Background: Kcnh1 is a voltage-gated potassium channel that is primarily expressed in brain.
Results: Knockdown of kcnh1 in zebrafish delays neural development and causes embryonic lethality.
Conclusion: Kcnh1 is involved in cell proliferation during early zebrafish development.
Significance: The finding that Kcnh1 has basic functions beyond neural signaling will help to elucidate its roles in physiology and cancer formation.
Keywords: Development, Electrophysiology, Neurodevelopment, Potassium Channels, Zebra Fish, EAG1 Channel, Morpholino
Abstract
The Kcnh1 gene encodes a voltage-gated potassium channel highly expressed in neurons and involved in tumor cell proliferation, yet its physiological roles remain unclear. We have used the zebrafish as a model to analyze Kcnh1 function in vitro and in vivo. We found that the kcnh1 gene is duplicated in teleost fish (i.e. kcnh1a and kcnh1b) and that both genes are maternally expressed during early development. In adult zebrafish, kcnh1a and kcnh1b have distinct expression patterns but share expression in brain and testis. Heterologous expression of both genes in Xenopus oocytes revealed a strong conservation of characteristic functional properties between human and fish channels, including a unique sensitivity to intracellular Ca2+/calmodulin and modulation of voltage-dependent gating by extracellular Mg2+. Using a morpholino antisense approach, we demonstrate a strong kcnh1 loss-of-function phenotype in developing zebrafish, characterized by growth retardation, delayed hindbrain formation, and embryonic lethality. This late phenotype was preceded by transcriptional up-regulation of known cell-cycle inhibitors (p21, p27, cdh2) and down-regulation of pro-proliferative factors, including cyclin D1, at 70% epiboly. These results reveal an unanticipated basic activity of kcnh1 that is crucial for early embryonic development and patterning.
Introduction
Kcnh1 is a voltage-gated potassium channel with canonical tetrameric structure and six-transmembrane domain topology. The Kcnh1 gene was originally named ether à go-go (eag) as its mutation in Drosophila melanogaster induced a rhythmic leg-shaking phenotype under ether anesthesia (1); it is also referred to as eag1 or Kv10.1. Kcnh1 mutant flies exhibit a high frequency of spontaneous action potentials and enhanced transmitter release in motor neurons (2). In mammals Kcnh1 expression is almost completely restricted to the brain (3–5), but no specific neuronal function could yet be described. The only physiological function ascribed to vertebrate Kcnh1 channels thus far is a promoting role at the onset of myoblast fusion (6, 7).
An important pathophysiological role for Kcnh1 in cancer formation has been proposed (8) because the human KCNH1 gene is overexpressed in a broad spectrum of cancers and channel inhibition can reduce cell proliferation (8–11). However, neither the gene expression profile nor the deduced functional properties of Kcnh1 channels provided insight into mechanisms underlying the channel oncogenic potential. Many oncogenes are involved in development, and cancer formation often recapitulates key processes of embryogenesis (12). Thus, it would be instructive to study Kcnh1 during embryonic development, but thus far no suitable vertebrate models have been established.
Recently it became evident that Danio rerio (zebrafish) is a valuable model for investigating Kcnh2 (Erg1, Kv11.1) channel diseases of the heart (13–18). With respect to physiological functions, Kcnh2 is the best-characterized member of the Kcnh channel family. It is involved in the control of cardiac action potentials, and mutations in KCNH2 can cause life-threatening cardiac arrhythmias in humans (19–21). Kcnh2 knockdown in zebrafish was found to mimic the known cardiac phenotype without systemic disturbance of embryo development (13–18).
We evaluated zebrafish as a model organism to study physiological functions of Kcnh1 potassium channels in vertebrates and their potential role in embryogenesis. We identified and cloned two fish orthologs of KCNH1, both sharing typical functional properties with human KCNH1 channels. The kcnh1 genes in zebrafish showed maternal expression, and morpholino-mediated knockdown caused severe and specific developmental anomalies. Our results provide evidence for a novel role of Kcnh1 voltage-gated potassium channels during embryo development and establish the zebrafish as a valuable model to study such functions.
EXPERIMENTAL PROCEDURES
Maintenance of Fish
Zebrafish embryos were obtained from matings of wild-type fish of the TüAB strain that had been kept in laboratory stocks in Jena for many generations, according to the local animal care program. Embryos were raised at 28 °C and staged according to Kimmel et al. (22).
Bioinformatics
The kcnh1 genes in zebrafish genome databases were identified using the GenBankTM (www.ncbi.nlm.nih.gov), JGI, Ensembl, and UCSC Genome browsers. Protein and nucleotide databases were searched using BLAT and BLAST algorithms (blastn and tblastn; E-values = e−30 as a threshold) using as a reference the human KCNH1 sequence NM_002238 (GenBankTM). For phylogenetic and synteny analyses, DNA sequences were edited and analyzed using the DNA-STAR package (Lasergene 5.02). Amino acid sequences were aligned using ClustalW and the core matrix PAM-45. Phylogenetic analyses were performed using distance-based methods as neighbor-joining and minimum-evolution incorporated in the MEGA software (23). Reliability of tree topologies was assessed using bootstrapping after 1000 iterations.
Cloning of kcnh1a and kcnh1b Coding Sequences
Total RNA isolated from 24 h post-fertilization (hpf)3 embryos was treated with DNase I and used as a template for cDNA synthesis using the SuperScript III RT kit (Invitrogen). Gene-specific PCR reactions were set up for three overlapping fragments of each zebrafish kcnh1 gene (primer information upon request). Resulting overlapping amplicons were used in second and third PCR reactions as templates with the respective primers of the distal ends. Finally, full-length cDNAs were ligated into the pGEM-T vector (Promega) and fully sequenced. Selected clones were digested with BamHI/XbaI and ligated into pGEM-HE (24).
Dominant negative mutants of zebrafish channels (Kcnh1a G438C; Kcnh1b G438C) were constructed using site-directed mutagenesis (QuikChange kit, Stratagene). PCR reactions were performed using the pGEM-HE clones of the corresponding genes as a template and primers carrying the desired mutation. Clones were fully sequenced to confirm the correct mutation. The dominant negative mutant of the human channel (KCNH1 G440C) was as described previously (25).
Whole-mount in Situ Hybridization
Gene-specific probes were amplified by PCR using 24-hpf zebrafish cDNA as the template and the following primers: kcnh1a probe forward (5′-CTG GAA AGA AGT AAC ACT AGC TCA GG-3′), kcnh1a probe reverse (5′-GTG TGT TCG GGA ATG GTT GG-3′), kcnh1b probe forward (5′-GGA CAC TTC TCA CGC AAT CTG G-3′), kcnh1b probe reverse (5′-GAT ATC CCC CTG CAG ATC TTG C-3′). Amplicons (819 bp for kcnh1a and 738 bp for kcnh1b) were ligated into pGEM-T vector (Promega) and sequenced. Digoxigenin-labeled antisense riboprobes were generated from linearized vectors (SacII) using the SP6 RNA polymerase (sense riboprobes: NotI and T7 RNA polymerase). Whole-mount in situ hybridization was performed essentially as described (26). Probes were detected using anti-digoxigenin alkaline phosphatase and nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate (Roche Applied Science). For characterization of kcnh1 morphants, the following markers were used: ntl (27), myoD (28), krox20 (29), atp1a3b (30), and foxd3, wnt11, and fgf8 (31). Photographs were taken using a Zeiss Axio Imager microscope (Carl Zeiss, Oberkochen, Germany) after embedding stained embryos in 1% methylcellulose or glycerol.
Morpholino Treatment of Zebrafish Embryos
The sequences of the used morpholinos (MOs) are: kcnh1a ATG (5′-CAC GGC GGG TCA TGC GCT CCA CTG A-3′), kcnh1b ATG (5′-GCG CTG AAG AGC CTC CTG CTA CA-3′), kcnh1 pan-ATG (5′-CTA GTC CTC TGC GTC CCC CGG CCA T-3′), kcnh1a e1i1 (5′-AGG TGC GTC TTA CCG TTA GAC CGT-3′), kcnh1a e2i2 (5′-GCA CAA TAT ATC TGT TAC CTG CAA G-3′), kcnh1b e1i1 (5′-AGT GTA CAA GGC TTT TCT CAC CGT T-3′), kcnh1b e2i2 (5′-ACA GAA ACA TGA AAT CAC CTG CAT G-3′). One to two-cell stage embryos were injected (∼5 nl) with different MO concentrations (ranging from 0.1 to 1 mm) and raised at 28.5 °C. As controls, we used a MO (5′-GCG CCA TTG CTT TGC AAG AAT TG-3′) to knock down the Δ113 transcript of p53 (not expressed before 48 hpf) (32) and a published kcnh2 ATG-MO (5′-GCG GCG CAC GGG CAT TTT TCA CGC G-3′) (14). Images were generated using the AxioVision software (Zeiss). To test the specificity of ATG-MO, we generated modified pCS2+GFP and pCS2+RFP mRNAs (kindly provided by J. Wittbrodt, Heidelberg) that contained the MO target sequences of kcnh1a and kcnh1b MO directly upstream of the corresponding start codon (Fig. 6, A and B). We performed serial injections into 1–2-cell-stage embryos, injecting MO first (0.5 mm final concentration) and then a mix of modified mRNAs (10 ng final concentration each) as described in Table 1. Live embryos (24 hpf) were scored for GFP and RFP signal using epifluorescence Stereo Discovery V8 microscope (Zeiss).
