Background: Polysialic acid is a developmentally regulated posttranslational modification.
Results: Loss of the polysialyltransferase ST8SiaII but not ST8SiaIV abolished polysialylation of SynCAM 1 in the mouse brain.
Conclusion: Polysialylation of SynCAM 1 is mediated by ST8SiaII throughout postnatal mouse brain development.
Significance: Studying the molecular requirements for protein polysialylation is crucial for understanding how this process is regulated.
Keywords: Alternative Splicing, Brain, Glycosylation, Posttranslational Modification, Sialyltransferase, Polysialic Acid
Abstract
Polysialic acid is a unique carbohydrate polymer specifically attached to a limited number of glycoproteins. Among them is synaptic cell adhesion molecule 1 (SynCAM 1), a member of the immunoglobulin (Ig) superfamily composed of three extracellular Ig-like domains. Polysialylation of SynCAM 1 is cell type-specific and was exclusively found in NG2 cells, a class of multifunctional progenitor cells that form specialized synapses with neurons. Here, we studied the molecular requirements for SynCAM 1 polysialylation. Analysis of mice lacking one of the two polysialyltransferases, ST8SiaII or ST8SiaIV, revealed that polysialylation of SynCAM 1 is exclusively mediated by ST8SiaII throughout postnatal brain development. Alternative splicing of the three variable exons 8a, 8b, and 8c can theoretically give rise to eight transmembrane isoforms of SynCAM 1. We detected seven transcript variants in the developing mouse brain, including three variants containing exon 8c, which was so far regarded as a cryptic exon in mice. Polysialylation of SynCAM 1 was restricted to four isoforms in perinatal brain. However, cell culture experiments demonstrated that all transmembrane isoforms of SynCAM 1 can be polysialylated by ST8SiaII. Moreover, analysis of domain deletion constructs revealed that Ig1, which harbors the polysialylation site, is not sufficient as an acceptor for ST8SiaII. The minimal polypeptide required for polysialylation contained Ig1 and Ig2, suggesting an important role for Ig2 as a docking site for ST8SiaII.
Introduction
Synaptic cell adhesion molecule 1 (SynCAM 1)3 (also known as Cadm1, Necl-2 (nectin-like molecule 2), or TSLC1 (tumor suppressor in lung carcinoma 1)) is a member of the immunoglobulin superfamily that was identified in the nervous system as a potent inducer of synapse formation (1). SynCAM 1 is prominently expressed in the developing and mature brain, mediating Ca2+-independent homo- and heterophilic interactions across the nascent and mature synaptic cleft (1–3). In developing neurons, SynCAM 1 shapes migrating growth cones, assembles at axo-dendritic contacts, and participates in adhesive trans interactions that induce presynaptic specializations (4, 5). Moreover, studies on genetic mouse models with increased or no SynCAM 1 expression demonstrated a crucial role of this synapse-organizing molecule in regulating the number and plasticity of excitatory synapses (6).
SynCAM 1 is a single-spanning membrane protein with three extracellular Ig-like domains and a short cytosolic part (1). The first Ig-like domain provides the binding interface for homo- and heterophilic trans interactions, whereas Ig2 and Ig3 were shown to drive cis oligomerization of SynCAM 1 (5, 7). The three Ig-like domains contain six potential N-glycosylation sites, and the presence of N-glycans at Asn-80 and Asn-104 in Ig1 was demonstrated to be essential for synapse induction by promoting adhesive trans interactions of SynCAM 1 (7).
Genetic and bioinformatic characterization of the human and murine SynCAM 1 gene revealed that they are composed of 12 and 11 exons, respectively. Alternative pre-mRNA splicing results in the formation of several transmembrane isoforms and a secreted form that encompasses only the Ig-like domains of SynCAM 1 (8–11). In the case of human SynCAM 1, differential usage of three alternative exons, here termed exons 8a, 8b, and 8c, can theoretically lead to eight membrane-bound isoforms, which differ only in a short juxtamembranous extracellular stem region. In mice, however, the variable exon 8c has been described as cryptic exon, and expression has been reported only for transcript variants lacking this exon (8, 10, 11). Notably, the peptide sequences encoded by the variable exons contain a high number of potential O-glycosylation sites (8), and developmental changes in SynCAM 1 glycosylation that are unique among synaptic adhesion molecules have been observed (12).
Using a glycoproteomics approach, we recently identified SynCAM 1 as a novel target for polysialylation (13). Polysialic acid (polySia) is an unusual carbohydrate structure, composed of α2,8-linked 5-N-acetylneuraminic acid (Neu5Ac), that is predominantly present in the developing brain of vertebrates. PolySia was first discovered as a dynamically regulated posttranslational modification of the neural cell adhesion molecule NCAM, a member of the Ig superfamily that is composed of five Ig-like and two fibronectin type III repeats (14–16). In the nervous system, NCAM represents by far the major polySia carrier, and biological roles of polySia have been almost exclusively studied in relationship to NCAM. Due to its biophysical properties, the polyanionic and highly hydrated glycan polySia disrupts the adhesive properties of NCAM and acts as a key regulator of NCAM-mediated interactions (17–19). As a modulator of cell interactions, polySia impacts dynamic processes, such as migration of neural precursor cells, axon targeting and fasciculation, and synaptic plasticity (20, 21).
Synthesis of polySia is mediated by the two Golgi-resident polysialyltransferases (polySTs) ST8SiaII and ST8SiaIV (22, 23). They are type II transmembrane proteins and share 59% sequence identity on the amino acid level. During brain development, both enzymes reach peak level in the perinatal phase, followed by a postnatal down-regulation (24–27).
In contrast to NCAM, which is fully polysialylated in the perinatal brain, only a small fraction of SynCAM 1 serves as polySia carrier (13). Moreover, this fraction of polySia-SynCAM 1 was found to be restricted to a particular cell population called NG2 cells (13). These multifunctional progenitor cells form unique synaptic associations with neurons through which they receive excitatory synaptic inputs (28–30). On SynCAM 1, polySia is attached selectively to N-glycans at Asn116 located in the first Ig-like domain (13). Polysialylation completely abolished homophilic SynCAM 1 binding in vitro, suggesting a functional role of polySia in regulating SynCAM 1-mediated interactions of NG2 cells (13).
In the present study, we investigated the molecular requirements for SynCAM 1 polysialylation by determining (i) the enzyme responsible for SynCAM 1 polysialylation during postnatal brain development, (ii) the SynCAM 1 isoforms that are targets for polysialylation, and (iii) the protein parts of SynCAM 1 that serve as minimal acceptor structure.
EXPERIMENTAL PROCEDURES
Antibodies, Enzymes, and Reagents
The anti-polySia monoclonal antibody (mAb) 735 (IgG2a)(31) and endosialidase (endoN) were purified as described previously (32, 33). The rabbit polyclonal antibody (pAb) directed against the C-terminal domain of SynCAM 1 was purchased from Sigma-Aldrich. Chicken anti-SynCAM 1 mAb 3E1 (IgY) was obtained from MBL, and mouse anti-SynCAM 1 mAb L45/30 (IgG1) was obtained from the University of California Davis/National Institutes of Health NeuroMab Facility. Rabbit anti-human IgG-Fc pAb was purchased from Bethyl Laboratories. Horseradish peroxidase-conjugated antibodies were from Southern Biotech. Peptide-N-glycosidase F (PNGase F) was purchased from Roche Applied Science.
Animals
St8sia2−/−, St8sia4−/−, and Ncam−/− mice (22, 23, 34) have been backcrossed to the C57BL/6J genetic background for six generations. St8sia2−/− mice were provided by Jamey Marth (University of California, Santa Barbara, CA), and Ncam−/− mice were provided by Harold Cremer (Developmental Biology Institute of Marseille Luminy, Campus de Luminy, Marseille, France). All protocols for animal use were in compliance with the German law for the protection of animals and approved by the local authorities.
Reverse Transcription-PCR Analysis
Total RNA from mouse brains was extracted using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. 2.5 μg of total RNA of C57BL/6J mouse brain was reverse transcribed using RevertAid reverse transcriptase (Fermentas) according to the manufacturer's recommendations. Amplification of SynCAM 1-specific cDNA was performed by PCR with the primers SK4s (5′-GTGACCAGTCAGCTGATGCTG-3′) and SK5as (5′-AAAATAGCGGCCCAGAATGATGAGCAA-3′). PCR products were separated on 3% agarose gels and stained with ethidium bromide. To verify the identity of the PCR products, individual bands were excised from the gel. Extracted DNA was subcloned into the TOPO-TA cloning vector (Invitrogen) and analyzed by sequencing.
Expression Plasmids
Full-length cDNA of murine SynCAM 1 was transcribed from 1 μg of total RNA of perinatal C57BL/6J mouse brain using the SuperScript first-strand synthesis system (Invitrogen). SynCAM 1 cDNAs of the transcript variants 8−, 8a, 8ab, 8b, 8bc, and 8abc were amplified by PCR with the primer pair MR68s (5′-GATCGGTACCATGGCGAGTGCTGTGCTG-3′) and MR69as (5′-GCCATGCGGCCGCCTAGATGAAGTACTCTTTCTTTTCTTCG-3′). After digestion with KpnI and NotI, the obtained PCR products were ligated into the corresponding sites of pcDNA3.1-Zeo (Invitrogen). The identity of each PCR product was confirmed by sequencing and sequences of variants 8-, 8a, 8ab, 8b, and 8bc were identical to the reported sequences with GenBankTM accession numbers NM_001025600, NM_018770, NM_207675, NM_207676, and AB377192, respectively. To generate a cDNA encoding isoform 8ac, which was not expressed in the mouse brain, the sequence of exon 8b was deleted in pcDNA3.1-Zeo-SynCAM 1–8abc by a PCR-based approach using the primers MH37s (5′-CCTTACCATCATCACAGGCCTTACTCAGTTGC-3′) and MH37as (5′-GCAACTGAGTAAGGCCTGTGATGATGGTAAGG-3′). Based on the obtained plasmid pcDNA3.1-Zeo-SynCAM 1–8ac, a full-length construct encoding isoform 8c was generated by deletion of the exon 8a sequence using the primers MH38s (5′-GCTGTATGTATACGGCCTTACTCAGTTGC-3′) and MH38as (5′-GCAACTGAGTAAGGCCGTATACATACAGC-3′).
