Background: Mechanisms underlying the regulation of DOHH expression are unclear.
Results: DOHH expression is inversely associated with miR-331-3p and miR-642-5p expression in PCa cells. Transfection with miR-331-3p and miR-642-5p inhibited DOHH expression, cell proliferation and enhanced the cytotoxic effect of mimosine.
Conclusion: miR-331-3p and miR-642-5p regulate DOHH expression in prostate.
Significance: Restoring miR-331-3p and/or miR-642-5p to PCa may be beneficial.
Keywords: Cell Proliferation, Gene Expression, Gene Regulation, MicroRNA, Prostate Cancer, DOHH, eIF5A, Mimosine
Abstract
The enzyme deoxyhypusine hydroxylase (DOHH) catalyzes the activation of eukaryotic translation initiation factor (eIF5A), a protein essential for cell growth. Using bioinformatic predictions and reporter gene assays, we have identified a 182-nt element within the DOHH 3′-untranslated region (3′-UTR) that contains a number of target sites for miR-331-3p and miR-642-5p. Quantitative RT-PCR studies demonstrated overexpression of DOHH mRNA and underexpression of miR-331-3p and miR-642-5p in several prostate cancer cell lines compared with normal prostate epithelial cells. Transient overexpression of miR-331-3p and/or miR-642-5p in DU145 prostate cancer cells reduced DOHH mRNA and protein expression and inhibited cell proliferation. We observed synergistic growth inhibition with the combination of miR-331-3p and miR-642-5p and mimosine, a pharmacological DOHH inhibitor. Finally, we identified a significant inverse relationship between the expression of miR-331-3p or miR-642-5p and DOHH in a cohort of human prostate cancer tissues. Our results suggest a novel role for miR-331-3p and miR-642-5p in the control of prostate cancer cell growth via the regulation of DOHH expression and eIF5A activity.
Introduction
Cell proliferation is tightly controlled by the eukaryotic translation initiation factor 5A (eIF5A), a small protein with two isoforms (eIF5A-1 and -2), which is highly conserved among eukaryotes, particularly around a unique hypusine residue (1, 2). Hypusine is a polyamine-derived amino acid that is formed in eIF5A by a post-translational enzymatic modification that occurs in two steps, the first of which involves cleavage of the polyamine spermidine and transfer of its 4-aminobutyl group to a specific lysine residue of the eIF5A precursor by deoxyhypusine synthase, forming a deoxyhypusine residue (3, 4). Subsequent hydroxylation of the eIF5A intermediate by deoxyhypusine hydroxylase (DOHH)3 produces the hypusine residue and a mature, active form of eIF5A. Although eIF5A was thought to be a general translation initiation factor (5, 6), recent studies do not support this, with depletion of eIF5A in yeast resulting only a small decrease in overall protein synthesis (7), but a decrease in cell proliferation and cell cycle arrest at the G1/S boundary. Therefore, eIF5A may be important for translation of proteins critical for cell cycle progression (8). Interestingly, eIF5A expression is elevated in intraepithelial neoplasia of the vulva (9) and colorectal and ovarian cancer cell lines (10).
The essential role of hypusine in cell proliferation has been confirmed in studies in which DOHH inhibitors cause G1/S cell cycle arrest (11) and block cell growth (12). A recent study by Balabanov and co-workers (13) demonstrated synergistic growth inhibition of leukemia cells treated with the chemotherapy drug imatinib and the deoxyhypusine synthase inhibitor GC7 or the DOHH inhibitor ciclopirox. Of particular interest, the DOHH inhibitor mimosine has been reported to inhibit cell cycle progression and proliferation of PC3, LNCaP, and DU145 prostate cancer (PCa) cells (14, 15) and breast cancer cells (16) to retard the growth of subcutaneous lung and pancreatic cancer xenografts in mice (17, 18) and to sensitize lung cancer cells to radiation (19). Thus, inhibition of DOHH expression may represent a therapeutic strategy to decrease tumor growth by blocking cell cycle progression and consequently cellular proliferation.
MicroRNAs (miRNAs) are a class of short, endogenous, non-coding RNA molecules that bind with imperfect complementarity to the 3′-untranslated regions (3′-UTRs) of target mRNAs, causing translational repression or message degradation (20, 21). miRNAs have important roles in normal cellular development and function (22, 23), including cell cycle regulation (24), and the altered expression of miRNAs is associated with cancer (25). Some miRNAs act as oncogenes or tumor suppressor genes (25, 26). For example, a decrease in the expression of the let-7 miRNA family members is associated with RAS oncogene overexpression and reduced survival in non-small cell lung cancer (27, 28). Conversely, increased miR-21 expression in a range of cancers, including those of the breast, prostate, lung, colon, pancreas, and stomach (29), is associated with reduced apoptosis, chemoresistance, and increased tumor growth (30).
Previously, we identified miR-331-3p as a putative tumor suppressor that is down-regulated in PCa (31). miR-331-3p regulates ERBB-2 expression and signaling (31), a process that involves an interplay between miR-331-3p and the RNA-binding protein HuR (32). In this study, we demonstrate that the DOHH mRNA 3′-UTR contains a 182-nt element that is a specific and direct target of miR-331-3p and miR-642-5p. RT-qPCR studies indicate that DOHH mRNA expression is increased, whereas miR-331-3p/miR-642-5p expression is decreased in PCa cell lines relative to normal prostate epithelial cells. Transfection of DU145 cells with miR-331-3p and/or miR-642-5p decreased DOHH mRNA and protein expression and reduced cell proliferation. Combining miR-331-3p and/or miR-642-5p overexpression with mimosine treatment produced synergistic growth inhibition. Finally, analysis of nine matched PCa and normal adjacent tissue samples demonstrated an inverse association between DOHH mRNA expression and miR-331-3p or miR-642-5p. Taken together, our results support a role for miR-331-3p and miR-642-5p as mediators of eIF5A activity and prostate epithelial cell proliferation via their modulation of DOHH expression.
EXPERIMENTAL PROCEDURES
Cell Culture, Plasmid DNA, miRNA Precursor Molecules, and DOHH Inhibitor
RWPE-1, LNCaP, C4–2B, DU145, PC3, and 22RV1 PCa cells were obtained from the American Type Culture Collection (ATCC) and cultured at 37 °C in 5% CO2 with RPMI 1640 supplemented with 10% fetal bovine serum. DOHH 3′-UTR reporter clones were generated by GenScript, Inc. (Piscataway) and consisted of a firefly luciferase reporter gene vector backbone (pmiR-REPORT; Ambion) to which was fused (i) full-length DOHH 3′-UTR (nt 1072–1761) of GenBankTM accession no. (NM_031303.4), (ii) a 182-nt DOHH 3′-UTR element (nt 1343–1525) of GenBankTM accession no. (NM_031303.4), or (iii) full-length DOHH 3′-UTR (nt 1072–1761) with deletion of the 182-nt element (nt 1343–1525) (see Fig. 2A). All plasmids were verified by DNA sequencing prior to use. Synthetic miRNA precursor molecules corresponding to human miR-331-3p (pre-miR miRNA precursor product ID PM10881), human miR-642-5p (pre-miR miRNA precursor product ID PM11477) and a negative control miRNA (miR-NC; pre-miR miRNA precursor negative control 1, product ID AM17110) were obtained from Ambion. The DOHH inhibitor mimosine was purchased from Sigma-Aldrich (M0253) and prepared as per manufacturer's instructions.
