structured core of SpaI is oriented with a very negatively charged surface toward the membrane (Fig. 4, A, upper right, and E). This is remarkable because the liposomes are also negatively charged due to an excess of phospholipids (DOPG and CL) carrying negatively charged head groups relative to DOPE, which contains a positively charged head group. However, because many lipoproteins possess a flexible linker preceding or succeeding the membrane anchoring residue(s) the overall surface charge of the structured core of these proteins is likely more related to their function than to their membrane interaction. One example is the lipoprotein LolB, which is part of the sorting and membrane localization complex in Gram-negative bacteria. LolB is thought to receive lipoproteins from the periplasmic chaperone LolA and transfer them to the inner leaflet of the outer membrane by swinging through its flexible N terminus (69). Its structure reveals a well defined hydrophobic cavity on the surface of LolB for accommodating the acyl chain of lipoproteins. In contrast, the distribution of positively and negatively charged residues on the protein surface does not result in a pattern that would allow the definition of a membrane interaction site. Correspondingly, the observed orientation of the structured core of SpaI with its negatively charged surface toward the membrane might be functionally important for its possible interactions with membrane-associated positively charged subtilin (see below).
A challenging question is the location of the interaction site between SpaI and subtilin. Cross-linking experiments clearly suggested a direct in vitro interaction between subtilin and SpaI (28) as well as nisin and NisI (32). However, we were unable to investigate this interaction by NMR spectroscopy because due to their highly hydrophobic nature neither subtilin nor entianin are soluble in our NMR buffers or related buffers. This unfortunately precludes NMR-based titration experiments of SpaI with subtilin or entianin and the observation of chemical shift perturbations.
Nevertheless, the structure of SpaI described here and the comparisons with the surface properties of the homologous structures EtnI and EriI suggest possible locations for a subtilin binding site. Subtilin harbors one positive charge at the N terminus and two positive charges at the C terminus but is otherwise very hydrophobic (Fig. 1A). Thus, the binding of subtilin to SpaI might be based on a combination of electrostatic and hydrophobic interactions. In agreement with such a hypothetical binding mode the surface of SpaI, which according to our NMR experiments points toward the periplasm (Fig. 4A, upper left, and supplemental Fig. S9, left) exhibits a pronounced central hydrophobic patch surrounded by a rim of negative charges. Therefore, this surface is a candidate for a binding site of subtilin and SpaI might thereby protect the membrane from subtilin insertion. The surface properties of this potential lantibiotic binding site are largely conserved in EtnI and in agreement with the very similar biophysical properties and sequences of subtilin and entianin (supplemental Fig. S9, left). In contrast, EriI shows less negative charges on this surface in accordance with the reduced positive charge of ericin S (supplemental Fig. S9, left).
Alternatively, the highly negatively charged membrane proximal surface of SpaI (Fig. 4A, upper right, and supplemental Fig. S9, right) might act by preventing lipid II-mediated subtilin oligomerization prior to pore formation by simply competing with the negatively charged pyrophosphate moiety of lipid II for subtilin binding. Interestingly, of all LanI proteins identified so far, only SpaI, EtnI, EriI, and NisI have an acidic surface charge and their corresponding lantibiotics have an overall positive charge. A model for LanI-mediated immunity by a mechanism based on competition was suggested, e.g. for PepI (70). In contrast to SpaI and its relatives, PepI is a basic protein conferring immunity to the highly positively charged lantibiotic Pep5 (70). Additionally, the strain expressing Pep5 does not code for an immunity mediating ABC transporter LanFEG. Thus, a direct binding of Pep5 to PepI seems unlikely. Instead it was suggested that PepI competes with Pep5 for binding to an anionic cellular target such as teichoic or lipoteichoic acids (70).
Another conceivable mechanism is that upon subtilin binding to lipid II mediated by its first two lanthionine rings, the positive charges at the C termini of subtilin in subtilin-lipid II oligomeric complexes are free to serve as binding site(s) for the highly negatively charged membrane proximal surface of SpaI (Fig. 4A, upper right, and supplemental Fig. S9, right). Such an interaction could either prevent the membrane insertion of these complexes or at a later stage close the membrane pores formed by subtilin-lipid II oligomers.