For generation of the plasmid encoding a SynCAM 1-Fc chimera composed of all three Ig-like domains of SynCAM 1 fused to the Fc part of human IgG1, the region encoding the corresponding extracellular domain of SynCAM 1 (amino acids 1–349) was amplified by PCR with primers MR91s (5′-GACTGCTAGCATGGCGAGTGCTGTGCTC-3′) and MR106as (5′-GATCAGATCTACTTACCTGTGTGGTCCACTGCCCCAATG-3′) using pcDNA3.1-Zeo-SynCAM 1-8− as template. The resulting PCR product was digested with NheI and BglII and ligated into the NheI/BamHI sites of pcDNA3.1-Ig upstream of the DNA sequence encoding the Fc part of human IgG1 (35). Constructs encoding the SynCAM 1-Fc chimera containing Ig1 and Ig2 (amino acids 1–242) and Ig1 (amino acids 1–145) were generated accordingly with primer pairs MR91s/MR107as (5′-GATCAGATCTACTTACCTGTCACTTCTAGATAGCGCTGGG-3′) and MR91s/MR92as (5′-GACTAGATCTACTTACCTGTAACCAGGACTGTGATGGTGG-3′), respectively. The plasmid pFlagHA-ST8SiaII encoding the full-length cDNA of murine ST8SiaII with an N-terminal FLAG-HA tag was constructed as described previously (36). The identity of all constructs was confirmed by sequencing.
Cell Culture and Transfection
CHO-K1, CHO-2A10, and LM-TK− cells were maintained in DMEM/Ham's F-12 medium (1:1) (Biochrom) supplemented with 5% FCS and 1 mm sodium pyruvate in a 37 °C, 5% CO2 incubator. CHO-2A10 cells are polySia-negative due to a defect in the ST8SiaIV gene (36, 37). Transfections were performed with Lipofectamine (Invitrogen) as described (32). For co-transfections, 2.5 × 105 cells were seeded per well of a 6-well plate. After 24 h in culture, cells were transfected using 6 μl of Lipofectamine per well together with a total of 1 μg of DNA containing 100 ng of plasmid encoding SynCAM 1, 100 ng of plasmid encoding ST8SiaII, and 800 ng of empty pcDNA3.1-Zeo vector. LM-TK− cells stably expressing FLAG-HA-tagged ST8SiaII were generated as described previously (32).
Immunoprecipitation
Mouse brains and cells were homogenized in 50 mm Tris-HCl, pH 8.0, containing 150 mm NaCl, 5 mm EDTA, 1% (v/v) Triton-X-100, 0.5% (w/v) sodium deoxycholate, 200 units/ml aprotinin, 2 mm phenylmethylsulfonyl fluoride, and 10 μg/ml leupeptin. For SynCAM 1 isolation, the supernatants of the obtained homogenates were mixed with anti-SynCAM 1 pAb (Sigma) and Protein G-Sepharose beads followed by incubation for 2 h on a rotating platform at 4 °C. For immunoprecipitation of polySia-SynCAM 1, mAb 735 coupled to M-280 tosyl-activated Dynabeads (Invitrogen) was used. Immunoprecipitates were washed three times with PBS and resuspended in sample buffer. For specific removal of polySia, one part of the isolated material was treated with 50 μg/ml endoN for 30 min at 37 °C.
Peptide-N-Glycosidase F Digestion
Immunoprecipitates were resuspended in 40 mm DTT, 0.5% SDS and boiled for 10 min. After adjusting the samples to a final concentration of 50 mm sodium phosphate, pH 7.5, 40 mm DTT, 0.5% SDS, and 1% Nonidet P-40, immunoprecipitates were incubated with PNGase F (50 units/ml) for 16 h at 37 °C.
In Vitro Polysialylation Assay
In vitro polysialylation of SynCAM 1-Fc was performed as described previously (13). Briefly, soluble SynCAM 1-Fc chimeras were produced in CHO-2A10 cells and immunoprecipitated from the cell culture supernatant with Protein A-Sepharose. After washing twice with PBS and twice with reaction buffer (10 mm MES, pH 6.7, 10 mm MnCl2), immunoprecipitates were incubated with 1 mm CMP-Neu5Ac (Nacalai Tesque) for 15 h at room temperature in the presence of 40 μg/ml of soluble murine ST8SiaII, lacking the transmembrane domain. After washing three times with PBS, beads were divided into two aliquots and resuspended in Laemmli sample buffer. For specific degradation of polySia, one aliquot was treated with 50 μg/ml SDS-resistant endoN for 30 min at 37 °C.
SDS-PAGE and Western Blot Analysis
SDS-PAGE and Western blotting was carried out as described (13) using the following primary antibodies: mAb 735 (5 μg/ml), mAb 3E1 (1 μg/ml), mAb L45/30 (1 μg/ml), and anti-SynCAM 1 pAb (0.5 μg/ml).
RESULTS
Polysialylation of SynCAM 1 during Postnatal Mouse Brain Development Depends Strictly on ST8SiaII
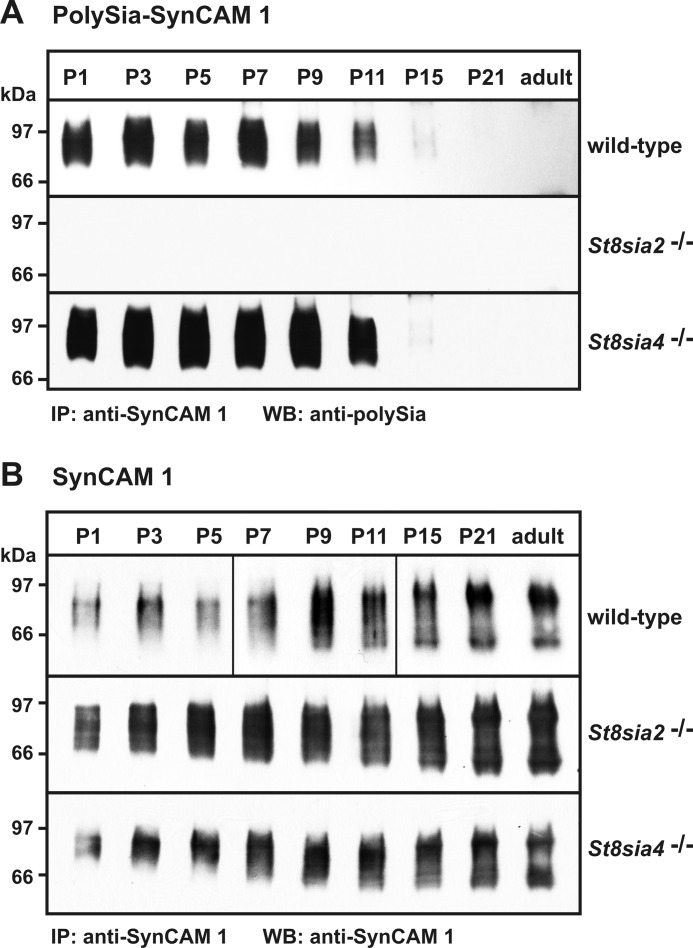
To dissect the contribution of the two polySTs ST8SiaII and ST8SiaIV to SynCAM 1 polysialylation, we compared the polysialylation level of SynCAM 1 in the postnatal brain of wild-type, St8sia2−/−, and St8sia4−/− mice. For each genotype, whole brains were collected at eight different time points between postnatal day 1 (P1) and P21 and from adult mice. SynCAM 1 was immunoprecipitated from brain lysates by a pAb directed against the intracellular C-terminal domain of SynCAM 1. The isolated protein was analyzed by Western blotting either with the polySia-specific mAb 735 to detect polySia-SynCAM 1 (Fig. 1A) or anti-SynCAM 1 mAb 3E1 to visualize the complete fraction of immunopreciptated SynCAM 1 (Fig. 1B). In brain homogenates of wild-type mice, analysis with mAb 735 revealed a broad band with an apparent molecular mass ranging from ∼80 to 110 kDa (Fig. 1A, top), representing the polysialylated form of SynCAM 1 (13). PolySia-SynCAM 1 was expressed at an almost constant level from P1 to P9, followed by a slight decrease toward P11. Between P11 and P15, however, a sharp drop in the polysialylation of SynCAM 1 was observed. At P15, minimal amounts of polySia-SynCAM 1 were still visible, but the signal reached the detection limit thereafter. In contrast, the overall expression level of SynCAM 1 stayed nearly constant (see top of Fig. 1B for SynCAM 1 analyzed after immunoprecipitation and supplemental Fig. S1A for total SynCAM 1 expression monitored by direct Western blot analysis of brain lysates). The total fraction of SynCAM 1 appeared as a diffuse band centering at 85 kDa throughout the first postnatal week. From P7 on, the SynCAM 1 signal started to extend toward the lower molecular mass range, and from P15 to adulthood, two prominent signals at ∼95 and ∼50 kDa were observed. Age-dependent variations in the SynCAM 1 protein pattern have been described previously and may reflect developmental changes in the iso- and glycoform pattern (1, 10).
FIGURE 1.