FIGURE 2.
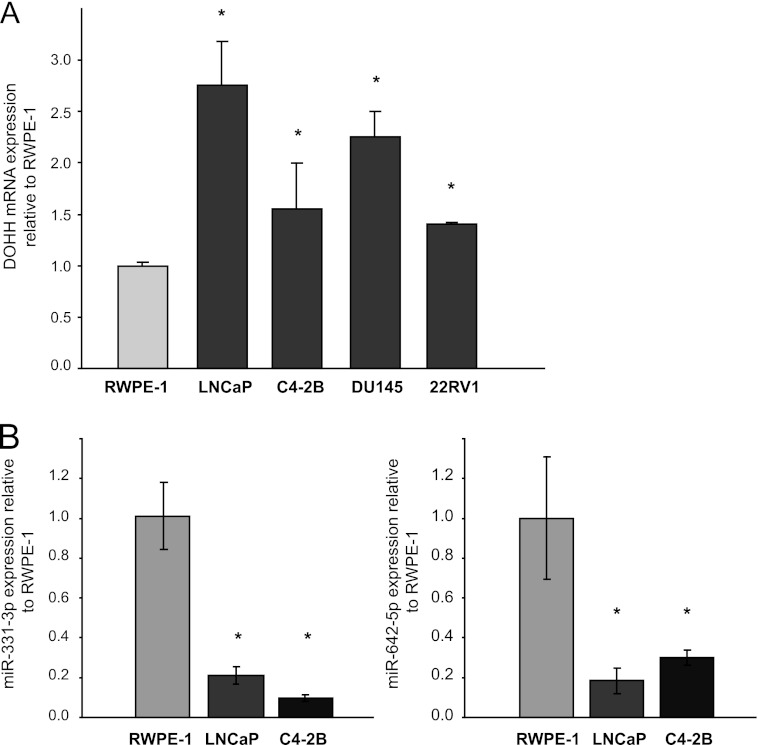
Inverse association between DOHH overexpression and miR-331-3p and miR-642-5p levels in PCa cell lines compared with an immortalized prostate epithelial cell line. A, RT-qPCR analysis of DOHH mRNA expression in RWPE-1 normal prostate epithelial cells and LNCaP, C4–2B, DU145, and 22RV1 PCa cells. Data are expressed relative to RWPE-1 cells, where DOHH mRNA expression was normalized to β-actin and GAPDH using GENEX software. Error bars represent S.D. *, p < 0.05. B, miR-331-3p and miR-642-5p expression in LNCaP and C4–2B PCa cell lines compared with RWPE-1 cells. Error bars represent S.D. *, p < 0.005.
RNA Extraction, Reverse Transcription, and Quantitative Polymerase Chain Reaction (RT-qPCR)
Total RNA was extracted from cell lines and tissue samples using Qiazol reagent (Qiagen). For RNA extractions from tissue, samples were first homogenized in Qiazol by 2 × 45-s pulses using 2.8-mm ceramic beads in a Precellys 24 Homogenizer (Bertin Technologies). For RT-qPCR analysis of RNA expression, 125 ng of total RNA was reverse-transcribed to cDNA using a Quantitect reverse transcription Kit (Qiagen). Quantitative PCR was performed on a Corbett 6000 Rotor-Gene thermocycler (Corbett Research) using Quantitect SYBR Mix (Qiagen) and validated Quantitect primer assays (Qiagen) for ACTB (catalog no. QT01680476), GAPDH (catalog no. QT00060746), and DOHH (catalog no. QT00235536) and primers for luciferase (Luc-F, 5′-TAC TGG GAC GAA GAC GAA CAC-3′; Luc-R, 5′-GTT CAC CGG CGT CAT CGT CG-3′). Expression of DOHH mRNA or luciferase mRNA expression relative to GAPDH and/or β-actin mRNA was determined using the 2−ΔΔCt method (33), and statistical analyses of RT-qPCR data were performed using GENEX software (MultiD).
For miRNA detection, TaqMan® miRNA assays (Invitrogen) for hsa-miR-331-3p, hsa-miR-642-5p and RNU6B small nuclear RNA (Invitrogen; Assay IDs 000545, 001592, and 001093, respectively), were used with 10 ng of total RNA and a Corbett 6000 Rotor-Gene thermocycler. Expression of mature miR-331-3p and miR-642-5p miRNAs relative to RNU6B small nuclear RNA was determined using the 2−ΔΔCt method (33). Statistical analysis of RT-qPCR data were performed using GenEx software (MultiD).
Transfection of miRNA Precursor Molecules and Reporter Gene Assays
PCa cells were seeded into six-well or 12-well plates or 10-cm2 dishes and transfected using Lipofectamine 2000 (Invitrogen) and precursor miRNA molecules at a final concentration of 30 nm, unless stated. Cells were harvested after 24 h for RNA isolation and 3 days for protein extraction.
Reporter gene assays were performed as described (34). Briefly, PCa cells were seeded in 12-well plates and co-transfected with 100 ng of firefly luciferase reporter plasmid DNA and 5 ng of control (Renilla luciferase; pRL-SV40) plasmid DNA and 1–30 nm final concentration of pre-miRNA (Ambion; pre-miR-331-3p, pre-miR-642-5p, pre-miR-NC, using Lipofectamine 2000. After 24 h, lysates were assayed for firefly and Renilla luciferase activities using the Dual-Luciferase Reporter Assay System (Promega) and a Fluostar OPTIMA microplate reader (BMG Labtech). Firefly luciferase activity for each sample was normalized to Renilla luciferase activity to yield a relative luciferase activity.
Protein Extraction and Western Blotting
Cytoplasmic protein extracts were prepared, and Western blotting was performed as described (34). Briefly, protein samples were resolved on NuPAGE 4–12% Bis Tris gels (Invitrogen) and transferred to PVDF membranes (Roche Diagnostics). Membranes were blocked in 5% skim milk/Tris Buffered Saline Tween and probed with anti-tubulin rat polyclonal antibody (1:1000, Abcam, ab6161-100) and anti-DOHH (C-19) goat polyclonal antibody (1:1000, Santa Cruz Biotechnology, sc-55157). Detection was performed using horseradish peroxidise-linked anti-rat-IgG (ab6734-1; Abcam) and anti-sheep/goat-IgG (AB324P; Chemicon) secondary antibodies with ECL Plus detection reagent and ECL-Hyperfilm (GE Healthcare).
Cell Proliferation Assays
For cell proliferation assays, DU145 cells were transfected with miRNA precursor molecules (see “Transfection of miRNA precursor molecules and reporter gene assays” above) and treated with or without mimosine (75 μm) at 24 h post transfection. Cell proliferation was assessed at 5 days post transfection with the CellTiter 96 Aqueous One Solution Cell Proliferation System (Promega) and a Fluostar Optima plate reader (BMG Scientific).
Matched Prostate Tumor and Normal Adjacent Tissue (NAT) Samples
Matched prostate tumor and normal adjacent tissue samples were obtained from Dr. Ronald Cohen (Uropath, Perth, Western Australia). Clinical characteristics of patient tumor samples are described in Table 1. All samples were verified to contain >80% tumor by a pathologist. Tissue samples were obtained with consent (institutional human ethics application number EC 2008/118).
TABLE 1.