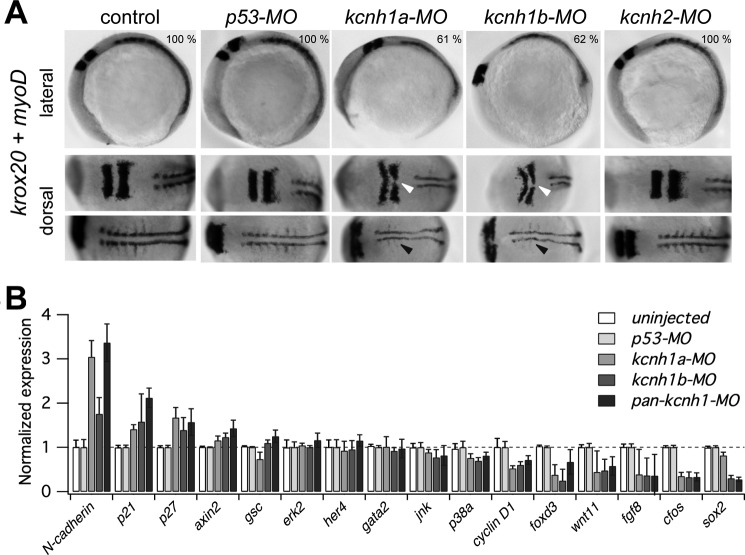
FIGURE 6.
Developmental delay of kcnh1 morphants affects hindbrain formation and somitogenesis. A, shown are representative whole-mount in situ hybridizations of zebrafish embryos with digoxigenin-labeled riboprobes for markers of hindbrain (krox20) and somites (myoD). Uninjected embryos (control) are compared with the indicated morphants. In lateral images (upper row), the dark-stained hindbrain rhombomeres 3 and 5 (krox20) and an extended stretch of myoD-stained somites are visible in the upper left and right region of the images, respectively. In the bottom row of the dorsal images the embryos are rotated to the left (anterior) by about 70 degrees. Note that in kcnh1a and kcnh1b morphants the rhombomeres 3 and 5 (white arrows) are thinner and that midline fusion of rhombomere 5 is incomplete. The lateral expansion and segmentation of somitic and presomitic mesoderm (black arrows) is reduced in both kcnh1 morphants. The percentage of embryos showing the given phenotype in two independent experiments is indicated in the upper images. B, early gene expression is modulated by kcnh1. Expression of the indicated genes in zebrafish morphants was analyzed by quantitative RT-PCR at 70% epiboly stage (7.5 hpf). Gene-specific quantitative PCR signals in uninjected controls as well as in kcnh1a, kcnh1b, and pan-kcnh1 morphants were normalized to p53-MO-injected controls, and ef1a served as the housekeeping gene for all samples. Genes are ordered according to the effect of pan-kcnh1-MO injection. Data are the mean ± S.D. from two independent experiments with each three reactions.
TABLE 1.
Specific effects of ATG morpholinos on the translation of modified GFP and RFP mRNAs
Numbers of embryos show detectable GFP and RFP signal (GFP/RFP) at 24 hpf, after the co-injection of the indicated morpholinos and mRNAs.
| Morpholino-target sequence mRNA | None | kcnh1a | kcnh1b | kcnh1 pan |
|---|---|---|---|---|
| GFP/RFP | 13/13 | 15/15 | 12/12 | 12/12 |
| kcnh1a MO wt-GFPa/kcnh1a MO mm-RFPb | 12/12 | 4/17 | 16/16 | 18/18 |
| kcnh1b MO wt-GFP/kcnh1b MO mm-RFP | 14/14 | 18/18 | 2/16 | 13/13 |
| kcnh1 pan-MO wt-GFP/kcnh1 pan-MO mm-RFP | 12/12 | 16/16 | 20/20 | 3/15 |
a wt indicates that the morpholino target sequence has the wild-type sequence as indicated in supplemental Fig. S1, A and B.
b mm indicates that the morpholino target sequence has five mismatches as shown in supplemental Fig. S1B.
RT-PCR Analysis
Total RNA was isolated from zebrafish embryos, morphants, or zebrafish organs using the RNeasy Mini kit (Qiagen) and treated with DNase. cDNA synthesis was performed using SuperScript III and oligo(dT) primers (Invitrogen). PCR analysis of cDNA was carried out with the Expand High Fidelity PCR system (Roche Applied Science) using gene-specific primers as follows: kcnh1a forward (5′-TTT TCT GGA AAA TAT TGT CCG ACG G-3′), kcnh1a reverse (5′-GTG GGC TGC AGT TGC TGA AGG-3′), kcnh1b forward (5′-GAG AAT ATA GTG CGG CGC TCC-3′), kcnh1b reverse (5′-TTT AAG CAC GTT TTC TCC CCG-3′), kcnh2 forward (5′-ATG CCC GTG CGC CGC G-3′), kcnh2 reverse (5′-CGA TGT CGT GCA GCG AGG ATG C-3′). Annealing temperatures were: kcnh1a primer pair, 60 °C; kcnh1b primer pair, 58 °C; kcnh2 primer pair, 62 °C. For loading control the expression of bactin was tested using forward primer (5′-GAG AAG ATC TGG CAT CAC ACC-3′) and reverse primer (5′-AGC TTC TCC TTG ATG TCA CG-3′) at an annealing temperature of 60 °C. For quantitative RT-PCR, total RNA was isolated from 40–50 zebrafish embryos at 70% epiboly stage. 800 ng of RNA was used to synthesize cDNA. PCR analysis was carried out using the SYBR GreenER quantitative PCR super mix for iCycler (Invitrogen) in an iCycler device (96-well format; Bio-Rad). All samples were measured as triplicates and normalized to the corresponding amounts of the housekeeping gene ef1a (33), which was measured within the same plate. Relative expression levels where calculated using the 2−ΔΔCT method (34) and are based on two independent experiments. The gene-specific primer set is listed in supplemental Table 1.
Electrophysiological Measurements
Capped mRNA was synthesized in vitro using the mMessage mMachine kit (Ambion) and NheI-linearized pGEM-HE constructs containing the respective channel-coding sequence as template. Stage-V Xenopus laevis oocytes were surgically obtained under ice water/tricaine anesthesia according to the local animal care program. Manually defolliculated oocytes were microinjected with channel-coding mRNA (<50 nl). For patch clamp measurements the vitelline membrane was removed by enzymatic treatment as described (35). Currents were measured 1–4 days after mRNA injection.
Inside-out Patch Clamp Measurements
Aluminum silicate glass pipettes with resistances of 1–2 megaohms were used for electrodes. An EPC-9 amplifier operated with PatchMaster software (both HEKA Elektronik, Lambrecht, Germany) was used as the patch clamp amplifier. For the I-V measurements in symmetrical K+, the bath solution contained: 100 mm potassium aspartate, 10 mm EGTA, 15 mm KCl, 10 mm HEPES (pH 8.0 with KOH). The pipette solution was composed of 103.6 mm potassium aspartate, 11.4 mm KCl, 1.8 mm CaCl2, 10 mm HEPES (pH 7.2 with KOH). For the measurement of Ca2+/CaM effects, the solutions used were “1 μm Ca2+” (115 mm potassium aspartate, 10 mm HEDTA, 5 mm CaCl2, 10 mm HEPES (pH 8.0 with KOH)) or “Ca2+-free” (100 mm potassium aspartate, 10 mm EGTA, 15 mm KCl, 10 mm HEPES (pH 8.0 with KOH)). The change of solutions was performed using a multichannel perfusion system, and the pipette tip was placed directly in the middle of streaming solution. His6-tagged human CaM was prepared as previously described (36).
Non-stationary Noise Analysis from On-cell Membrane Patches
Non-stationary noise analysis was performed from sets of at least 200 successive current sweeps containing responses to depolarizations to −20, 0, 20, 40, and 80 mV. Ensemble variances were compiled from differences of neighboring records (37), and variance-current plots of the activating current phases were simultaneously fitted for all voltages (V) as described (38) to yield the single-channel current i(V), the maximal open probability Po(V), and the number of active channels.
Two-electrode Voltage Clamp
Currents were recorded at 24.5 ± 0.5 °C using a Turbo-TEC 10CD amplifier (NPI electronic, Tamm, Germany) operated with PatchMaster software (HEKA Elektronik). Filament glass pipettes (Hilgenberg, Malsfeld, Germany) filled with 2 m KCl and having resistances between 0.6 and 0.8 megaohms were used for electrodes. The bath solution (normal frog Ringer) contained 115 mm NaCl, 2.5 mm KCl, 1.8 mm CaCl2, 10 mm HEPES (pH 7.2, NaOH). The addition of 5 mm MgCl2 to this solution is indicated in the figures of the respective experiments. Co-expression of wild-type and dominant negative mutant channels was obtained by microinjection of a 1:1 mix of the corresponding mRNAs. For these experiments 3 independent injections of each RNA mix were performed with 5–10 oocytes per injection.
Data analysis was performed with FitMaster software (HEKA Elektronik) and IgorPro software (WaveMetrics, Lake Oswego, OR). Pooled data are represented as the means ± S.E. (n = number of independent experiments). p values from unpaired Student's t tests are indicated as * (<0.05), ** (<0.01), and *** (<0.001).