Expression of SynCAM 1 and polySia-SynCAM 1 in postnatal mouse brain of wild-type, St8sia2−/−, and St8sia4−/− mice. A, expression of polySia-SynCAM 1. Using an anti-SynCAM 1 pAb, SynCAM 1 was immunoprecipitated (IP) from whole brain lysates obtained at P1, P3, P5, P7, P9, P11, P21, and adult stage. Samples were separated by 8% SDS-PAGE and analyzed by Western blotting (WB) using the polySia-specific mAb 735. B, expression of SynCAM 1. To monitor SynCAM 1 expression in all three genotypes and to control the immunoprecipitation step performed in A, one aliquot of each sample analyzed in A was separated by 8% SDS-PAGE and analyzed by Western blotting using the anti-SynCAM 1 mAb 3E1. IP, immunoprecipitation; WB, Western blot analysis.
Compared with wild-type animals, polysialylation and overall expression level of SynCAM 1 were unchanged in St8sia4−/− mice. Moreover, down-regulation of polySia-SynCAM 1 followed the same time course (bottom panels in Fig. 1, A and B, respectively). Conversely, loss of ST8SiaII resulted in a complete absence of polySia-SynCAM 1 at all time points analyzed (see Fig. 1A, middle, and supplemental Fig. S1B for data obtained after prolonged exposure time). In contrast, no changes in the overall expression of SynCAM 1 were observed (middle panel of Fig. 1B and supplemental Fig. S1A). These data highlight that throughout postnatal brain development, ST8SiaII is essential for the polysialylation of SynCAM 1, whereas ST8SiaIV is dispensable for this process.
SynCAM 1 Transcript Variants Expressed during Postnatal Mouse Brain Development
In the perinatal phase, both ST8SiaII and ST8SiaIV reach peak levels and an almost ubiquitous expression pattern throughout the brain (24–26). At this stage, NCAM is completely polysialylated (27), whereas only a small subfraction of SynCAM 1 is modified by polySia (13). This raises the question whether polysialylation of SynCAM 1 is restricted to particular iso- and/or glycoforms in vivo. To address this point, we first analyzed the SynCAM 1 isoform pattern during postnatal mouse brain development.
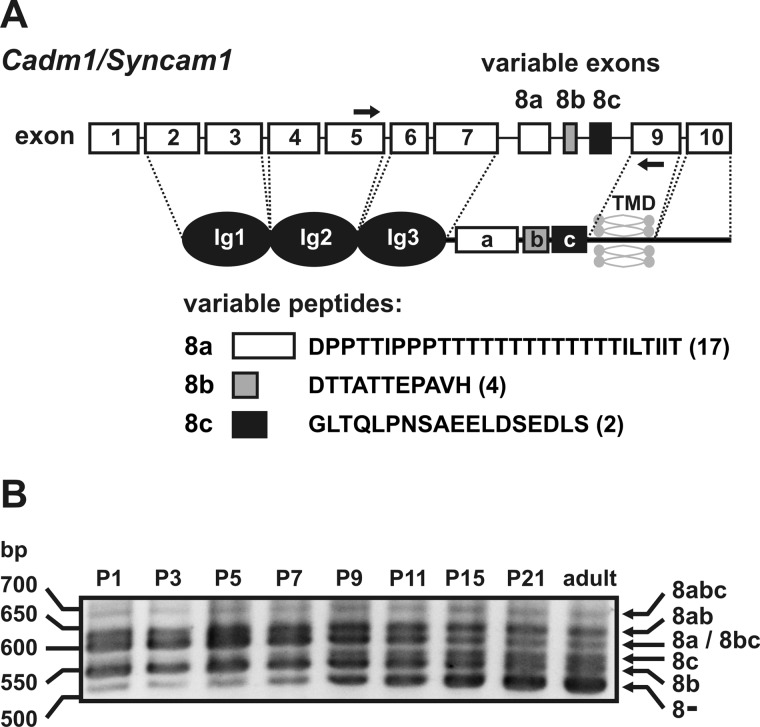
For informative designation of SynCAM 1 transcripts and isoforms, we named exons and variable peptides as shown in Fig. 2A. The N-terminal signal peptide of SynCAM 1 is encoded by exon 1, and the three Ig-like domains are encoded by exons 2–7. The three variable exons that encode short peptides located in the juxtamembranous extracellular stem region were designated as 8a, 8b, and 8c. The transmembrane region is encoded by exon 9 and the C-terminal intracellular domain by exon 10. For unequivocal designation of each transcript variant, we named them according to the absence (variant 8−) or presence of variable exons (e.g. variant 8b contains only exon 8b, whereas variant 8abc contains all variable exons). Consistent with the current view that the variable exon 8c is a cryptic exon in mouse (8, 10), we found no exon 8c-containing murine sequences in the databases when we searched specifically for expressed sequence tags. However, by general BLAST search analysis, we found partial and/or complete coding sequences for the exon 8c-containing murine transcript variants 8abc (GenBankTM accession number EU541476), 8bc (accession numbers EU541477 and AB37719), and 8c (accession number EU541478).
FIGURE 2.
SynCAM 1 transcript variants expressed during postnatal mouse brain development. A, schematic representation of the murine SynCAM 1 gene (Cadm1). Exons that are subject to alternative splicing were designated as 8a, 8b, and 8c. The dashed lines indicate which protein parts are encoded by particular exons. Peptides encoded by the variably spliced exons 8a, 8b, and 8c are shown as white, gray, and black boxes, respectively. Their peptide sequences are given together with the number of predicted O-glycosylation sites in parentheses. The arrows indicate the binding sites of the exon 5-specific forward primer SK4 and the exon 9-specific reverse primer SK5 used for RT-PCR in B. B, RT-PCR analysis of SynCAM 1 transcript variants during postnatal mouse brain development. Transcript variants encoding membrane-bound isoforms of SynCAM 1 were amplified by RT-PCR from total RNA of mouse brains collected at P1, P3, P5, P7, P9, P15, P21, and adult stage. PCR products were separated on a 3% agarose gel and assigned to individual transcript variants according to size (see arrows on the right). TMD, transmembrane domain.
To investigate which transcript variants of SynCAM 1 are expressed during postnatal mouse brain development, we performed RT-PCR analysis with an exon 5-specific forward and an exon 9-specific reverse primer as described by Hagiyama et al. (10). Based on the calculated fragment size, five of the obtained PCR products could be attributed to transcript variants 8−, 8b, 8c, 8ab, and 8abc (see Fig. 2B). Transcript variants 8a and 8bc differ only by 3 bp, and the corresponding PCR products could not be separated. However, sequencing of the PCR products demonstrated that during postnatal brain development, all possible combinations of the three variable exons were expressed with the single exception of variant 8ac, which was not found at any developmental stage.
Notably, transcript variant 8abc showed a constantly low expression level throughout postnatal brain development, which might have hampered the identification of this variant in previous studies (10, 11). Transcript variant 8c was hardly detectable before P7/P9 but was constantly expressed thereafter. The most prominent age-dependent changes were seen for variant 8−, which was strongly up-regulated between P1 and P21 and appeared as the predominant transcript variant in the adult mouse brain.
Polysialylation of SynCAM 1 in Perinatal Mouse Brain Was Restricted to Four Isoforms
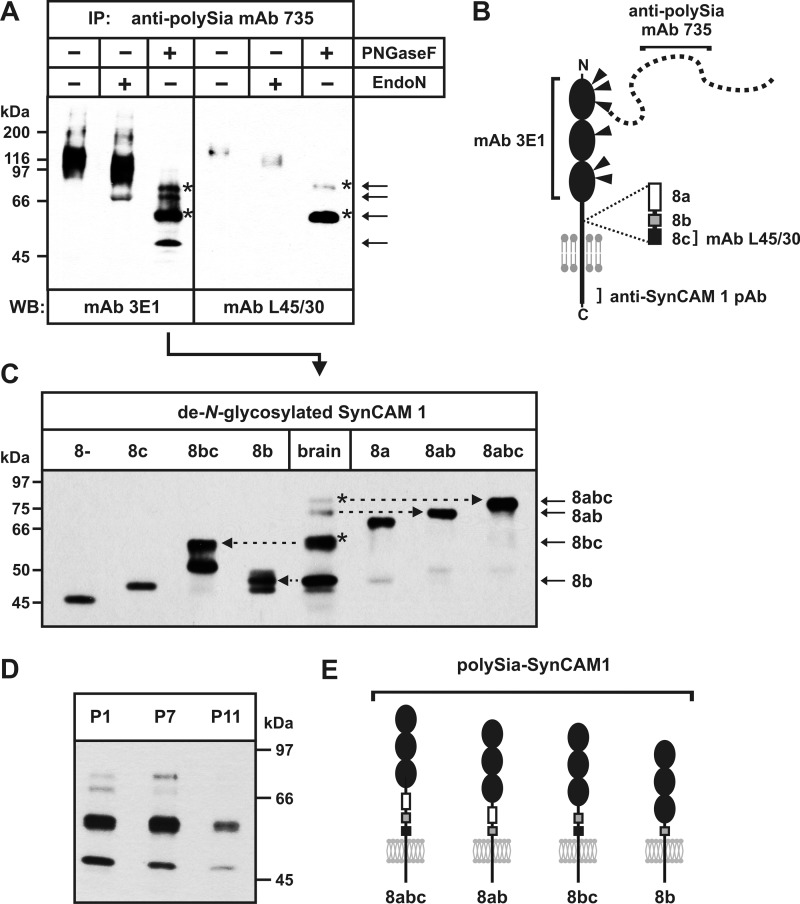
To analyze which SynCAM 1 isoforms serve as targets for polysialylation in vivo, we isolated the polysialylated fraction of SynCAM 1 from P1 brain by immunoprecipitation with the anti-polySia mAb 735. Because more than 95% of all polySia is attached to NCAM in the perinatal brain of wild-type mice (13), we used brains of Ncam−/− mice for efficient isolation of polySia-SynCAM 1. As shown previously, polysialylation of SynCAM 1 occurs independently of NCAM polysialylation and is identical in Ncam−/− and wild-type mice. The isolated polysialylated fraction of SynCAM 1 was characterized before and after specific removal of polySia by endoN or after de-N-glycosylation by PNGase F. After separation by SDS-PAGE, the material was analyzed by Western blotting using two different anti-SynCAM 1 antibodies: mAb 3E1 directed against the extracellular part of SynCAM 1 shared by all isoforms and mAb L45/30 directed against the variable peptide encoded by exon 8c (see Fig. 3B for schematic representation of the antibody binding sites).