Characteristics of NAT versus tumor prostate tissues with DOHH mRNA overexpression (n = 5) used in this study
T, tumor.
| Matched T vs. NAT | Age | Gleason score | Stage | Extra-prostatic spread | Grade % 4/5 |
|---|---|---|---|---|---|
| 1 | 63 | 7 | T2c | No | 60 |
| 2 | 59 | 7 | T2c | No | 10 |
| 3 | 63 | 9 | T3a | Yes | 100 |
| 4 | 59 | 8 | T3a | Yes | 100 |
| 5 | 65 | 7 | T3b | No | 70 |
Statistical Analysis
Statistical analysis of RT-qPCR data were performed using GENEX software (MultiD). All analyses were performed at a minimum confidence interval of 95% (confidence interval = 0.95), and normality of data were confirmed by Kolmogorov-Smirnoff test.
For NAT versus tumor data, the expression of miR-331-3p, miR-642-5p, and DOHH between NAT versus tumor pairs was determined using Box-Whisker plots and Wilcoxon matched pairs signed rank testing using GraphPad Prism software (version 5, GraphPad Software, Inc.).
Statistical analysis of reporter gene assay data were performed using Student's t test, where p < 0.05 represented a significant difference. Error bars represent S.D. Synergy in combined miRNA and mimosine experiments was assessed according to the method of Bliss (35), where synergy occurs when EBliss < EObserved.
RESULTS
A 182-nt Element within the DOHH 3′-UTR Is a Target for miR-331-3p and miR-642-5p
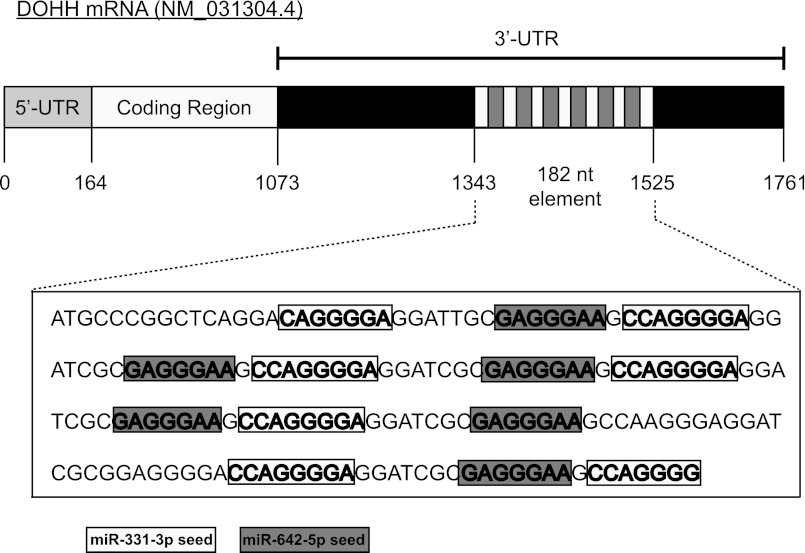
We are interested in understanding the action of the tumor suppressor miR-331-3p in cancer (31, 32). To identify new putative targets, we utilized TargetScan (36) analysis (release 6.2, June 2012), which revealed that the highest confidence predicted target of miR-331-3p is DOHH, an enzyme involved in the hypusination and activation of eIF5A. We identified a 182-nt element within the DOHH 3′-UTR, unusual in that it contains seven putative miR-331-3p binding sites (Fig. 1), with TargetScan context scores (37) ranging from 34–91%. Of interest, the 182-nt DOHH 3′-UTR element also contains six putative miR-642-5p binding sites (Fig. 1), with TargetScan context scores ranging from 29–35%. Although the element is not conserved between human, rat, and mouse, the presence of multiple putative miR-331-3p and miR-642-5p binding sites within this DOHH 3′-UTR fragment warranted further investigation. We analyzed DOHH mRNA expression in a panel of PCa cell lines (LNCaP, C4–2B, DU145, PC3, and 22RV1) compared with a normal prostate epithelial cell line (RWPE-1) and found DOHH mRNA to be significantly up-regulated in all PCa cell lines (Fig. 2A). Next, we used TaqMan miRNA RT-qPCR assays to detect miR-331-3p and miR-642-5p expression in LNCaP and C4–2B PCa cells compared with RWPE-1 cells and found both miRNAs to be significantly down-regulated in the PCa cell lines (Fig. 2B). The inverse relationship between DOHH mRNA and miR-331-3p and miR-642-5p in PCa cell lines suggested the potential for these miRNAs to regulate DOHH expression in the prostate.
FIGURE 1.
The DOHH 3′-UTR contains a 182-nt element with multiple predicted miR-331-3p and miR-642-5p target sites. Shown is a schematic representation of the DOHH mRNA with its 182-nt 3′-UTR element containing seven miR-331-3p and six miR-642-5p binding sites predicted by TargetScan (version 6.2). miR-331-3p seed sequences are shown in boxes in boldface type, and miR-642-5p seed sequences are shown in boldface type within shaded boxes.
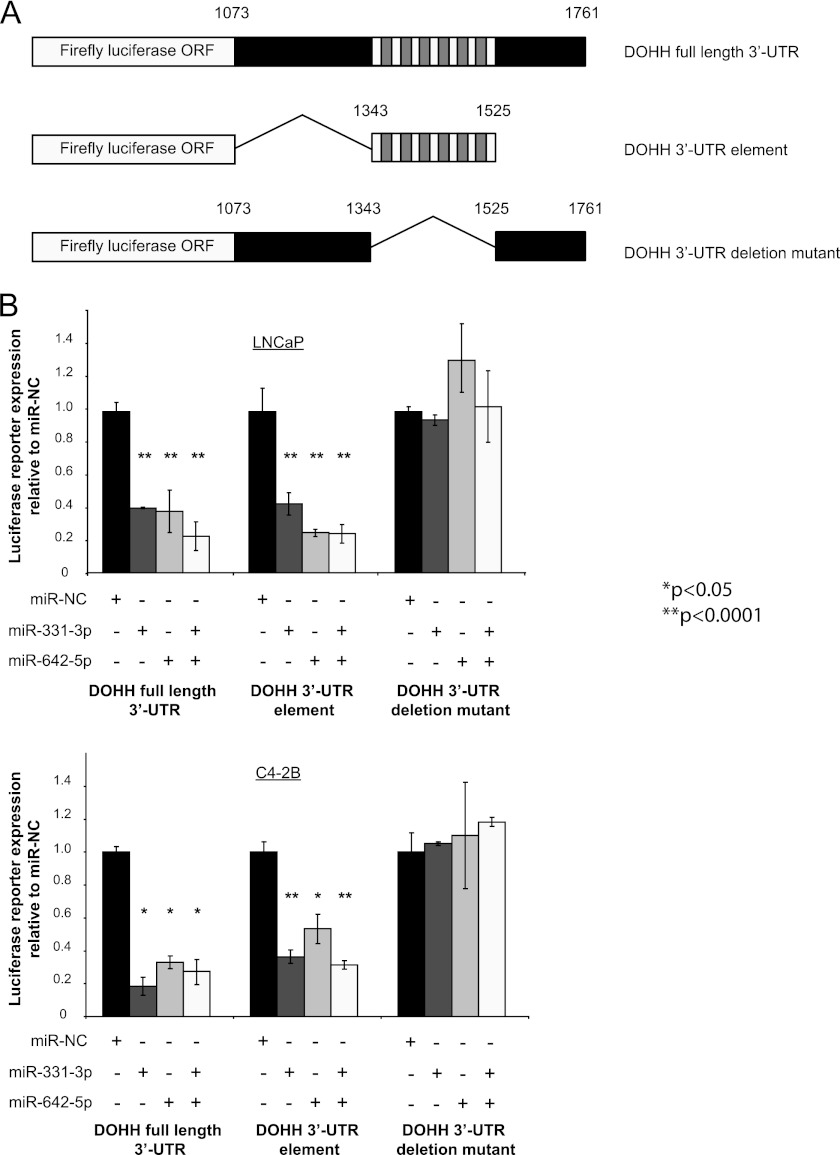
We performed assays using LNCaP and C4–2B cells with miR-331-3p and/or miR-642-5p and luciferase reporter constructs that contained the full-length DOHH 3′-UTR, the 182-nt DOHH 3′-UTR element, or a DOHH 3′-UTR deletion mutant lacking the 182-nt element (Fig. 3A). Transfection with miR-331-3p and/or miR-642-5p significantly down-regulated activity of the full-length DOHH 3′-UTR luciferase reporter (Fig. 3B). miR-331-3p and/or miR-642-5p produced a similar down-regulation of reporter activity with the 182-nt DOHH 3′-UTR element alone; an effect that was not observed with the 182-nt DOHH 3′-UTR element deletion mutant reporter (Fig. 3B). Taken together, these data indicate that the 182-nt element within the 3′-UTR of DOHH is a direct target of both miR-331-3p and miR-642-5p.