RESULTS
Two kcnh1 Genes in Zebrafish
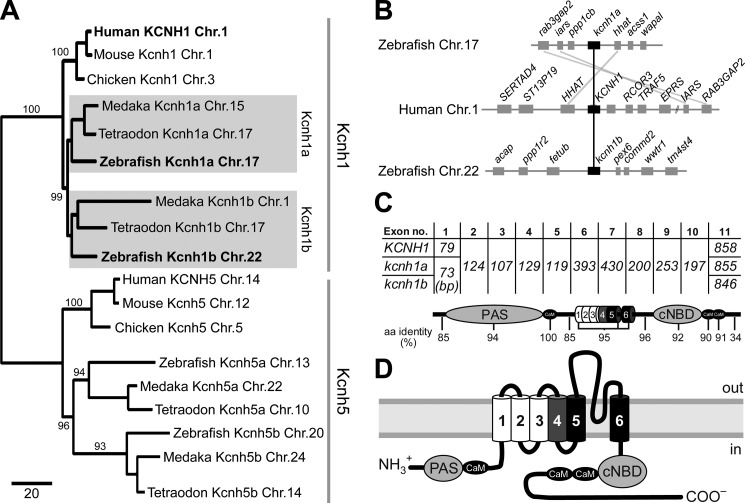
An in silico search for putative KCNH1 (EAG1) orthologs in the zebrafish genome yielded four putative candidates. Two of those, located on chromosomes 17 and 22, encode proteins with more than 90% amino acid identity to human KCNH1. The gene on chromosome 17 had already been predicted as kcnh1 in the Ensembl database (39). The second two hits scored at 86–88% amino acid identity to KCNH1 and represent the orthologs of the closest gene family member KCNH5 (EAG2), located on chromosomes 20 and 13. A phylogenetic analysis comparing human, mouse, and chicken Kcnh1 and Kcnh5 genes to the corresponding genes in the fish genomes from medaka, puffer fish, and zebrafish supported this assignment (Fig. 1A). These findings are in accordance with the suggested whole-genome duplication that occurred in a common ancestor of teleost fish (40–42). We named the kcnh1 genes in zebrafish kcnh1a (chromosome 17, GenBankTM accession number HQ703597) and kcnh1b (chromosome 22, accession number HQ703598). Synteny analysis of the chromosome regions encoding Kcnh1 loci in human and zebrafish genomes revealed similarities in gene organization for human KCNH1 and zebrafish kcnh1a, whereas the kcnh1b locus on zebrafish chromosome 22 showed no obvious overlaps in gene organization (Fig. 1B).
FIGURE 1.
Conserved Kcnh1 channels in zebrafish and man. A, shown is a distance-based phylogenetic tree of kcnh1 and kcnh5 genes in three fish genomes and their human, mouse, and chicken orthologs. Fish kcnh1a and kcnh1b paralogs are highlighted with gray boxes. The chromosome (Chr.) number associated with each gene is provided. Numbers at internodes indicate supporting bootstrap values after 1000 iterations. Branch lengths represent the relative amount of molecular divergence as indicated in the scale bar (standardized rate of amino acid replacements per site). B, synteny analysis of human and zebrafish Kcnh1 loci reveal conservation of gene clusters in fish and man. C, shown is exon distribution and amino acid identities among Kcnh1 proteins. Exon lengths in base pairs are indicated for human (KCNH1) and zebrafish (kcnh1a/b) genes. Conserved structural domains of the Kcnh1 proteins are assigned to the corresponding coding exons, and the identity of amino acid sequences between all three proteins is indicated below. D, membrane topology and domain structure of a single Kcnh1 subunit is shown. The positively charged voltage-sensor helix and the two pore helices are indicated in gray and black, respectively. PAS, Per-ARNT-Sim domain; cNBD, cyclic nucleotide-binding domain. Binding sites for Ca2+/CaM are shown as black ovals.
Cloning of the zebrafish KCNH1 orthologs, both being assembled from 11 exons, revealed that all exon borders are conserved between fish and human (Fig. 1C). The only two exons with differences in length are exons 1 and 11. Conservation at the level of amino acids is lowest (34% identity) in the second half of the cytosolic carboxyl termini of the predicted channel proteins (Fig. 1C). By contrast, all six membrane-spanning segments, the amino-terminal Per-ARNT-Sim domain and the carboxyl-terminal cyclic nucleotide binding domain as well as three CaM binding motifs are highly conserved between human and zebrafish channel proteins (Fig. 1, C and D).
Kcnh1a and kcnh1b Code for Functional Voltage-gated Potassium Channels with Typical KCNH1-like Characteristics
We expressed both zebrafish kcnh1 genes in X. laevis oocytes by injection of in vitro transcribed mRNA. Electrophysiological recordings revealed the formation of potassium channels with functional properties similar to human KCNH1 (Fig. 2). In inside-out patch clamp experiments, stepwise depolarization of the membrane potential elicited K+ currents very similar in amplitude as well as in activation and deactivation kinetics (Fig. 2A). Overall, the steady-state current-voltage dependences of the three channels largely superimposed in the voltage range explored (Fig. 2B). The currents measured at the ends of depolarizing pulses and peak tail currents were plotted versus depolarization voltages and analyzed by fits with 4th-order Boltzmann functions. The half-maximal gate activation voltages were similar (KCNH1, −65.1 ± 2.3 mV; Kcnh1a, −54.0 ± 2.4 mV; Kcnh1b, −55.3 ± 4.5 mV), whereas the corresponding slope factor was lower for Kcnh1b (17.2 ± 0.5 mV) in comparison to KCNH1 (24.7 ± 1.3 mV) and Kcnh1a (21.0 ± 0.6 mV) (Fig. 2C). Single-channel characteristics were estimated by non-stationary noise analysis applied to on-cell patch clamp recordings at various voltages (Fig. 2D). The single-channel conductance was estimated by linear data fits to the single-channel current as a function of voltage. Both zebrafish channels had a higher single-channel conductance than human KCNH1 (11.1 ± 0.5 picosiemens (pS); n = 5), with 13.5 ± 0.6 pS (n = 4) for Kcnh1a and 18.1 ± 0.9 pS (n = 7) for Kcnh1b. The extrapolated reversal potentials, between −45 and −50 mV in all cases, indicated a strong selectivity of all three channels for K+ over Na+ ions.
FIGURE 2.
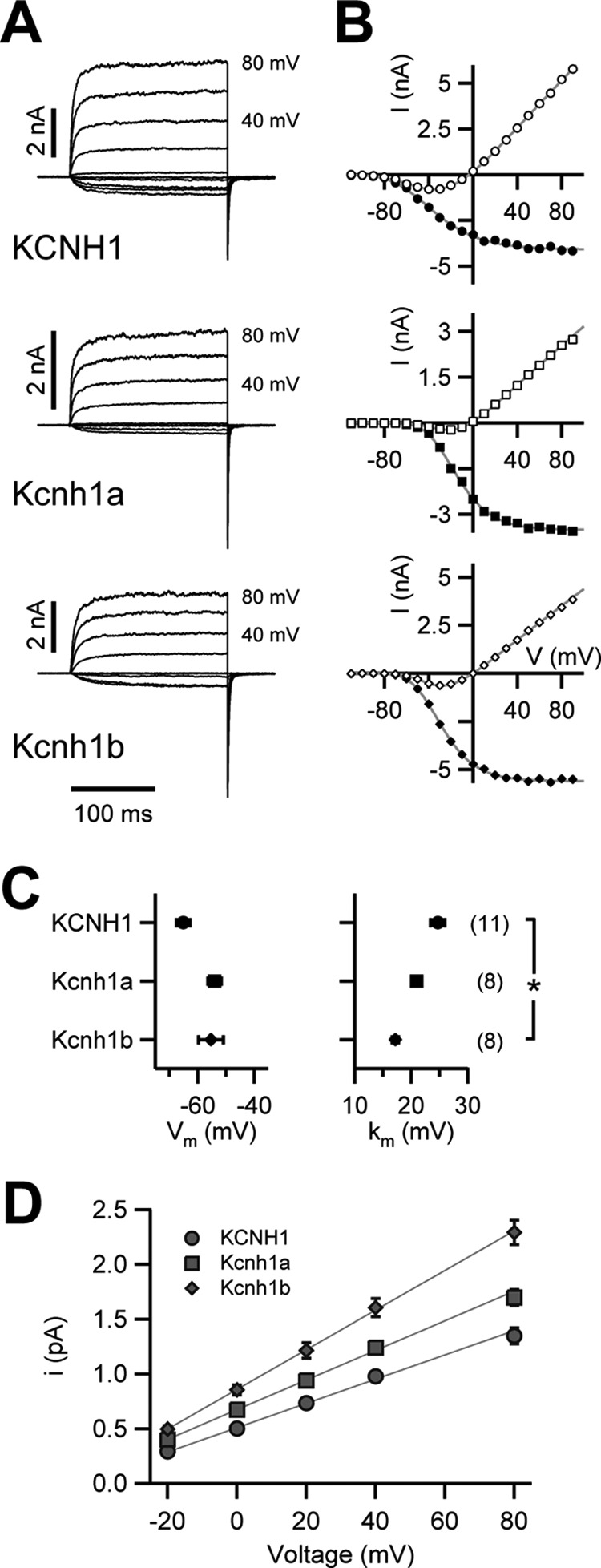
Electrophysiological properties of Kcnh1 channels expressed in Xenopus oocytes. A, current-voltage relationships were determined in inside-out patch configuration. Shown are superimposed current traces for depolarizing 200-ms steps ranging from −110 to +100 mV (from a holding potential of −120 mV) in steps of 10 mV at an interval of 5 s (only traces between −100 and +80 mV are shown, step size 20 mV; +80- and +40-mV traces are labeled). B, shown is mean current within the last 50 ms of the depolarizing steps (open symbols) and peak tail currents at −120 mV (filled symbols) as a function of the test voltage. Superimposed gray curves are fits according to a 4th-order Boltzmann function (tail currents) or a 4th-order Boltzmann function with consideration of a linear conductance (mean currents), yielding a half-maximal voltage of gate activation (Vm) and a corresponding slope factor (km). C, shown are the results of the analysis of steady-state voltage dependence of activation; Vm and km as the mean ± S.E. The numbers of experiments for A–C are shown in parentheses; * denotes statistical difference to the value obtained for KCNH1 with p < 0.05 (Student's t test). D, single-channel conductance determination for zebrafish Kcnh1 channels and human KCNH1 are shown. Single-channel current amplitudes are plotted as a function of test voltage determined with non-stationary noise analysis from on-cell membrane patches. Error bars denote S.E. values (n = 4–7). The straight lines are linear fits to estimate the single-channel conductance.