FIGURE 3.
Isoform pattern of polysialylated SynCAM 1. A, analysis of polySia-SynCAM 1 immunoprecipitated (IP) from postnatal day 1 Ncam−/− mouse brain using the anti-polySia mAb 735. Immunoprecipitated polySia-SynCAM 1 was separated by 10% SDS-PAGE before and after the removal of polySia by endoN treatment or after de-N-glycosylation by PNGase F. Western blot analysis (WB) was performed with anti-SynCAM 1 antibodies against the extracellular part shared by all SynCAM 1 isoforms (mAb 3E1), or against the variable peptide encoded by exon 8c (mAb L45/30). The positions of the four protein bands obtained after de-N-glycosylation and staining with mAb 3E1 are indicated by arrows. Bands obtained after de-N-glycosylation that were stained by both mAb 3E1 and mAb L45/30 are marked by asterisks. B, schematic representation of anti-SynCAM 1 and anti-polySia antibody binding sites. mAb 3E1 is directed against the extracellular part of SynCAM 1 encompassing the Ig domains, mAb L45/30 detects specifically the variable peptide encoded by exon 8c, and the anti-SynCAM 1 pAb used in this study is directed against the C-terminal intracellular part of SynCAM 1. mAb 735 recognizes polySia composed of at least eight α2,8-linked sialic acids. C, assignment of SynCAM 1 isoforms that serve as polySia carriers in P1 mouse brain. PolySia-SynCAM 1 was immunoisolated from postnatal day 1 Ncam−/− mouse brain followed by de-N-glycosylation as described in A. In parallel, each of the indicated SynCAM 1 isoforms was transiently expressed in CHO cells, immunoprecipitated with the anti-SynCAM 1 pAb, and de-N-glycosylated by PNGase F. Proteins were separated by 8% SDS-PAGE and analyzed by Western blotting using anti-SynCAM 1 mAb 3E1. The four bands detected after de-N-glycosylation of mouse-derived polySia-SynCAM 1 (brain) are indicated by arrows on the right and can be assigned to the four isoforms 8abc, 8ab, 8bc, and 8b. Asterisks highlight those isoforms that contain the peptide encoded by exon 8c as demonstrated by staining with mAb L45/30 in A. D, isoform pattern of polySia-SynCAM 1 at different time points of postnatal brain development. Using the anti-polySia mAb 735, polySia-SynCAM 1 was immunoisolated from Ncam−/− mouse brain obtained at the indicated time points. The immunoprecipitated material was de-N-glycosylated and analyzed by Western blotting using anti-SynCAM 1 mAb 3E1. E, schematic representation of the four SynCAM 1 isoforms that serve as targets for polysialylation in postnatal mouse brain.
In the case of mAb 3E1 (Fig. 3A, left), a broad signal with an apparent molecular mass of 90–125 kDa was detected. Removal of polySia by endoN resulted in a moderate increase in electrophoretic mobility, whereas four discrete bands appeared after de-N-glycosylation by PNGase F. Their apparent molecular masses of about 50, 60, 70, and 80 kDa exceeded the masses of 42–48 kDa calculated for the unmodified SynCAM 1 isoforms, suggesting the presence of O-glycans. Moreover, the bands at 60 and 80 kDa were also detected by mAb L45/30 (Fig. 3A, right) which is specific for the peptide encoded by exon 8c (supplemental Fig. S2, bottom).
To assign the four bands obtained after de-N-glycosylation to particular isoforms, we compared their electrophoretic mobilities with the mobility of individual de-N-glycosylated SynCAM 1 isoforms. To obtain these marker proteins, we generated full-length cDNAs for each transmembrane isoform and expressed them individually in CHO cells. After immunoprecipitation by an anti-SynCAM 1 pAb recognizing the C-terminal domain, all isoforms were analyzed by Western blotting before and after PNGase F treatment (supplemental Fig. S2). All isoforms were modified by N-glycans as demonstrated by a significant increase in electrophoretic mobility after de-N-glycosylation. With the exception of isoforms 8− and 8c, the apparent molecular masses of the de-N-glycosylated isoforms exceeded the mass calculated for the unmodified polypeptide. This probably reflects the presence of O-glycans. The most prominent mass difference was seen for the peptide 8a-containing isoforms (supplemental Fig. S2), which is in agreement with 17, 4, and 2 O-glycosylation sites predicted for peptide 8a, 8b, and 8c, respectively.
Defined, de-N-glycosylated isoforms were then used as markers to assign the isoforms of SynCAM 1 that served as polySia carrier in P1 mouse brain. PolySia-SynCAM 1 was again immunoisolated from perinatal Ncam−/− brain using the anti-polySia mAb 735. After de-N-glycosylation, the electrophoretic mobilities of the four bands were directly compared with the mobility of the de-N-glycosylated forms of each individually expressed isoform (Fig. 3C). The bands at 80 and 70 kDa co-migrated with isoforms 8abc and 8ab, respectively. The band at 60 kDa co-migrated with the upper band of isoform 8bc, which represents most likely the fully O-glycosylated form. The fourth band at ∼48 kDa showed an electrophoretic mobility similar to that of isoforms 8b and 8c. However, due to the fact that this band was not recognized by the peptide 8c-specific mAb L45/30 (Fig. 3A, right), it was assigned to isoform 8b. Based on this approach, the isoforms underlying the polysialylated fraction of SynCAM 1 at P1 were identified as 8abc, 8ab, 8bc, and 8b, all characterized by the presence of an exon 8b-encoded peptide (Fig. 3E). For polySia-SynCAM 1 expressed at P7, we observed the same set of four isoforms, with 8b and 8bc being the major polySia carriers (Fig. 3D). At P11, when polysialylation of SynCAM 1 starts to decrease (Fig. 1A), less SynCAM 1 was pulled out by anti-polySia mAb 735 (Fig. 3D). For this material, only isoforms 8b and 8bc were detected as underlying polySia carriers (Fig. 3D). Isoforms 8ab and 8abc, which had been observed as minor polySia carriers at P1, might have been below the detection limit at P11 due to the observed overall decrease in SynCAM 1 polysialylation at this time point.
ST8SiaII Can Polysialylate All Transmembrane-bound Isoforms of SynCAM 1
Aimed at understanding the impact of the variable peptides for efficient polysialylation of membrane-bound SynCAM 1, we studied polysialylation of individual isoforms in cell culture experiments. However, all of our initial attempts to generate polysialylated SynCAM 1 by transient or stable co-expression of ST8SiaII and SynCAM 1 failed. After we had established which isoforms of SynCAM 1 were polysialylated in vivo, we selected one of them, namely isoform 8abc, and optimized co-expression conditions in the murine cell line LMTK−. This fibroblast cell line lacks endogenous expression of SynCAM 1 (supplemental Fig. S3) and is negative for NCAM and both polySTs (32). Under standard transfection conditions, SynCAM 1 was extremely well expressed, whereas only small amounts of polySia-SynCAM 1 were generated (supplemental Fig. S3). Decreasing the amount of SynCAM 1 plasmid resulted in decreased expression of SynCAM 1 but in parallel resulted in increased levels of polySia-SynCAM 1, suggesting a detrimental effect of increased SynCAM 1 expression on the polysialylation of this acceptor molecule. Optimal polysialylation of SynCAM 1 was achieved by transient co-transfection of low but equal amounts of SynCAM 1 and ST8SiaII cDNA (see supplemental Fig. S3, right).
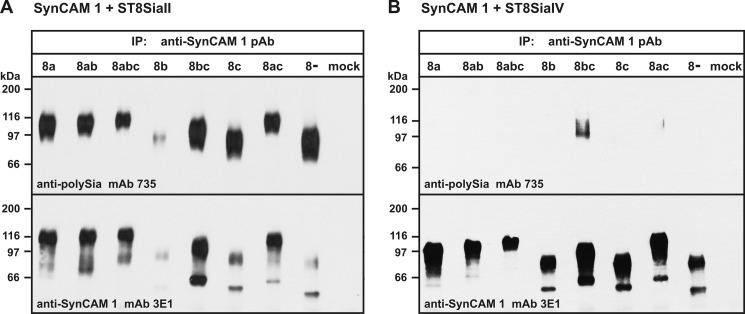
Under these optimized conditions, each SynCAM 1 isoform was then individually co-expressed with ST8SiaII (Fig. 4A). After immunoprecipitation of SynCAM 1 with anti-SynCAM 1 pAb, the polysialylation status and overall expression level of SynCAM 1 was compared by Western blot analysis using anti-polySia mAb 735 (Fig. 4A, top) and anti-SynCAM 1 mAb 3E1 (Fig. 4A, bottom), respectively. Notably, polysialylated SynCAM 1 was detected for all SynCAM 1 isoforms, including isoform 8− lacking all variable peptides. Thus, ST8SiaII has the ability to polysialylate all transmembrane isoforms of SynCAM 1 independent of the presence or absence of variable peptides.
FIGURE 4.