FIGURE 3.
A 182-nt element within the DOHH mRNA 3′-UTR is a target for miR-331-3p and miR-642-5p in PCa cells. A, schematic representation of firefly luciferase reporter constructs containing either the full-length DOHH 3′-UTR, the 182-nt DOHH 3′-UTR element, or a DOHH 3′-UTR deletion mutant lacking the 182-nt element. nt positions of the DOHH 3′-UTR sequences are indicated (GenBank accession no. NM_031304.4). B, luciferase reporter assays using LNCaP and C4–2B cells co-transfected with miR-331-3p and/or miR-642-5p and the reporter gene constructs described above. For each sample, firefly luciferase activity was normalized to Renilla luciferase activity. Data for each reporter construct are expressed relative to miR-NC transfected cells. Error bars represent S.D. *, p < 0.05; **, p < 0.0001.
miR-331-3p and miR-642-5p Regulate DOHH Expression in DU145 PCa Cells
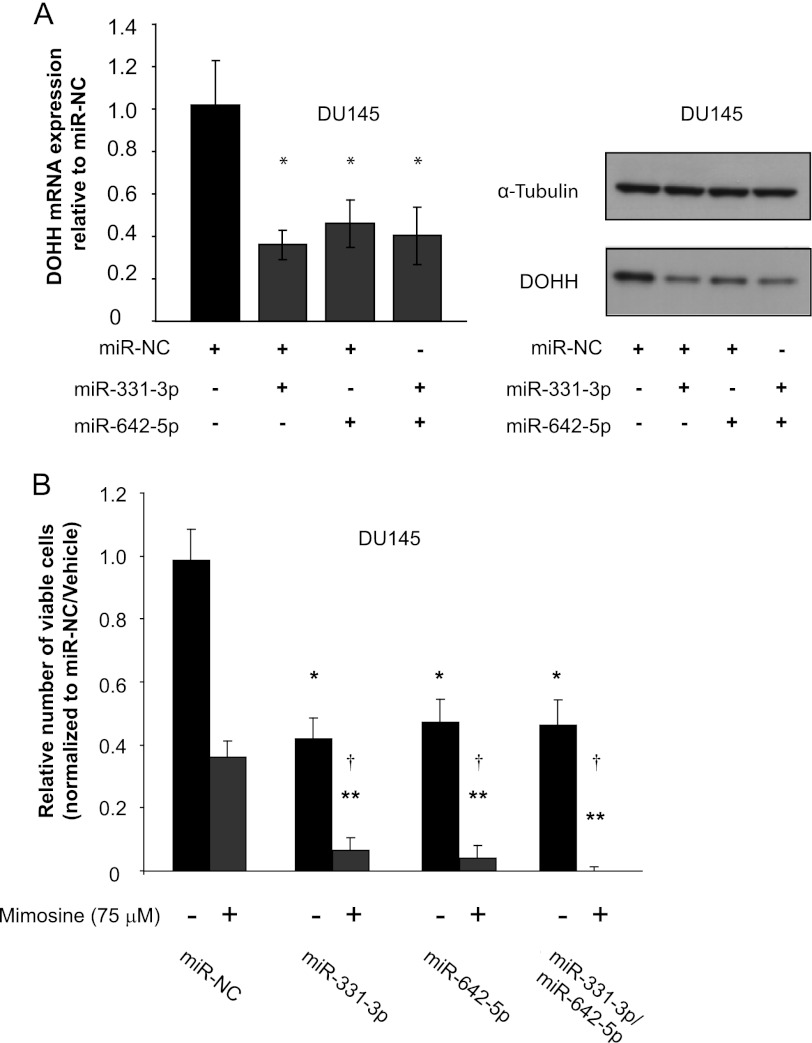
We next investigated the capacity for miR-331-3p and miR-642-5p to regulate endogenous DOHH expression. Transfection of DU145 cells with miR-331-3p and/or miR-642-5p significantly reduced expression of DOHH mRNA (Fig. 4A). We also found reduced luciferase RNA levels in DU145 cells transfected with the luciferase reporter carrying the DOHH 3′-UTR element (Fig. 3A) and miR-331-3p/miR-642-5p (supplemental Fig. S1), consistent with a post-transcriptional effect of these miRNAs. In parallel Western blotting experiments, transient transfection of miR-331-3p and/or miR-642-5p inhibited expression of DOHH protein (Fig. 4A). Taken together, these results suggest that miR-331-3p and miR-642-5p regulate DOHH expression in the prostate by inducing decay of its mRNA, resulting in reduced levels of DOHH protein.
FIGURE 4.
miR-331-3p and miR-642-5p down-regulate DOHH mRNA and protein expression in DU145 PCa cells. A, RT-qPCR and Western blotting analysis of DOHH mRNA and protein expression 24 and 48 h, respectively, after transfection of DU145 cells with miR-331-3p and/or miR-642-5p or miR-NC. α-Tubulin is included as a loading control. *, p < 0.005. B, cell proliferation assay 5 days post transfection of DU145 PCa cells with miR-331-3p and/or miR-642-5p, or miR-NC. Error bars represent S.D., and data are expressed relative to miR-NC/vehicle. *, p < 0.05. In some instances, mimosine (75 μm) was added 48 h post transfection, and cell proliferation was analyzed at 3 days later. Error bars represent S.D., and data are expressed relative to miR-NC/mimosine. **, p < 0.001. †, synergy according to the Bliss Additivity model (35).
Overexpression of miR-331-3p and/or miR-642-5p Inhibits Growth of DU145 PCa Cells and Produces a Synergistic Effect on Proliferation with the DOHH inhibitor Mimosine
We investigated the functional significance of miR-331-3p and miR-642-5p overexpression in DU145 cells. Cells were transfected with synthetic precursor molecules for miR-331-3p and/or miR-642-5p or with the negative control miRNA (miR-NC). Cell proliferation was assessed 5 days post transfection using a cell titer assay. miR-331-3p and/or miR-642-5p significantly reduced DU145 cell proliferation when compared with miR-NC (Fig. 4B), an effect likely to be due in part to decreased DOHH expression and hence eIF5A activity.