Hallmarks of mammalian Kcnh1 channels are two unique features; 1) extracellular Mg2+ ions markedly slow down activation (43, 44), and 2) intracellular Ca2+/CaM inhibit current conduction (36). Two-electrode voltage clamp experiments shown in Fig. 3A demonstrate that 5 mm Mg2+ in the external solution slows the activation of all three channel types with similar potency. Normalized current values, reached after 20 ms, revealed a slightly stronger effect of external Mg2+ on both zebrafish channels compared with human KCNH1. The current ratios (I [Mg2+]/I [noMg2+]) at 20 ms were 0.09 ± 0.01 (n = 5) and 0.06 ± 0.02 (n = 5) for Kcnh1a and Kcnh1b, respectively, and 0.21 ± 0.07 (n = 5) for human KCNH1.
FIGURE 3.
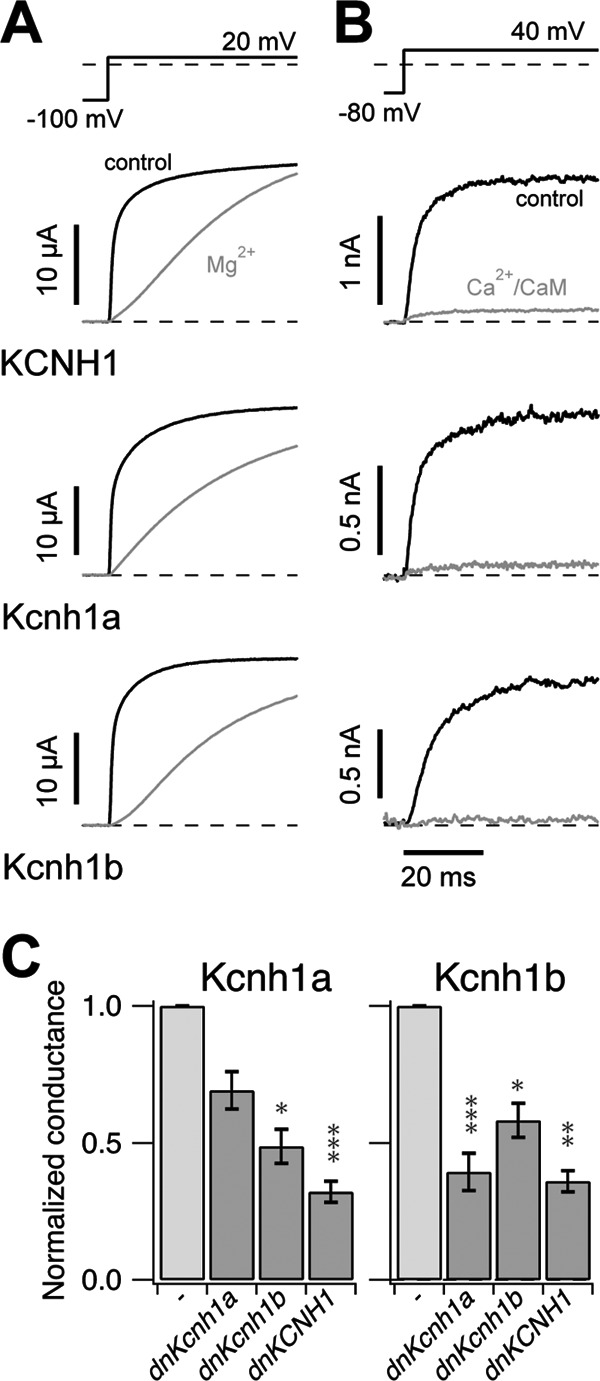
Kcnh1-specific characteristics of channel activation and inhibition. A, shown is dependence of channel activation on prepulse potential and Mg2+ ions. Current traces elicited by depolarizing 300-ms steps to 20 mV from holding potentials as indicated in the corresponding pulse protocol (upper panel) were measured in the absence (black curves) and presence (gray curves) of external 5 mm Mg2+ ions. Measurements were performed using two-electrode voltage clamp. B, channel inhibition by Ca2+/CaM is shown. Superimposition of current traces before (black) and 100 s after (gray) internal application of 100 nm CaM + 1 μm Ca2+ was measured from inside-out oocyte patches. Normalized residual currents in Ca2+/CaM were 12.6 ± 4.3, 18.9 ± 7.1, and 8.8 ± 5.7% for KCNH1 (n = 4), Kcnh1a (n = 5), and Kcnh1b (n = 5), respectively. C, Kcnh1 channels can form heteromers. mRNA encoding dominant-negative (dn) mutants of Kcnh1a (G438C), Kcnh1b (G438C), or human KCNH1 (G440C) were co-injected at 1:1 ratio with wild-type mRNA of Kcnh1a or Kcnh1b, respectively. Using two-electrode voltage clamp, K+ currents were elicited by stepwise depolarization (as in Fig. 2A), and the maximal conductances (Gmax) were determined. The bars show the mean ± S.E. from three independent experiments (n = 7–15). All values are normalized to Gmax measured after injection of wild-type channel mRNA alone. Student's t tests are indicated as * (<0.05), ** (<0.01), and *** (<0.001).
Channel regulation by Ca2+/CaM was analyzed using single-step depolarizations of inside-out membrane patches to 40 mV. Fig. 3B shows current traces from representative experiments before and after application of 1 μm Ca2+ and 100 nm CaM to the cytosolic membrane face. Analysis of the remaining currents after Ca2+/CaM application did not show significant differences between KCNH1 and the zebrafish channels. In summary, cloning and expression of two kcnh1 genes from zebrafish revealed that both genes encode functional voltage-gated potassium channels, sharing the characteristic biophysical features of human KCNH1 channels.
Given the functional and structural similarities between both zebrafish channel paralogs, we wondered if Kcnh1a and Kcnh1b are able to form heteromeric channels. After a previously described experimental approach (25, 45), we generated dominant negative mutants of both channels by single amino acid replacements (G438C) in the selectivity filter. mRNA encoding the mutant zebrafish channels as well as a dominant negative human KCNH1 variant (25) were each co-injected with wild-type mRNA for the zebrafish channels (Fig. 3C). All three mutant constructs led to reduced current amplitudes of Kcnh1a and Kcnh1b channels, strongly suggesting that both zebrafish paralogs can form hetero-tetrameric channels with each other as well as with the human KCNH1 channel. A nonspecific current reduction via an excess of mRNA could be excluded, as comparable high amounts of the respective wild-type mRNAs resulted in increased current amplitudes (not shown).
Both Zebrafish kcnh1 Genes Are Expressed in Neural Tissue
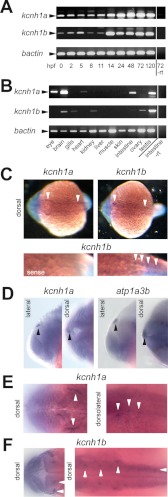
We analyzed the temporal gene expression patterns of kcnh1a and kcnh1b by RT-PCR at different developmental stages. Both paralogs were expressed maternally and could be detected in all developmental stages analyzed (Fig. 4A). The expression levels of both genes were stable until 11 hpf and increased significantly thereafter, displaying constantly high levels until 5 days post-fertilization. Interestingly, the time point around 14 hpf marks the start of neurulation in zebrafish.
FIGURE 4.
Expression of kcnh1a and kcnh1b during early development and in the adult zebrafish. A, temporal mRNA expression patterns of kcnh1a and kcnh1b in zebrafish embryos at the specified stages (in hours post fertilization, hpf) were analyzed by RT-PCR. One sample without reverse transcriptase (−rt) was used as a negative control. bactin was used as a housekeeping gene. Arrowheads indicate the expected amplicon sizes. B, shown is the expression pattern of kcnh1a/b in adult zebrafish tissues. mRNA was isolated from the specified organs, and analysis was performed as in A. C–F, dorsal and lateral views, anterior to the left, of zebrafish embryos at 19 hpf (C) and 24 hpf (D–F) were stained by whole-mount in situ hybridization. Arrowheads point to specific expression domains. C, upper panels show whole-mount staining for kcnh1a (left) and kcnh1b (right); the lower panels show enlarged posterior sections after kcnh1b staining (right) versus control staining with a kcnh1b sense probe (left). D, the two left panels (lateral and dorsal view) show an expression domain for kcnh1a (arrowheads) that was absent in kcnh1b staining. For comparison, the right-hand images show equivalent staining with an atp1a3b-specific probe that was previously shown to mark the epiphysis (30). E, image details show kcnh1a expression domains (arrowheads) in anterior (left, dorsal) and central to posterior parts (right, dorsolateral) of the embryo. F, dorsal view images in two focal planes show kcnh1b expression domains (arrowheads) in anterior (left) and central to posterior regions (right) of the embryo.