Polysialylation of SynCAM 1 isoforms. A, polysialylation of SynCAM 1 isoforms by ST8SiaII. Each transmembrane isoform of SynCAM 1 was co-expressed with ST8SiaII in the murine fibroblast cell line LM-TK−, and SynCAM 1 was immunoprecipitated (IP) with an anti-SynCAM 1 pAb directed against the C-terminal intracellular domain. Each isoform was then analyzed by SDS-PAGE and Western blotting using anti-polySia mAb 735 to detect polySia-SynCAM 1 (top) or anti-SynCAM 1 mAb 3E1 to control the overall expression of SynCAM 1 (bottom). Exposure time for both blots was 15 s. B, polysialylation of SynCAM 1 isoforms by ST8SiaIV. Each transmembrane isoform of SynCAM 1 was co-expressed with ST8SiaIV in LM-TK− cells, and SynCAM 1 expression was analyzed as in A. Exposure times were 1 min and 20 s for the top and bottom, respectively.
ST8SiaII Acts More Efficiently on SynCAM 1 than ST8SiaIV
The finding that polysialylation of SynCAM 1 relies exclusively on ST8SiaII in vivo (Fig. 1A) prompted us to compare the ability of ST8SiaII and ST8SiaIV to polysialylate this acceptor molecule in a cellular context. The different SynCAM 1 isoforms were now co-expressed with ST8SiaIV (Fig. 4B). After an exposure time of 1 min, polysialylation was only seen for isoforms 8bc and 8ac (Fig. 4B, top), and prolonged exposure times of ≥10 min were required to detect faint signals for all isoforms (data not shown). By contrast, co-expression with ST8SiaII resulted in strong polySia-SynCAM 1 signals that appeared already after an exposure time of 15 s (Fig. 4A). Moreover, analysis with an anti-SynCAM 1 antibody revealed that SynCAM 1 that was co-expressed with ST8SiaII showed reduced electrophoretic mobility compared with SynCAM 1 expressed in the presence of ST8SiaIV (Fig. 4, A and B, compare bottom panels). The observed shift in mobility indicates that ST8SiaII had converted the majority of SynCAM 1 to the polysialylated form, whereas ST8SiaIV had modified only a limited portion of the SynCAM 1 pool.
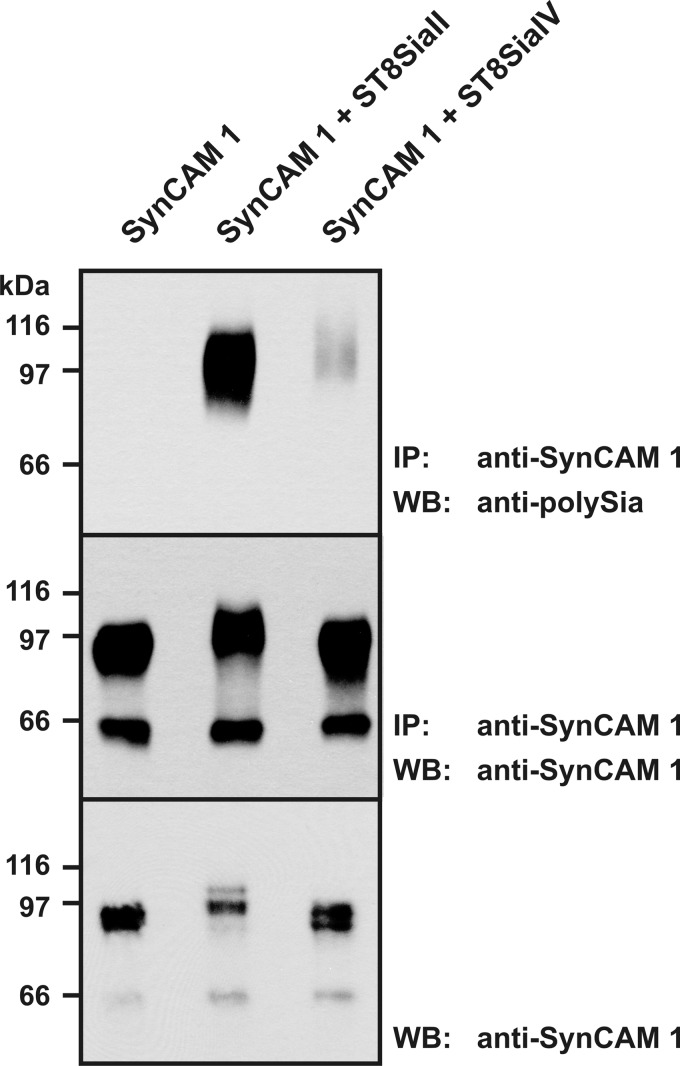
To visualize these differences on the same blot, SynCAM 1 of the isoform 8bc was expressed either alone or together with ST8SiaII or ST8SiaIV (Fig. 5). Although similar amounts of SynCAM 1 were expressed and immunoprecipitated in all three settings (Fig. 5, bottom and middle, respectively), clear differences in the extent of polysialylation were observed. Although co-expression of ST8SiaII resulted in a strong polySia-SynCAM 1 signal, only tiny amounts of polysialylated SynCAM 1 were detected in the presence of ST8SiaIV (Fig. 5, top). Analysis of the total fraction of SynCAM 1 (Fig. 5, bottom) revealed that co-expression with ST8SiaII but not ST8SiaIV resulted in a shift in electrophoretic mobility. Together, these experiments provide the first evidence that ST8SiaII and ST8SiaIV are distinct in their ability to use SynCAM 1 as an acceptor substrate.
FIGURE 5.
Comparison of the SynCAM 1 polysialylation capacity of ST8SiaII and ST8SiaIV. SynCAM 1 of the transmembrane isoform 8bc was transiently expressed in LM-TK− cells, either alone or together with ST8SiaII or ST8SiaIV. After immunoprecipitation (IP) with an anti-SynCAM 1 pAb directed against the C-terminal intracellular domain, SynCAM 1 was analyzed by Western blotting (WB) using anti-polySia mAb 735 to detect polySia-SynCAM 1 (top) or anti-SynCAM 1 mAb 3E1 to control the immunoprecipitation step (middle). In parallel, equal amounts of lysate were analyzed directly by Western blotting using anti-SynCAM 1 mAb 3E1 (bottom).
The Ig-like Domains Ig1 and Ig2 Are Sufficient for in Vitro Polysialylation of SynCAM 1
In order to identify the minimal structural requirements for SynCAM 1 polysialylation, soluble SynCAM 1-Fc chimera were generated encompassing (i) all three Ig-like domains, (ii) Ig1 and Ig2, or (iii) exclusively Ig1, the domain that carries the polysialylation site (13). After secretory expression in CHO-2A10 cells, which lack endogenous polyST activity (36, 37), SynCAM 1-Fc chimeras were adsorbed to Protein A-Sepharose and incubated with purified soluble ST8SiaII and the donor substrate CMP-Neu5Ac. Reaction products were analyzed before and after endoN treatment by Western blotting. Polysialylated SynCAM 1 was visualized by the polySia-specific mAb 735 (Fig. 6, top), whereas overall expression of the different Fc chimera was controlled by additional staining with an anti-Fc antibody (Fig. 6, bottom).
FIGURE 6.
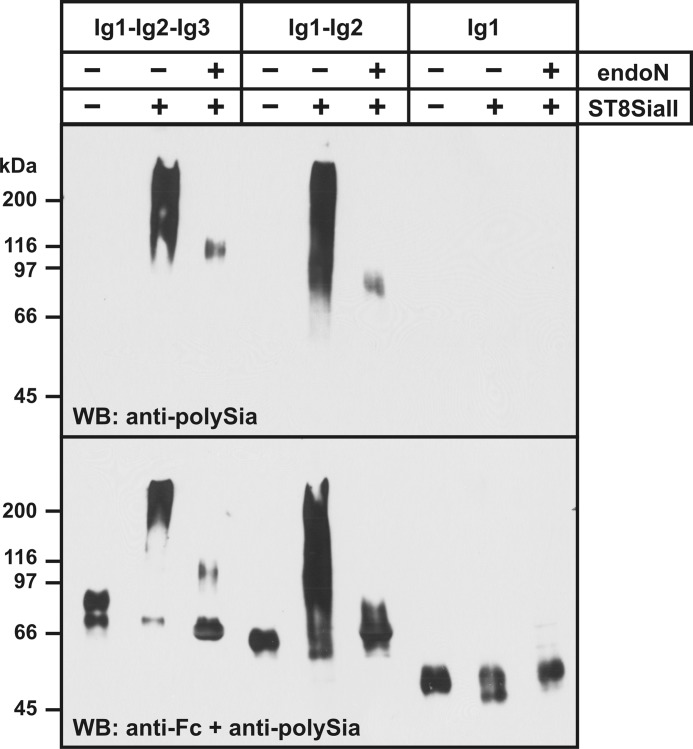
In vitro polysialylation of SynCAM 1 domain deletion constructs. Soluble SynCAM 1-Fc chimeras containing all three Ig domains of SynCAM 1 (Ig1-Ig2-Ig3), the first two Ig domains (Ig1-Ig2), or only Ig1 were expressed in CHO-2A10 cells. After adsorption to Protein A-Sepharose beads, SynCAM 1-Fc was incubated with ST8SiaII in the presence of CMP-Neu5Ac. Reaction products were separated by 8% SDS-PAGE before and after removal of polySia by endoN treatment and were analyzed by Western blotting (WB) with anti-polySia mAb 735 (top) or a combination of anti-polySia and anti-Fc antibodies (bottom).
For the SynCAM 1-Fc chimeras encompassing all three Ig domains or exclusively Ig1 and Ig2, endoN-sensitive signals were obtained with mAb 735, demonstrating that these variants were polysialylated by ST8SiaII. By contrast, no polysialylation was observed for SynCAM 1-Fc composed of only Ig1, although similar amounts of this chimera were adsorbed to Protein A-Sepharose (Fig. 6, bottom). This finding demonstrates that Ig1, the domain harboring the polysialylation site, is not sufficient for polysialylation in vitro and suggests an important role of Ig2 as recognition site for ST8SiaII.