We next used cell titer assays to assess the combination of miR-331-3p and/or miR-642-5p and the DOHH inhibitor mimosine, which has been shown to inhibit the growth of multiple cancer cell lines in vitro and in vivo (14–18). We hypothesized that the combined effect of DOHH down-regulation and inhibition might produce a synergistic growth inhibitory effect on cancer cell proliferation. DU145 cells were transiently transfected with synthetic miRNA precursor molecules, and after 2 days were treated with mimosine (75 μm) or vehicle for a further 3 days. The combination of miR-331-3p and/or miR-642-5p, and mimosine produced a synergistic inhibition of DU145 cell growth (Fig. 4B). Using the Bliss model (35), synergy occurs when the observed fractional inhibition for the combination of miR-331-3p and/or miR-642-5p and mimosine (defined as Eobserved) exceeds the sum of the combined miRNA effect and the mimosine effect (defined as EBliss) (Fig. 4B; Table 2). Taken together, these results indicate that down-regulation of DOHH expression by miR-331-3p and/or miR-642-5p is associated with reduced DU145 PCa cell proliferation and that this decrease in DOHH levels results in a synergistic growth inhibition when combined with the DOHH inhibitor mimosine.
TABLE 2.
Growth inhibition values for synergy calculations
| E values | Growth inhibition miR-331-3p/Mimosine | Growth inhibition miR-642-5p/Mimosine | Growth inhibition miR-331-3p/miR-642-5p/Mimosine |
|---|---|---|---|
| Ea (miRNA) | 0.572 | 0.523 | 0.532 |
| Eb (mimosine) | 0.632 | 0.632 | 0.632 |
| EBliss | 0.842 | 0.825 | 0.828 |
| Eobserved | 0.933a | 0.961a | 1.014a |
a Synergy according to the Bliss Additivity model (35).
miR-331-3p and miR-642-5p Are Down-regulated in a Subset of PCa Samples, Which Overexpress DOHH
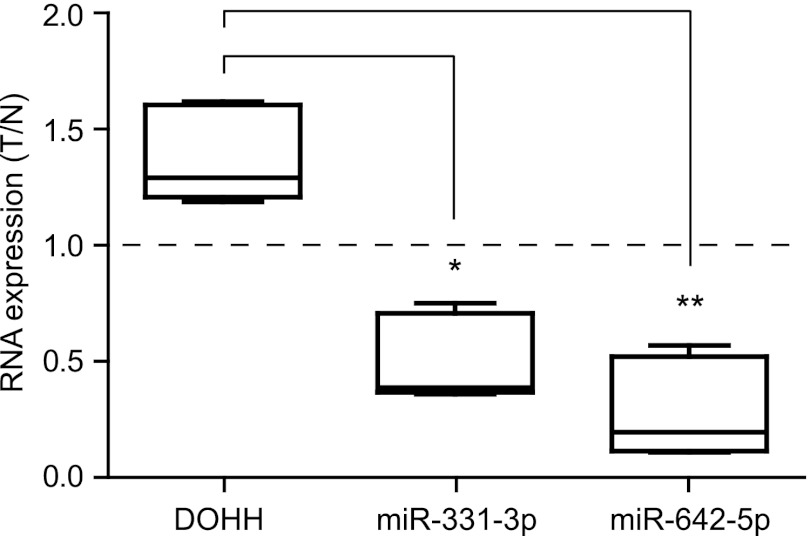
We and others (31, 32, 38–40) have identified a putative role for miR-331-3p as a tumor suppressor in PCa. To further investigate its role and in particular the functional association between miR-331-3p, miR-642-5p and DOHH in PCa, we examined RNA expression levels in human PCa tissue samples. We used RT-qPCR to determine expression of DOHH mRNA in a cohort of nine prostate tumor samples with matched NAT. We observed DOHH mRNA expression was elevated in five of nine (56%) tumor samples compared with matched normal adjacent tissues (data not shown). As we observed an inverse association between DOHH mRNA expression and miR-331-3p and miR-642-5p in PCa cell lines (Fig. 2), we focused on the subset of five tumor samples with increased DOHH expression and using TaqMan miRNA RT-qPCR, we found expression of miR-331-3p and miR-642-5p was significantly reduced in each instance versus its matched NAT (Fig. 5). miR-331-3p and miR-642-5p expression was also reduced in the subset of four tumor samples lacking DOHH overexpression (supplemental Fig. S2). These findings suggest that loss of miR-331-3p and miR-642-5p may facilitate the elevated expression of DOHH and subsequent cellular growth, in part via eIF5A activation, in PCa.
FIGURE 5.
Reduced miR-331-3p and miR-642-5p expression in a subset of PCa with elevated DOHH mRNA expression. RT-qPCR analysis of DOHH mRNA, miR-331-3p, and miR-642-5p expression in a cohort of five matched tumor versus NAT (N) pairs, with expression of each RNA shown in a box plot as a ratio of tumor/normal (T/N), where >1 indicates higher expression in tumor than normal tissue, and <1 indicates lower expression in tumor than normal.
DISCUSSION
Previously, we reported that miR-331-3p is down-regulated in PCa. We demonstrated that by restoring miR-331-3p to PCa cells, ERBB2 expression and signaling was blocked (31). Others have since confirmed the reduced expression of miR-331-3p in PCa (39, 41). The present study sought to extend our understanding of the functional significance of miR-331-3p in PCa cells. Bioinformatic analysis identified DOHH, an enzyme critical for activation of eIF5A and cell cycle progression, as a predicted target of miR-331-3p. Within the DOHH 3′-UTR is a 182-nt element that contains seven putative miR-331-3p and six putative miR-642-5p binding sites. There is an inverse association between DOHH mRNA expression and the levels of miR-331-3p and miR-642-5p in PCa cell lines and a normal prostate epithelial cell line. The capacity of miR-331-3p and miR-642-5p to regulate DOHH expression was confirmed at the mRNA and protein levels, and miR-331-3p and miR-642-5p inhibited proliferation of DU145 cells and synergistically repressed cell proliferation by the DOHH inhibitor mimosine. Finally, we have demonstrated the inverse relationship between DOHH mRNA and miR-331-3p and miR-642-5p in a subset of PCa patients.
A major limitation in understanding the functional effects of specific miRNAs is the paucity of validated target genes. Many studies rely on exhaustive lists of predicted miRNA target genes generated by algorithms such as TargetScan (36), miRanda (42), or PicTar (43), followed by experimental confirmation of the regulation of a target gene by a specific miRNA (44). Alternative approaches to identify miRNA target genes typically involve microarray analysis of cells in which activity of a specific miRNA has been modulated, allowing the identification of a series of candidate miRNA target genes (45). Underpinning this approach is the observation that a majority of endogenous miRNA targets are regulated at the mRNA level (46). More recently, immunoprecipitation of cross-linked Argonaute (Ago) protein-RNA complexes (HITS-CLIP) and RNA sequencing has allowed the identification of genome-wide miRNA and target mRNA interaction maps, expanding our knowledge of miRNA target genes and understanding of the nature of miRNA-mRNA interactions (47).