Analysis of the spatial distribution of mRNA levels in organs of adult zebrafish revealed distinct, but overlapping expression patterns of both paralogs (Fig. 4B). For kcnh1a, expression was highest in testis and brain, moderate in the eye and the gastro-intestinal tract, and low in the heart. Strong expression of kcnh1b was observed in testis, whereas lower levels were detected in brain and ovary. Kcnh1b was also expressed in the gills, the kidney, and the intestine.
To analyze the spatial expression patterns during development, we performed whole-mount in situ hybridization experiments in embryos of different developmental stages using sequences within the least conserved last exons of both genes as probes. At 19 hpf, both paralogs showed weak but specific staining patterns; transcripts were detected in discrete domains of the hindbrain and trunk (Fig. 4C). Hindbrain expression was detectable in the rostrolateral part, anterior of the otic placode, most likely reflecting trigeminal ganglia (Fig. 4C, upper panels, left arrowheads). Expression of both paralogs in the trunk was confined to discrete spots arranged in bilateral rows (top, right arrowheads and bottom, right panel). This staining pattern likely reflects primary neurons such as Rohon-Beard neurons or motor neurons.
At the 24-hpf stage, a domain was detected in the dorsal midline of the brain expressing specifically kcnh1a (Fig. 4D, left), but not kcnh1b (not shown). This kcnh1a-specific domain closely resembled a staining of the epiphysis using the marker atp1a3b (30) as shown in Fig. 4D, right, for comparison.
As already seen at 19 hpf, kcnh1a expression domains at 24 hpf comprised a distinct, curved area in the hindbrain (Fig. 4E, left). In addition, three areas dorsal of the otic vesicle showed positive signals (Fig. 4E, right). In contrast, kcnh1b expression at 24 hpf was observed bilateral at two confined areas in the cerebellum (Fig. 4F, left panel) and in three prominently stained bilateral domains, two of which were located dorsomedial and another caudolateral from the otic vesicle (Fig. 4F, right panel, upright arrowheads). The dorsolateral kcnh1b expression domains along the trunk were less pronounced but still evident (Fig. 4F, right panel, right arrowhead). In summary, the expression patterns for kcnh1a and kcnh1b were overlapping but not identical.
Knockdown of kcnh1a or kcnh1b Induces Severe Developmental Disturbances
The maternal expression of kcnh1a and kcnh1b suggested a physiological function during early development (46). Because formation of neuronal structures is one of the earliest developmental processes, we hypothesized that neuronal expression of kcnh1 genes might be of general importance for the growing embryo. To test this hypothesis we used a morpholino-mediated gene knockdown approach. MOs are antisense oligonucleotides that can prevent gene expression on two different levels (47). ATG morpholinos (ATG-MO) inhibit the translation of mature mRNAs. Alternatively, splice morpholinos (splice-MO) that are directed against specific splice donor or acceptor sites result in inclusion of an intron, exclusion of an exon, or usage of an alternative splice site. To rule out potential unspecific effects, we employed three different morpholinos for each kcnh1 paralog, two splice-MO and one ATG-MO. In addition, one ATG-MO was designed to interfere with both paralogs (panATG-MO). As controls we used previously described ATG-MOs targeting the closely related kcnh2 potassium channel gene or the p53 tumor suppressor gene. Expression of kcnh2 is confined to the heart, and knockdown of this gene has been reported to cause pericardial edemas, the absence of circulation, and atrioventricular block (13, 14). Knockdown of p53 was reported to have no early phenotype, as expression of p53 in zebrafish does not start before 48 hpf (32).
The most dramatic effect of all kcnh1-directed MO injections was a dose-dependent mortality of early embryos. Fig. 5A summarizes the survival rates at 24 h. Although uninjected controls and embryos injected with the p53-targeting morpholino had almost 100% survival rates, all morpholinos targeting kcnh1 genes caused a dramatic reduction of survival irrespective of the mode of interference of the morpholino. At the lowest morpholino concentration (0.1 mm) about 50% of the morphants survived, and at the maximum concentration of 1 mm, the survival dropped below 20%. Apparently, both kcnh1 paralogs must be involved in vital functions during development of the first 24 h. Knockdown of the kcnh2 potassium channel gene had a milder phenotype and even at the highest concentration at least 50% of the embryos survived.
FIGURE 5.
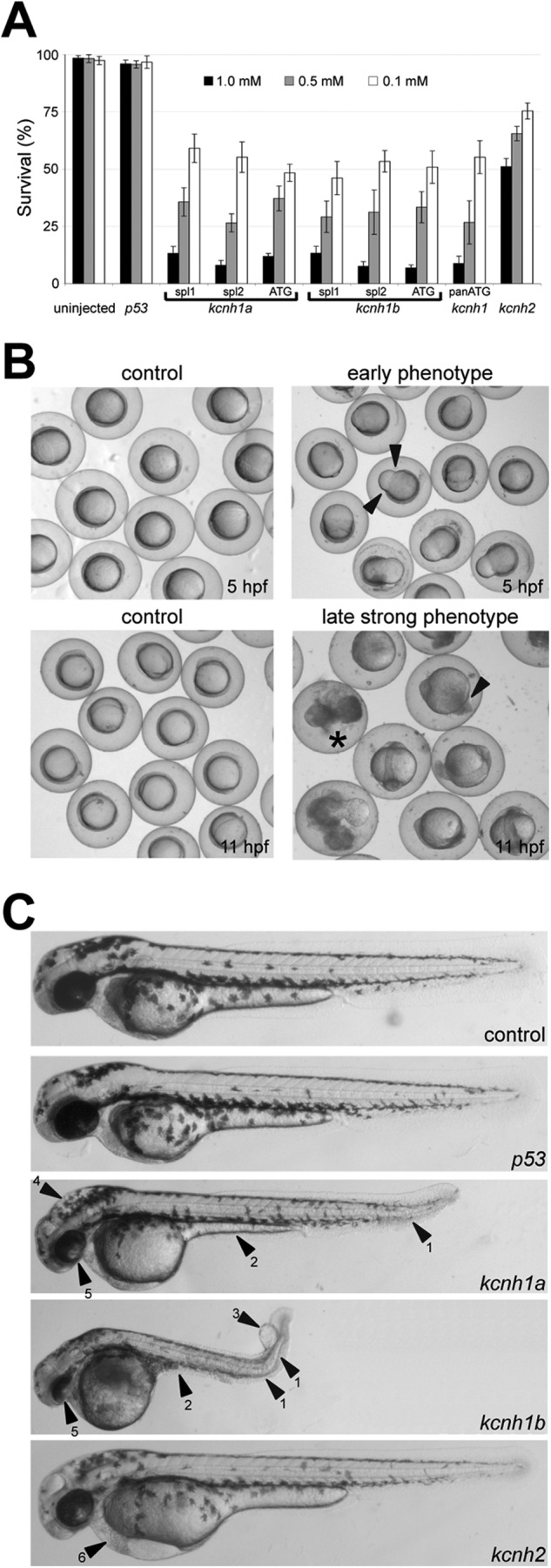
Phenotype of kcnh1a and kcnh1b zebrafish morphants. A, shown are survival rates of morphants at 24 h post-injection of the indicated MO concentrations. For both kcnh1a and kcnh1b, three different gene-specific MOs, affecting either splicing (spl1, spl2) or translation (ATG), were used to knock down gene function. A panATG MO was directed against both kcnh1 paralogs. Uninjected embryos and embryos injected with MOs targeting p53 or kcnh2 genes served as controls. Results are based on four independent injections (mean ± S.E., n = 20–50). B, shown are two representative phenotypes of kcnh1a morphants after injection of ATG-interfering MOs (0.5 mm) at the indicated time points. Arrowheads in the upper right image indicate a typical constriction of the marginal enveloping cell layer at 5 hpf. At later stages (11 hpf), a strong phenotype was characterized by progressive cell detachment at the tail region (lower right panel, arrowhead), and subsequent necrosis (asterisk). C, shown is a phenotype of kcnh1 morphants at 48 hpf in comparison to uninjected controls and p53 or kcnh2 morphants. All injections were performed with ATG-MO at 0.5 mm. Note the general growth retardation of kcnh1 morphants and frequent characteristics, such as kinked tails (arrowhead 1), yolk-sac extension defects (2), tail edemas (3), hydrocephaly (4), and poorly developed eyes (5). The bottom image shows a kcnh2 morphant with typical heart edema (arrowhead 6).