DISCUSSION
In contrast to most other glycosyltransferases, which modify glycan structures irrespective of the underlying protein, the two polySTs ST8SiaII and ST8SiaIV are highly protein-specific and polysialylate only a restricted set of acceptor molecules. In addition to the major polySia carrier NCAM, only a few mammalian glycoproteins have been identified that can be modified by polysialylation: the α-subunit of the voltage-gated sodium channel in adult rat brain (38); the scavenger receptor CD36 in human milk (39); the cell surface glycoprotein neuropilin-2 on human dendritic cells (40); SynCAM 1 on NG2 cells of the developing mouse brain (13); and the two polySTs themselves, which can polysialylate their own N-glycans in a process called autopolysialylation (41, 42). Genetic mouse models lacking either ST8SiaII or ST8SiaIV were generated to unravel the impact of each polyST in vivo (22, 23). So far, these models have been used exclusively to study the function of the two polySTs during polysialylation of the most prevalent polySia carrier NCAM, which can be polysialylated by ST8SiaII and ST8SiaIV (22, 23, 43–45). In accordance with the observed differences in expression patterns, ST8SiaII was found to be the predominant enzyme during early postnatal development, important to keep the complete brain NCAM pool in the polysialylated state, whereas in the adult stage, NCAM polysialylation depends almost exclusively on ST8SiaIV (22–24, 27, 46). During development, both enzymes contribute to NCAM polysialylation, and in the absence of ST8SiaII, more than half of the NCAM pool is kept in the polysialylated state (26). Moreover, direct comparison of polySia-NCAM from perinatal brain of wild-type, St8sia2−/−, and St8sia4−/− mice revealed only minor differences, highlighting that both ST8SiaII and ST8SiaIV have the capacity to synthesize polySia of comparable quality on NCAM (26, 47).
In contrast to NCAM polysialylation, the present study demonstrates that throughout postnatal brain development, polysialylation of SynCAM 1 is catalyzed exclusively by ST8SiaII. Loss of ST8SiaII completely abolished SynCAM 1 polysialylation, whereas no significant changes were observed in the absence of ST8SiaIV. Thus, our data highlight a unique role of ST8SiaII in polysialylation of SynCAM 1. During postnatal brain development, prominent signals for polySia-SynCAM 1 were observed between P1 and P11, whereas polysialylation of SynCAM 1 was drastically down-regulated thereafter, resulting in a sharp drop between P11 and P15. This time course followed the previously described down-regulation of ST8SiaII transcript level, which dropped by 95% during postnatal brain development, with the strongest reduction between P5 and P11 (27).
Previously, we have shown that in vitro, both polySTs, ST8SiaII and ST8SiaIV, were able to polysialylate SynCAM 1. These in vitro polysialylation experiments were performed with truncated, soluble proteins that lack their transmembrane domains. In vivo, however, polySTs and SynCAM 1 are transmembrane proteins that are fixed to the Golgi membrane during the polysialylation step. The observation that ST8SiaIV cannot compensate for ST8SiaII deficiency in vivo suggests that SynCAM 1 is a poor substrate for ST8SiaIV under in vivo conditions. This was further substantiated by our co-expression experiments, demonstrating that in the cellular context, ST8SiaII is much more efficient in polysialylating SynCAM 1 than ST8SiaIV. Although ST8SiaIV was able to recognize SynCAM 1 as an acceptor molecule, the enzyme modified only a limited portion of the SynCAM 1 pool, resulting in only minute amounts of polySia-SynCAM 1. By contrast, ST8SiaII clearly outperformed ST8SiaIV in its ability to modify almost the complete pool of SynCAM 1.
Alternative splicing, representing a versatile mechanism for generating a large pool of functionally unique proteins, is particularly frequent in the nervous system (48, 49). In the case of SynCAM 1, alternative splicing of the short variable exons 8a, 8b, and 8c can, theoretically, give rise to eight transmembrane isoforms of SynCAM 1. Here, we show that all of them with the single exception of isoform 8ac were expressed during postnatal mouse brain development. So far, exon 8c has been regarded as cryptic exon in mice (8). However, based on RT-PCR and sequencing of the obtained products, we demonstrated the occurrence of three exon 8c-containing transcript variants (i.e. 8abc, 8bc, and 8c), which were not identified in similar approaches performed previously to study developmental changes in the SynCAM 1 transcript pattern in the mouse brain (10, 11).
Notably, the variable peptides encoded by exon 8a, 8b, and 8c contain 17, 4, and 2 putative O-glycosylation sites, respectively, resulting in numerous glycoforms. Stretches of O-glycosylation sites are known to influence protein conformation, leading to an elongated overall structure, as described for P-selectin glycoprotein ligand-1 and ovine submaxillary mucin (50, 51). Accordingly, a highly O-glycosylated stem region in SynCAM 1 was suggested to position the Ig-like domains away from the plasma membrane, thereby regulating their accessibility for trans interactions (8). Moreover, O-glycosylation may affect cis oligomerization of SynCAM 1, which was shown to precede synapse assembly and to promote trans adhesion (5).
On the transcript level, we observed the most striking developmental changes for isoform 8−, which lacks an O-glycosylated stem region. Although expressed at a low level during the first postnatal week, transcript variant 8− became the most prevalent variant in the adult brain (Fig. 2B). This suggests that SynCAM 1 isoforms with high numbers of potential O-glycosylation sites may dominate in early brain development, whereas isoform 8−, lacking any O-glycosylation sites, becomes the major isoform in the adult stage. These developmental changes might regulate the distinct functions of SynCAM 1 in the developing and mature brain (4–6, 12).
Precise knowledge on the SynCAM 1 isoform pattern and recombinant expression of individual isoforms enabled us to identify the isoforms underlying polySia-SynCAM 1 as 8abc, 8ab, 8bc, and 8b. Although polysialylation of SynCAM 1 was restricted to four particular isoforms in vivo, our data obtained by cell culture experiments demonstrated that ST8SiaII is able to polysialylate all transmembrane isoforms of SynCAM 1 in vitro. Thus, the four isoforms that serve as targets for polysialylation in vivo represent most likely the SynCAM 1 isoforms expressed in NG2 cells, the cells in which polysialylation of SynCAM 1 occurs. In vivo, polysialylation of SynCAM 1 is restricted to these cells, suggesting that they provide the particular environment required for SynCAM 1 polysialylation. The key prerequisite might be a critical balance between SynCAM 1 and ST8SiaII. High expression levels of the acceptor SynCAM 1 prevented its efficient polysialylation by ST8SiaII, as demonstrated by our co-expression experiments. SynCAM 1 has been shown to undergo excessive lateral self-assembly, a process that precedes the engagement of SynCAM 1 in trans interactions and is mediated by the membrane proximal Ig domains 2 and 3 (5). Thus, elevated SynCAM 1 levels may favor cis oligomerization, and the resulting clustering of SynCAM 1 in the membrane might reduce its accessibility for ST8SiaII.
Previously, we demonstrated that polySia is selectively attached to SynCAM 1 via N-glycans at Asn-116 located in the first Ig-like domain (13). By analysis of domain deletion variants of soluble SynCAM 1 variants, we now established the minimal structural requirements for SynCAM 1 polysialylation. Although Ig1 harbors the polysialylation site, this domain alone did not serve as an acceptor substrate for ST8SiaII. However, a variant encompassing Ig1 and Ig2 was efficiently polysialylated, and no significant differences in polysialylation were observed compared with a variant containing all three Ig-like domains. These findings indicate that Ig2 provides the critical interface for the recognition of SynCAM 1 by ST8SiaII.
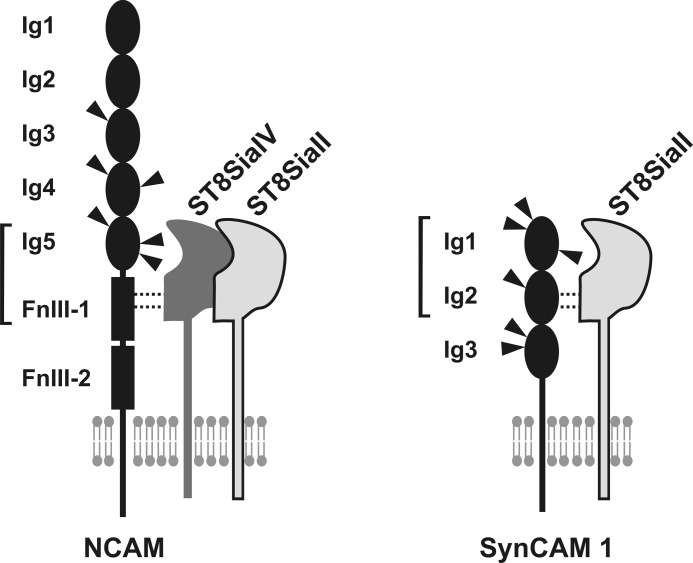
As summarized in Fig. 7, the minimal structural requirements for polysialylation of SynCAM 1 are highly reminiscent of what was observed for NCAM. The extracellular part of NCAM is composed of five Ig-like domains followed by two fibronectin type III repeats (FnIII-1 and -2) (52). Among the six N-glycosylation sites of NCAM, only two particular sites located in Ig5 are used for polysialylation (53–55). Nevertheless, Ig5 alone did not serve as an acceptor substrate for the polySTs, whereas a truncated NCAM variant encompassing the tandem domains Ig5 and FnIII-1 was efficiently polysialylated (56). The importance of the FnIII-1 domain as a recognition site was further substantiated by polyST-NCAM interaction studies (57). Subsequent structural analysis of FnIII-1 revealed the presence of an α-helix, an unusual feature among FnIII repeats, and an acidic surface patch essentially required for NCAM polysialylation (58, 59).