We investigated DOHH as a putative miR-331-3p and miR-642-5p target based upon its TargetScan prediction and the presence of multiple putative target sites for each miRNA within a 182-nt DOHH 3′-UTR element. This element is unusual due to its concentration of seed binding sites for miR-331-3p and miR-642-5p. It is thought that the presence of multiple sites for a single miRNA within the 3′-UTR of a target gene confers tighter regulatory control over that mRNA (48). Using experimental techniques, we confirmed DOHH as a target gene of miR-331-3p and miR-642-5p in PCa cells. We are unaware of any reported targets for miR-642-5p to date; however, a number of studies have implicated miR-331-3p in the regulation of cell cycle progression and cell growth. miR-331-3p expression is significantly elevated in human fibroblasts induced to enter quiescence by serum starvation (49), suggesting it might regulate a network of genes involved in controlling growth and cell cycle. Comparing miRNA and mRNA expression profiles of immortalized lymphoblastoid cell lines, Wang and co-workers (50) identified a significant inverse correlation in expression between miR-331-3p and genes involved in DNA replication, DNA metabolism, and cell cycle. It was subsequently shown that miR-331-3p expression was significantly decreased in aggressive PCa, where it regulates the cell cycle control genes KIF23 (kinesin family member 23) and CDCA5 (cell division cycle-associated 5) (39). Interestingly, this study also reported decreased proliferation and increased apoptosis in PCa cell lines transfected with miR-331-3p, a finding that is consistent with our data showing that DU145 cell proliferation is inhibited by ectopic expression of miR-331-3p. Another report identified loss of miR-331-3p in gastric cancer and demonstrated that transfection of gastric cancer cell lines with miR-331-3p reduced expression of its direct target E2F1 (38), a protein with a crucial role in controlling cell cycle progression (51). Furthermore, miR-331-3p transfection decreased proliferation of gastric cancer cell lines and induced G0/G1 phase cell cycle arrest. Of interest, Lee and co-workers (52) demonstrated that miR-330 acts as a tumor suppressor in PCa via suppression of E2F1, further supporting a role for miRNAs in cell cycle regulation. Our data suggest that miR-331-3p, in conjunction with miR-642-5p, acts as a PCa tumor suppressor to regulate proliferation not only via decreased ERBB-2/Akt signaling and E2F1 expression, but also through control of DOHH expression, which in turn alters the hypusination and activation of eIF5A (Fig. 6).
FIGURE 6.
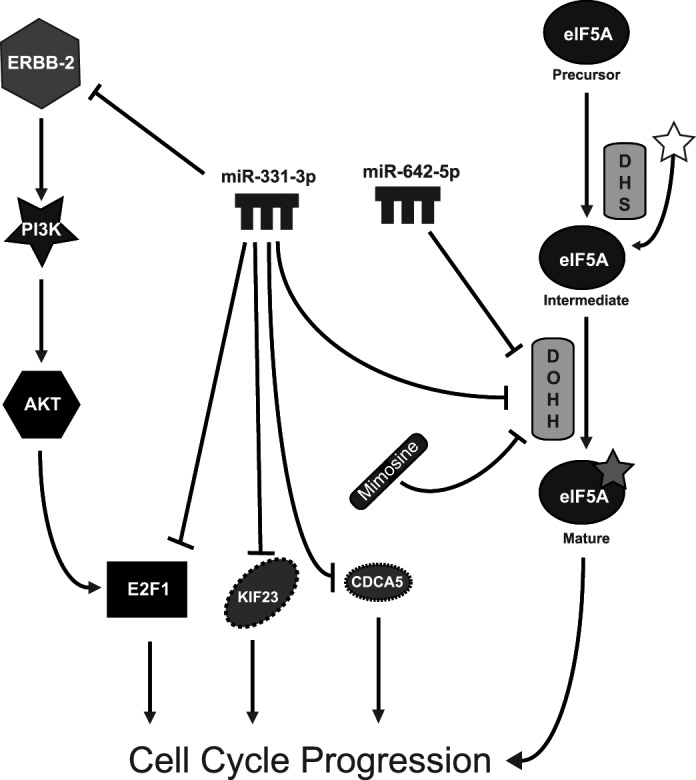
miR-331-3p and miR-642-5p regulate PCa cell proliferation via down-regulation of multiple target mRNAs. miR-331-3p directly regulates ERBB-2 expression and downstream signaling (31) and also targets cell cycle molecules, including E2F1, KIF23, and CDCA5 (38, 39). Both miR-331-3p and miR-642-5p target DOHH, an enzyme critical for the activation of eIF5A via hypusination, a process essential for cell cycle progression and cell growth. Synergistic growth inhibition is observed with the combination of miR-331-3p, miR-642-5p, and the DOHH inhibitor mimosine. DHS, deoxyhypusine synthase.
eIF5A is of considerable interest as a potential therapeutic target in human disease. Inhibition of eIF5A hypusination with the drugs ciclopirox and deferiprone decreases HIV-1 gene expression (53), whereas eIF5A and DOHH promote inflammation and pancreatic β-cell dysfunction, with inhibition of DOHH or depletion of eIF5A protecting against glucose intolerance in murine inflammatory models of diabetes (54). Overexpression of eIF5A has been shown in ovarian and colorectal cancers, and eIF5A promotes anchorage-independent growth in vitro and tumor formation in vivo (55, 56). A recent report demonstrated overexpression of eIF5A and its hypusine-forming enzymes DOHH and deoxyhypusine synthase in glioblastoma patient samples (57). Interestingly, deoxyhypusine synthase and DOHH expression was significantly higher in glioblastoma (grade IV) samples compared with grade I–III tumors, and the deoxyhypusine synthase inhibitor GC7 reduced proliferation of glioblastoma cell lines but not normal human astrocytes. We observed elevated DOHH expression in five of nine prostate tumor samples relative to matched NAT, and in each of these cases, there was a significant down-regulation of miR-331-3p and miR-642-5p. We propose a model in which the decreased expression of miR-331-3p and miR-642-5p in PCa permits the up-regulation of DOHH and other pro-proliferative gene targets, such as E2F1 (Fig. 6). Therefore, restoration of miR-331-3p and miR-642-5p may have therapeutic potential in the treatment of PCa. Furthermore, the synergistic action of miR-331-3p and miR-642-5p with the DOHH inhibitor mimosine suggests that therapeutic up-regulation of these miRNAs could augment the efficacy of DOHH/eIF5A inhibitors in the treatment of PCa.
Supplementary Material
Acknowledgments
We thank Dianne Beveridge and Dr. Scott Bringans (Proteomics International) for helpful discussions regarding this article.
This work was supported by the National Health and Medical Research Council of Australia and the Prostate Cancer Foundation of Australia.

This article contains supplemental Figs. S1 and S2.
- DOHH
- deoxyhypusine hydroxylase monooxygenase
- miRNA
- microRNA
- PCa
- prostate cancer
- nt
- nucleotide(s)
- RT-qPCR
- quantitative RT-PCR
- pre-miR
- precursor miRNA
- miR-NC
- negative control miRNA
- NAT
- normal adjacent tissue.