The earliest morphologic phenotype of kcnh1 morphants was a constriction of the marginal enveloping cell layer (Fig. 5B, upper right panel). In seven independent injections with three different morpholinos for each of the two kcnh1 paralogs as well as with the pan-ATG MO (all at 0.5 mm), this phenotype was detectable in 15- 30% of the injected embryos (n ≥ 75) at 5 hpf. This constriction did not occur in any of the three controls. Later, at 11 hpf, 16–37% of the kcnh1 morphants showed a strong phenotype with progressive cell detachment at the tails and eventually necrosis (Fig. 5B, lower right image). At 18 hpf most embryos had not developed further, and only 7–21% of the injected embryos in the seven groups of kcnh1 morphants showed no severe malformations but were clearly retarded in growth compared with uninjected embryos. Given the severity of the phenotype in early development, we assume that these embryos are hypomorphic, most likely due to incomplete kcnh1 knockdown.
Inspection of surviving morphants at 48 hpf revealed an overall growth retardation in all kcnh1 morphants, irrespective of morpholino type (Fig. 5C). Beyond this general phenotype, frequently observed characteristics of kcnh1 morphants were kinked tails (Fig. 5C, arrowhead 1), yolk sac extension defects (2), tail edemas (3), hydrocephaly (4), and poorly developed eyes (5). By contrast, p53 morphants showed no differences in comparison to uninjected controls. The kcnh2 morphants showed no growth retardation, but more than 50% of these embryos displayed typical heart edema (Fig. 5C, bottom, arrowhead 6).
To confirm efficiency and specificity of the morpholinos targeting kcnh1a and kcnh1b, we employed two strategies. First, we compared the efficiency of ATG-MOs on the protein level using modified GFP and RFP constructs harboring the morpholino target sequences upstream of the respective start codon supplemental Fig. S1, A–C; Table 1). Second, we examined the specificity and efficiency of kcnh1 splice-MOs in knocking down the respective wild-type mRNAs using RT-PCR (supplemental Fig. S1D). The kcnh1 ATG-MOs specifically blocked their target sequences supplemental Fig. S1C and Table 1), and the kcnh1splice-MOs specifically knocked down the levels of their target mRNAs in a dose-dependent manner supplemental Fig. S1D). The RT-PCR analyses also indicated lack of transcriptional compensation between kcnh1 paralogs, as the knockdown of one kcnh1 gene did not affect the mRNA levels of the other.
Knockdown of kcnh1a and kcnh1b Delays Hindbrain Formation and Somitogenesis
To gain insight into the events underlying the developmental defects in kcnh1 morphants, we analyzed the expression pattern of developmental markers via whole-mount in situ hybridization in morphants with a mild phenotype. Analysis of early fate specification during gastrulation (5 hpf) using the axial mesoderm marker no tail (ntl) did not reveal any differences between kcnh1a and kcnh1b morphants and uninjected controls supplemental Fig. S2). However, during embryonic segmentation (12 hpf), kcnh1a and kcnh1b morphants consistently displayed a lateral expansion of hindbrain territories labeled by krox20 (rhombomeres 3 and 5, Fig. 6, white arrowheads). In kcnh1 morphants, the rhombomeres 3 and 5 were thinner and wider than in controls, and rhombomere boundaries were less sharp. In kcnh1b morphants, often no krox20-positive cells had reached the midline axis of rhombomere 5 at this time point (Fig. 6, white arrowheads in the upper dorsal panel). Together these findings suggest that hindbrain development in kcnh1 morphants is delayed by several hours.
Labeling of somites in the axial mesoderm with a myoD probe was also indicative of a slower development of kcnh1 morphants. At 12 hpf the number of clearly distinguishable somites was reduced, and they failed to expand laterally as in the controls (Fig. 6, black arrowheads in the lower dorsal panel). These results indicate that kcnh1 MO disturbed the normal extension of the embryonic anterioposterior axis (myoD) and affected hindbrain patterning as visualized by the neural fate marker krox20.
Knockdown of kcnh1 Alters the Expression of Genes Involved in Cell-cycle Control
To gain an insight into potential mechanisms underlying the described phenotype in kcnh1 morphants, we used quantitative PCR and monitored the mRNA expression of genes required for zebrafish development. This gene set comprises transcription factors, cell-cycle regulators, protein kinases, and adhesion proteins. We assumed that the delay in hindbrain formation and somitogenesis at 12 hpf must be preceded by more subtle changes at earlier time points and chose the onset of constriction of the enveloping layer (70% epiboly, 7.5 hpf) for expression analysis. Consistent with our viability data (Fig. 5A), the knockdown of p53 had no significant impact compared with uninjected controls. Fig. 6B shows the changes of mRNA levels, ordered from up-regulated to down-regulated genes. At this time point, the mRNA levels of several genes showed no or only minor changes upon kcnh1-MO injection. Among the non-responsive genes are the protein kinase genes erk2, her4, jnk, and the transcription factor gata2. This demonstrates that gene expression in kcnh1 morphants is not generally disturbed. By contrast, the expression of three genes (N-cadherin, p21, p27) was up-regulated by more than 50% in at least one of the morphant groups. The products of p21 and p27 are known for their function as cyclin-dependent kinase inhibitors and, thus, inhibitors of cell proliferation. Also N-cadherin (cdh2) has recently been shown to restrict cell proliferation in zebrafish embryos (48). The up-regulation of N-cadherin was more pronounced in kcnh1a morphants and in embryos injected with the pan-kcnh1-MO than in kcnh1b morphants. A third group of genes was found down-regulated in kcnh1 morphants. The respective gene products have diverse functions, including secreted signaling proteins (wnt11, fgf8), transcription factors (foxD3, cfos, sox2), and cell-cycle regulators (cyclin D1). Of note, the down-regulation of cyclinD1 is in agreement with previous cell-based studies, reporting that siRNA targeting kcnh1 down-regulates cyclinD1 in human breast cancer cells (49). All the down-regulated genes are in one way or another associated with proliferation. Morpholino-induced gene repression was largely independent of the targeted kcnh1 paralog, with exception of sox2, which was not significantly altered in kcnh1a morphants, but was strongly reduced by kcnh1b-MO and pan-kcnh1-MO (Fig. 6B). Taken together, the quantitative PCR data reveal that delayed patterning and growth reduction seen at 12 hpf in knch1-depleted zebrafish embryos are preceded by specific alterations in the transcription levels of genes involved in the control of cell proliferation. We were interested to see whether the impact of kcnh1 knockdown on regulatory genes is predominantly quantitative or if the spatial expression of responsive genes is also disturbed. To address this question we selected three down-regulated genes (fgf8, wnt11, foxd3) and analyzed their expression domains by in situ hybridization at the gastrulation stage (90% epiboly) and during early somitogenesis supplemental Fig. S3). We did not observe prominent changes of the spatial expression profiles. However, a moderate reduction of signal intensity upon pan-kcnh1-MO injection was typically observed for fgf8 and foxd3 during gastrulation supplemental Fig. S3A).
DISCUSSION
The physiological and pathophysiological roles of the voltage-gated potassium channel Kcnh1 in humans and other mammals are far from being understood. Further investigations require suitable model organisms, allowing a faithful comparison to the situation in humans. We used zebrafish as a model to study expression and function of kcnh1 genes during vertebrate embryogenesis. We found that fish kcnh1 genes are duplicated (so called kcnh1a and kcnh1b) and highly conserved compared with the human ortholog. In adult fish we observed expression of both genes in the brain and expression of kcnh1a in the eye, which is consistent with the expression patterns found in mammals (3, 5). Kcnh1 inactivation led to severe and lethal phenotypes. Molecular analysis of kcnh1 loss-of-function phenotypes revealed a general delay of development in kcnh1 morphants. The normal anterio-posterior patterning of zebrafish embryos is impaired, with evident loss of neural fate markers in the hindbrain region and reduced expression of muscle fate markers during segmentation. Our results thus suggest a crucial role of Kcnh1 channels during embryogenesis.
Electrophysiological characterization of heterologously expressed Kcnh1a and Kcnh1b channels revealed a functional behavior similar to that of the human channel but with slightly increased single-channel conductance of the fish channels. The typical modulation of channel activation kinetics by prepulse potential and external Mg2+ ions is also conserved between human and fish channels. This may be explained by almost complete conservation of the transmembrane segments S2 through S4, harboring the binding site for external Mg2+ ions (50). Similarly, the CaM binding motifs identified in the human KCNH1 channel (36, 51) are well conserved in the fish orthologs, and Ca2+ sensitivity of both fish channels could be confirmed in Xenopus oocytes.
After a whole-genome duplication event, a common fate of duplicated genes is the loss of function of one gene copy by inactivating mutations (52). Alternatively, preservation of two functional gene copies may occur if one gene adapts a new function (neofunctionalization) or if both genes experience a partial loss of subfunctions (53). In the case of the kcnh1 genes in zebrafish, our heterologous expression data do not support neofunctionalization or loss of subfunctions. The analysis of mRNA expression of kcnh1a and kcnh1b in the developing embryo did not reveal evidence for temporal subfunctionalization, and both mRNA species were already present in the fertilized egg. We hypothesize that the two kcnh1 paralogs underwent at least in part a spatial subfunction partitioning, as we observed overlapping but not identical expression domains for the two genes. This is exemplified by the exclusive detection of mRNA for kcnh1a in the epiphysis at 24 hpf, whereas at the same developmental stage three bilateral domains exhibited prominent and exclusive staining for kcnh1b. For domains that coexpress both paralogs (e.g. primary neurons along the body axis), the formation of heteromeric channels must be assumed, as simultaneous expression in Xenopus oocytes provided evidence for mixed channel formation. This is in agreement with the previous observation that human channels of the EAG subfamily (KCNH1, KCNH5) also form heteromers (25). Functional coassembly of closely related channels has also been described for human eag-related gene (ERG) channels (45).