The structure of SpaI reported here provides an important step toward the molecular understanding of the immunity mechanism of B. subtilis in particular and of other lantibiotic producing strains in general. Understanding the immunity mechanism of lantibiotic producing strains will help to design and express novel lantibiotics to cope with the emerging resistance of bacterial strains against common antibiotics.
Supplementary Material
Acknowledgments
We are grateful to Dr. Christian Richter for help with the NMR experiments and Dr. Jürgen Graf for conducting initial buffer screens. We thank Prof. Volker Dötsch for the kind gift of the TEV-Protease construct and Björn Meyer and Prof. Michael Karas for MALDI-MS analysis of the C-terminal SpaI fragment.
This work was supported by an Aventis Foundation professorship (to J. W.), a Lichtenberg professorship of the Volkswagen Foundation (to P. G.), the Deutsche Forschungsgemeinschaft (to P. G.), the Center of Biomolecular Magnetic Resonance (BMRZ), the Cluster of Excellence ”Macromolecular Complexes“ (to K.-D. E. and J. W.), and the Förderfonds of the Goethe University Frankfurt am Main.

This article contains supplemental ”Experimental Procedures,“ Tables S1 and S2, and Figs. S1–S10.
The atomic coordinates and structure factors (code 2lvl) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
NMR assignments have been deposited in the BioMagResBank (BMRB) under accession number 17534.
- DOPG
- 1,2-dioleoyl-sn-glycero-3-phospho-(1′-rac-glycerol)
- DOPE
- 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine
- CL
- cardiolipin
- HSQC
- heteronuclear single quantum coherence
- TROSY
- transverse relaxation-optimized spectroscopy
- r.m.s.
- root mean square
- RDC
- residual dipolar coupling.
REFERENCES
- 1. Schnell N., Entian K. D., Schneider U., Götz F., Zähner H., Kellner R., Jung G. (1988) Prepeptide sequence of epidermin, a ribosomally synthesized antibiotic with four sulfide rings. Nature 333, 276–278 [DOI] [PubMed] [Google Scholar]
- 2. Breukink E., de Kruijff B. (2006) Lipid II as a target for antibiotics. Nat. Rev. Drug Discov. 5, 321–332 [DOI] [PubMed] [Google Scholar]
- 3. Piper C., Cotter P. D., Ross R. P., Hill C. (2009) Discovery of medically significant lantibiotics. Curr. Drug Discov. Technol. 6, 1–18 [DOI] [PubMed] [Google Scholar]
- 4. Chatterjee C., Paul M., Xie L., van der Donk W. A. (2005) Biosynthesis and mode of action of lantibiotics. Chem. Rev. 105, 633–684 [DOI] [PubMed] [Google Scholar]
- 5. Klein C., Kaletta C., Schnell N., Entian K. D. (1992) Analysis of genes involved in biosynthesis of the lantibiotic subtilin. Appl. Environ. Microbiol. 58, 132–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Engelke G., Gutowski-Eckel Z., Hammelmann M., Entian K. D. (1992) Biosynthesis of the lantibiotic nisin. Genomic organization and membrane localization of the NisB protein. Appl. Environ. Microbiol. 58, 3730–3743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Siegers K., Heinzmann S., Entian K. D. (1996) Biosynthesis of lantibiotic nisin. Post-translational modification of its prepeptide occurs at a multimeric membrane-associated lanthionine synthetase complex. J. Biol. Chem. 271, 12294–12301 [DOI] [PubMed] [Google Scholar]
- 8. Kiesau P., Eikmanns U., Gutowski-Eckel Z., Weber S., Hammelmann M., Entian K. D. (1997) Evidence for a multimeric subtilin synthetase complex. J. Bacteriol. 179, 1475–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gross E., Morell J. L. (1971) The structure of nisin. J. Am. Chem. Soc. 93, 4634–4635 [DOI] [PubMed] [Google Scholar]