REFERENCES
- 1. Caraglia M., Park M. H., Wolff E. C., Marra M., Abbruzzese A. (2011) Amino Acids [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Park M. H., Nishimura K., Zanelli C. F., Valentini S. R. (2010) Functional significance of eIF5A and its hypusine modification in eukaryotes. Amino Acids 38, 491–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen K. Y., Liu A. Y. (1997) Biochemistry and function of hypusine formation on eukaryotic initiation factor 5A. Biol. Signals 6, 105–109 [DOI] [PubMed] [Google Scholar]
- 4. Park M. H., Wolff E. C., Folk J. E. (1993) Hypusine: Its post-translational formation in eukaryotic initiation factor 5A and its potential role in cellular regulation. Biofactors 4, 95–104 [PubMed] [Google Scholar]
- 5. Kemper W. M., Merrick W. C., Redfield B., Liu C. K., Weissbach H. (1976) Purification and properties of rabbit reticulocyte elongation factor 1. Arch. Biochem. Biophys. 174, 603–612 [DOI] [PubMed] [Google Scholar]
- 6. Benne R., Hershey J. W. (1978) The mechanism of action of protein synthesis initiation factors from rabbit reticulocytes. J. Biol. Chem. 253, 3078–3087 [PubMed] [Google Scholar]
- 7. Kang H. A., Hershey J. W. (1994) Effect of initiation factor eIF-5A depletion on protein synthesis and proliferation of Saccharomyces cerevisiae. J. Biol. Chem. 269, 3934–3940 [PubMed] [Google Scholar]
- 8. Hanauske-Abel H. M., Slowinska B., Zagulska S., Wilson R. C., Staiano-Coico L., Hanauske A. R., McCaffrey T., Szabo P. (1995) Detection of a subset of polysomal mRNAs associated with modulation of hypusine formation at the G1-S boundary. Proposal of a role for eIF-5A in onset of DNA replication. FEBS Lett. 366, 92–98 [DOI] [PubMed] [Google Scholar]
- 9. Cracchiolo B. M., Heller D. S., Clement P. M., Wolff E. C., Park M. H., Hanauske-Abel H. M. (2004) Eukaryotic initiation factor 5A-1 (eIF5A-1) as a diagnostic marker for aberrant proliferation in intraepithelial neoplasia of the vulva. Gynecol Oncol. 94, 217–222 [DOI] [PubMed] [Google Scholar]
- 10. Park J. H., Aravind L., Wolff E. C., Kaevel J., Kim Y. S., Park M. H. (2006) Molecular cloning, expression, and structural prediction of deoxyhypusine hydroxylase: A HEAT-repeat-containing metalloenzyme. Proc. Natl. Acad. Sci. U.S.A. 103, 51–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hanauske-Abel H. M., Park M. H., Hanauske A. R., Popowicz A. M., Lalande M., Folk J. E. (1994) Inhibition of the G1-S transition of the cell cycle by inhibitors of deoxyhypusine hydroxylation. Biochim. Biophys. Acta 1221, 115–124 [DOI] [PubMed] [Google Scholar]
- 12. Park M. H., Lee Y. B., Joe Y. A. (1997) Hypusine is essential for eukaryotic cell proliferation. Biol. Signals 6, 115–123 [DOI] [PubMed] [Google Scholar]
- 13. Balabanov S., Gontarewicz A., Ziegler P., Hartmann U., Kammer W., Copland M., Brassat U., Priemer M., Hauber I., Wilhelm T., Schwarz G., Kanz L., Bokemeyer C., Hauber J., Holyoake T. L., Nordheim A., Brümmendorf T. H. (2007) Hypusination of eukaryotic initiation factor 5A (eIF5A): A novel therapeutic target in BCR-ABL-positive leukemias identified by a proteomics approach. Blood 109, 1701–1711 [DOI] [PubMed] [Google Scholar]
- 14. Chung L. C., Tsui K. H., Feng T. H., Lee S. L., Chang P. L., Juang H. H. (2012) l-Mimosine blocks cell proliferation via upregulation of B-cell translocation gene 2 and N-myc downstream regulated gene 1 in prostate carcinoma cells. Am. J. Physiol. Cell Physiol. 302, C676–685 [DOI] [PubMed] [Google Scholar]
- 15. Wartenberg M., Fischer K., Hescheler J., Sauer H. (2002) Modulation of intrinsic P-glycoprotein expression in multicellular prostate tumor spheroids by cell cycle inhibitors. Biochim. Biophys. Acta 1589, 49–62 [DOI] [PubMed] [Google Scholar]
- 16. Kulp K. S., Vulliet P. R. (1996) Mimosine blocks cell cycle progression by chelating iron in asynchronous human breast cancer cells. Toxicol. Appl. Pharmacol. 139, 356–364 [DOI] [PubMed] [Google Scholar]
- 17. Chang H. C., Weng C. F., Yen M. H., Chuang L. Y., Hung W. C. (2000) Modulation of cell cycle regulatory protein expression and suppression of tumor growth by mimosine in nude mice. Int. J. Oncol 17, 659–665 [DOI] [PubMed] [Google Scholar]
- 18. Zalatnai A. (2005) P-glycoprotein expression is induced in human pancreatic cancer xenografts during treatment with a cell cycle regulator, mimosine. Pathol Oncol Res. 11, 164–169 [DOI] [PubMed] [Google Scholar]
- 19. Cook J. (2001) Radiation sensitization of mammalian cells by metal chelators. Radiat. Res. 155, 304–310 [DOI] [PubMed] [Google Scholar]
- 20. Bartel D. P. (2004) MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116, 281–297 [DOI] [PubMed] [Google Scholar]
- 21. Humphreys D. T., Westman B. J., Martin D. I., Preiss T. (2005) MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc. Natl. Acad. Sci. U.S.A. 102, 16961–16966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen J. F., Mandel E. M., Thomson J. M., Wu Q., Callis T. E., Hammond S. M., Conlon F. L., Wang D. Z. (2006) The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 38, 228–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cheng A. M., Byrom M. W., Shelton J., Ford L. P. (2005) Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 33, 1290–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bueno M. J., Malumbres M. (2011) MicroRNAs and the cell cycle. Biochim. Biophys. Acta 1812, 592–601 [DOI] [PubMed] [Google Scholar]
- 25. Zhang B., Pan X., Cobb G. P., Anderson T. A. (2007) microRNAs as oncogenes and tumor suppressors. Dev. Biol. 302, 1–12 [DOI] [PubMed] [Google Scholar]
- 26. Esquela-Kerscher A., Slack F. J. (2006) Oncomirs: microRNAs with a role in cancer. Nat. Rev. Cancer 6, 259–269 [DOI] [PubMed] [Google Scholar]
- 27. Johnson S. M., Grosshans H., Shingara J., Byrom M., Jarvis R., Cheng A., Labourier E., Reinert K. L., Brown D., Slack F. J. (2005) RAS is regulated by the let-7 microRNA family. Cell 120, 635–647 [DOI] [PubMed] [Google Scholar]
- 28. Takamizawa J., Konishi H., Yanagisawa K., Tomida S., Osada H., Endoh H., Harano T., Yatabe Y., Nagino M., Nimura Y., Mitsudomi T., Takahashi T. (2004) Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 64, 3753–3756 [DOI] [PubMed] [Google Scholar]
- 29. Volinia S., Calin G. A., Liu C. G., Ambs S., Cimmino A., Petrocca F., Visone R., Iorio M., Roldo C., Ferracin M., Prueitt R. L., Yanaihara N., Lanza G., Scarpa A., Vecchione A., Negrini M., Harris C. C., Croce C. M. (2006) A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. U.S.A. 103, 2257–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Si M. L., Zhu S., Wu H., Lu Z., Wu F., Mo Y. Y. (2007) miR-21-mediated tumor growth. Oncogene 26, 2799–2803 [DOI] [PubMed] [Google Scholar]