FIGURE 7.
Minimal structural requirements for the polysialylation of NCAM and SynCAM 1. Tandem domains representing the minimal structural requirement for polysialylation of NCAM and SynCAM 1 are highlighted by brackets.
For SynCAM 1, structural information is so far only available for the C-terminal intracellular domain (60). Future work focused on the structural analysis of the extracellular part will help to identify structural features in Ig2 that provide the docking sites for ST8SiaII and define SynCAM 1 as a target for polysialylation.
Supplementary Material
Acknowledgments
We are grateful to Rita Gerardy-Schahn for helpful discussions and continuous support. We thank Françoise Routier and Herbert Hildebrandt for critical remarks on the manuscript.
This work was supported by Deutsche Forschungsgemeinschaft Grants GE 527/5 (to H. G.) and MU 1774/3 (to M. M.).

This article contains supplemental Figs. S1–S3.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s)JQ782219.
- SynCAM 1
- synaptic cell adhesion molecule 1
- Cadm1
- cell adhesion molecule 1
- endoN
- endosialidase
- FnIII
- fibronectin type III domain
- NCAM
- neural cell adhesion molecule 1
- Neu5Ac
- 5-N-acetylneuraminic acid
- pAb
- polyclonal antibody
- Pn
- postnatal day n
- PNGase F
- peptide-N-glycosidase F
- polySia
- polysialic acid
- polyST
- polysialyltransferase.
REFERENCES
- 1. Biederer T., Sara Y., Mozhayeva M., Atasoy D., Liu X., Kavalali E. T., Südhof T. C. (2002) SynCAM, a synaptic adhesion molecule that drives synapse assembly. Science 297, 1525–1531 [DOI] [PubMed] [Google Scholar]
- 2. Sara Y., Biederer T., Atasoy D., Chubykin A., Mozhayeva M. G., Südhof T. C., Kavalali E. T. (2005) Selective capability of SynCAM and neuroligin for functional synapse assembly. J. Neurosci. 25, 260–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Thomas L. A., Akins M. R., Biederer T. (2008) Expression and adhesion profiles of SynCAM molecules indicate distinct neuronal functions. J. Comp. Neurol. 510, 47–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Stagi M., Fogel A. I., Biederer T. (2010) SynCAM 1 participates in axo-dendritic contact assembly and shapes neuronal growth cones. Proc. Natl. Acad. Sci. U.S.A. 107, 7568–7573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fogel A. I., Stagi M., Perez de Arce K., Biederer T. (2011) Lateral assembly of the immunoglobulin protein SynCAM 1 controls its adhesive function and instructs synapse formation. EMBO J. 30, 4728–4738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Robbins E. M., Krupp A. J., Perez de Arce K., Ghosh A. K., Fogel A. I., Boucard A., Südhof T. C., Stein V., Biederer T. (2010) SynCAM 1 adhesion dynamically regulates synapse number and impacts plasticity and learning. Neuron 68, 894–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fogel A. I., Li Y., Giza J., Wang Q., Lam T. T., Modis Y., Biederer T. (2010) N-Glycosylation at the SynCAM (synaptic cell adhesion molecule) immunoglobulin interface modulates synaptic adhesion. J. Biol. Chem. 285, 34864–34874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Biederer T. (2006) Bioinformatic characterization of the SynCAM family of immunoglobulin-like domain-containing adhesion molecules. Genomics 87, 139–150 [DOI] [PubMed] [Google Scholar]
- 9. Koma Y., Ito A., Wakayama T., Watabe K., Okada M., Tsubota N., Iseki S., Kitamura Y. (2004) Cloning of a soluble isoform of the SgIGSF adhesion molecule that binds the extracellular domain of the membrane-bound isoform. Oncogene 23, 5687–5692 [DOI] [PubMed] [Google Scholar]
- 10. Hagiyama M., Ichiyanagi N., Kimura K. B., Murakami Y., Ito A. (2009) Expression of a soluble isoform of cell adhesion molecule 1 in the brain and its involvement in directional neurite outgrowth. Am. J. Pathol. 174, 2278–2289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hagiyama M., Furuno T., Hosokawa Y., Iino T., Ito T., Inoue T., Nakanishi M., Murakami Y., Ito A. (2011) Enhanced nerve-mast cell interaction by a neuronal short isoform of cell adhesion molecule-1. J. Immunol. 186, 5983–5992 [DOI] [PubMed] [Google Scholar]
- 12. Fogel A. I., Akins M. R., Krupp A. J., Stagi M., Stein V., Biederer T. (2007) SynCAMs organize synapses through heterophilic adhesion. J. Neurosci. 27, 12516–12530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Galuska S. P., Rollenhagen M., Kaup M., Eggers K., Oltmann-Norden I., Schiff M., Hartmann M., Weinhold B., Hildebrandt H., Geyer R., Mühlenhoff M., Geyer H. (2010) Synaptic cell adhesion molecule SynCAM 1 is a target for polysialylation in postnatal mouse brain. Proc. Natl. Acad. Sci. U.S.A. 107, 10250–10255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Edelman G. M., Chuong C. M. (1982) Embryonic to adult conversion of neural cell adhesion molecules in normal and staggerer mice. Proc. Natl. Acad. Sci. U.S.A. 79, 7036–7040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rothbard J. B., Brackenbury R., Cunningham B. A., Edelman G. M. (1982) Differences in the carbohydrate structures of neural cell adhesion molecules from adult and embryonic chicken brains. J. Biol. Chem. 257, 11064–11069 [PubMed] [Google Scholar]
- 16. Finne J., Finne U., Deagostini-Bazin H., Goridis C. (1983) Occurrence of α2–8-linked polysialosyl units in a neural cell adhesion molecule. Biochem. Biophys. Res. Commun. 112, 482–487 [DOI] [PubMed] [Google Scholar]
- 17. Fujimoto I., Bruses J. L., Rutishauser U. (2001) Regulation of cell adhesion by polysialic acid. Effects on cadherin, immunoglobulin cell adhesion molecule, and integrin function and independence from neural cell adhesion molecule binding or signaling activity. J. Biol. Chem. 276, 31745–31751 [DOI] [PubMed] [Google Scholar]
- 18. Weinhold B., Seidenfaden R., Röckle I., Mühlenhoff M., Schertzinger F., Conzelmann S., Marth J. D., Gerardy-Schahn R., Hildebrandt H. (2005) Genetic ablation of polysialic acid causes severe neurodevelopmental defects rescued by deletion of the neural cell adhesion molecule. J. Biol. Chem. 280, 42971–42977 [DOI] [PubMed] [Google Scholar]
- 19. Hildebrandt H., Mühlenhoff M., Oltmann-Norden I., Röckle I., Burkhardt H., Weinhold B., Gerardy-Schahn R. (2009) Imbalance of neural cell adhesion molecule and polysialyltransferase alleles causes defective brain connectivity. Brain 132, 2831–2838 [DOI] [PubMed] [Google Scholar]
- 20. Mühlenhoff M., Oltmann-Norden I., Weinhold B., Hildebrandt H., Gerardy-Schahn R. (2009) Brain development needs sugar. The role of polysialic acid in controlling NCAM functions. Biol. Chem. 390, 567–574 [DOI] [PubMed] [Google Scholar]
- 21. Rutishauser U. (2008) Polysialic acid in the plasticity of the developing and adult vertebrate nervous system. Nat. Rev. Neurosci. 9, 26–35 [DOI] [PubMed] [Google Scholar]
- 22. Eckhardt M., Bukalo O., Chazal G., Wang L., Goridis C., Schachner M., Gerardy-Schahn R., Cremer H., Dityatev A. (2000) Mice deficient in the polysialyltransferase ST8SiaIV/PST-1 allow discrimination of the roles of neural cell adhesion molecule protein and polysialic acid in neural development and synaptic plasticity. J. Neurosci. 20, 5234–5244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Angata K., Long J. M., Bukalo O., Lee W., Dityatev A., Wynshaw-Boris A., Schachner M., Fukuda M., Marth J. D. (2004) Sialyltransferase ST8Sia-II assembles a subset of polysialic acid that directs hippocampal axonal targeting and promotes fear behavior. J. Biol. Chem. 279, 32603–32613 [DOI] [PubMed] [Google Scholar]
- 24. Ong E., Nakayama J., Angata K., Reyes L., Katsuyama T., Arai Y., Fukuda M. (1998) Developmental regulation of polysialic acid synthesis in mouse directed by two polysialyltransferases, PST and STX. Glycobiology 8, 415–424 [DOI] [PubMed] [Google Scholar]
- 25. Hildebrandt H., Becker C., Mürau M., Gerardy-Schahn R., Rahmann H. (1998) Heterogeneous expression of the polysialyltransferases ST8Sia II and ST8Sia IV during postnatal rat brain development. J. Neurochem. 71, 2339–2348 [DOI] [PubMed] [Google Scholar]
- 26. Galuska S. P., Oltmann-Norden I., Geyer H., Weinhold B., Kuchelmeister K., Hildebrandt H., Gerardy-Schahn R., Geyer R., Mühlenhoff M. (2006) Polysialic acid profiles of mice expressing variant allelic combinations of the polysialyltransferases ST8SiaII and ST8SiaIV. J. Biol. Chem. 281, 31605–31615 [DOI] [PubMed] [Google Scholar]