To date, the developmental activity of kcnh1 genes has not been reported in mammalian models nor have embryonic phenotypes been described in Drosophila eag mutants. Early development is regulated by a combination of maternal factors (deposited in the egg during embryogenesis) and zygotic factors generated by the embryo itself (46). Our data indicate that zebrafish kcnh1a and kcnh1b are two of such maternal factors crucial for development. The knockdown of either one of the kcnh1 genes led to severe phenotypes including early mortality and cell detachment.
In line with the predominantly neuronal expression of both kcnh1 genes, an obvious late effect of morpholino treatment was an initial retardation of the developing brain accompanied by the loss of hindbrain markers. With ongoing development, these effects extended to other parts of the body including the tail, and the whole embryos did not reach normal length. In this context it is important to note that expression of both kcnh1 paralogs was not restricted to the brain but was also detectable along the body axis in primary neurons of the spinal cord. Thus, the observed fundamental role of both kcnh1 paralogs is in line with the temporal and spatial expression patterns of both genes.
Morpholino antisense technology provides a powerful tool to study gene function in vivo, but off-target effects have also been described to cause neural cell death and even general retardation (54). To exclude misinterpretations, we controlled our MO experiments in two ways, 1) by comparing our kcnh1 morphant phenotypes with two independent MO and 2) by analyzing the extent of interference with normal kcnh1 pre mRNA splicing via RT-PCR and expression of modified GFP/RFP mRNAs.
Although the morpholino-induced phenotypic alterations became apparent with the development of early brain structures and the onset of segmentation, changes in the transcriptional profile of distinct genes were already evident at 7.5 hpf (70% epiboly). Some of the observed changes like the up-regulation of cell-cycle inhibitors p21 and p27 immediately suggest a mechanistic basis for the developmental delay at later time points. For the cell adhesion protein N-cadherin (cdh2), a causal link of up-regulation to the phenotypic changes is less intuitive, but a specific role for N-cadherin in zebrafish neural development has recently been demonstrated (48). N-cadherin was found to restrict cell proliferation in the dorsal neural tube, a mechanism that might well be involved in the impaired neural development of kcnh1 morphants. Principally, the impact of kcnh1 knockdown on responsive genes might also involve a spatial disorganization of expression, in addition to quantitative effects. However, our analysis of fgf8, wnt11, and foxd3 at later developmental stages did not reveal prominent changes in the respective expression patterns.
It remains to be determined how Kcnh1 proteins can influence gene expression in the developing embryo. The unique dependence of Kcnh1 channels on intracellular Ca2+, distinguishing it from Kcnh2 and other voltage-gated potassium channels, may be of special interest because Ca2+-dependent signals have a fundamental role in cellular signaling and a role for Ca2+ in patterning of the early zebrafish embryo is well established (for review, see Webb and Miller (55)). A recent report showing that human and rat Kcnh1 channels can also localize to the inner nuclear membrane (56) suggests that direct interactions with nuclear regulators and even with chromatin might also occur. Interestingly, of nine genes that we found regulated by kcnh1 depletion, two genes were affected specifically by kcnh1a (N-cadherin) or by kcnh1b (sox2), suggesting that some regulatory functions of the paralogs must be unique. Given that critical players in embryogenesis are often re-employed in tumorigenesis, the disclosed role of Kcnh1 in ontogenetic processes will also have consequences for the future understanding of Kcnh1 channels in cell proliferation and cancer.
Supplementary Material
Acknowledgments
We thank Wolfgang Driever for help in interpreting expression patterns, Birgit Perner for sharing expertise in microscopy and Angela Roβner, Steffi Arend as well as Christin Hahn for excellent technical support. We thank Ulrike Gausmann for valuable help with bioinformatic analysis.
This work was supported in part by a grant from the Deutsche Forschungsgemeinschaft (SFB 604) (to C. E. and to S. H. H.).

This article contains supplemental Table S1 and Figs. S1–3.
- hpf
- hours post fertilization
- MO
- morpholino
- HEDTA
- N-(2-hydroxyethyl)ethylenediaminetriacetic acid
- CaM
- calmodulin
- RFP
- red fluorescent protein.
REFERENCES
- 1. Kaplan W. D., Trout W. E. (1969) The behavior of four neurological mutants of Drosophila. Genetics 61, 399–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ganetzky B., Wu C. F. (1983) Neurogenetic analysis of potassium currents in Drosophila. Synergistic effects on neuromuscular transmission in double mutants. J. Neurogenet. 1, 17–28 [DOI] [PubMed] [Google Scholar]
- 3. Ludwig J., Weseloh R., Karschin C., Liu Q., Netzer R., Engeland B., Stansfeld C., Pongs O. (2000) Cloning and functional expression of rat eag2, a new member of the ether-à-go-go family of potassium channels and comparison of its distribution with that of eag1. Mol. Cell. Neurosci. 16, 59–70 [DOI] [PubMed] [Google Scholar]
- 4. Martin S., Lino de Oliveira C., Mello de Queiroz F., Pardo L. A., Stühmer W., Del Bel E. (2008) Eag1 potassium channel immunohistochemistry in the CNS of adult rat and selected regions of human brain. Neuroscience 155, 833–844 [DOI] [PubMed] [Google Scholar]
- 5. Saganich M. J., Machado E., Rudy B. (2001) Differential expression of genes encoding subthreshold-operating voltage-gated K+ channels in brain. J. Neurosci. 21, 4609–4624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bijlenga P., Occhiodoro T., Liu J. H., Bader C. R., Bernheim L., Fischer-Lougheed J. (1998) An ether -à-go-go K+ current, Ih-eag, contributes to the hyperpolarization of human fusion-competent myoblasts. J. Physiol. 512, 317–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Occhiodoro T., Bernheim L., Liu J. H., Bijlenga P., Sinnreich M., Bader C. R., Fischer-Lougheed J. (1998) Cloning of a human ether-a-go-go potassium channel expressed in myoblasts at the onset of fusion. FEBS Lett. 434, 177–182 [DOI] [PubMed] [Google Scholar]
- 8. Pardo L. A., del Camino D., Sánchez A., Alves F., Brüggemann A., Beckh S., Stühmer W. (1999) Oncogenic potential of EAG K+ channels. EMBO J. 18, 5540–5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gavrilova-Ruch O., Schönherr K., Gessner G., Schönherr R., Klapperstück T., Wohlrab W., Heinemann S. H. (2002) Effects of imipramine on ion channels and proliferation of IGR1 melanoma cells. J. Membr. Biol. 188, 137–149 [DOI] [PubMed] [Google Scholar]
- 10. Hemmerlein B., Weseloh R. M., Mello de Queiroz F., Knötgen H., Sánchez A., Rubio M. E., Martin S., Schliephacke T., Jenke M., Heinz-Joachim-Radzun, Stühmer W., Pardo L. A. (2006) Overexpression of Eag1 potassium channels in clinical tumors. Mol. Cancer 5, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Spitzner M., Ousingsawat J., Scheidt K., Kunzelmann K., Schreiber R. (2007) Voltage-gated K+ channels support proliferation of colonic carcinoma cells. FASEB J. 21, 35–44 [DOI] [PubMed] [Google Scholar]
- 12. Kalluri R., Weinberg R. A. (2009) The basics of epithelial-mesenchymal transition. J. Clin. Invest. 119, 1420–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Arnaout R., Ferrer T., Huisken J., Spitzer K., Stainier D. Y., Tristani-Firouzi M., Chi N. C. (2007) Zebrafish model for human long QT syndrome. Proc. Natl. Acad. Sci. U.S.A. 104, 11316–11321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hassel D., Scholz E. P., Trano N., Friedrich O., Just S., Meder B., Weiss D. L., Zitron E., Marquart S., Vogel B., Karle C. A., Seemann G., Fishman M. C., Katus H. A., Rottbauer W. (2008) Deficient zebrafish ether-à-go-go-related gene channel gating causes short-QT syndrome in zebrafish reggae mutants. Circulation 117, 866–875 [DOI] [PubMed] [Google Scholar]
- 15. Langheinrich U., Vacun G., Wagner T. (2003) Zebrafish embryos express an orthologue of HERG and are sensitive toward a range of QT-prolonging drugs inducing severe arrhythmia. Toxicol. Appl. Pharmacol. 193, 370–382 [DOI] [PubMed] [Google Scholar]
- 16. Milan D. J., Peterson T. A., Ruskin J. N., Peterson R. T., MacRae C. A. (2003) Drugs that induce repolarization abnormalities cause bradycardia in zebrafish. Circulation 107, 1355–1358 [DOI] [PubMed] [Google Scholar]