- 31. Epis M. R., Giles K. M., Barker A., Kendrick T. S., Leedman P. J. (2009) miR-331-3p regulates ERBB-2 expression and androgen receptor signaling in prostate cancer. J. Biol. Chem. 284, 24696–24704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Epis M. R., Barker A., Giles K. M., Beveridge D. J., Leedman P. J. (2011) The RNA-binding protein HuR opposes the repression of ERBB-2 gene expression by microRNA miR-331-3p in prostate cancer cells. J. Biol. Chem. 286, 41442–41454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Livak K. J., Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)) method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 34. Giles K. M., Barker A., Zhang P. M., Epis M. R., Leedman P. J. (2011) MicroRNA regulation of growth factor receptor signaling in human cancer cells. Methods Mol. Biol. 676, 147–163 [DOI] [PubMed] [Google Scholar]
- 35. Bliss C. (1939) The toxicity of poisons applied jointly. Ann. Appl. Biol. 26, 585–615 [Google Scholar]
- 36. Lewis B. P., Burge C. B., Bartel D. P. (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20 [DOI] [PubMed] [Google Scholar]
- 37. Grimson A., Farh K. K., Johnston W. K., Garrett-Engele P., Lim L. P., Bartel D. P. (2007) MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell 27, 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Guo X., Guo L., Ji J., Zhang J., Zhang J., Chen X., Cai Q., Li J., Gu Q., Liu B., Zhu Z., Yu Y. (2010) miRNA-331-3p directly targets E2F1 and induces growth arrest in human gastric cancer. Biochem. Biophys. Res. Commun. 398, 1–6 [DOI] [PubMed] [Google Scholar]
- 39. Wang L., Tang H., Thayanithy V., Subramanian S., Oberg A. L., Cunningham J. M., Cerhan J. R., Steer C. J., Thibodeau S. N. (2009) Gene networks and microRNAs implicated in aggressive prostate cancer. Cancer Res. 69, 9490–9497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. White N. M., Youssef Y. M., Fendler A., Stephan C., Jung K., Yousef G. M. (2012) The miRNA-kallikrein axis of interaction: A new dimension in the pathogenesis of prostate cancer. Biol. Chem. 393, 379–389 [DOI] [PubMed] [Google Scholar]
- 41. Xu G., Wu J., Zhou L., Chen B., Sun Z., Zhao F., Tao Z. (2010) Characterization of the small RNA transcriptomes of androgen-dependent and -independent prostate cancer cell line by deep sequencing. PLoS One 5, e15519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. John B., Enright A. J., Aravin A., Tuschl T., Sander C., Marks D. S. (2004) Human MicroRNA targets. PLoS Biol. 2, e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Krek A., Grün D., Poy M. N., Wolf R., Rosenberg L., Epstein E. J., MacMenamin P., da Piedade I., Gunsalus K. C., Stoffel M., Rajewsky N. (2005) Combinatorial microRNA target predictions. Nat. Genet. 37, 495–500 [DOI] [PubMed] [Google Scholar]
- 44. Witkos T. M., Koscianska E., Krzyzosiak W. J. (2011) Practical aspects of microRNA target prediction. Curr. Mol. Med. 11, 93–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Webster R. J., Giles K. M., Price K. J., Zhang P. M., Mattick J. S., Leedman P. J. (2009) Regulation of epidermal growth factor receptor signaling in human cancer cells by microRNA-7. J. Biol. Chem. 284, 5731–5741 [DOI] [PubMed] [Google Scholar]
- 46. Guo H., Ingolia N. T., Weissman J. S., Bartel D. P. (2010) Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466, 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chi S. W., Zang J. B., Mele A., Darnell R. B. (2009) Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 460, 479–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jackson R. J., Standart N. (2007) How do microRNAs regulate gene expression? Sci. STKE 2007, re1. [DOI] [PubMed] [Google Scholar]
- 49. Maes O. C., Sarojini H., Wang E. (2009) Stepwise up-regulation of microRNA expression levels from replicating to reversible and irreversible growth arrest states in WI-38 human fibroblasts. J. Cell. Physiol. 221, 109–119 [DOI] [PubMed] [Google Scholar]
- 50. Wang L., Oberg A. L., Asmann Y. W., Sicotte H., McDonnell S. K., Riska S. M., Liu W., Steer C. J., Subramanian S., Cunningham J. M., Cerhan J. R., Thibodeau S. N. (2009) Genome-wide transcriptional profiling reveals microRNA-correlated genes and biological processes in human lymphoblastoid cell lines. PLoS One 4, e5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Engelmann D., Pützer B. M. (2012) The dark side of E2F1: In transit beyond apoptosis. Cancer Res. 72, 571–575 [DOI] [PubMed] [Google Scholar]
- 52. Lee K. H., Chen Y. L., Yeh S. D., Hsiao M., Lin J. T., Goan Y. G., Lu P. J. (2009) MicroRNA-330 acts as tumor suppressor and induces apoptosis of prostate cancer cells through E2F1-mediated suppression of Akt phosphorylation. Oncogene 28, 3360–3370 [DOI] [PubMed] [Google Scholar]
- 53. Hoque M., Hanauske-Abel H. M., Palumbo P., Saxena D., D'Alliessi Gandolfi D., Park M. H., Pe'ery T., Mathews M. B. (2009) Inhibition of HIV-1 gene expression by Ciclopirox and Deferiprone, drugs that prevent hypusination of eukaryotic initiation factor 5A. Retrovirology 6, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Maier B., Ogihara T., Trace A. P., Tersey S. A., Robbins R. D., Chakrabarti S. K., Nunemaker C. S., Stull N. D., Taylor C. A., Thompson J. E., Dondero R. S., Lewis E. C., Dinarello C. A., Nadler J. L., Mirmira R. G. (2010) The unique hypusine modification of eIF5A promotes islet beta cell inflammation and dysfunction in mice. J. Clin. Invest. 120, 2156–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Guan X. Y., Fung J. M., Ma N. F., Lau S. H., Tai L. S., Xie D., Zhang Y., Hu L., Wu Q. L., Fang Y., Sham J. S. (2004) Oncogenic role of eIF-5A2 in the development of ovarian cancer. Cancer Res. 64, 4197–4200 [DOI] [PubMed] [Google Scholar]
- 56. Xie D., Ma N. F., Pan Z. Z., Wu H. X., Liu Y. D., Wu G. Q., Kung H. F., Guan X. Y. (2008) Overexpression of EIF-5A2 is associated with metastasis of human colorectal carcinoma. Hum. Pathol. 39, 80–86 [DOI] [PubMed] [Google Scholar]
- 57. Preukschas M. H., Schulte A., Lamszus K., Sievert H., Bokemeyer C., Balabanov S. (2011) Overexpression of eukaryotic initiation factor 5a (eIF-5A) and hypusine-forming enzymes in glioblastoma patient samples and therapeutic potential of hypusine modification for treatment of glioblastoma multiforme in 2011 European Multidisciplinary Cancer Congress, Vol. 47, Suppl. 1, Sept. 2011, p. 157, Elsevier [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.