- 10. Buchman G. W., Banerjee S., Hansen J. N. (1988) Structure, expression, and evolution of a gene encoding the precursor of nisin, a small protein antibiotic. J. Biol. Chem. 263, 16260–16266 [PubMed] [Google Scholar]
- 11. Banerjee S., Hansen J. N. (1988) Structure and expression of a gene encoding the precursor of subtilin, a small protein antibiotic. J. Biol. Chem. 263, 9508–9514 [PubMed] [Google Scholar]
- 12. Stein T., Borchert S., Conrad B., Feesche J., Hofemeister B., Hofemeister J., Entian K. D. (2002) Two different lantibiotic-like peptides originate from the ericin gene cluster of Bacillus subtilis A1/3. J. Bacteriol. 184, 1703–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fuchs S. W., Jaskolla T. W., Bochmann S., Kötter P., Wichelhaus T., Karas M., Stein T., Entian K. D. (2011) Entianin, a novel subtilin-like lantibiotic from Bacillus subtilis subsp. spizizenii DSM 15029T with high antimicrobial activity. Appl. Environ. Microbiol. 77, 1698–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bierbaum G., Sahl H. G. (2009) Lantibiotics. Mode of action, biosynthesis, and bioengineering. Curr. Pharm. Biotechnol. 10, 2–18 [DOI] [PubMed] [Google Scholar]
- 15. Brötz H., Josten M., Wiedemann I., Schneider U., Götz F., Bierbaum G., Sahl H. G. (1998) Role of lipid-bound peptidoglycan precursors in the formation of pores by nisin, epidermin, and other lantibiotics. Mol. Microbiol. 30, 317–327 [DOI] [PubMed] [Google Scholar]
- 16. Hasper H. E., Kramer N. E., Smith J. L., Hillman J. D., Zachariah C., Kuipers O. P., de Kruijff B., Breukink E. (2006) An alternative bactericidal mechanism of action for lantibiotic peptides that target lipid II. Science 313, 1636–1637 [DOI] [PubMed] [Google Scholar]
- 17. Schüller F., Benz R., Sahl H. G. (1989) The peptide antibiotic subtilin acts by formation of voltage-dependent multistate pores in bacterial and artificial membranes. Eur. J. Biochem. 182, 181–186 [DOI] [PubMed] [Google Scholar]
- 18. Breukink E., Wiedemann I., van Kraaij C., Kuipers O. P., Sahl H., de Kruijff B. (1999) Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science 286, 2361–2364 [DOI] [PubMed] [Google Scholar]
- 19. Parisot J., Carey S., Breukink E., Chan W. C., Narbad A., Bonev B. (2008) Molecular mechanism of target recognition by subtilin, a class I lanthionine antibiotic. Antimicrob. Agents Chemother. 52, 612–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hsu S. T., Breukink E., Tischenko E., Lutters M. A., de Kruijff B., Kaptein R., Bonvin A. M., van Nuland N. A. (2004) The nisin-lipid II complex reveals a pyrophosphate cage that provides a blueprint for novel antibiotics. Nat. Struct. Mol Biol. 11, 963–967 [DOI] [PubMed] [Google Scholar]
- 21. Breukink E., van Heusden H. E., Vollmerhaus P. J., Swiezewska E., Brunner L., Walker S., Heck A. J., de Kruijff B. (2003) Lipid II is an intrinsic component of the pore induced by nisin in bacterial membranes. J. Biol. Chem. 278, 19898–19903 [DOI] [PubMed] [Google Scholar]
- 22. Hasper H. E., de Kruijff B., Breukink E. (2004) Assembly and stability of nisin-lipid II pores. Biochemistry 43, 11567–11575 [DOI] [PubMed] [Google Scholar]
- 23. Draper L. A., Ross R. P., Hill C., Cotter P. D. (2008) Lantibiotic immunity. Curr. Protein Pept. Sci. 9, 39–49 [DOI] [PubMed] [Google Scholar]