- 17. Mittelstadt S. W., Hemenway C. L., Craig M. P., Hove J. R. (2008) Evaluation of zebrafish embryos as a model for assessing inhibition of hERG. J. Pharmacol. Toxicol. Methods 57, 100–105 [DOI] [PubMed] [Google Scholar]
- 18. Scholz E. P., Niemer N., Hassel D., Zitron E., Bürgers H. F., Bloehs R., Seyler C., Scherer D., Thomas D., Kathöfer S., Katus H. A., Rottbauer W. A., Karle C. A. (2009) Biophysical properties of zebrafish ether-à-go-go-related gene potassium channels. Biochem. Biophys. Res. Commun. 381, 159–164 [DOI] [PubMed] [Google Scholar]
- 19. Brugada R., Hong K., Dumaine R., Cordeiro J., Gaita F., Borggrefe M., Menendez T. M., Brugada J., Pollevick G. D., Wolpert C., Burashnikov E., Matsuo K., Wu Y. S., Guerchicoff A., Bianchi F., Giustetto C., Schimpf R., Brugada P., Antzelevitch C. (2004) Sudden death associated with short-QT syndrome linked to mutations in HERG. Circulation 109, 30–35 [DOI] [PubMed] [Google Scholar]
- 20. Sanguinetti M. C., Jiang C., Curran M. E., Keating M. T. (1995) A mechanistic link between an inherited and an acquired cardiac arrhythmia. HERG encodes the IKr potassium channel. Cell 81, 299–307 [DOI] [PubMed] [Google Scholar]
- 21. Sanguinetti M. C., Tristani-Firouzi M. (2006) hERG potassium channels and cardiac arrhythmia. Nature 440, 463–469 [DOI] [PubMed] [Google Scholar]
- 22. Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B., Schilling T. F. (1995) Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310 [DOI] [PubMed] [Google Scholar]
- 23. Kumar S., Tamura K., Nei M. (1994) MEGA. Molecular evolutionary genetics analysis software for microcomputers. Comput. Appl. Biosci. 10, 189–191 [DOI] [PubMed] [Google Scholar]
- 24. Liman E. R., Tytgat J., Hess P. (1992) Subunit stoichiometry of a mammalian K+ channel determined by construction of multimeric cDNAs. Neuron 9, 861–871 [DOI] [PubMed] [Google Scholar]
- 25. Schönherr R., Gessner G., Löber K., Heinemann S. H. (2002) Functional distinction of human EAG1 and EAG2 potassium channels. FEBS Lett. 514, 204–208 [DOI] [PubMed] [Google Scholar]
- 26. Hauptmann G., Gerster T. (1994) Two-color whole-mount in situ hybridization to vertebrate and Drosophila embryos. Trends Genet. 10, 266. [DOI] [PubMed] [Google Scholar]
- 27. Schulte-Merker S., Ho R. K., Herrmann B. G., Nüsslein-Volhard C. (1992) The protein product of the zebrafish homologue of the mouse T gene is expressed in nuclei of the germ ring and the notochord of the early embryo. Development 116, 1021–1032 [DOI] [PubMed] [Google Scholar]
- 28. Weinberg E. S., Allende M. L., Kelly C. S., Abdelhamid A., Murakami T., Andermann P., Doerre O. G., Grunwald D. J., Riggleman B. (1996) Developmental regulation of zebrafish MyoD in wild-type, no tail, and spadetail embryos. Development 122, 271–280 [DOI] [PubMed] [Google Scholar]
- 29. Oxtoby E., Jowett T. (1993) Cloning of the zebrafish krox-20 gene (krx-20) and its expression during hindbrain development. Nucleic Acids Res. 21, 1087–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Thisse B., Pflumio S., Fürthauer M., Loppin B., Heyer V., Degrave A., Woehl R., Lux A., Steffan T., Charbonnier X., Thisse C. (2001) ZFIN Direct Data Submission, available online [Google Scholar]
- 31. Reifers F., Böhli H., Walsh E. C., Crossley P. H., Stainier D. Y., Brand M. (1998) Fgf8 is mutated in zebrafish acerebellar (ace) mutants and is required for maintenance of midbrain-hindbrain boundary development and somitogenesis. Development 125, 2381–2395 [DOI] [PubMed] [Google Scholar]
- 32. Robu M. E., Larson J. D., Nasevicius A., Beiraghi S., Brenner C., Farber S. A., Ekker S. C. (2007) p53 activation by knockdown technologies. PLoS Genet 3, e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McCurley A. T., Callard G. V. (2010) Time course analysis of gene expression patterns in zebrafish eye during optic nerve regeneration. J. Exp. Neurosci. 2010, 17–33 [PMC free article] [PubMed] [Google Scholar]
- 34. Livak K. J., Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 35. Wang M. H. (2004) A technical consideration concerning the removal of oocyte vitelline membranes for patch clamp recording. Biochem. Biophys. Res. Commun. 324, 971–972 [DOI] [PubMed] [Google Scholar]
- 36. Schönherr R., Löber K., Heinemann S. H. (2000) Inhibition of human ether à go-go potassium channels by Ca2+/calmodulin. EMBO J. 19, 3263–3271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Heinemann S. H., Conti F. (1992) Nonstationary noise analysis and application to patch clamp recordings. Methods Enzymol. 207, 131–148 [DOI] [PubMed] [Google Scholar]
- 38. Starkus J. G., Varga Z., Schönherr R., Heinemann S. H. (2003) Mechanisms of the inhibition of Shaker potassium channels by protons. Pflügers Arch. 447, 44–54 [DOI] [PubMed] [Google Scholar]
- 39. Kohn M., Högel J., Vogel W., Minich P., Kehrer-Sawatzki H., Graves J. A., Hameister H. (2006) Reconstruction of a 450-My-old ancestral vertebrate protokaryotype. Trends Genet 22, 203–210 [DOI] [PubMed] [Google Scholar]
- 40. Amores A., Force A., Yan Y. L., Joly L., Amemiya C., Fritz A., Ho R. K., Langeland J., Prince V., Wang Y. L., Westerfield M., Ekker M., Postlethwait J. H. (1998) Zebrafish hox clusters and vertebrate genome evolution. Science 282, 1711–1714 [DOI] [PubMed] [Google Scholar]
- 41. Taylor J. S., Braasch I., Frickey T., Meyer A., Van de Peer Y. (2003) Genome duplication, a trait shared by 22000 species of ray-finned fish. Genome Res 13, 382–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Venkatesh B. (2003) Evolution and diversity of fish genomes. Curr. Opin. Genet. Dev. 13, 588–592 [DOI] [PubMed] [Google Scholar]
- 43. Ludwig J., Terlau H., Wunder F., Brüggemann A., Pardo L. A., Marquardt A., Stühmer W., Pongs O. (1994) Functional expression of a rat homologue of the voltage gated either á go-go potassium channel reveals differences in selectivity and activation kinetics between the Drosophila channel and its mammalian counterpart. EMBO J. 13, 4451–4458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schönherr R., Mannuzzu L. M., Isacoff E. Y., Heinemann S. H. (2002) Conformational switch between slow and fast gating modes. Allosteric regulation of voltage sensor mobility in the EAG K+ channel. Neuron 35, 935–949 [DOI] [PubMed] [Google Scholar]
- 45. Wimmers S., Wulfsen I., Bauer C. K., Schwarz J. R. (2001) Erg1, erg2 and erg3 K+ channel subunits are able to form heteromultimers. Pflügers Arch. 441, 450–455 [DOI] [PubMed] [Google Scholar]
- 46. Pelegri F. (2003) Maternal factors in zebrafish development. Dev. Dyn. 228, 535–554 [DOI] [PubMed] [Google Scholar]
- 47. Summerton J. (1999) Morpholino antisense oligomers. The case for an RNase H-independent structural type. Biochim. Biophys. Acta 1489, 141–158 [DOI] [PubMed] [Google Scholar]
- 48. Chalasani K., Brewster R. M. (2011) N-cadherin-mediated cell adhesion restricts cell proliferation in the dorsal neural tube. Mol. Biol. Cell 22, 1505–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Borowiec A. S., Hague F., Gouilleux-Gruart V., Lassoued K., Ouadid-Ahidouch H. (2011) Regulation of IGF-1-dependent cyclin D1 and E expression by hEag1 channels in MCF-7 cells. The critical role of hEag1 channels in G1 phase progression. Biochim. Biophys. Acta 1813, 723–730 [DOI] [PubMed] [Google Scholar]
- 50. Silverman W. R., Tang C. Y., Mock A. F., Huh K. B., Papazian D. M. (2000) Mg2+ modulates voltage-dependent activation in ether-à-go-go potassium channels by binding between transmembrane segments S2 and S3. J. Gen. Physiol. 116, 663–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ziechner U., Schönherr R., Born A. K., Gavrilova-Ruch O., Glaser R. W., Malesevic M., Küllertz G., Heinemann S. H. (2006) Inhibition of human ether à go-go potassium channels by Ca2+/calmodulin binding to the cytosolic N and C termini. FEBS J. 273, 1074–1086 [DOI] [PubMed] [Google Scholar]
- 52. Ohno S. (1970) Evolution by Gene Duplication, Springer-Verlag, Berlin, New York [Google Scholar]
- 53. Force A., Lynch M., Pickett F. B., Amores A., Yan Y. L., Postlethwait J. (1999) Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151, 1531–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Heasman J. (2002) Morpholino oligos. Making sense of antisense? Dev. Biol. 243, 209–214 [DOI] [PubMed] [Google Scholar]
- 55. Webb S. E., Miller A. L. (2007) Ca2+ signaling and early embryonic patterning during zebrafish development. Clin. Exp. Pharmacol. Physiol. 34, 897–904 [DOI] [PubMed] [Google Scholar]
- 56. Chen Y., Sánchez A., Rubio M. E., Kohl T., Pardo L. A., Stühmer W. (2011) Functional K(v)10.1 channels localize to the inner nuclear membrane. PLoS One 6, e19257. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.