- 27. Oltmann-Norden I., Galuska S. P., Hildebrandt H., Geyer R., Gerardy-Schahn R., Geyer H., Mühlenhoff M. (2008) Impact of the polysialyltransferases ST8SiaII and ST8SiaIV on polysialic acid synthesis during postnatal mouse brain development. J. Biol. Chem. 283, 1463–1471 [DOI] [PubMed] [Google Scholar]
- 28. Paukert M., Bergles D. E. (2006) Synaptic communication between neurons and NG2+ cells. Curr. Opin. Neurobiol. 16, 515–521 [DOI] [PubMed] [Google Scholar]
- 29. Nishiyama A., Komitova M., Suzuki R., Zhu X. (2009) Polydendrocytes (NG2 cells). Multifunctional cells with lineage plasticity. Nat. Rev. Neurosci. 10, 9–22 [DOI] [PubMed] [Google Scholar]
- 30. Trotter J., Karram K., Nishiyama A. (2010) NG2 cells. Properties, progeny, and origin. Brain Res. Rev. 63, 72–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Frosch M., Görgen I., Boulnois G. J., Timmis K. N., Bitter-Suermann D. (1985) NZB mouse system for production of monoclonal antibodies to weak bacterial antigens. Isolation of an IgG antibody to the polysaccharide capsules of Escherichia coli K1 and group B meningococci. Proc. Natl. Acad. Sci. U.S.A. 82, 1194–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mühlenhoff M., Manegold A., Windfuhr M., Gotza B., Gerardy-Schahn R. (2001) The impact of N-glycosylation on the functions of polysialyltransferases. J. Biol. Chem. 276, 34066–34073 [DOI] [PubMed] [Google Scholar]
- 33. Stummeyer K., Dickmanns A., Mühlenhoff M., Gerardy-Schahn R., Ficner R. (2005) Crystal structure of the polysialic acid-degrading endosialidase of bacteriophage K1F. Nat. Struct. Mol. Biol. 12, 90–96 [DOI] [PubMed] [Google Scholar]
- 34. Cremer H., Lange R., Christoph A., Plomann M., Vopper G., Roes J., Brown R., Baldwin S., Kraemer P., Scheff S. (1994) Inactivation of the N-CAM gene in mice results in size reduction of the olfactory bulb and deficits in spatial learning. Nature 367, 455–459 [DOI] [PubMed] [Google Scholar]
- 35. Röckle I., Seidenfaden R., Weinhold B., Mühlenhoff M., Gerardy-Schahn R., Hildebrandt H. (2008) Polysialic acid controls NCAM-induced differentiation of neuronal precursors into calretinin-positive olfactory bulb interneurons. Dev. Neurobiol. 68, 1170–1184 [DOI] [PubMed] [Google Scholar]
- 36. Windfuhr M., Manegold A., Muhlenhoff M., Eckhardt M., Gerardy-Schahn R. (2000) Molecular defects that cause loss of polysialic acid in the complementation group 2A10. J. Biol. Chem. 275, 32861–32870 [DOI] [PubMed] [Google Scholar]
- 37. Eckhardt M., Mühlenhoff M., Bethe A., Koopman J., Frosch M., Gerardy-Schahn R. (1995) Molecular characterization of eukaryotic polysialyltransferase-1. Nature 373, 715–718 [DOI] [PubMed] [Google Scholar]
- 38. Zuber C., Lackie P. M., Catterall W. A., Roth J. (1992) Polysialic acid is associated with sodium channels and the neural cell adhesion molecule N-CAM in adult rat brain. J. Biol. Chem. 267, 9965–9971 [PubMed] [Google Scholar]
- 39. Yabe U., Sato C., Matsuda T., Kitajima K. (2003) Polysialic acid in human milk. CD36 is a new member of mammalian polysialic acid-containing glycoprotein. J. Biol. Chem. 278, 13875–13880 [DOI] [PubMed] [Google Scholar]
- 40. Curreli S., Arany Z., Gerardy-Schahn R., Mann D., Stamatos N. M. (2007) Polysialylated neuropilin-2 is expressed on the surface of human dendritic cells and modulates dendritic cell-T lymphocyte interactions. J. Biol. Chem. 282, 30346–30356 [DOI] [PubMed] [Google Scholar]
- 41. Mühlenhoff M., Eckhardt M., Bethe A., Frosch M., Gerardy-Schahn R. (1996) Autocatalytic polysialylation of polysialyltransferase-1. EMBO J. 15, 6943–6950 [PMC free article] [PubMed] [Google Scholar]
- 42. Close B. E., Colley K. J. (1998) In vivo autopolysialylation and localization of the polysialyltransferases PST and STX. J. Biol. Chem. 273, 34586–34593 [DOI] [PubMed] [Google Scholar]
- 43. Kojima N., Tachida Y., Yoshida Y., Tsuji S. (1996) Characterization of mouse ST8Sia II (STX) as a neural cell adhesion molecule-specific polysialic acid synthase. Requirement of core α1,6-linked fucose and a polypeptide chain for polysialylation. J. Biol. Chem. 271, 19457–19463 [DOI] [PubMed] [Google Scholar]
- 44. Mühlenhoff M., Eckhardt M., Bethe A., Frosch M., Gerardy-Schahn R. (1996) Polysialylation of NCAM by a single enzyme. Curr. Biol. 6, 1188–1191 [DOI] [PubMed] [Google Scholar]
- 45. Angata K., Suzuki M., Fukuda M. (1998) Differential and cooperative polysialylation of the neural cell adhesion molecule by two polysialyltransferases, PST and STX. J. Biol. Chem. 273, 28524–28532 [DOI] [PubMed] [Google Scholar]
- 46. Kojima N., Yoshida Y., Tsuji S. (1995) A developmentally regulated member of the sialyltransferase family (ST8Sia II, STX) is a polysialic acid synthase. FEBS Lett. 373, 119–122 [DOI] [PubMed] [Google Scholar]
- 47. Galuska S. P., Geyer R., Gerardy-Schahn R., Mühlenhoff M., Geyer H. (2008) Enzyme-dependent variations in the polysialylation of the neural cell adhesion molecule (NCAM) in vivo. J. Biol. Chem. 283, 17–28 [DOI] [PubMed] [Google Scholar]
- 48. Muotri A. R., Gage F. H. (2006) Generation of neuronal variability and complexity. Nature 441, 1087–1093 [DOI] [PubMed] [Google Scholar]
- 49. Li Q., Lee J. A., Black D. L. (2007) Neuronal regulation of alternative pre-mRNA splicing. Nat. Rev. Neurosci. 8, 819–831 [DOI] [PubMed] [Google Scholar]
- 50. Rose M. C., Voter W. A., Sage H., Brown C. F., Kaufman B. (1984) Effects of deglycosylation on the architecture of ovine submaxillary mucin glycoprotein. J. Biol. Chem. 259, 3167–3172 [PubMed] [Google Scholar]
- 51. Li F., Erickson H. P., James J. A., Moore K. L., Cummings R. D., McEver R. P. (1996) Visualization of P-selectin glycoprotein ligand-1 as a highly extended molecule and mapping of protein epitopes for monoclonal antibodies. J. Biol. Chem. 271, 6342–6348 [DOI] [PubMed] [Google Scholar]
- 52. Cunningham B. A., Hemperly J. J., Murray B. A., Prediger E. A., Brackenbury R., Edelman G. M. (1987) Neural cell adhesion molecule. Structure, immunoglobulin-like domains, cell surface modulation, and alternative RNA splicing. Science 236, 799–806 [DOI] [PubMed] [Google Scholar]
- 53. Nelson R. W., Bates P. A., Rutishauser U. (1995) Protein determinants for specific polysialylation of the neural cell adhesion molecule. J. Biol. Chem. 270, 17171–17179 [DOI] [PubMed] [Google Scholar]
- 54. von Der Ohe M., Wheeler S. F., Wuhrer M., Harvey D. J., Liedtke S., Mühlenhoff M., Gerardy-Schahn R., Geyer H., Dwek R. A., Geyer R., Wing D. R., Schachner M. (2002) Localization and characterization of polysialic acid-containing N-linked glycans from bovine NCAM. Glycobiology 12, 47–63 [DOI] [PubMed] [Google Scholar]
- 55. Liedtke S., Geyer H., Wuhrer M., Geyer R., Frank G., Gerardy-Schahn R., Zähringer U., Schachner M. (2001) Characterization of N-glycans from mouse brain neural cell adhesion molecule. Glycobiology 11, 373–384 [DOI] [PubMed] [Google Scholar]
- 56. Close B. E., Mendiratta S. S., Geiger K. M., Broom L. J., Ho L. L., Colley K. J. (2003) The minimal structural domains required for neural cell adhesion molecule polysialylation by PST/ST8Sia IV and STX/ST8Sia II. J. Biol. Chem. 278, 30796–30805 [DOI] [PubMed] [Google Scholar]
- 57. Thompson M. G., Foley D. A., Swartzentruber K. G., Colley K. J. (2011) Sequences at the interface of the fifth immunoglobulin domain and first fibronectin type III repeat of the neural cell adhesion molecule are critical for its polysialylation. J. Biol. Chem. 286, 4525–4534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mendiratta S. S., Sekulic N., Hernandez-Guzman F. G., Close B. E., Lavie A., Colley K. J. (2006) A novel α-helix in the first fibronectin type III repeat of the neural cell adhesion molecule is critical for N-glycan polysialylation. J. Biol. Chem. 281, 36052–36059 [DOI] [PubMed] [Google Scholar]
- 59. Mendiratta S. S., Sekulic N., Lavie A., Colley K. J. (2005) Specific amino acids in the first fibronectin type III repeat of the neural cell adhesion molecule play a role in its recognition and polysialylation by the polysialyltransferase ST8Sia IV/PST. J. Biol. Chem. 280, 32340–32348 [DOI] [PubMed] [Google Scholar]
- 60. Busam R. D., Thorsell A. G., Flores A., Hammarström M., Persson C., Öbrink B., Hallberg B. M. (2011) Structural basis of tumor suppressor in lung cancer 1 (TSLC1) binding to differentially expressed in adenocarcinoma of the lung (DAL-1/4.1B). J. Biol. Chem. 286, 4511–4516 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.