- 24. Alkhatib Z., Abts A., Mavaro A., Schmitt L., Smits S. H. (2012) Lantibiotics. How do producers become self-protected? J. Biotechnol. 159, 145–154 [DOI] [PubMed] [Google Scholar]
- 25. Kuipers O. P., Beerthuyzen M. M., Siezen R. J., De Vos W. M. (1993) Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis. Requirement of expression of the nisA and nisI genes for development of immunity. Eur. J. Biochem. 216, 281–291 [DOI] [PubMed] [Google Scholar]
- 26. Klein C., Entian K. D. (1994) Genes involved in self-protection against the lantibiotic subtilin produced by Bacillus subtilis ATCC 6633. Appl. Environ. Microbiol. 60, 2793–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Engelke G., Gutowski-Eckel Z., Kiesau P., Siegers K., Hammelmann M., Entian K. D. (1994) Regulation of nisin biosynthesis and immunity in Lactococcus lactis 6F3. Appl. Environ. Microbiol. 60, 814–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stein T., Heinzmann S., Düsterhus S., Borchert S., Entian K. D. (2005) Expression and functional analysis of the subtilin immunity genes spaIFEG in the subtilin-sensitive host Bacillus subtilis MO1099. J. Bacteriol. 187, 822–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Halami P. M., Stein T., Chandrashekar A., Entian K. D. (2010) Maturation and processing of SpaI, the lipoprotein involved in subtilin immunity in Bacillus subtilis ATCC 6633. Microbiol. Res. 165, 183–189 [DOI] [PubMed] [Google Scholar]
- 30. Siegers K., Entian K. D. (1995) Genes involved in immunity to the lantibiotic nisin produced by Lactococcus lactis 6F3. Appl. Environ. Microbiol. 61, 1082–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ra R., Beerthuyzen M. M., de Vos W. M., Saris P. E., Kuipers O. P. (1999) Effects of gene disruptions in the nisin gene cluster of Lactococcus lactis on nisin production and producer immunity. Microbiology 145, 1227–1233 [DOI] [PubMed] [Google Scholar]
- 32. Stein T., Heinzmann S., Solovieva I., Entian K. D. (2003) Function of Lactococcus lactis nisin immunity genes nisI and nisFEG after coordinated expression in the surrogate host Bacillus subtilis. J. Biol. Chem. 278, 89–94 [DOI] [PubMed] [Google Scholar]
- 33. Takala T. M., Koponen O., Qiao M., Saris P. E. (2004) Lipid-free NisI. Interaction with nisin and contribution to nisin immunity via secretion. FEMS Microbiol. Lett. 237, 171–177 [DOI] [PubMed] [Google Scholar]
- 34. Entian K. D., de Vos W. M. (1996) Genetics of subtilin and nisin biosyntheses. Biosynthesis of lantibiotics. Antonie Leeuwenhoek 69, 109–117 [DOI] [PubMed] [Google Scholar]
- 35. Willey J. M., van der Donk W. A. (2007) Lantibiotics. Peptides of diverse structure and function. Annu. Rev. Microbiol. 61, 477–501 [DOI] [PubMed] [Google Scholar]
- 36. Christ N. A., Duchardt-Ferner E., Düsterhus S., Kötter P., Entian K. D., Wöhnert J. (2012) NMR resonance assignment of the autoimmunity protein SpaI from Bacillus subtilis ATCC 6633. Biomol. NMR Assign. 6, 9–13 [DOI] [PubMed] [Google Scholar]
- 37. Markley J. L., Bax A., Arata Y., Hilbers C. W., Kaptein R., Sykes B. D., Wright P. E., Wüthrich K. (1998) Recommendations for the presentation of NMR structures of proteins and nucleic acids. IUPAC-IUBMB-IUPAB Inter-Union Task Group on the standardization of data bases of protein and nucleic acid structures determined by NMR spectroscopy. J. Biomol. NMR. 12, 1–23 [DOI] [PubMed] [Google Scholar]
- 38. Sattler M., Schleucher J., Griesinger C. (1999) Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog. Nuclear Mag. Reson. Spectrosc. 34, 93–158 [Google Scholar]
- 39. Kay L. E., Nicholson L. K., Delaglio F., Bax A., Torchia D. A. (1992) Pulse sequences for removal of the effects of cross correlation between dipolar and chemical shift anisotropy relaxation mechanisms on the measurement of heteronuclear T1 and T2 values in proteins. J. Magn. Res. 97, 359–375 [Google Scholar]
- 40. Farrow N. A., Muhandiram R., Singer A. U., Pascal S. M., Kay C. M., Gish G., Shoelson S. E., Pawson T., Forman-Kay J. D., Kay L. E. (1994) Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry 33, 5984–6003 [DOI] [PubMed] [Google Scholar]
- 41. Cordier F., Dingley A. J., Grzesiek S. (1999) A doublet-separated sensitivity-enhanced HSQC for the determination of scalar and dipolar one-bond J-couplings. J. Biomol. NMR 13, 175–180 [DOI] [PubMed] [Google Scholar]
- 42. Hansen M. R., Mueller L., Pardi A. (1998) Tunable alignment of macromolecules by filamentous phage yields dipolar coupling interactions. Nat. Struct. Biol. 5, 1065–1074 [DOI] [PubMed] [Google Scholar]
- 43. Vranken W. F., Boucher W., Stevens T. J., Fogh R. H., Pajon A., Llinas M., Ulrich E. L., Markley J. L., Ionides J., Laue E. D. (2005) The CCPN data model for NMR spectroscopy. Development of a software pipeline. Proteins 59, 687–696 [DOI] [PubMed] [Google Scholar]
- 44. Dosset P., Hus J. C., Marion D., Blackledge M. (2001) A novel interactive tool for rigid-body modeling of multidomain macromolecules using residual dipolar couplings. J. Biomol. NMR. 20, 223–231 [DOI] [PubMed] [Google Scholar]
- 45. Güntert P. (2009) Automated structure determination from NMR spectra. Eur. Biophys. J. 38, 129–143 [DOI] [PubMed] [Google Scholar]
- 46. Cordier F., Grzesiek S. (1999) Direct observation of hydrogen bonds in proteins by interresidue 3H HNC' scalar couplings. J. Am. Chem. Soc. 121, 1601–1602 [DOI] [PubMed] [Google Scholar]
- 47. Keller R. (2004) The Computer-aided Resonance Tutorial, CANTINA Verlag, Goldau [Google Scholar]
- 48. Herrmann T., Güntert P., Wüthrich K. (2002) Protein NMR structure determination with automated NOE identification in the NOESY spectra using the new software ATNOS. J. Biomol. NMR 24, 171–189 [DOI] [PubMed] [Google Scholar]
- 49. Herrmann T., Güntert P., Wüthrich K. (2002) Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 319, 209–227 [DOI] [PubMed] [Google Scholar]
- 50. Shen Y., Delaglio F., Cornilescu G., Bax A. (2009) TALOS+. A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR. 44, 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Güntert P., Mumenthaler C., Wüthrich K. (1997) Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 273, 283–298 [DOI] [PubMed] [Google Scholar]
- 52. Gottstein D., Kirchner D. K., Güntert P. (2012) Simultaneous single-structure and bundle representation of protein NMR structures in torsion angle space. J. Biomol. NMR. 52, 351–364 [DOI] [PubMed] [Google Scholar]
- 53. Koradi R., Billeter M., Güntert P. (2000) Point-centered domain decomposition for parallel molecular dynamics simulation. Comp. Phys. Commun. 124, 139–147 [Google Scholar]
- 54. Ponder J. W., Case D. A. (2003) in Advances in Protein Chemistry, Vol. 66, pp. 27–85, Academic Press, San Diego, CA: [DOI] [PubMed] [Google Scholar]
- 55. Bhattacharya A., Tejero R., Montelione G. T. (2007) Evaluating protein structures determined by structural genomics consortia. Proteins 66, 778–795 [DOI] [PubMed] [Google Scholar]
- 56. Dolinsky T. J., Nielsen J. E., McCammon J. A., Baker N. A. (2004) PDB2PQR. An automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 32, W665–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Electrostatics of nanosystems. Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Delano W. (2010) The PyMOL Molecular Graphics System, version 1.3 Schrödinger, LLC [Google Scholar]
- 59. Burkard M., Stein T. (2008) Microtiter plate bioassay to monitor the interference of antibiotics with the lipid II cycle essential for peptidoglycan biosynthesis. J. Microbiol. Methods 75, 70–74 [DOI] [PubMed] [Google Scholar]
- 60. García de la Torre J., Huertas M. L., Carrasco B. (2000) HYDRONMR. Prediction of NMR relaxation of globular proteins from atomic level structures and hydrodynamic calculations. J. Magn. Reson. 147, 138–146 [DOI] [PubMed] [Google Scholar]
- 61. Holm L., Rosenström P. (2010) Dali server. Conservation mapping in three-dimension. Nucleic Acids Res. 38, W545-W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. O'Leary W. M., Wilkinson S. G. (1988) in Microbial Lipids (Ratledge C., Wilkinson S. G., eds) 1st Ed., pp. 117–201, Academic Press, London [Google Scholar]
- 63. Seydlová G., Svobodová J. (2008) Development of membrane lipids in the surfactin producer Bacillus subtilis. Folia Microbiol. 53, 303–307 [DOI] [PubMed] [Google Scholar]
- 64. Takala T. M., Saris P. E. (2006) C terminus of NisI provides specificity to nisin. Microbiology 152, 3543–3549 [DOI] [PubMed] [Google Scholar]
- 65. Hancock J. F. (2003) Ras proteins. Different signals from different locations. Nat. Rev. Mol. Cell Biol. 4, 373–384 [DOI] [PubMed] [Google Scholar]
- 66. Brunsveld L., Kuhlmann J., Alexandrov K., Wittinghofer A., Goody R. S., Waldmann H. (2006) Lipidated ras and rab peptides and proteins-synthesis, structure, and function. Angew. Chem. Int. Ed. Engl. 45, 6622–6646 [DOI] [PubMed] [Google Scholar]
- 67. Kim K. H., Kang H. S., Okon M., Escobar-Cabrera E., McIntosh L. P., Paetzel M. (2011) Structural characterization of Escherichia coli BamE, a lipoprotein component of the β-barrel assembly machinery complex. Biochemistry 50, 1081–1090 [DOI] [PubMed] [Google Scholar]
- 68. Leverrier P., Declercq J. P., Denoncin K., Vertommen D., Hiniker A., Cho S. H., Collet J. F. (2011) Crystal structure of the outer membrane protein RcsF, a new substrate for the periplasmic protein-disulfide isomerase DsbC. J. Biol. Chem. 286, 16734–16742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Takeda K., Miyatake H., Yokota N., Matsuyama S., Tokuda H., Miki K. (2003) Crystal structures of bacterial lipoprotein localization factors, LolA and LolB. EMBO J. 22, 3199–3209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hoffmann A., Schneider T., Pag U., Sahl H. G. (2004) Localization and functional analysis of PepI, the immunity peptide of Pep5-producing Staphylococcus epidermidis strain 5. Appl. Environ. Microbiol. 70, 3263–3271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., Thornton J. M. (1996) AQUA and PROCHECK-NMR. Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 8, 477–486 [DOI] [PubMed] [Google Scholar]
- 72. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 73. Mulder F. A., Schipper D., Bott R., Boelens R. (1999) Altered flexibility in the substrate-binding site of related native and engineered high-alkaline Bacillus subtilisins. J. Mol. Biol. 292, 111–123 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.